Abstract
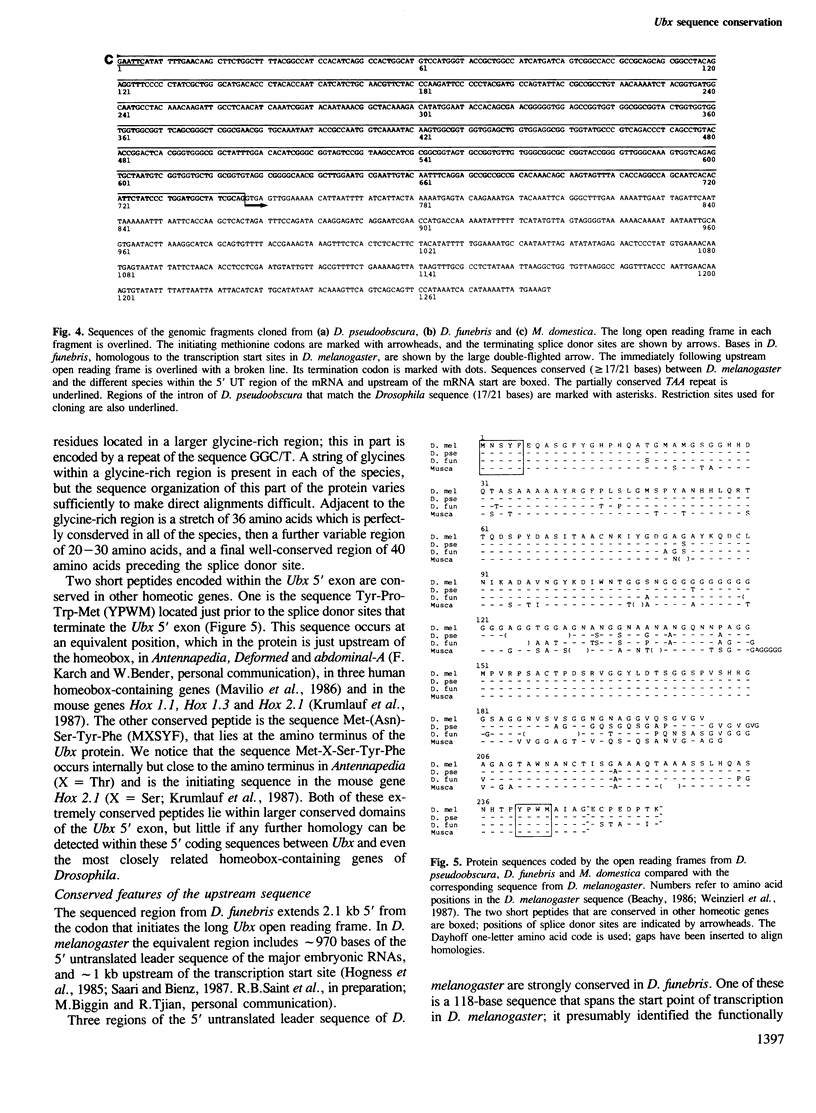
Clones homologous to the 5' region of the Ultrabithorax gene of Drosophila melanogaster have been isolated from D. pseudoobscura, D. funebris and Musca domestica. Regions that encode most of the Ubx protein have been sequenced in all three of these species, and the 5' upstream region has been sequenced in D. funebris to a point ˜1000 bases upstream of the probable mRNA start site. Here we compare these sequences with those described elsewhere for D. melanogaster. Deduced amino acid sequences of the Ubx protein show 8% (D. pseudoobscura), 15% (D. funebris) and 22% (M. domestica) divergence from D. melanogaster. However, these figures mask very different rates of evolution in different regions of the protein. A glycine-rich (`hinge') region is conserved in each of these species, although its length is variable. Comparison of D. funebris and D. melanogaster sequences in the long 5' untranslated leader region of the mRNA, and in the region immediately upstream of the start point of transcription, reveals tightly conserved elements embedded in an otherwise non-homologous sequence. These conserved elements include a 118-bp region that spans the mRNA start site, an internally repetitive (TAA)n region in the untranslated leader and a short repeated motif immediately upstream of the ATG codon that initiates the major open reading frame of the Ubx protein. Two other conserved elements were identified upstream of the transcription start site; both elements have structural features consistent with a role as recognition sites for regulatory proteins.
Keywords: bithorax complex, diptera, evolution, upstream elements
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. E., Martinez-Arias A. The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 1985 Jul;4(7):1689–1700. doi: 10.1002/j.1460-2075.1985.tb03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M. E., Martinez-Arias A., Weinzierl R., Wilde C. D. Function and expression of ultrabithorax in the Drosophila embryo. Cold Spring Harb Symp Quant Biol. 1985;50:195–200. doi: 10.1101/sqb.1985.050.01.025. [DOI] [PubMed] [Google Scholar]
- Akam M. E. The location of Ultrabithorax transcripts in Drosophila tissue sections. EMBO J. 1983;2(11):2075–2084. doi: 10.1002/j.1460-2075.1983.tb01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy P. A., Helfand S. L., Hogness D. S. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985 Feb 14;313(6003):545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Bender W., Weiffenbach B., Karch F., Peifer M. Domains of cis-interaction in the bithorax complex. Cold Spring Harb Symp Quant Biol. 1985;50:173–180. doi: 10.1101/sqb.1985.050.01.023. [DOI] [PubMed] [Google Scholar]
- Beverley S. M., Wilson A. C. Molecular evolution in Drosophila and the higher Diptera II. A time scale for fly evolution. J Mol Evol. 1984;21(1):1–13. doi: 10.1007/BF02100622. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman R. K., Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986 Apr 20;188(4):499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Casanova J., Sánchez-Herrero E., Morata G. Prothoracic transformation and functional structure of the Ultrabithorax gene of Drosophila. Cell. 1985 Sep;42(2):663–669. doi: 10.1016/0092-8674(85)90123-0. [DOI] [PubMed] [Google Scholar]
- Cavener D. R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987 Feb 25;15(4):1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E., Levine M., Gehring W. J. Regulation of Antennapedia transcript distribution by the bithorax complex in Drosophila. Nature. 1984 Jan 19;307(5948):287–289. doi: 10.1038/307287a0. [DOI] [PubMed] [Google Scholar]
- Harding K., Wedeen C., McGinnis W., Levine M. Spatially regulated expression of homeotic genes in Drosophila. Science. 1985 Sep 20;229(4719):1236–1242. doi: 10.1126/science.3898362. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromi Y., Kuroiwa A., Gehring W. J. Control elements of the Drosophila segmentation gene fushi tarazu. Cell. 1985 Dec;43(3 Pt 2):603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- Hogness D. S., Lipshitz H. D., Beachy P. A., Peattie D. A., Saint R. B., Goldschmidt-Clermont M., Harte P. J., Gavis E. R., Helfand S. L. Regulation and products of the Ubx domain of the bithorax complex. Cold Spring Harb Symp Quant Biol. 1985;50:181–194. doi: 10.1101/sqb.1985.050.01.024. [DOI] [PubMed] [Google Scholar]
- Holmgren R., Corces V., Morimoto R., Blackman R., Meselson M. Sequence homologies in the 5' regions of four Drosophila heat-shock genes. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3775–3778. doi: 10.1073/pnas.78.6.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T. False starts in translational control of gene expression. Nature. 1985 Aug 15;316(6029):580–581. doi: 10.1038/316580a0. [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Ish-Horowicz D., Howard K. R. Correlative changes in homoeotic and segmentation gene expression in Krüppel mutant embryos of Drosophila. EMBO J. 1986 Jul;5(7):1659–1665. doi: 10.1002/j.1460-2075.1986.tb04409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W., Martinez-Arias A. The correct activation of Antennapedia and bithorax complex genes requires the fushi tarazu gene. Nature. 1986 Dec 11;324(6097):592–597. doi: 10.1038/324592a0. [DOI] [PubMed] [Google Scholar]
- Johansen H., Schümperli D., Rosenberg M. Affecting gene expression by altering the length and sequence of the 5' leader. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7698–7702. doi: 10.1073/pnas.81.24.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Herskowitz I. A repressor (MAT alpha 2 Product) and its operator control expression of a set of cell type specific genes in yeast. Cell. 1985 Aug;42(1):237–247. doi: 10.1016/s0092-8674(85)80119-7. [DOI] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Wong M. L., O'Farrell P. H. Electron microscopic heteroduplex mapping identifies regions of the engrailed locus that are conserved between Drosophila melanogaster and Drosophila virilis. Mol Cell Biol. 1985 Dec;5(12):3600–3609. doi: 10.1128/mcb.5.12.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R., Hultmark D., Gehring W. J. Selective translation of heat shock mRNA in Drosophila melanogaster depends on sequence information in the leader. EMBO J. 1985 Aug;4(8):2053–2060. doi: 10.1002/j.1460-2075.1985.tb03891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984 Jan 25;12(2):857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R., Holland P. W., McVey J. H., Hogan B. L. Developmental and spatial patterns of expression of the mouse homeobox gene, Hox 2.1. Development. 1987 Apr;99(4):603–617. doi: 10.1242/dev.99.4.603. [DOI] [PubMed] [Google Scholar]
- LEWIS E. B. Pseudoallelism and gene evolution. Cold Spring Harb Symp Quant Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- Laughon A., Carroll S. B., Storfer F. A., Riley P. D., Scott M. P. Common properties of proteins encoded by the Antennapedia complex genes of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:253–262. doi: 10.1101/sqb.1985.050.01.032. [DOI] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Liu C. C., Simonsen C. C., Levinson A. D. Initiation of translation at internal AUG codons in mammalian cells. Nature. 1984 May 3;309(5963):82–85. doi: 10.1038/309082a0. [DOI] [PubMed] [Google Scholar]
- Lomedico P. T., McAndrew S. J. Eukaryotic ribosomes can recognize preproinsulin initiation codons irrespective of their position relative to the 5' end of mRNA. Nature. 1982 Sep 16;299(5880):221–226. doi: 10.1038/299221a0. [DOI] [PubMed] [Google Scholar]
- Mavilio F., Simeone A., Giampaolo A., Faiella A., Zappavigna V., Acampora D., Poiana G., Russo G., Peschle C., Boncinelli E. Differential and stage-related expression in embryonic tissues of a new human homoeobox gene. Nature. 1986 Dec 18;324(6098):664–668. doi: 10.1038/324664a0. [DOI] [PubMed] [Google Scholar]
- McGarry T. J., Lindquist S. The preferential translation of Drosophila hsp70 mRNA requires sequences in the untranslated leader. Cell. 1985 Oct;42(3):903–911. doi: 10.1016/0092-8674(85)90286-7. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Miller A. M., MacKay V. L., Nasmyth K. A. Identification and comparison of two sequence elements that confer cell-type specific transcription in yeast. Nature. 1985 Apr 18;314(6012):598–603. doi: 10.1038/314598a0. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell. 1984 May;37(1):273–283. doi: 10.1016/0092-8674(84)90323-4. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Pastorcic M., Wang H., Elbrecht A., Tsai S. Y., Tsai M. J., O'Malley B. W. Control of transcription initiation in vitro requires binding of a transcription factor to the distal promoter of the ovalbumin gene. Mol Cell Biol. 1986 Aug;6(8):2784–2791. doi: 10.1128/mcb.6.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachat F. H., Hogness D. S. Repetitive sequences in isolated Thomas circles from Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1974;38:371–381. doi: 10.1101/sqb.1974.038.01.040. [DOI] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Baumgartner P., Gehring W. J. Structural organization and sequence of the homeotic gene Antennapedia of Drosophila melanogaster. EMBO J. 1986 Apr;5(4):733–739. doi: 10.1002/j.1460-2075.1986.tb04275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C., McGinnis W., Carrasco A. E., De Robertis E. M., Gehring W. J. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984 Jul 5;310(5972):70–71. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. A gene product required for correct initiation of segmental determination in Drosophila. Nature. 1981 Sep 3;293(5827):36–41. doi: 10.1038/293036a0. [DOI] [PubMed] [Google Scholar]
- Struhl G., Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985 Dec 1;4(12):3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G., White R. A. Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell. 1985 Dec;43(2 Pt 1):507–519. doi: 10.1016/0092-8674(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E., Vernós I., Marco R., Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985 Jan 10;313(5998):108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Tautz D., Trick M., Dover G. A. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986 Aug 14;322(6080):652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- White R. A., Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986 Oct 24;47(2):311–321. doi: 10.1016/0092-8674(86)90453-8. [DOI] [PubMed] [Google Scholar]
- White R. A., Wilcox M. Distribution of Ultrabithorax proteins in Drosophila. EMBO J. 1985 Aug;4(8):2035–2043. doi: 10.1002/j.1460-2075.1985.tb03889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. A., Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984 Nov;39(1):163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]
- Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984 May 17;309(5965):229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]



