Abstract
Pediatric diarrhea is a common cause of death among children under 5 years of age. In the current study, we investigated the frequency of intestinal parasites among 580 pediatric patients with chronic diarrhea. Parasitic protozoa (all species combined) were detected by molecular tools in 22.9% of the children and the most common parasite was Cryptosporidium spp. (15.1%). Blastocystis hominis was detected in 4.7%, Dientamoeba fragilis in 4%, Giardia duodenalis in 1.7%, and Entamoeba histolytica in 0.17%. Protozoan infections were observed among all regional groups, but prevalence was highest among Qatari subjects and during the winter season. Typing of Cryptosporidium spp. revealed a predominance of Cryptosporidium parvum in 92% of cases with mostly the IIdA20G1 subtype. Subtypes IIdA19G2, IIdA18G2, IIdA18G1, IIdA17G1, IIdA16G1, and IIdA14G1 were also detected. For Cryptosporidium hominis, IbA10G2 and IbA9G3 subtypes were identified. This study provides supplementary information for implementing prevention and control strategies to reduce the burden of these pediatric protozoan infections. Further analyses are required to better understand the local epidemiology and transmission of Cryptosporidium spp. in Qatar.
Keywords: protozoa, diarrhea, pediatrics, Qatar, RT-PCR, statistical analysis, Cryptosporidium, genotyping
Introduction
Diarrhoeal disease is the world's second leading cause of death among young children (UNICEF, 2013). The primary cause of diarrhea is the ingestion of infectious agents, bacterial, viral, or protozoan, from human or animal feces. Transmission occurs through anthroponotic, zoonotic, foodborne, and waterborne routes (Moore et al., 2016). It consists of ingestion of contaminated food or water, direct contact with infected feces, person-to-person contact, and poor personal hygiene (Boithias et al., 2016).
Intestinal protozoa that are most commonly associated with diarrhea in children include Giardia duodenalis, Blastocystis hominis, Entamoeba histolytica, Cryptosporidium spp., and Dientamoeba fragilis (Maas et al., 2014). Symptoms of infection follow an incubation period of approximately 1 week and include watery diarrhea, cramp, and nausea that may lead to inappetence and undernutrition, as observed in children in West Africa and South America (Quihui-Cota et al., 2015). The World Health Organization (WHO) has identified Cryptosporidium spp. as globally the most common diarrhea-causing protozoan (Pedersen et al., 2014), and the Center for Disease Control and Prevention has reported that giardiasis is experienced by approximately 33% of people in developing countries (CDC, 2015). Accordingly, due to their significant public health and socioeconomic implications, both parasites Cryptosporidium spp. and G. duodenalis were included in the WHO's “Neglected disease initiative” in 2004 (Osman et al., 2016). A wide diversity of Cryptosporidium spp. and subtypes infect humans and molecular analysis of the GP60 marker has been useful for identifying contamination sources and thus inferring their transmission routes (Cacciò and Chalmers, 2016). The cosmopolitan intestinal protozoa B. hominis and D. fragilis are both also of relevance in this context (Stark et al., 2016). D. fragilis is frequently detected as a co-infection with multiple protozoa (Garcia, 2016) especially with B. hominis, which is highly prevalent in Qatar (Abu-Madi et al., 2016a).
Conventional diagnosis of these protozoa relies widely on microscopical examination of stool samples. However, coproscopic identification is limited by a low sensitivity as evidenced by earlier studies reporting a marked difference in the prevalences of B. hominis and G. duodenalis when comparing the results of coproscopic and molecular analyses (Abu-Madi et al., 2015; Gotfred-Rasmussen et al., 2016). Moreover, coproscopic analysis is unable to distinguish between the pathogenic and non-pathogenic species of Entamoeba (Ali, 2015), between different species of Cryptosporidium and different assemblages of Giardia (Haque et al., 2009) as these are morphologically identical within genera. Hence there is an urgent need to adopt more sensitive and specific methods for species identification (Boughattas and Salehi, 2014), as accurate diagnosis will lead to early and more appropriate treatment (Maçin et al., 2016). By employing real-time PCR (RT-PCR), the aim of the current study was to investigate the frequency of intestinal parasites in pediatric patients with chronic diarrhea and to clarify the importance of individual species and relevant risk factors in such cases.
Materials and methods
Sample collection
A total of 580 stool samples were collected in sterile containers from diarrhoeal hospitalized pediatric patients during a whole year (March 2015–March 2016). Samples were transported to the Biomedical Research Center at Qatar University by ice box and stored at −20°C. The study was carried out in accordance with the recommendations of the Medical Research Centre and the Research Committee at Hamad Medical Corporation, Qatar (Research protocol # 16367/16). As the patients were young children, family members gave written informed consent in accordance with the Declaration of Helsinki. DNA was extracted using the QIAamp DNA stool minikit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RT-PCR detection
For the diagnosis of Cryptosporidium spp., D. fragilis, G. duodenalis, and E. histolytica, different sets of primers and probes were used as listed in Table 1. For B. hominis, SyberGreen chemistry was used for parasite identification (Abu-Madi et al., 2015).
Table 1.
Primers and probes used in this study.
| Parasite | Target gene | Primers/Probes | Sequence 5′−3′ |
|---|---|---|---|
| Blastocystis hominis* | SSU rDNA | FwdS1 | GGTCCGGTGAACACTTTGGATTT |
| RvsS2 | CCTACGGAAACCTTGTTACGACTTCA | ||
| Giardia duodenalis | SSU rRNA | Gd-80F | GACGGCTCAGGACAACGGTT |
| Gd-127R | TTGCCAGCGGTGTCCG | ||
| 105T | FAM-CCCGCGGCG/ZEN/GTCCCTGCTAG | ||
| Dientamoeba fragilis | SSU rDNA | DF3 | GTTGAATACGTCCCTGCCCTTT |
| DF4 | TGATCCAATGATTTCACCGAGTCA | ||
| Probe | FAM-CACACCGCCCGTCGCTCCTA | ||
| Entamoeba histolytica | 18S rRNA | Edh-239F | ATTGTCGTGGCATCCTAACTCA |
| Edh-88R | GCGGACGGCTCATTATAACA | ||
| 96T | HEX-TCATTGAAT/ZEN/GAATTGGCCATTT | ||
| Cryptosporidium spp. | 18S rRNA | SCL2 | CAGTTATAGTTTACTTGATAATC |
| SCR2 | CAATACCCTACCGTCTAAAG | ||
| CrySB | FAM/CCGTGGTAATTCTAGAGCTA/BHQ |
No probe was used in the detection of this protozoan but SyberGreen chemistry.
A unidirectional workflow pre- to post-PCR was enforced, and preparation of PCR reaction mixture, DNA preparations and PCR were carried out in facilities physically separate from each other (Tellevik et al., 2016). Sample analysis was done using Biosystems Cycler 7500 with a total reactional volume of 20 and 3 μl of DNA as described elsewhere for other protozoa with duplicates of positive and negative controls included in each run (Abu-Madi et al., 2015). PCR cycling conditions for Cryptosporidum spp. were 95°C for 10 min, followed by 50 cycles each cycle comprising 95°C for 15 s, 50°C for 30 s, and 72°C for 45 s each, and a final extension at 72°C for 2 min.
Cryptosporidium geno/sub-typing
The RT-PCR positive products for Cryptosporidium spp. were subjected to restriction fragment length polymorphism (RFLP) analysis to identify the Cryptosporidium species (Coupe et al., 2005). Specimens that contained Cryptosporidium parvum or Cryptosporidium hominis were further subtyped by DNA sequencing of the PCR product of the gp60 gene (Sulaiman et al., 2005). Samples' sequences were aligned with reference sequences using the MAFFT program (EMBL) and BLAST searches were conducted with the edited sequences to determine the similarity of the samples with already published sequences. To determine the taxonomic positions, phylogenetic trees were constructed using the neighbor joining method by SplitsTree software, with robustness of groupings assessed using 1,000 bootstrap replicates of the data. The GP60 nucleotide sequences of the different subtypes were deposited in the GenBank database and the accession numbers are provided in the Results Section.
Statistical analysis
Prevalence data are shown with 95% confidence limits (CL95), calculated as described by Rohlf and Sokal (1995) employing bespoke software. Prevalence was analyzed by maximum likelihood techniques based on log-linear analysis of contingency tables using the software package IBM SPSS Statistics (Version 23). Initially, full factorial models were fitted, incorporating as factors sex (SEX at 2 levels, males and females), age (AGE CLASS at 5 levels: Age class 1 (1 day–10 months), Age class 2 (11–20 months), Age class 3 (21–30 months), Age class 4 (31–60 months), and Age class 5 (>60 months); season [SEASON at 2 levels, winter (November–April) and summer (May–October)]; region of origin (REGION at 5 levels, as defined below). The children (n = 580) came from 37 countries which we allocated to five geographical regions for ease of initial analysis. These were Qatar (n = 168); Middle East (n = 92) comprising Bahrain (n = 3), Iraq (n = 6), Jordan (n = 23), Lebanon (n = 15), Oman (n = 2), Palestine (n = 3), Saudi Arabia (n = 6), and Syria (n = 17); and Yemen (n = 17); Africa (n = 119) comprising Djibouti (n = 1), Egypt (n = 88), Eritrea (n = 1), Libya (n = 1), Mauritania (n = 1), Morocco (n = 3), Nigeria (n = 1), South Africa (n = 1) Sudan (n = 10), and Tunisia (n = 12); Asia (n = 178) comprising Bangladesh (n = 6), China (n = 1), India (n = 87), Nepal (n = 3), Pakistan (n = 56), Philippines (n = 16), and Sri Lanka (n = 9); Other (n = 23) comprising children from other continents North America (n = 9), Great Britain (n = 3), Canada (n = 1), The Netherlands (n = 1), France (n = 3), Greece (n = 1), Italy (n = 1), Poland (n = 1), Spain (n = 2), and Venezuela (n = 1).
For each species of parasite, and for higher taxa, the presence/absence of parasites (INFECTION) was coded as a binary factor. The four explanatory factors listed above were fitted initially to all models that were evaluated. For each level of analysis in turn, beginning with the most complex model, involving all possible main effects and interactions, those combinations that did not contribute significantly to explaining variation in the data were eliminated in a stepwise fashion beginning with the highest-level interaction (the backward selection procedure). A minimum sufficient model was then obtained, for which the likelihood ratio of χ2 was not significant, indicating that the model was sufficient in explaining the data. The importance of each term (i.e., interactions involving INFECTION) in the final model was assessed by the probability that its exclusion would alter the model significantly and these values relating to interactions that included INFECTION are given in the text. Where relevant we also fitted in turn models with just one of the factors and INFECTION. A 2 × 2 χ2-test was implemented in some instances based on (Barnard et al., 2007).
Species richness (number of species harbored by each subject) was analyzed by generalized linear models (GLM) in SPSS 23, based on Poisson errors and we report the value of Wald χ2.
Results
Table 2 shows the distribution of subjects by region of origin and sex. Numbers in each age class are given in Figure 1. As can be seen, the largest category was children from Asia, and the smallest category was that referred to as “other” representing subjects from Europe and North and South America.
Table 2.
Number of children in the study by country/region of origin and sex.
| Country/Region of origin | Sex | Number | Total by region |
|---|---|---|---|
| Qatar | Male | 93 | |
| Female | 75 | 168 | |
| Middle East | Male | 51 | |
| Female | 41 | 92 | |
| Africa | Male | 72 | |
| Female | 47 | 119 | |
| Asia | Male | 93 | |
| Female | 85 | 178 | |
| Other* | Male | 13 | |
| Female | 10 | 23 |
This category included subjects from Europe, North and South America.
Figure 1.
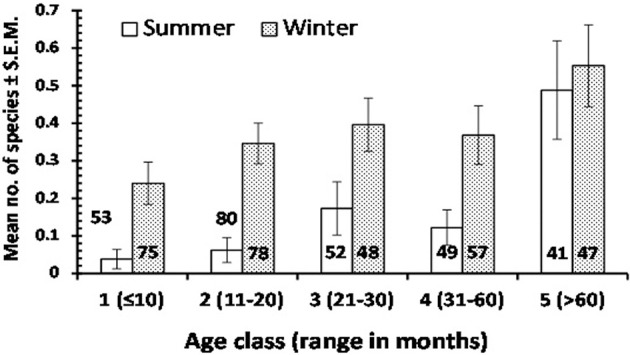
The mean no. of enteric protozoan species detected in children of varying age in summer and winter seasons. The sample size of each subset of data is given on the column or above the column to avoid obscuring the error bars. For statistical analysis see text.
Overall prevalence
The prevalence of infection with each of the detected species is shown in Table 3. Overall, parasitic protozoa were detected in 22.9% of the children, and the most common species was C. parvum. Only two subjects harbored Cryptosporidium meleagridis and only one E. histolytica.
Table 3.
Prevalence of protozoan parasites.
| Species/Taxon | % | 95% CL |
|---|---|---|
| Blastocystis hominis | 4.7 | 3.43–6.26 |
| Giardia duodenalis | 1.7 | 1.05–2.83 |
| Dientamoeba fragilis | 4.0 | 2.85–5.49 |
| Entamoeba histolytica | 0.17 | 0.090–0.690 |
| Cryptosporidium (species combined) | 15.1 | 13.25–18.07 |
| C. parvum | 14.7 | 12.44–17.15 |
| C. hominis | 0.86 | 0.450–1.710 |
| C. meleagridis | 0.34 | 0.180–0.940 |
| All species combined | 22.9 | 20.25–25.85 |
Effect of region of origin
The prevalence of combined protozoan infections varied significantly between regions (REGION × INFECTION, = 19.3, P = 0.001). Highest prevalence was recorded among Qatari children and the lowest among those from Asia (Table 4). An even stronger regional effect was found for C. parvum (REGION × INFECTION, = 40.5, P < 0.001), and again prevalence was highest among the Qataris. However, B. hominis showed similar prevalence among children from each of the five regions, varying only in the range 3.4–5.6%. For D. fragilis prevalence was relatively high among children from Europe and the Americas (Table 4), but there was no significant difference between the regions when tested in models that also took into account SEX, AGE, and SEASON.
Table 4.
Prevalence of enteric protozoa in children, by region of their origin, age, sex, and season.
| Factor | Combined | C.parvum | B. hominis | D. fragilis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean | 95%CL | Mean | 95%CL | Mean | 95%CL | Mean | 95%CL | ||
| REGION | ||||||||||
| Qatar | 168 | 33.9 | 25.98–42.94 | 26.8 | 19.44–35.44 | 4.8 | 2.06–10.23 | 6.0 | 2.86–11.62 | |
| Middle East | 92 | 23.9 | 14.07–37.10 | 19.6 | 10.76–32.45 | 4.4 | 1.00–13.60 | 2.2 | 0.21–10.36 | |
| Africa | 119 | 17.7 | 12.51–24.16 | 10.1 | 6.27–15.52 | 3.4 | 1.43–7.25 | 1.7 | 0.48–5.04 | |
| Asia | 178 | 15.2 | 9.43–22.98 | 4.5 | 1.82–10.08 | 5.6 | 2.56–11.48 | 3.4 | 1.14–8.47 | |
| Other | 23 | 26.1 | 12.03–47.78 | 8.7 | 1.57–27.81 | 4.4 | 0.23–21.25 | 13.0 | 3.66–32.35 | |
| SEX | ||||||||||
| Males | 322 | 19.6 | 15.57–24.30 | 13.7 | 10.29–17.86 | 3.1 | 1.66–5.63 | 4.4 | 2.57–7.15 | |
| Females | 258 | 27.1 | 22.97–31.72 | 15.9 | 12.59–19.79 | 6.6 | 4.5–9.47 | 3.5 | 2.05–5.76 | |
| AGE | ||||||||||
| Age Class 1 | 128 | 14.1 | 9.36–20.35 | 10.2 | 6.22–15.83 | 0.78 | 0.110–3.740 | 1.6 | 0.40–5.03 | |
| Age Class 2 | 158 | 19.6 | 13.48–27.40 | 15.2 | 9.79–22.45 | 0.63 | 0.070–4.010 | 1.9 | 0.46–6.20 | |
| Age Class 3 | 100 | 26.0 | 18.07–35.45 | 18.0 | 11.35–26.91 | 4.0 | 1.38–9.85 | 4.0 | 1.38–9.85 | |
| Age Class 4 | 106 | 22.6 | 17.1–29.20 | 14.2 | 9.74–19.81 | 5.7 | 3.12–9.89 | 1.9 | 0.61–5.10 | |
| Age Class 5 | 88 | 38.6 | 26.73–51.69 | 17.1 | 9.04–28.95 | 17.1 | 9.04–28.95 | 13.6 | 6.83–25.20 | |
| SEASON | ||||||||||
| Summer | 275 | 12.0 | 9.05–15.68 | 4.0 | 2.40–6.47 | 5.1 | 3.28–7.80 | 4.0 | 2.40–6.47 | |
| Winter | 305 | 32.8 | 27.98–38.01 | 24.3 | 19.97–29.11 | 4.3 | 2.54–6.95 | 3.9 | 2.28–6.55 | |
The highest value in each data-subset is in bold.
Effect of host age
There was a highly significant difference in prevalence of combined protozoa across the age classes (AGE CLASS × INFECTION, = 18.8, P = 0.001), with the highest prevalence recorded among the oldest age class (61–174 month old; Table 4). The species with the greatest contribution to this age effect were B. hominis (Table 4; AGE CLASS × INFECTION, = 34.5, P < 0.001) and D. fragilis (AGE CLASS × INFECTION, = 19.7, P = 0.001). In contrast the prevalence of C. parvum varied only in the range from 10.0 to 18.0% (Table 4) among the 5 age classes.
Effect of host sex
Prevalence of combined protozoa was significantly higher among female children (Table 4; SEX × INFECTION, = 4.6, P = 0.032), and this was to some extend attributable to the sex bias arising from B. hominis (SEX × INFECTION, = 3.9, P = 0.048) for which prevalence was twice as high among female compared with male children. There was no evidence of any sex bias for C. parvum or D. fragilis (Table 4).
Effect of season
Prevalence of combined protozoan infection differed markedly and significantly between the seasons (Table 4; SEASON × INFECTION, = 36.9, P < 0.001), being 2.7 × higher in winter months compared with summer. A much greater seasonal effect was found for C. parvum (SEASON × INFECTION, = 5 3.0, P < 0.001) with prevalence in the winter months being 6 × higher than that in the summer (Table 4). However, no seasonal effect was evident for the prevalence of B. hominis or D. fragilis (Table 4).
Other species
The other enteric protozoa recorded were too rare to analyze reliably. G. duodenalis was detected in ten children, five of whom were Asians, two African, two Qatari, and one from the Yemen in the Middle East. Six were females, seven of these infections were detected in the winter season and they were spread across all the age classes except the very youngest. Only one case of E. histolytica was recorded in a 71-month old (Age Class 5) Asian boy in the summer period. C. hominis was recorded in five subjects, three of whom were Asians, four were females, and three cases were in the winter. There were only two cases of C. meleagridis, both among age class 1 boys, both recorded in the winter, one from Africa and the other Qatari.
Species richness
Table 5 summarizes mean values for species richness by each factor taken into consideration. Mean species richness was significantly affected by region of origin of the children (Wald = 11.1, P = 0.026), the highest value being derived from Qatari children and the lowest from African children. Additionally, mean species richness was affected by season (Wald = 22.1, P < 0.001) and by host age (Wald = 29.5, P < 0.001), but there was also a significant 2-way interaction between these latter two factors (SEASON × AGE CLASS on species richness, Wald = 12.7, P = 0.013), which is illustrated in Figure 1. This shows clearly that the age related increase in mean species richness was more rapid in the winter compared to in the summer months.
Table 5.
Mean enteric protozoan species richness of children by nationality, age, sex, and season.
| Factor | Mean | SEM | |
|---|---|---|---|
| Region | Qatar | 0.40 | 0.047 |
| Middle East | 0.27 | 0.056 | |
| Africa | 0.18 | 0.035 | |
| Asia | 0.19 | 0.038 | |
| Other | 0.30 | 0.117 | |
| Age class | 1 | 0.16 | 0.036 |
| 2 | 0.20 | 0.033 | |
| 3 | 0.28 | 0.051 | |
| 4 | 0.26 | 0.048 | |
| 5 | 0.52 | 0.084 | |
| Season | Summer | 0.15 | 0.029 |
| Winter | 0.36 | 0.032 | |
| Sex | Male | 0.24 | 0.03 |
| Female | 0.30 | 0.032 |
The highest value in each data-subset is in bold.
Polyparasitism
Among the 133 children infected with enteric protozoa, concurrent species infections were detected in only 16. The maximum number of species harbored by a single individual was 4 (one Asian boy, aged 71 months and tested in summer months, who was positive for B. hominis, D. fragilis, E. histolytica, and C. hominis). Two children harbored three species and 13 two species. The remaining 117 infected children were positive for only one species. Based on the prevalence of the seven individual species recorded (Table 3) in the entire study group, these numbers are much as predicted (Janovy et al., 1995) in the absence of interactions between species ( = 3.06, P = 0.38; prediction = 438.9, 129.2, 11.5, 0.4, and 0, for uninfected, and those carrying 1, 2, 3, and 4 species, respectively).
However, when focusing on the two dominant species (B. hominis and C. parvum) among the 133 infected children (i.e., positive for at least one of the seven enteric protozoa detected in the study), some evidence for a negative association between these two species was evident, as illustrated in Figure 2. Based on a prevalence of 20.3% for B. hominis among the 133 children infected with at least one protozona species, prevalence among C. parvum infected children was markedly lower than expected, and accordingly higher among those without C. parvum infection (2 × 2 χ2-test, = 35.4, P < 0.0001). In a log-linear model, with AGE, SEX, REGION, and SEASON taken into account, there was also a significant association between prevalence of B. hominis and C. parvum in this subsample of children (INFECTION with B. hominis × INFECTION with C. parvum, = 27.3, P < 0.001), that was independent of any of other factors that were fitted to the model. However, when the whole data-set was analyzed (n = 580), prevalence of B. hominis among subjects with C. parvum [4.7% (1.28–13.59)] and those without [4.6% (2.41–8.46)] was almost identical.
Figure 2.
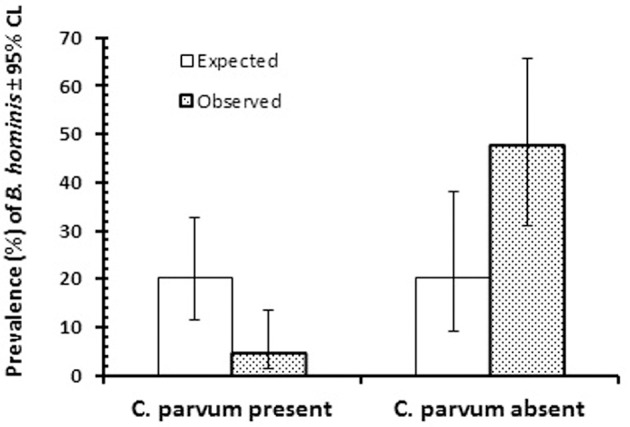
Prevalence of B. hominis among the 133 infected children that were infected with at least one of the enteric protozoa in the study. The figure shows prevalence of B. hominis among those in which C. parvum was also detected (n = 85) or was absent (n = 48). For statistical analysis see text.
Our data also revealed a significant positive relationship between B. hominis and D. fragilis (INFECTION with B. hominis × INFECTION with D. fragilis, = 22.9, P < 0.001) in the whole data-set (n = 580), and this retained significance when we controlled for REGION, AGE, SEX and SEASON ( = 13.2, P < 0.001). Prevalence of D. fragilis was only 2.7% [1.83–4.00] among subjects without B. hominis, but more than 10-fold higher, 29.6 [14.77–50.00] among those with B. hominis. When analysis was confined to the 133 subjects that carried at least one protozoan species, prevalence of D. fragilis was also more than twice higher among B. hominis infected subjects [29.6% (14.77–50.00)] compared to those without B. hominis [14.2% (9.74–19.81)] but in this case the difference was not significant.
Cryptosporidium geno/sub-typing
A total of 90 samples, out of 580 that were positive by RT-PCR, were successfully genotyped. The 18S rRNA PCR-RFLP analysis revealed distinctive banding patterns. The majority of isolates [83/90 (92.22%)], were identified as C. parvum, making this the dominant species identified in the study population. Four samples (4.44%) were positive for C. hominis, one (1.11%) for C. meleagridis, one mixed (1.11%) C. parvum + C. hominis, and one mixed (1.11%) C. parvum + C. meleagridis. The GP60 gene was successfully sequenced in a total of 35 isolates, of which 31/35 (88.6%) were C. parvum, while 4/35 (11.4%) were C. hominis (Figure 3).
Figure 3.

Phylogenetic analysis of some of the C. hominis and C. parvum subtypes using neighbor-joining analysis of the gylcoprotein 60 (gp60) gene. Reference sequences are preceded by indication of the country of origin.
Alignments with reference sequences classified all C. parvum isolates into one family: IId. Further sub-classification led to 10 different subtypes of which the most frequently observed types were A20G1 (n = 22/ KY709197), A17G1 (n = 3; samples Qa342, Qa530, and Qa577/ Acc.num: KY709201), and A18G1 (n = 2; samples Qa522 and Qa 558/Acc.num: KY709200). A unique isolate was identified for each of the following sub-type: A19G2 (sample Qa574/Acc.num: KY709198), A18G2 (sample Qa58/Acc.num: KY709199), A16G1 (sample Qa249/Acc.num: KY709202), and A14G1 (sample Qa482/Acc.num: KY709203). The C. hominis subtype family identified among the four isolates was Ib. Within this subtype family, only two subtypes were identified with subtype IbA9G3 being the most common, present in three cases (samples Qa468, Qa471, and Qa479/Acc.num: KY709204, and KY709205) and subtype IbA10G2 identified in just one case (sample Qa208/Acc.num: KY709206).
Discussion
While a range of different pathogenic organisms can cause pediatric diarrhea in Qatar, including rotaviruses and adenoviruses (Al-Thani et al., 2013) and foodborne bacteria such as, Eschirichia coli (Weam et al., 2016), in the current study we have shown that a significant percentage of cases of pediatric diarrhea (22.9%) is also associated with the presence of intestinal protozoan parasites. The most common among these was C. parvum, accounting for 14.7% of cases.
Cryptosporidiosis is globally a widespread disease with prevalence in humans reported to range between 1.1% in Iran (Tahvildar-Biderouni and Salehi, 2014) to more than 60% (El Shazly et al., 2007; Abdel-Hafeez et al., 2012). Prevalence of B. hominis in humans has been found to vary from 0.5 to 76% (Abu-Madi et al., 2015) and even as high as 100% among Senegalese children (El Safadi et al., 2014) and that of D. fragilis from 0.3 to 89% (Garcia, 2016). In our current work, we observed a prevalence of 4% for D. fragilis which is similar to prevalence values reported from Tunisia 5·5% (Ayadi and Bahri, 1999) and Australia 5% (Stark et al., 2016). Moreover, we also found evidence for a positive association between this species and B. hominis and this is consistent with reports on pediatric populations in Lebanon (Osman et al., 2016), and Portugal (Júlio et al., 2015).
Host intrinsic factors including age and sex have been assessed to identify potential factors that may influence the risk of infection with B. hominis in pediatric patients elsewhere (Nithyamathi et al., 2016) and in agreement with earlier studies, prevalence of infection with this species in our study was also found to increase significantly with host age. An age-effect was also detected for D. fragilis, however, there was no evidence in our data for an age effect in the case of C. parvum. The increase of infection prevalence with host age that we detected in the cases of B. hominis and D. fragilis may be related to changes in breast feeding practices with a reduction in exclusive breastfeeding as the children grow older. Although we did not find a significant increase in prevalence with host age in the case of Cryptosporidium spp., failure to breast feed is considered to be a risk factor for cryptosporidiosis (Aldeyarbi et al., 2016). Indeed mothers provide protective passive immunity against different parasites infection (Abdel-Hafeez et al., 2013; Pedersen et al., 2014), as reflected for example in the high levels of anti-Cryptosporidium IgA in maternal breast milk (Korpe et al., 2013). G. duodenalis infection has also been reported to be significantly less frequent among breastfed children (1.9%) compared with those not on breastfeeding (6.5%) in Egypt as well as among Rwandan children (Tellevik et al., 2016), but in our study the number of cases was too few to allow statistical analysis of any age effect in this species.
Prevalence of B. hominis was found to differ significantly between the sexes being twice as high among female children in contrast to observations elsewhere reporting higher prevalence among male subjects (Nithyamathi et al., 2016; Osman et al., 2016). The contrasting findings suggest that the distribution of intestinal protozoan infections is spatially heterogeneous with respect to host sex, and differences between studies are most likely linked with variation in local environmental conditions in the communities, and as a consequence with physiological properties intrinsic locally to hosts.
Intestinal protozoan infections are considered to be transmitted via the fecal-oral route, contaminated water/food (Anvari-Tafti et al., 2016), close contact with animals, poor personal hygiene and inadequate sanitation, and are facilitated by the environmentally resistant cyst stage of the parasites. Drinking contaminated water, especially surface water, is a known and significant risk factor for B. hominis infection (Abdulsalam et al., 2013). Cryptosporidiosis is also primarily spread by ingestion of contaminated water, and was ranked fifth among the 24 most important food-borne parasites (FAO, 2012). Among gastroenteritis outbreaks reported in England and Wales during the 1990s, Cryptosporidium spp. were identified as the probable cause in 14 outbreaks associated with public water supplies and swimming pools (Insulander et al., 2005; Suppes et al., 2016) as these species are resistant to chlorine at the levels usually used in water treatment (Maçin et al., 2016). The usage of surface water for irrigation can lead also to human infections through the consumption of contaminated fresh raw vegetables (FDA, 2015). Indeed, Cryptosporidium spp. have been detected on Cilantro leaves in Costa Rita and on lettuce in Norway (Sunnotel et al., 2006) and outbreaks of cryptosporidiosis have been observed to be associated with the consumption of fresh pre-cut salad leaves in England (McKerr et al., 2015) and fresh parsley in Sweden (Insulander et al., 2008). Green leafy vegetables such as, lettuce, rocket, and parsley have uneven surfaces and this provides a convenient substrate for the attachment of parasite eggs, cysts, and oocysts, either at the farms where they are grown or subsequently when washed with contaminated water (Said, 2012).
Our analysis has shown that prevalence of combined protozoan infections, and C. parvum in particular, was highest among the Qatari, despite Qatari nationals representing only 12% of the total population in the country (Qatar's Ministry of Development Planning and Statistics; Abu-Madi et al., 2016b). The most plausible routes for infection are via day-care centers (Clavel et al., 1996), swimming pools (Suppes et al., 2016), and food (as the consumption of salads is frequent in the country even in fast food restaurants). As children in Qatar have little contact with farm animals and the country relies on desalinated sea water that is piped to houses and used for drinking, these two potential routes for transmission are unlikely (Nazemalhosseini-Mojarad et al., 2012). A Portuguese study reported that the risk of acquiring this infection was 12 times greater for children who attended school, suggesting probable human-to-human transmission (Júlio et al., 2015). Foreign workers may also play a role in the transmission of intestinal parasites. In Saudi Arabia, pediatric parasitic infections (including Cryptosporidium spp.) have been suggested to be associated with the widespread custom of employing foreign maids who usually have direct contact with children. A high prevalence (46.5%) of intestinal parasites among female Asian housekeepers in that study, suggests that members of families with foreign housekeepers may be at some risk of parasitic infections (Ghenghesh et al., 2012).
As the etiological agents of diarrhoeal diseases occur naturally in both temperate and tropical areas, epidemics arising from the different agents may show specific seasonality. Meteorological conditions are known to have different effects on the transport, spread, reproduction, and persistence of a range of pathogens (Boithias et al., 2016). Concurring with our results, a recent pediatric study in Thailand, found a higher prevalence of intestinal parasites in the cool season compared with the hot season (Tellevik et al., 2016). In neighboring countries, such as, Kuwait, where high temperatures and a very dry atmosphere are typically experienced during the summer, and are followed by a short cool winter as observed in Qatar (Boughattas et al., 2017), prevalence of cryptosporidiosis also shows seasonality with highest prevalence during winter months (Iqbal et al., 2001). There is no regular rainy season in Kuwait or in Qatar, and thus the possibility of water-borne transmission attributable to natural sources of water is minimal. However, most studies relate seasonality of outbreaks of diarrhea positively with relative humidity (Jagai et al., 2009), whereas others report an opposite effect (Kiani et al., 2016) with correlation to water shortage and to greater reliance on less safe water sources (e.g., stored water) at certain times of the year. Interestingly, the period of high prevalence of infection in Kuwait has been reported to correspond to the “winter desert camping” season in the country when a large number of families spend an extended period of time in desert camps supplied with hundreds of overhead water tanks (Iqbal et al., 2011). A Spanish study reported marked seasonal variation in diarrhea, with peak prevalence in winter among 1-to 3-year-olds, which seemed to be related to attendance at child care centers, as the attendance was lowest in summer due to holidays (Clavel et al., 1996). Thus, this lack of consistency among seasonality studies and the absence of a straightforward relationship between ambient temperature, water quality and incidence of diarrhea highlight the complex relationship between hydro-meteorological and ecological factors and the transmission of waterborne diseases.
The distribution of Cryptosporidium spp. in humans is known to vary with geographic region and also with the socioeconomic conditions of the study population. Our findings are consistent with previous reports from other Middle Eastern countries (Nazemalhosseini-Mojarad et al., 2012). In Saudi Arabia, C. parvum was identified in 43 of 53 examined samples (Alyousefi et al., 2013) and in Kuwait C. parvum was the predominant causative agent of cryptosporidiosis in children (Sulaiman et al., 2005; Iqbal et al., 2011). Gp60 subtyping has shown that not all C. parvum infections in humans are of zoonotic transmission (Xiao, 2010). Within each subtype family, one subtype is frequently seen to dominate in specific communities/countries. For example, in industrialized countries most of C. hominis infections are caused by the Ib subtype family (Waldron et al., 2009), which appears to be the most virulent as reflected by the numerous outbreaks with which it has been linked. Worldwide there are two common subtypes within the subtype family Ib: IbA9G3 and IbA10G2. IbA9G3 is commonly seen in Malawi, Kenya, and India, whereas IbA10G2 is commonly seen in South Africa, Botswana, Jamaica, and Peru (Xiao, 2010). Using GenBank BLASTN, a similar IbA9G3 subtype to that identified in our isolates has been observed among children in North India (Gatei et al., 2007) and Bangladesh (Hira et al., 2011). The IbA10G2 subtype was reported to be involved in waterborne outbreaks especially in Sweden (Gherasim et al., 2012) and almost half of the C. hominis outbreaks in USA (Cacciò and Chalmers, 2016). It has been also associated with swimming pool outbreaks in Australia (Ng-Hublin et al., 2015).
Sequence analysis of the GP60 locus in the present study has identified only C. parvum subtype family IId. The source of the IId subtype in humans is not yet clear. It is the dominant C. parvum subtype family in humans in Middle Eastern countries such as, Iran, Syria (Kassouha et al., 2016), and Saudi Arabia (Nazemalhosseini-Mojarad et al., 2012). The most frequent subtype observed in our samples was the IIdA20G1 as identified predominantly in Egypt, Jordan (Helmy et al., 2013; Hijjawi et al., 2016), and among Kuwaiti children (Sulaiman et al., 2005; Iqbal et al., 2011). Recurrently identified in our population, subtype IIdA17G1, was reported to be the cause of an outbreak in Finland in 2012, with more than 250 individuals involved and it was associated with consumption of fries and salads (Cacciò and Chalmers, 2016), whereas subtype IIdA18G1 has been widely identified in Europe (e.g., Czech Republic, Spain) and in the Middle East (e.g., Turkey, Iran, Kuwait, etc.). Less frequently observed, subtype IIdA19G2 has been detected in France as a foodborne etiological agent (GenBank number KT716847.1), IIdA14G1 in Spain (Plutzer and Karanis, 2009), and IIdA16G1 in Egypt and Tunisia (Rahmouni et al., 2014). C. meleagridis, mainly isolated from poultry and birds, has been frequently observed in Africa (Aldeyarbi et al., 2016) as well as within mixed cryptosporidium infections (Ng-Hublin et al., 2013). Although, as stated earlier, Qatari children have limited contact with wildlife, such animal subtypes may infect them indirectly through housemaids who themselves may have been exposed to animals back home and/or through their presence in the food supply via remnants of fertilizers of animal origin on salad crops.
Conclusion
In our previous work, we suggested that transmission of some protozoa does occur in Qatar and our current results support this hypothesis based on the high rate of infection among Qatari subjects. Cryptosporidiosis has been shown to be the most frequent pediatric diarrheic infection. Food borne infective stages and a human-to-human role appear to constitute the most probable routes of transmission. In order to better understand this parasite's circulation among the population of Qatar, further studies targeting housemaids and food handlers are required.
Author contributions
SB carried out the experiments and drafted the manuscript; JB carried out the statistical analysis and participated in the preparation and refinement of the manuscript; KA collected the samples and their background information; AS contributed to the molecular detection; WA contributed in sequencing investigation; AA contributed to the interpretation of the results and participated in refinement of the manuscript; MA conceived the study, collected the data and revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript. The contents of this report are solely the responsibility of the authors and do not necessarily represent the official views of Qatar University and QRNF.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Our grateful thanks go to Sondos Khader and Monira Fouladi for their technical assistance and to the Biomedical Research Center at Qatar University for providing the facilities for this work.
Footnotes
Funding. This study was funded by the Qatar National Research Fund (QRNF) at Qatar Foundation through the National Priorities Research Program (Project No. NPRP8-1556-3-313).
References
- Abdel-Hafeez E. H., Ahmad A. K., Ali B. A., Moslam F. A. (2012). Opportunistic parasites among immunosuppressed children in Minia District, Egypt. Korean J. Parasitol. 50, 57–62. 10.3347/kjp.2012.50.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Hafeez E. H., Belal U. S., Abdellatif M. Z. M., Naoi K., Norose K. (2013). Breast-feeding protects infantile diarrhea caused by intestinal protozoan infections. Korean J. Parasitol. 51, 519–524. 10.3347/kjp.2013.51.5.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdulsalam A. M., Ithoi I., Al-Mekhlafi H. M., Khan A. H., Ahmed A., Surin J., et al. (2013). Prevalence, predictors and clinical significance of Blastocystis sp. in Sebha, Libya. Parasit. Vectors 6:86. 10.1186/1756-3305-6-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Madi M. A., Behnke J. M., Boughattas S., Al-Thani A., Doiphode S. H. (2016a). A decade of intestinal protozoan epidemiology among settled immigrants in Qatar. BMC Infect. Dis. 16:370. 10.1186/s12879-016-1728-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Madi M. A., Behnke J. M., Boughattas S., Al-Thani A., Doiphode S. H., Deshmukh A. (2016b). Helminth infections among long-term-residents and settled immigrants in Qatar in the decade from 2005 to 2014: temporal trends and varying prevalence among subjects from different regional origins. Parasit. Vectors 9:153. 10.1186/s13071-016-1433-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Madi M., Aly M., Behnke J. M., Clark C. G., Balkhy H. (2015). The distribution of Blastocystis subtypes in isolates from Qatar. Parasit. Vectors 8:465. 10.1186/s13071-015-1071-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeyarbi H. M., Abu El-Ezz N. M., Karanis P. (2016). Cryptosporidium and Cryptosporidiosis: the African perspective. Environ. Sci. Pollut. Res. Int. 23, 13811–13821. 10.1007/s11356-016-6746-6 [DOI] [PubMed] [Google Scholar]
- Ali I. K. (2015). Intestinal amebae. Clin. Lab. Med. 35, 393–422. 10.1016/j.cll.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Al-Thani A., Baris M., Al-Lawati N., Al-Dhahry S. (2013). Characterising the aetiology of severe acute gastroenteritis among patients visiting a hospital in Qatar using real-time polymerase chain reaction. BMC Infect. Dis. 13:329. 10.1186/1471-2334-13-329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alyousefi N. A., Mahdy M. A., Lim Y. A., Xiao L., Mahmud R. (2013). First molecular characterization of Cryptosporidium in Yemen. Parasitology 140, 729–734. 10.1017/S0031182012001953 [DOI] [PubMed] [Google Scholar]
- Anvari-Tafti M. H., Eslami G., Teimourzadeh-Baboli A., Ghafourzadeh M. (2016). Food-borne protozoan infection in HIV+/AIDS patients and healthy individuals: a case-control study in Iran. J. Food Qual. Hazards Control 3, 93–96. [Google Scholar]
- Ayadi A., Bahri I. (1999). Dientamoeba fragilis: pathogenic flagellate? Bull. Soc. Pathol. Exot. 92, 299–301. [PubMed] [Google Scholar]
- Barnard C., Gilbert F., McGregor P. (2007). Asking Questions in Biology, 3rd Edn. London: Prentice Hall. [Google Scholar]
- Boithias L., Choisy M., Souliyaseng N., Jourdren M., Quet F., Buisson Y., et al. (2016). Hydrological regime and water shortage as drivers of the seasonal incidence of diarrheal diseases in a topical montane environment. PLoS Negl. Trop. Dis. 10:e0005195 10.1371/journal.pntd.0005195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughattas S., Behnke J., Sharma A., Abu-Madi M. (2017). Seroprevalence of Toxoplasma gondii infection in feral cats in Qatar. BMC Vet. Res. 13:26. 10.1186/s12917-017-0952-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughattas S., Salehi R. (2014). Molecular approaches for detection and identification of foodborne pathogens. J. Food Qual. Hazards Control 1, 1–6. [Google Scholar]
- Cacciò S. M., Chalmers R. M. (2016). Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. 22, 471–480. 10.1016/j.cmi.2016.04.021 [DOI] [PubMed] [Google Scholar]
- CDC (2015). Available online at: https://www.cdc.gov/parasites/giardia/infection-sources.html
- Clavel A., Olivares J. L., Fleta J., Castillo J., Varea M., Ramos F. J., et al. (1996). Seasonality of cryptosporidiosis in children. Eur. J. Clin. Microbiol. Infect. Dis. 15, 77–79. 10.1007/BF01586190 [DOI] [PubMed] [Google Scholar]
- Coupe S., Sarfati C., Hamane S., Derouin F. (2005). Detection of Cryptosporidium and identification to the species level by nested PCR and restriction fragment length polymorphism. J. Clin. Microbiol. 43, 1017–1023. 10.1128/JCM.43.3.1017-1023.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Safadi D., Gaayeb L., Meloni D., Cian A., Poirier P., Wawrzyniak I., et al. (2014). Children of Senegal river basin show the highest prevalence of Blastocystis sp. ever observed worldwide. BMC Infect. Dis. 14:164. 10.1186/1471-2334-14-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Shazly A. M., Soltan D. M., El-Sheikha H. M., Sadek G. S., Morsy A. T. (2007). Correlation of ELISA copro-antigen and oocysts count to the severity of cryptosporidiosis parvum in children. J. Egypt. Soc. Parasitol. 37, 107–120. [PubMed] [Google Scholar]
- FAO (2012). Available online at: http://www.fao.org/food/food-safety-quality/a-z-index/foodborne-parasites/en/
- FDA (2015). Available online at: http://www.fda.gov/Food/FoodScienceResearch/SafePracticesforFoodProcesses/ucm091265.htm
- Garcia L. S. (2016). Dientamoeba fragilis, one of the neglected intestinal Protozoa. J. Clin. Microbiol. 54, 2243–2250. 10.1128/JCM.00400-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei W., Das P., Dutta P., Sen A., Cama V., Lal A. A., et al. (2007). Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7, 197–205. 10.1016/j.meegid.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Ghenghesh K. S., Ghanghish K., El-Mohammady H., Franka E. (2012). Cryptosporidium in countries of the Arab world: the past decade (2002–2011). Libyan J. Med. 7. 10.3402/ljm.v7i0.19852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherasim A., Lebbad M., Insulander M., Decraene V., Kling A., Hjertqvist M., et al. (2012). Two geographically separated food-borne outbreaks in Sweden linked by an unusual Cryptosporidium parvum subtype. Euro Surveill. 17:20318. [DOI] [PubMed] [Google Scholar]
- Gotfred-Rasmussen H., Lund M., Enemark H. L., Erlandsen M., Petersen E. (2016). Comparison of sensitivity and specificity of 4 methods for detection of Giardia duodenalis in feces: immunofluorescence and PCR are superior to microscopy of concentrated iodine-stained samples. Diagn. Microbiol. Infect. Dis. 84, 187–190. 10.1016/j.diagmicrobio.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Haque R., Mondal D., Karim A., Molla I. H., Rahim A., Faruque A. S. G., et al. (2009). Prospective case-control study of the association between common enteric protozoal parasites and diarrhea in Bangladesh. Clin. Infect. Dis. 48, 1191–1197. 10.1086/597580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmy Y. A., Krücken J., Nöckler K., von Samson-Himmelstjerna G., Zessin K. H. (2013). Molecular epidemiology of Cryptosporidium in livestock animals and humans in the Ismailia province of Egypt. Vet. Parasitol. 193, 15–24. 10.1016/j.vetpar.2012.12.015 [DOI] [PubMed] [Google Scholar]
- Hijjawi N., Mukbel R., Yang R., Ryan U. (2016). Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet. Parasitol. 228, 116–120. 10.1016/j.vetpar.2016.08.015 [DOI] [PubMed] [Google Scholar]
- Hira K. G., Mackay M. R., Hempstead A. D., Ahmed S., Karim M. M., O'Connor R. M., et al. (2011). Genetic diversity of Cryptosporidium spp. from Bangladeshi children. J. Clin. Microbiol. 49, 2307–2310. 10.1128/JCM.00164-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insulander M., de Jong B., Svenungsson B. (2008). A food-borne outbreak of cryptosporidiosis among guests and staff at a hotel restaurant in Stockholm county, Sweden, September 2008. Euro Surveill. 13:19071. [PubMed] [Google Scholar]
- Insulander M., Lebbad M., Stenström T. A., Svenungsson B. (2005). An outbreak of cryptosporidiosis associated with exposure to swimming pool water. Scand. J. Infect. Dis. 37, 354–360. 10.1080/00365540410021072 [DOI] [PubMed] [Google Scholar]
- Iqbal J., Hira P. R., Al-Ali F., Philip R. (2001). Cryptosporidiosis in Kuwaiti children: seasonality and endemicity. Clin. Microbiol. Infect. 7, 261–266. 10.1046/j.1198-743x.2001.00254.x [DOI] [PubMed] [Google Scholar]
- Iqbal J., Khalid N., Hira P. R. (2011). Cryptosporidiosis in Kuwaiti children: association of clinical characteristics with Cryptosporidium species and subtypes. J. Med. Microbiol. 60, 647–652. 10.1099/jmm.0.028001-0 [DOI] [PubMed] [Google Scholar]
- Janovy J., Jr., Clopton R. E., Clopton D. A., Snyder S. D., Efting A., Krebs L. (1995). Species density distributions as null models for ecologically significant interactions of parasite species in an assemblage. Ecol. Modell. 77, 189–196. [Google Scholar]
- Jagai J. S., Castronovo D. A., Monchak J., Naumova E. N. (2009). Seasonality of cryptosporidiosis: a meta-analysis approach. Environ. Res. 109, 465–478. 10.1016/j.envres.2009.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Júlio C., Furtado C., Rocha R., Escobar C., Brito M. J., Oleastro M. (2015). Detection of Dientamoeba fragilis in portuguese children with acute gastroenteritis between 2011 and 2013. Parasitology 142, 1398–1403. 10.1016/0304-3800(93)E0087-J [DOI] [PubMed] [Google Scholar]
- Kassouha M., Soukkarieh C., Alkhaled A. (2016). First genotyping of Cryptosporidium spp. in pre-weaned calves, broiler chickens and children in Syria by PCR-RFLP analysis. Vet. Parasitol. 225, 86–90. 10.1016/j.vetpar.2016.06.009 [DOI] [PubMed] [Google Scholar]
- Kiani H., Haghighi A., Rostami A., Azargashb E., Tabaei S. J., Solgi A., et al. (2016). Prevalence, risk factors and symptoms associated to intestinal parasite infections among patients with gastrointestinal disorders in Nahavand, Western Iran. Rev. Inst. Med. Trop. Sao Paulo 58:42. 10.1590/S1678-9946201658042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpe P. S., Liu Y., Siddique A., Kabir M., Ralston K., Ma J. Z., et al. (2013). Breast milk parasite-specific antibodies and protection from amebiasis and cryptosporidiosis in Bangladeshi infants: a prospective cohort study. Clin. Infect. Dis. 56, 988–992. 10.1093/cid/cis1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas L., Dorigo-Zetsma J. W., de Groot C. J., Bouter S., Plötz F. B., van Ewijk B. E. (2014). Detection of intestinal protozoa in paediatric patients with gastrointestinal symptoms by multiplex real-time PCR. Clin. Microbiol. Infect. 20, 545–550. 10.1111/1469-0691.12386 [DOI] [PubMed] [Google Scholar]
- Maçin S., Kaya F., Çaǧdaş D., Hizarcioglu-Gulsen H., Saltik-Temizel I. N., Tezcan İ., et al. (2016). Detection of parasites in children with chronic diarrhea. Pediatr. Int. 58, 531–533. 10.1111/ped.12959 [DOI] [PubMed] [Google Scholar]
- McKerr C., Adak G. K., Nichols G., Gorton R., Chalmers R. M., Kafatos G., et al. (2015). An outbreak of Cryptosporidium parvum across England & Scotland associated with consumption of fresh pre-cut salad leaves, May 2012. PLoS ONE 10:e0125955. 10.1371/journal.pone.0125955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. E., Elwin K., Phot N., Seng C., Mao S., Suy K., et al. (2016). Molecular characterization of Cryptosporidium species and Giardia duodenalis from symptomatic cambodian children. PLoS Negl. Trop. Dis. 10:e0004822. 10.1371/journal.pntd.0004822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazemalhosseini-Mojarad E., Feng Y., Xiao L. (2012). The importance of subtype analysis of Cryptosporidium spp. in epidemiological investigations of human cryptosporidiosis in Iran and other Mideast countries. Gastroenterol. Hepatol. Bed Bench 5, 67–70. [PMC free article] [PubMed] [Google Scholar]
- Ng-Hublin J. S., Combs B., Mackenzie B., Ryan U. (2013). Human cryptosporidiosis diagnosed in Western Australia: a mixed infection with Cryptosporidium meleagridis, the Cryptosporidium mink genotype, and an unknown Cryptosporidium species. J. Clin. Microbiol. 51, 2463–2465. 10.1128/JCM.00424-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng-Hublin J. S., Hargrave D., Combs B., Ryan U. (2015). Investigation of a swimming pool-associated cryptosporidiosis outbreak in the Kimberley region of Western Australia. Epidemiol. Infect. 143, 1037–1041. 10.1017/S095026881400106X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithyamathi K., Chandramathi S., Kumar S. (2016). Predominance of Blastocystis sp. infection among school children in Peninsular Malaysia. PLoS ONE 11:e0136709. 10.1371/journal.pone.0136709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M., El Safadi D., Cian A., Benamrouz S., Nourrisson C., Poirier P., et al. (2016). Prevalence and risk factors for intestinal protozoan infections with Cryptosporidium, Giardia, Blastocystis and Dientamoeba among schoolchildren in Tripoli, Lebanon. PLoS Negl. Trop. Dis. 10:e0004496 10.1371/journal.pntd.0004496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen S. H., Wilkinson A. L., Andreasen A., Warhurst D. C., Kinunghi S. M., Urassa M., et al. (2014). Cryptosporidium prevalence and risk factors among mothers and infants 0 to 6 months in rural and semi-rural Northwest Tanzania: a prospective cohort study. PLoS Negl. Trop. Dis. 8:e3072. 10.1371/journal.pntd.0003072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutzer J., Karanis P. (2009). Genetic polymorphism in Cryptosporidium species: an update. Vet. Parasitol. 165, 187–199. 10.1016/j.vetpar.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Quihui-Cota L., Lugo-Flores C. M., Ponce-Martínez J. A., Morales-Figueroa G. G. (2015). Cryptosporidiosis: a neglected infection and its association with nutritional status in schoolchildren in northwestern Mexico. J. Infect. Dev. Ctries. 9, 878–883. 10.3855/jidc.6751 [DOI] [PubMed] [Google Scholar]
- Rahmouni I., Essid R., Aoun K., Bouratbine A. (2014). Glycoprotein 60 diversity in Cryptosporidium parvum causing human and cattle cryptosporidiosis in the rural region of Northern Tunisia. Am. J. Trop. Med. Hyg. 90, 346–350. 10.4269/ajtmh.13-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf F. J., Sokal R. R. (1995). Statistical Tables. San Francisco, CA: W. H. Freeman and Company. [Google Scholar]
- Said D. E. S. (2012). Detection of parasites in commonly consumed raw vegetables. Alex. Med. J. 48, 345–352. 10.1016/j.ajme.2012.05.005 [DOI] [Google Scholar]
- Stark D., Barratt J., Chan D., Ellis J. T. (2016). Dientamoeba fragilis, the neglected trichomonad of the human bowel. Clin. Microbiol. Rev. 29, 553–580. 10.1128/CMR.00076-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I. M., Hira P. R., Zhou L., Al-Ali F. M., Al-Shelahi F. A., Shweiki H. M., et al. (2005). Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43, 2805–2809. 10.1128/JCM.43.6.2805-2809.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnotel O., Lowery C. J., Moore J. E., Dooley J. S., Xiao L., Millar B. C., et al. (2006). Cryptosporidium. Lett. Appl. Microbiol. 43, 7–16. 10.1111/j.1472-765X.2006.01936.x [DOI] [PubMed] [Google Scholar]
- Suppes L. M., Canales R. A., Gerba C. P., Reynolds K. A. (2016). Cryptosporidium risk from swimming pool exposures. Int. J. Hyg. Environ. Health 219, 915–919. 10.1016/j.ijheh.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Tahvildar-Biderouni F., Salehi N. (2014). Detection of Cryptosporidium infection by modified ziehl-neelsen and PCR methods in children with diarrheal samples in pediatric hospitals in Tehran. Gastroenterol. Hepatol. Bed Bench 7, 125–130. [PMC free article] [PubMed] [Google Scholar]
- Tellevik M. G., Blomberg B., Kommedal Ø., Maselle S. Y., Langeland N., Moyo S. J. (2016). High prevalence of faecal carriage of ESBL-producing Enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS ONE 11:e0168024. 10.1371/journal.pone.0168024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNICEF (2013). Committing to Child Survival: a Promise Renewed—Progress Report 2013. New York, NY: UNICEF; Available online at: http://www.unicef.org/publications/index_70354.html [Google Scholar]
- Waldron L. S., Ferrari B. C., Gillings M. R., Power M. L. (2009). Terminal restriction fragment length polymorphism for identification of Cryptosporidium species in human feces. Appl. Environ. Microbiol. 75, 108–112. 10.1128/AEM.01341-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weam B., Abraham M., Doiphode S., Peters K., Ibrahim E., Sultan A., et al. (2016). Foodborne bacterial pathogens associated with the risk of gastroenteritis in the state of qatar. Int. J. Health Sci. 10, 197–207. [PMC free article] [PubMed] [Google Scholar]
- Xiao L. (2010). Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124, 80–89. 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]


