Abstract
The relationship between C-reactive protein (CRP) gene rs1205 polymorphism and the risk of colorectal cancer (CRC) has been investigated previously. However, the results were conflicting. In the present study, we assessed whether CRP gene rs1205 polymorphism was associated with the risk of CRC by meta-analysis. We searched in PubMed, Embase, and the CNKI databases. Pooled odds ratios (ORs) and 95% confidence intervals (CIs) were calculated. Seven original studies involving 4,181 cases and 10,601 controls analyzed the association between CRP gene rs1205 polymorphism and CRC risk. No significant association was found between CRP gene rs1205 polymorphism and CRC risk in this meta-analysis. Sensitivity analysis did not draw different findings. Stratification analyses of ethnicity, type of cancer, and genotype method also did not obtain any association between CRP gene rs1205 polymorphism and CRC risk. In conclusion, this meta-analysis indicates that CRP gene rs1205 polymorphism was not associated with the risk of CRC.
Keywords: CRP, CRC, meta-analysis, polymorphism, rs1205
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most common cancer cause of death globally [1]. The pathogenesis of cancer has not been completely elucidated. However, a significant correlation between inflammation and human cancer was first established almost 27 years ago [2], and inflammatory reactions have received widespread attention in cancer community ever since. Two hypotheses are studied regarding the association between inflammation and cancer. First, the induction hypothesis states that chronic inflammation results in excessive cell proliferation and activation of a cascade of cellular actions that can lead to induction of irreversible DNA damage. Persistent irritation and inflammation subsequently promote these initiated cells, resulting in tumor growth, progression of metastatic disease, and immunosuppression [3]. Second, the immune response of the host is studied as a consequence of tumor growth itself. In both hypotheses, products of inflammatory processes are believed to be biomarkers for cancer [4–6].
C-reactive protein (CRP) is the phenotype acute-phase protein induced by hepatocytes, known as an inflammatory biomarker. The CRP gene is located at chromosome 1q21–1q23 consisting of two exons and spans 1.9 kb in length, including 29 single nucleotide polymorphisms (SNPs). CRP is associated with a wide range of diseases, including atherosclerosis and diabetes mellitus [7,8]. Given that cancer is related to several forms of inflammation, CRP levels have also been implicated. Many studies demonstrated that elevated level of CRP was associated with the increased risk of multiple cancers, such as colorectal, esophageal, hepatic, breast, and pancreatic cancer [4,9–14]. Recent data from the European Prospective Investigation into Cancer and Nutrition (EPIC) study showed a positive association between circulating CRP and risk of colon cancer. Therefore, it is reasonable to hypothesize that the CRP may be a candidate gene for CRC susceptibility.
Recently, a lot of studies explored the relationship between CRP gene rs1205 polymorphism and CRC risk [15–20]. However, the results of these studies were conflicting and inconclusive because of the clinical heterogeneity, different ethnic populations, and small sample sizes. In order to precisely elucidate the genetic role for CRP gene rs1205 polymorphism in the development of CRC, we performed a comprehensive meta-analysis to clarify the association between this SNP and CRC risk.
Materials and methods
Identification of eligible studies and data extraction
We performed a comprehensive literature search throughout PubMed, Embase, and CNKI databases to retrieve the genetic association studies of CRC. The following terms were used in our searching strategies: ‘c-reactive protein’, ‘CRP’, ‘SNP’, ‘polymorphism’, ‘variant’, ‘cancer’, ‘carcinoma’, and ‘malignancy’. Additional potential omitted studies (such as reference lists of identified studies) have been identified by hand screening. All studies were carefully selected and were up to date as of May 1, 2017. The inclusion criteria for studies were as follows: (1) studies that evaluated the association between CRP gene rs1205 polymorphism and CRC risk; (2) studied on human beings; (3) contained genotype data for the calculation of odds ratios (ORs) and 95% confidence intervals (CIs). Related information was carefully extracted from all eligible studies. The following information was extracted from each study: author, year of publication, ethnicity based on the continent of origin of the study population, source of controls (SOC), numbers of cases and controls, and the genotype methods.
Evaluation of statistical associations
All statistical analyses were performed using the Stata 11.0 software (StataCorp, College Station, TX, U.S.A.). ORs and 95% CIs were used to assess the strength of associations between CRP gene rs1205 polymorphism and CRC risk. Stratification analyses were carried out by SOC. P<0.05 was considered statistically significant. Multivariate ORs and corresponding 95% CIs between extreme levels of annualized case volume (highest versus lowest) were pooled using a random-effects model, accounting for clinical heterogeneity. Heterogeneity across studies was assessed by using the Q statistic with its P value and I2 statistic [21,22]. Pooled ORs and 95% CIs were calculated in our meta-analysis that was performed using the following genetic models: (1) allele, (2) recessive, (3) homozygous, (4) heterozygous, and (5) dominant. The power of this meta-analysis was calculated with a significant value of 0.05 [23]. Two reviewers independently performed the extraction of data and assessed the quality of study based on the Newcastle–Ottawa Scale (NOS) scores [24]. All disagreements were discussed and resolved with consensus.
Evaluation of publication bias and heterogeneity
Potential publication bias was assessed by Begg’s and Egger’s linear regression test [25]. P<0.05 was considered to indicate statistically significant. We performed sensitivity analysis by omitting each study in turn to determine the effect on the test of heterogeneity and evaluated the stability of the overall results.
Results
Characteristics of the included studies
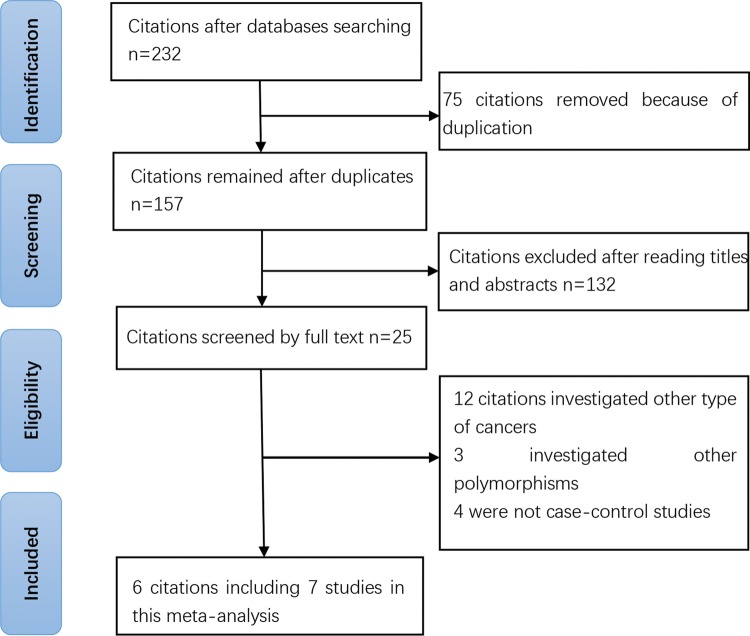
As showed in Figure 1, we derived 232 citations from the databases of PubMed, Embase, and CNKI. Seventy-five citations were removed due to duplication. Of the 157 remaining citations, 132 were excluded after reading titles and abstracts. Twenty-five citations were excluded after being screened by full text: 12 citations investigated other type of cancers; 3 investigated other polymorphisms; 4 were not case–control studies. The characteristics of included studies are summarized in Tables 1 and 2. The NOS of all included studies ranged from 6 to 8 stars, suggesting that these studies were of high methodological quality.
Figure 1. Selection for eligible citations included in this meta-analysis.
Table 1. Characteristics of included studies.
| Study | Year | Nationality | Type | Number of cases/controls | Genotype method |
|---|---|---|---|---|---|
| Nimptsch et al. | 2015 | Mixed | CRC | 727/727 | TaqMan |
| Yang et al. | 2011 | China | CRC | 421/218 | TaqMan |
| Slattery et al. (CC) | 2011 | U.S.A. | CRC | 1574/1970 | Golden Gate assay |
| Slattery et al. (RC) | 2011 | U.S.A. | CRC | 791/999 | Golden Gate assay |
| Ognjanovic et al. | 2010 | U.S.A. | CRC | 271/539 | TaqMan |
| Tsilidis et al. | 2009 | U.S.A. | CRC | 208/381 | TaqMan |
| Siemes et al. (CRC) | 2006 | Holland | CRC | 189/5767 | TaqMan |
Abbreviation: CC, colon cancer.
Table 2. Characteristics of included studies.
| Author and year | SOC | Ethnicity | Case | Control | NOS | ||||
|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | ||||
| Nimptsch (2015) | PB | Mixed | 358 | 292 | 71 | 302 | 342 | 81 | 8 |
| Yang (2011) | PB | Asians | 72 | 197 | 152 | 40 | 111 | 67 | 6 |
| Slattery (2011) (CC) | PB | Mixed | 700 | 659 | 163 | 882 | 845 | 157 | 6 |
| Slattery (2011) (RC) | PB | Mixed | 295 | 325 | 79 | 406 | 403 | 92 | 7 |
| Ognjanovic (2010) | PB | Mixed | 55 | 119 | 96 | 146 | 250 | 140 | 7 |
| Tsilidis (2009) | PB | Mixed | 99 | 83 | 24 | 167 | 156 | 51 | 6 |
| Siemes (2006) (CRC) | PB | Caucasians | 78 | 92 | 19 | 2584 | 2595 | 588 | 7 |
Abbreviations: CC, colon cancer; PB, population-based; RC, rectal cancer.
Meta-analysis of CRP gene rs1205 polymorphism
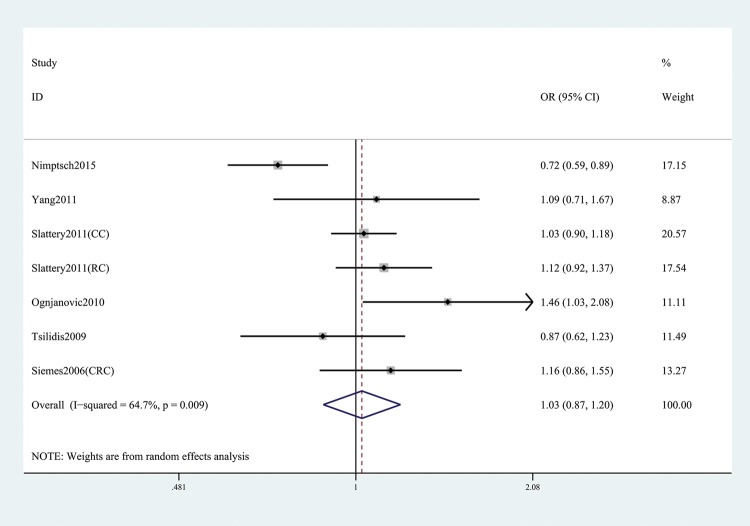
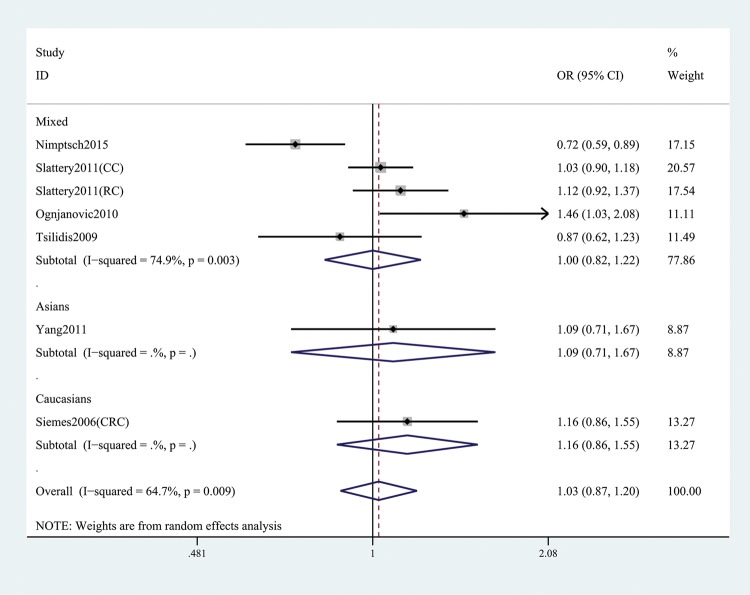
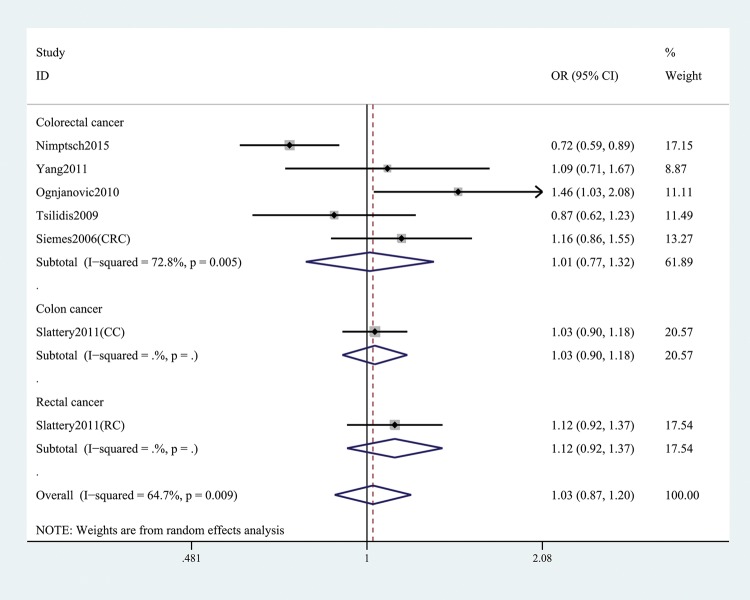
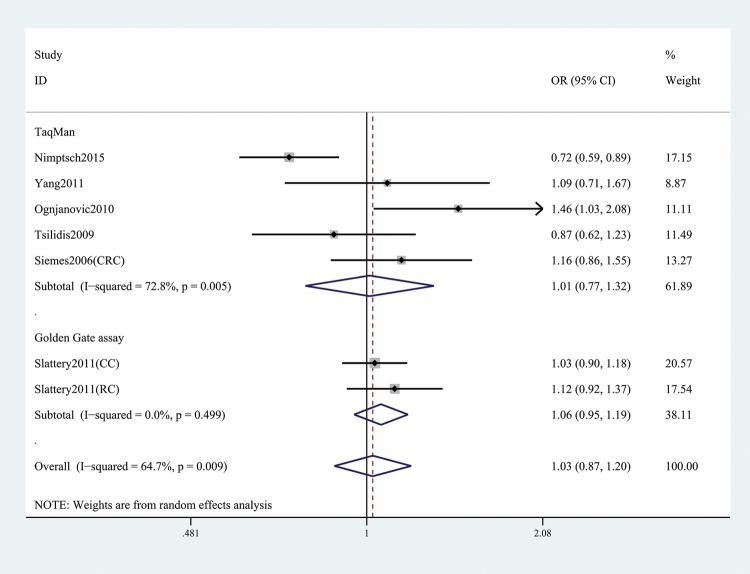
In the general analysis, we found that CRP gene rs1205 was not associated with CRC risk (T versus C: OR and 95% CI, 1.05 (0.93, 1.19), P=0.421; TT versus CC: OR and 95% CI, 1.14 (0.91, 1.43), P=0.257; TT + CT versus CC: OR and 95% CI, 1.03 (0.87, 1.20), P=0.758; TT versus CT+CC: OR and 95%CI, 1.16 (0.98, 1.37), P=0.078; TC versus CC: OR and 95% CI, 0.99 (0.86, 1.14), P=0.866, Table 3 and Figure 2). Stratification analyses of ethnicity (Figure 3), type of CRC (Figure 4), and genotype method (Figure 5) also did not obtain any association between this SNP and CRC risk (Table 3).
Table 3. Summary of results of the meta-analysis from different comparative genetic models.
| Comparison | OR (95% CI) | P-value | P for heterogeneity | I2 (%) | Model |
|---|---|---|---|---|---|
| T versus C | |||||
| Total | 1.05 (0.93, 1.19) | 0.421 | 0.002 | 70.8 | Random |
| Ethnicity | |||||
| Mixed | 1.03 (0.88, 1.21) | 0.695 | 0.001 | 79.8 | Random |
| Asians | 1.15 (0.91, 1.45) | 0.255 | |||
| Caucasians | 1.08 (0.87, 1.34) | 0.489 | |||
| Cancer type | |||||
| CRC | 1.04 (0.85, 1.28) | 0.718 | 0.001 | 79.3 | Random |
| CC | 1.08 (0.97, 1.19) | 0.158 | |||
| RC | 1.09 (0.94, 1.27) | 0.240 | |||
| Genotype method | |||||
| TaqMan | 1.04 (0.85, 1.28) | 0.718 | 0.001 | 79.3 | Random |
| Golden Gate assay | 1.08 (0.99, 1.18) | 0.064 | 0.875 | <0.001 | Random |
| TT versus CC | |||||
| Total | 1.14 (0.91, 1.43) | 0.257 | 0.029 | 57.3 | Random |
| Ethnicity | |||||
| Mixed | 1.13 (0.84, 1.52) | 0.435 | 0.008 | 71.1 | Random |
| Asians | 1.26 (0.78, 2.04) | 0.346 | |||
| Caucasians | 1.07 (0.64, 1.78) | 0.793 | |||
| Cancer type | |||||
| CRC | 1.08 (0.76, 1.54) | 0.674 | 0.014 | 67.8 | Random |
| CC | 1.31 (1.03, 1.66) | 0.029 | |||
| RC | 1.18 (0.84, 1.65) | 0.330 | |||
| Genotype method | |||||
| TaqMan | 1.08 (0.76, 1.54) | 0.674 | 0.014 | 67.8 | Random |
| Golden Gate assay | 1.26 (1.04, 1.54) | 0.019 | 0.630 | <0.001 | Random |
| TT + CT versus CC | |||||
| Total | 1.03 (0.87, 1.20) | 0.758 | 0.009 | 64.7 | Random |
| Ethnicity | |||||
| Mixed | 1.00 (0.82, 1.22) | 0.999 | 0.003 | 74.9 | Random |
| Asians | 1.09 (0.71, 1.67) | 0.694 | |||
| Caucasians | 1.16 (0.86, 1.55) | 0.336 | |||
| Cancer type | |||||
| CRC | 1.01 (0.77, 1.32) | 0.929 | 0.005 | 72.8 | Random |
| CC | 1.03 (0.90, 1.18) | 0.632 | |||
| RC | 1.12 (0.92, 1.37) | 0.253 | |||
| Genotype method | |||||
| TaqMan | 1.01 (0.77, 1.32) | 0.929 | 0.005 | 72.8 | Random |
| Golden Gate assay | 1.06 (0.95, 1.19) | 0.299 | 0.499 | <0.001 | Random |
| TT versus CT + CC | |||||
| Total | 1.16 (0.98, 1.37) | 0.078 | 0.140 | 37.8 | Random |
| Ethnicity | |||||
| Mixed | 1.15 (0.93, 1.43) | 0.205 | 0.063 | 55.1 | Random |
| Asians | 1.27 (0.90, 1.81) | 0.175 | |||
| Caucasians | 0.98 (0.61, 1.59) | 0.949 | |||
| Cancer type | |||||
| CRC | 1.10 (0.86, 1.42) | 0.78 | 0.076 | 52.7 | Random |
| CC | 1.32 (1.05, 1.66) | 2.36 | |||
| RC | 1.12 (0.81, 1.54) | 0.70 | |||
| Genotype method | |||||
| TaqMan | 1.10 (0.86, 1.42) | 0.78 | 0.076 | 52.7 | Random |
| Golden Gate assay | 1.25 (1.03, 1.50) | 2.32 | 0.415 | <0.001 | Random |
| TC versus CC | |||||
| Total | 0.99 (0.86, 1.14) | 0.866 | 0.056 | 51.1 | Random |
| Ethnicity | |||||
| Mixed | 0.96 (0.81, 1.15) | 0.668 | 0.030 | 62.7 | Random |
| Asians | 0.99 (0.63, 1.55) | 0.951 | |||
| Caucasians | 1.17 (0.86, 1.60) | 0.304 | |||
| Cancer type | |||||
| CRC | 0.97 (0.77, 1.22) | 0.789 | 0.040 | 60.2 | Random |
| CC | 0.98 (0.85, 1.13) | 0.809 | |||
| RC | 1.11 (0.90, 1.37) | 0.329 | |||
| Genotype method | |||||
| TaqMan | 0.97 (0.77, 1.22) | 0.789 | 0.040 | 60.2 | Random |
| Golden Gate assay | 1.02 (0.91, 1.15) | 0.727 | 0.056 | 51.1 | Random |
Figure 2. Forest plot shows OR for the associations between rs1205 polymorphism and CRC risk (TT + CT versus CC).
Figure 3. Stratification analysis by ethnicity shows OR for the association between rs1205 polymorphism and CRC risk (TT + CT versus CC).
Figure 4. Stratification analysis by type of cancer shows OR for the association between rs1205 polymorphism and CRC risk (TT + CT versus CC).
Figure 5. Stratification analysis by genotype method shows OR for the association between rs1205 polymorphism and CRC risk (TT + CT versus CC).
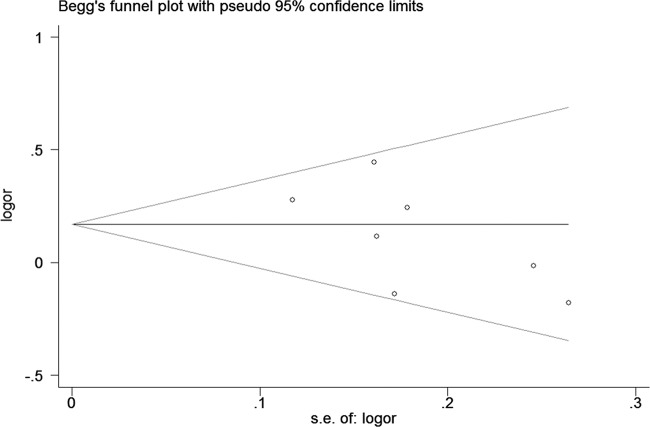
We assessed sensitivity analysis by omitting each study one at a time in every genetic model for rs1205 polymorphism. The pooled ORs for the effects of the SNP on the risk for CRC risk indicated that our data were stable and trustworthy. Both Egger’s and Begg’s tests were used to evaluated the publication bias of this meta-analysis. Our data revealed that there was no obvious publication bias for CRP rs1205 polymorphism (Figure 6).
Figure 6. Begg’s tests for publication bias about rs1205 polymorphism and CRC (TT versus CT + CC).
Discussion
To our best knowledge, this is the first quantitative assessment of the genetic association studies reporting on the relationship between CRP gene rs1205 polymorphism and CRC susceptibility. CRP is one of the most common acute-phase proteins induced by hepatocytes. Plasma CRP level may dramatically increase by up to 10,000-fold at the time of acute responses to severe tissue damage or serious infection [26]. Several previous studies reported the association between CRP gene rs1205 polymorphism and risk of CRC, but the results were inconsistent [15–20]. This meta-analysis summarized seven case–control studies with 4,181 cases and 10,601 controls, and provided evidence that CRP gene rs1205 polymorphism was not associated with CRC risk. Stratification analyses of ethnicity, type of cancer, and genotype method also did not obtain any association between this SNP and CRC risk.
A single study could be underpowered because of sample size, diversity inheritance of the heterogeneous, different ethnicities, clinical heterogeneity, and so on. For instance, Nimptsch et al. [15], Ognjanovic et al. [16], Tsilidis et al. [19], Slattery et al. [18], and Yang et al. [20] reported a significant association between CRP rs1205 polymorphism and CRC risk. However, Siemes et al. [17] failed to replicate this association in a study from Netherlands. To overcome these disaccords, we performed this comprehensive meta-analysis to evaluate the association of CRP rs1205 polymorphism with CRC risk and different ethnicities. Two meta-analyses [27,28] investigated CRP gene rs1205 polymorphism with cancer susceptibility previously. Zhang et al. [27] found no significant association between CRP rs1205 polymorphism and the risk of overall cancer. However, in subgroup analysis by cancer type, marginally increased risk was observed in CRC. Geng et al. [28] found that rs1205 polymorphism increased the risk of overall cancer. In addition, stratification analysis of cancer type in their meta-analysis [28] suggested that rs1205 polymorphism was also associated with an increased risk of CRC. We included additional studies and found that this SNP was not associated with CRC risk. Stratification analysis of ethnicity also did not obtain any association between this SNP and CRC risk. It is noteworthy that Zhang et al. [27] did not include two studies [15,20], while Geng et al. [28] did not include four studies [15,17,19,20]. Consequently, the reliability of their conclusions should be interpreted with caution. We believed our meta-analysis has some strengths over previous meta-analyses for the following reasons. First, the present study is the first systematical meta-analysis regarding the association between CRP gene rs1205 polymorphism and CRC risk. Second, we identified seven studies [15–20] with larger sample size, including 4,181 cases and 10,601 controls with regard to rs1205 polymorphism. Large sample and unbiased epidemiological studies of predisposition gene polymorphisms could provide insight into the association between candidate genes and diseases. Third, sensitivity analysis indicated that our data about rs1205 polymorphism were trustworthy and robust. Fourth, we conducted stratification analyses of ethnicity, type of cancer, and genotype method (previous meta-analyses did not perform), although no association was obtained. Fifth, the power analysis indicated that our study had a power of 93.2% to detect the effect of rs1205 polymorphism on CRC susceptibility with an OR of 1.14.
Several potential limitations should be addressed in this meta-analysis. First, the heterogeneity of this meta-analysis is high, so the data should be interpreted with caution. Second, due to limited data, we could not conduct further stratification analyses of other potential factors, such as age, gender, smoking, and alcohol consumption. Third, our results were based on unadjusted estimates for confounding factors, which might have affected the final results. Fourth, we could not assess potential gene–gene and gene–environment interactions because of the lack of relevant data. Fifth, the conclusions of some stratification analyses about rs1205 polymorphism should be interpreted with caution due to limited sample size. Sixth, the sample sizes of some stratification analyses were limited. Finally, we cannot examine the association between CRP gene rs1205 polymorphism and the clinical manifestations of CRC.
In conclusion, this meta-analysis confirms that CRP gene rs1205 polymorphism is not associated with the risk of CRC. Further studies with large sample size is necessary to validate whether CRP gene rs1205 polymorphism contribute to CRC susceptibility.
Abbreviations
- CI
confidence interval
- CRP
C-reactive protein
- NOS
Newcastle–Ottawa Scale
- OR
odds ratio
- SNP
single nucleotide polymorphism
- SOC
source of controls
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
Dilong Fang conceived and designed this work, and analyzed the data. Yu Ye wrote the paper.
References
- 1.Brenner H., Kloor M. and Pox C.P. (2014) Colorectal cancer. Lancet 383, 1490–1502 [DOI] [PubMed] [Google Scholar]
- 2.Virchow R. (1989) Cellular pathology. As based upon physiological and pathological histology. Lecture XVI–Atheromatous affection of arteries. 1858. Nutr. Rev. 47, 23–25 [DOI] [PubMed] [Google Scholar]
- 3.Coussens L.M. and Werb Z. (2002) Inflammation and cancer. Nature 420, 860–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto K., Ikeda Y., Korenaga D., Tanoue K., Hamatake M., Kawasaki K. et al. (2005) The impact of preoperative serum C-reactive protein on the prognosis of patients with hepatocellular carcinoma. Cancer 103, 1856–1864 [DOI] [PubMed] [Google Scholar]
- 5.Chung Y.C. and Chang Y.F. (2003) Serum C-reactive protein correlates with survival in colorectal cancer patients but is not an independent prognostic indicator. Eur. J. Gastroenterol. Hepatol. 15, 369–373 [DOI] [PubMed] [Google Scholar]
- 6.Nikiteas N.I., Tzanakis N., Gazouli M., Rallis G., Daniilidis K., Theodoropoulos G. et al. (2005) Serum IL-6, TNFalpha and CRP levels in Greek colorectal cancer patients: prognostic implications. World J. Gastroenterol. 11, 1639–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danesh J. and Pepys M.B. (2000) C-reactive protein in healthy and in sick populations. Eur. Heart J. 21, 1564–1565 [DOI] [PubMed] [Google Scholar]
- 8.Danesh J., Wheeler J.G., Hirschfield G.M., Eda S., Eiriksdottir G., Rumley A. et al. (2004) C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N. Engl. J. Med. 350, 1387–1397 [DOI] [PubMed] [Google Scholar]
- 9.Douglas J.B., Silverman D.T., Weinstein S.J., Graubard B.I., Pollak M.N., Tao Y. et al. (2011) Serum C-reactive protein and risk of pancreatic cancer in two nested, case-control studies. Cancer Epidemiol. Biomarkers Prev. 20, 359–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erlinger T.P., Platz E.A., Rifai N. and Helzlsouer K.J. (2004) C-reactive protein and the risk of incident colorectal cancer. J. Am. Med. Assoc. 291, 585–590 [DOI] [PubMed] [Google Scholar]
- 11.Pierce B.L., Ballard-Barbash R., Bernstein L., Baumgartner R.N., Neuhouser M.L., Wener M.H. et al. (2009) Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J. Clin. Oncol. 27, 3437–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gockel I., Dirksen K., Messow C.M. and Junginger T. (2006) Significance of preoperative C-reactive protein as a parameter of the perioperative course and long-term prognosis in squamous cell carcinoma and adenocarcinoma of the oesophagus. World J. Gastroenterol. 12, 3746–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Il'yasova D., Colbert L.H., Harris T.B., Newman A.B., Bauer D.C., Satterfield S. et al. (2005) Circulating levels of inflammatory markers and cancer risk in the health aging and body composition cohort. Cancer Epidemiol. Biomarkers Prev. 14, 2413–2418 [DOI] [PubMed] [Google Scholar]
- 14.Eklund C.M. (2009) Proinflammatory cytokines in CRP baseline regulation. Adv. Clin. Chem. 48, 111–136 [DOI] [PubMed] [Google Scholar]
- 15.Nimptsch K., Aleksandrova K., Boeing H., Janke J., Lee Y.A., Jenab M. et al. (2015) Association of CRP genetic variants with blood concentrations of C-reactive protein and colorectal cancer risk. Int. J. Cancer 136, 1181–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ognjanovic S., Yamamoto J., Saltzman B., Franke A., Ognjanovic M., Yokochi L. et al. (2010) Serum CRP and IL-6, genetic variants and risk of colorectal adenoma in a multiethnic population. Cancer Causes Control 21, 1131–1138 [DOI] [PubMed] [Google Scholar]
- 17.Siemes C., Visser L.E., Coebergh J.W., Splinter T.A., Witteman J.C., Uitterlinden A.G. et al. (2006) C-reactive protein levels, variation in the C-reactive protein gene, and cancer risk: the Rotterdam Study. J. Clin. Oncol. 24, 5216–5222 [DOI] [PubMed] [Google Scholar]
- 18.Slattery M.L., Curtin K., Poole E.M., Duggan D.J., Samowitz W.S., Peters U. et al. (2011) Genetic variation in C-reactive protein in relation to colon and rectal cancer risk and survival. Int. J. Cancer 128, 2726–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsilidis K.K., Helzlsouer K.J., Smith M.W., Grinberg V., Hoffman-Bolton J., Clipp S.L. et al. (2009) Association of common polymorphisms in IL10, and in other genes related to inflammatory response and obesity with colorectal cancer. Cancer Causes Control 20, 1739–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S.H., Huang C.J., Chang S.C. and Lin J.K. (2011) Association of C-reactive protein gene polymorphisms and colorectal cancer. Ann. Surg. Oncol. 18, 1907–1915 [DOI] [PubMed] [Google Scholar]
- 21.Higgins J.P., Thompson S.G., Deeks J.J. and Altman D.G. (2003) Measuring inconsistency in meta-analyses. Br. Med. J. 327, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins J.P. and Thompson S.G. (2002) Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 [DOI] [PubMed] [Google Scholar]
- 23.Hedges L.V. and Pigott T.D. (2001) The power of statistical tests in meta-analysis. Psychol. Methods 6, 203–217 [PubMed] [Google Scholar]
- 24.Stang A. (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 25, 603–605 [DOI] [PubMed] [Google Scholar]
- 25.Peters J.L., Sutton A.J., Jones D.R., Abrams K.R. and Rushton L. (2006) Comparison of two methods to detect publication bias in meta-analysis. J. Am. Med. Assoc. 295, 676–680 [DOI] [PubMed] [Google Scholar]
- 26.Gabay C. and Kushner I. (1999) Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340, 448–454 [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y. and Jiang L. (2014) CRP 1059 G/C and 1846G/A polymorphisms and cancer risk: a meta-analysis of 26,634 subjects. Clin. Res. Hepatol. Gastroenterol. 38, 607–612 [DOI] [PubMed] [Google Scholar]
- 28.Geng P., Sa R., Li J., Li H., Liu C., Liao Y. et al. (2016) Genetic polymorphisms in C-reactive protein increase cancer susceptibility. Sci. Rep. 6, 17161. [DOI] [PMC free article] [PubMed] [Google Scholar]