Abstract
The presence of mouse embryonic stem (ES) cells makes the mouse a powerful model organism for reverse genetics, gene function study through mutagenesis of specific genes. In contrast, forward genetics, identification of mutated genes responsible for specific phenotypes, has an advantage to uncover novel pathways and unknown genes because no a priori assumptions are made about the mutated genes. However, it has been hampered in mice because of the lack of a system in which a large-scale mutagenesis and subsequent isolation of mutated genes can be performed efficiently. Here, we demonstrate the efficient chromosomal transposition of a Tc1/mariner-like transposon, Sleeping Beauty, in mice. This system allows germ-line mutagenesis in vivo and will facilitate certain aspects of phenotype-driven genetic screening in mice.
The mouse has become an important model system for gene function studies. Use of embryonic stem (ES) cells in combination with the gene-targeting method allows the generation of mutant mice with desired alterations of the genome (1). However, the phenotype-driven approach has been limited in mice. A large-scale production of mutant mice by chemical mutagenesis has been reported recently (2, 3). This approach has the advantage of introducing mutations at high frequency and creating a large number of mutant mice with multiple mutations in the short term. However, no efficient method is available to identify the genes responsible for the phenotypic changes.
Transposons are the mobile elements that transpose in the genome and that have been used as unique tools for insertional mutagenesis. In contrast to chemical mutagenesis, the transposon system allows for the efficient isolation of causative genes by using the transposon sequence as a tag for the insertion site. The power of transposon-tagged mutagenesis has been proven in certain organisms such as Drosophila melanogaster (4, 5), Caenorhabditis elegans (6), and plants (7). The P element of Drosophila is among the best known transposons for insertional mutagenesis (4, 5); however, it is not effective for other species, probably because of the requirement of factor(s) produced by Drosophila. On the other hand, Tc1/mariner-like elements appear to be less dependent on host factor(s) for the following reasons. First, they exist in a wide range of species, from unicellular organisms to mammals (6). Second, recombinant transposase mediates transposition in vitro (8, 9). In a mouse culture system, chromosomal transposition was demonstrated for the first time by using Sleeping Beauty (SB), a Tc1/mariner-like transposable element reconstructed from fish in vitro (10), but the efficiency was extremely low (11). As an initial step to use the SB transposon as a tool to create mutant mice, we studied the efficiency of the SB transposition in mice.
Materials and Methods
Plasmid Construction and Generation of Transgenic Mice.
To make the pCX-SB construct, a blunt-ended SacII SB fragment from pSB10 (10) was inserted at the blunt-ended EcoRI site of an enhanced green fluorescent protein (EGFP) expression vector, pCX-EGFP (12), after removal of an EcoRI EGFP fragment. The SalI–BamHI fragment of pCX-SB was gel-purified and injected into fertilized eggs obtained from the mating of male and female mice, [C57BL/6 × C3H (BCF1)] × BCF1, to generate an SB transgenic mouse.
The transposon construct pTransCX-GFP:Neo was created by multiple steps. A 383-bp inverted repeats/direct repeats (IR/DR)-R fragment was isolated from pT/neo (10) by digesting with BamHI, filling in and cutting with EcoRI, and inserted into the HincII–EcoRI site of pBluescript II (Stratagene), resulting in pBS-IR/DR(R). A 363-bp IR/DR-L fragment was excised from pT/neo after digesting with SacI, blunting the end and cutting with BamHI, and inserted into the XbaI–BamHI site of pBS-IR/DR(R) after the XbaI site was filled in, resulting in pBS-IR/DR(R,L). A 1.3-kb SpeI–SacI fragment containing exon 2 and intron 2 of the phosphatidylinositol glycan class a (Piga) gene (13) was inserted at the SpeI–SacI site of pBluescript II to generate pPiga, and a 1.7-kb SalI–EcoRI fragment of pCX-EGFP containing the CAG promoter was inserted at the SalI–EcoRI site of pPiga, resulting in pCX-Piga. The EcoRI EGFP fragment from pCX-EGFP was inserted at the EcoRI site of pCX-Piga to generate pCX-EGFP-Piga. The SalI–SacI fragment of pCX-EGFP-Piga containing CAG promoter, EGFP, and Piga gene fragment was blunt-ended and inserted at the EcoRI–BamHI site of pBS-IR/DR(R,L) located between IR/DR-R and IR/DR-L after both EcoRI and BamHI sites were blunt-ended, resulting in pTransCX-GFP. Finally, a 2.3-kb NotI fragment of PGK-neo cassette flanked by two loxP sites (14) was inserted at the unique NotI site of pTransCX-GFP to generate pTransCX-GFP:Neo. We found that the Piga exon2–intron2 fragment has an unexpected activity of polyA addition signal, although there is no canonical polyA addition signal in this fragment (data not shown). Therefore, this region is designated as “pA” in all of the figures. Two versions of transgenic mice were generated by injecting a 4.5-kb Asp-718 fragment and a full-length SacI fragment into BCF1 × BCF1 fertilized eggs.
Mouse Breeding.
The transgenic lines were established by mating the founder transgenic mice with C57BL/6. The doubly transgenic mice, lTRA-GFP:SB, were obtained by mating the established transgenic mice. The progeny of the male and female doubly transgenic mice were generated by crossing with female ICR and male C57BL/6, respectively.
PCR Analysis.
Transposon excision was detected by PCR using the following primer set: TgTP-2L, 5′-ACACAGGAAACAGCTATGACCATGATTACG-3′ and TgTP-1U, 5′-GACCGCTTCCTCGTGCTTTACGGTATC-3′. Each primer is located outside of the IR/DR-R and IR/DR-L of the pTransCX-GFP:Neo, respectively. PCR was performed by using the HotStarTaq system (Qiagen) under the following conditions: 95°C for 15 min, 50 cycles of 94°C for 1 min, 59°C for 1 min, and 72°C for 1 min, followed by 1 cycle of 72°C for 10 min in the last step. PCR products were directly sequenced by using dye terminators and the ABI373A DNA sequencer (Applied Biosystems).
Genotypes of transgenic mice were determined by using the following primer pairs: EGFP-1U, 5′-CACCCTCGTGACCACCCTGACCTAC-3′ and EGFP-1L, 5′-CTTGATGCCGTTCTTCTGCTTGTCG-3′ for GFP gene; and SB-2U, 5′-TCCTAGAGATGAACGTACTTTGGTG-3′ and SB-1L, 5′-ATCCACATAATTTTCCTTCCTCATG-3′ for SB transgene. PCR condition was same as above except that the annealing temperature was 55°C and the cycle number was 30. The resulting PCR products were 313 bp for GFP gene and 466 bp for SB transgene.
Flanking sequences at the novel integration sites of transposon were amplified by means of ligation-mediated PCR (or splinkerette-PCR) as described (10, 15) and were directly sequenced. Genomic sequences of the wild-type loci corresponding to the novel integration sites in mice 1–8 and 1–9 were determined by PCR amplification. The following primer pairs were designed from the flanking sequences of each integration site: SI1–8L3, 5′-CAGAGAAAAAAATGTTTTACCTGTACTTTTAGG-3′ and SI1–8R3, 5′-AAGCTGTTTGGCCTAATGTTATGAATG-3′ for mouse 1–8; and SI1–9L2, 5′-ATCTTTGTATAGGGAAAGGCCATTGAC-3′ and SI1–9R2, 5′-GATCAAGGGGATACAAATTGGAAAAG-3′ for mouse 1–9. Genomic DNA from wild-type ES cells was used as a template, and the PCR products were directly sequenced.
Chromosome Preparation and Fluorescence in Situ Hybridization (FISH).
Chromosome preparation and FISH were performed as described (16, 17) from mitogen-stimulated splenocyte cultures. The 4.1-kb SalI–Asp-718 fragment of the pTransCX-GFP:Neo (Fig. 1b) subcloned into pBluescript II was used as a probe. FISH images were observed under a Nikon fluorescence microscope using Nikon filter sets B-2A and UV-2A.
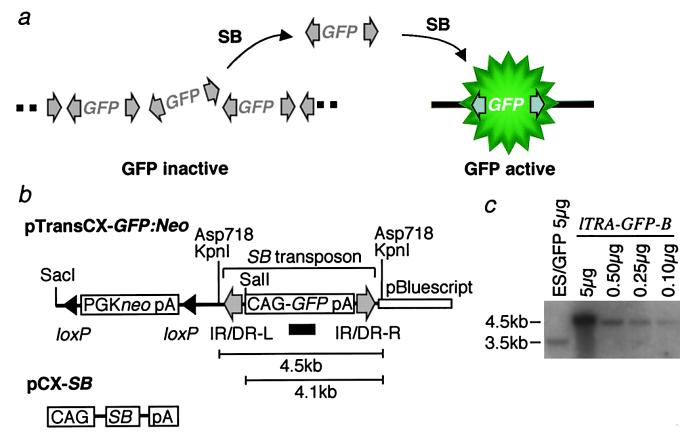
Figure 1.
An overview of the experimental system. (a) Strategy to identify transposition events. The multiply integrated GFP genes flanked by transposon elements (gray arrows) are epigenetically repressed at the original integration site. Excision of transposons by SB transposase releases GFP genes from repression status, and transposition events are detected by GFP activity at the novel site. (b) Constructs used to create transgenic mice. pTransCX-GFP:Neo contains a GFP expression cassette flanked by IR/DR-R and IR/DR-L, constituting an SB transposon element. Two fragments used for generating transgenic mice are described in the text. pCX-SB is the expression vector of SB transposase. Black arrowheads, loxP site; gray arrows, IR/DR-L and IR/DR-R; open bar, pBluescript II; CAG, CAG promoter; pA, polyA addition signal. The GFP probe used for Southern blot analysis in c is shown as a black bar. (c) Estimation of transposon copy number by Southern blot analysis. Genomic DNA from the lTRA-GFP-B transgenic mice was digested by KpnI, and indicated amounts of DNAs were loaded on a gel. As a control for copy number estimation, 5 μg of KpnI–XhoI digested genomic DNA from an ES cell line (ES/GFP) containing a single copy of GFP gene was included. The band sizes are shown on the left side.
Results
Strategy to Identify Transposition.
We developed a system using GFP as a reporter to monitor transposition efficiently. GFP has proven to be a sensitive marker that allows for the detection of gene expression in mice (12). The general strategy is as follows. From the founder mice containing the GFP gene flanked by the IR/DRs of transposon sequences, we select the ones in which GFP expression is repressed (Fig. 1a). These integration sites serve as donor sites for transposition. Transposition releases the GFP gene from its repressed status, and GFP expression is activated at the new locus (Fig. 1a). We generated transgenic mice with two versions of GFP constructs, a short version and a long version (Fig. 1b). The short version is a 4.5-kb Asp-718 fragment that contains only the transposon with the GFP gene driven by a ubiquitous strong promoter, CAG (18), whereas the long version is a SacI-linearized full-length plasmid that includes a PGK-neo cassette flanked by loxP sites and pBluescript vector backbone. The GFP signal was detected in all five transgenic lines generated from the short version of the construct, but it was undetectable in any of the three transgenic mice carrying the long version, probably owing to the presence of an element that suppressed GFP expression in the PGK-neo cassette or the pBluescript vector sequence or both. Two of the latter transgenic lines (lTRA-GFP-A and lTRA-GFP-B mice) were used for the experiment. The initial focus was on the lTRA-GFP-B line, and ≈20 transgene copies were identified in the lTRA-GFP-B mouse by means of Southern blot analysis (Fig. 1c). To mediate transposition, we also established another transgenic line expressing the SB transposase (SB mice) (Fig. 1b). lTRA-GFP-B mice and SB mice were mated to generate lTRA-GFP-B:SB doubly transgenic mice carrying both the SB transposon and transposase.
Detection of Transposon Excision Events.
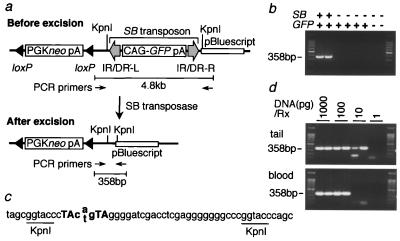
Transposons transpose in a “cut-and-paste” manner. We first examined the “cut” (excision) step by using tail DNA from the mice doubly positive for lTRA-GFP and SB genes. The excision was monitored by means of PCR by using primers located outside of the transposon (Fig. 2a), as excision of the transposon brings the two primers close enough for PCR amplification. Clear bands were detected in DNA from lTRA-GFP-B:SB doubly positive mice but not from lTRA-GFP-B singly positive mice (Fig. 2b), suggesting that SB transposase mediated the excision. Transposon leaves unique “footprints” at their excision sites consisting of a pair of TA dinucleotides spaced by three nucleotides (11). The footprint sequence was identified in the PCR product (Fig. 2c), confirming that SB transposase-mediated excision did occur.
Figure 2.
Transposon excision in lTRA-GFP-B:SB doubly transgenic mice. (a) Structure of vector DNA at the original integration site. PCR primers are located 4.8 kb apart before excision, which is beyond the PCR capacity. Transposon excision by SB transposase brings two primers close enough for PCR amplification. PCR primers are shown by thin arrows. Other symbols and abbreviations are as shown in the legend to Fig. 1. (b) Transposon excision specific for lTRA-GFP-B:SB doubly transgenic mice. Tail DNAs from two mice carrying both transposon and SB transposase gene (GFP+SB+), four mice carrying only transposon (GFP+SB−), and two mice without both transgenes (GFP−SB−) were analyzed. Size marker is a 100-bp ladder. (c) Sequence of the PCR product. Three nucleotides (cag or ctg) and TA dinucleotides repeat shown in bold are the unique footprints typically left at the excision sites (11). Both “a” and “t” were identified in the middle of the footprints by direct sequencing of the PCR product. The rest of the sequence completely matched the vector sequence. (d) High frequency of excision events in lTRA-GFP-B:SB doubly transgenic mice. Genomic DNAs from tail (Upper) and blood (Lower) of lTRA-GFP-B:SB doubly transgenic mice were serially diluted and analyzed for excision events by PCR. Amount of DNAs used per reaction is shown at the top of the panels.
Excision frequency was estimated by means of PCR by using serially diluted DNAs from tail and peripheral blood (Fig. 2d). The minimal amount of DNA per PCR reaction required to detect excision was 10 pg for the tail and 100 pg for the blood. This DNA amount corresponds to 1.5 cells in the tail and 15 cells in the blood, indicating that the excision frequency was as high as 1 in 1.5 cells in the tail and 1 in 15 cells in the blood.
Activation of GFP Expression by Transposition.
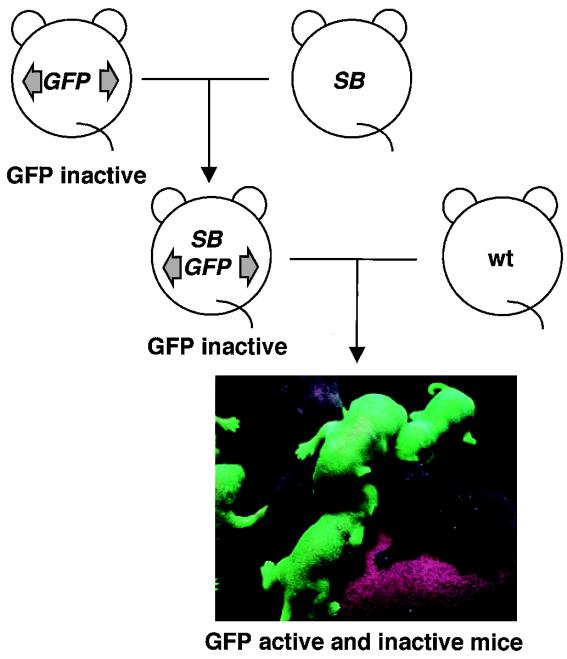
Because the frequency of the “cut” (excision) step was surprisingly high, we expected that the “paste” (integration) step would also be high. In that case, we could anticipate the GFP expression in lTRA-GFP-B:SB doubly positive mice to correspond to the model shown in Fig. 1a. However, no GFP signal was microscopically observed in the tail, nor was any GFP signal detected by FACS analysis of peripheral blood (data not shown), although the excision was definitely confirmed from the same sample as shown in Fig. 2d. We considered that the GFP signal was below the detection level or that repression of the GFP gene was maintained at the novel integration sites. Another possible explanation for this finding is that the “paste” (integration) step is extremely inefficient compared with the “cut” (excision) step. We therefore designed an experiment to detect integration irrespective of GFP expression. We mated the lTRA-GFP-B:SB doubly positive mice with wild-type mice to transmit the locus of the novel integration site to the next generation. Successful transmission would generate mice with a transposon at a specific novel integration site throughout the body and would enable us to detect integration with methods other than GFP, such as Southern blot analysis or ligation-mediated PCR amplification of novel flanking regions (15).
However, the mating unexpectedly resulted in GFP-active mice, that is, mice positive for the GFP signal (Fig. 3). When a male lTRA-GFP-B:SB doubly positive mouse was used for mating, the percentage of GFP-active mice of all of the mice bearing the GFP gene was as high as 63% (Table 1). Similar results were obtained when another line of the male lTRA-GFP mouse (lTRA-GFP-A) was used (Table 1). GFP-active mice were also born from a female lTRA-GFP-B:SB doubly positive mouse, although the frequency was lower than for the progeny from the male lTRA-GFP-B:SB doubly positive mouse (Table 1). The intensity of GFP fluorescence varied widely (data not shown). These results strongly suggest that SB transposase-mediated transposition occurred and transposons landed in different locations on the chromosomes.
Figure 3.
Activation of GFP expression in the progeny from lTRA-GFP:SB doubly transgenic mice. lTRA-GFP:SB doubly transgenic mice were obtained by the mating of lTRA-GFP mice and SB transgenic mice. Neither lTRA-GFP transgenic mice carrying GFP gene only or lTRA-GFP:SB doubly transgenic mice showed GFP signal. However, some of the progeny from lTRA-GFP:SB doubly transgenic mice were clearly GFP positive.
Table 1.
Summary of the mating between ITRA-GFP:SB doubly positive mice and wild-type mice
| Parents
|
Progeny
|
||||
|---|---|---|---|---|---|
| ITRA-GFP:SB | Sex | No. of total mice born | GFP gene-positive | GFP signal-positive | Frequency of GFP-active mice,* % |
| B | M | 61 | 36 | 23 | 63 |
| F | 21 | 13 | 3 | 23 | |
| A | M | 19 | 10 | 8 | 80 |
The number of GFP signal-positive mice was divided by the number of GFP gene-positive mice.
Molecular Characterization of Transposition.
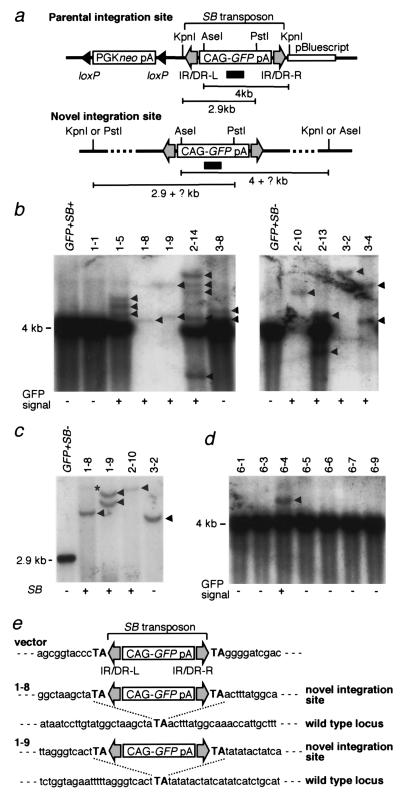
To demonstrate that transposition occurred, we performed Southern blot analysis using tail DNA (Fig. 4). We first analyzed the progeny of the male lTRA-GFP-B:SB doubly positive mouse. Tail DNA of the eight GFP-active mice and the four GFP-inactive mice including the lTRA-GFP-B:SB doubly positive mouse were double-digested with KpnI and AseI and hybridized with the GFP probe (Fig. 4 a and b). Bands longer than 4 kb were expected as a result of these transpositions. Intense 4-kb bands generated by the multiple-integrated transposons at the original integration site were not seen in 1-8, 1-9, 2-10, 3-2, and 3-4, which suggested that the original integration site was lost because of chromosome segregation in these mice. On the other hand, novel bands of more than 4 kb were seen in all of the GFP-active mice, indicating that transposition had occurred. Most of the bands were different in size; this means that transposon had integrated into various chromosomal sites. Mice 2-13 and 2-14 showed bands unexpectedly smaller than 4 kb, and this may have been the result of aberrant patterns of transposition. Mouse 1-1 did not show any novel band corresponding to the absence of GFP fluorescence, but mouse 3-8 did, although no GFP signal could be detected by microscopic observation of the tail or by FACS analysis of peripheral blood (data not shown). We speculate that in this case, the level of GFP expression from the novel integration sites was too weak for detection. To further confirm that the novel bands indeed reflected transposition events, we digested tail DNAs with KpnI and PstI and hybridized them with the GFP probe (Fig. 4c). Bands longer than 2.9 kb were expected from the novel integration sites as shown in Fig. 4a. We chose four mice that lacked the band of the multiple-integrated transposons as shown in Fig. 4b because the strong signal of this band could obscure the novel bands. To obtain clear bands, we loaded 20 μg of digested DNA per lane, which is three times more than that used for the results shown in Fig. 4b. As predicted, bands longer than 2.9 kb were obtained in all of the mice (Fig. 4c), and the number of bands in each mouse was the same as that resulting from KpnI and AseI digestion (Fig. 4 b and c) except the faint band in mouse 1-8 (indicated by an asterisk). PCR was used to examine the transmission of the SB transposase gene from the lTRA-GFP-B:SB doubly positive mouse, and it was found in all of the mice except mouse 3-2 (Fig. 4c). Expression of the SB transposase gene may induce mosaicism of the integration sites because the novel integration sites are continuously exposed to the chance of transposition. If this were the case, we could expect variations in hybridization intensities among the novel bands. The faint band in mouse 1-8 may represent an integration site present in much less than one copy per cell. However, other novel bands were discrete (indicated by arrowheads), and the intensities were similar irrespective of the presence or absence of the SB transposase gene. This result suggests that the mosaicism is not significant if present at all. The mice without the original integration site presented in Fig. 4c are suitable for estimating the number of new germ-line integrations per animal. We excluded the mice containing the original integration site because the transposition events may occur in somatic tissues. There were five new bands in the four animals shown in Fig. 4c. Therefore, the rate of the new germ-line integration events is 1.25 transpositions per GFP-positive mouse. The progeny from the female lTRA-GFP-B:SB doubly positive mouse were also examined (Fig. 4d). The entire first litter containing the GFP gene was analyzed by means of KpnI and AseI digestion. Only one mouse (6-4) showed the GFP signal, and the novel band was detected only in this mouse, consistent with the idea that GFP expression reflects transposition.
Figure 4.
Molecular characterization of transposition. (a) Restriction map at the parental and novel integration sites. Location of GFP probe used in b–d is shown by a black bar. Other symbols and abbreviations are as described in the legend to Fig. 1. (b) Southern blot analysis of the progeny from a male lTRA-GFP-B:SB doubly positive mouse by AseI–KpnI digestion. GFP+SB− is the lTRA-GFP-B mouse carrying the transposon only, GFP+SB+ is the lTRA-GFP-B:SB doubly positive mouse, and the rest of the mice are the progeny from the mating of a male lTRA-GFP-B:SB doubly positive mouse and a wild-type mouse (Fig. 3). Presence or absence of GFP signals is shown under the panel. Location of the strong 4-kb band coming from multiple copies of transposon at the original integration site is shown on the left side. Arrowheads indicate the extra bands. Seven micrograms of DNAs were loaded per lane. (c) Southern blot analysis of the same progeny by PstI–KpnI digestion. Twenty micrograms of DNAs were loaded per lane except in GFP+SB−, in which 3 μg were loaded. Location of the 2.9-kb band resulting from the original integration site is shown on the left side. A faint band in mouse 1-8 is indicated by an asterisk. Presence or absence of the SB transposase gene determined by PCR is shown under the panel. (d) Southern blot analysis of the progeny from a female lTRA-GFP-B:SB doubly positive mouse by AseI–KpnI digestion. Seven micrograms of DNAs were loaded per lane. (e) Flanking sequences at the novel integration sites in mice 1-8 and 1-9. The TA dinucleotides flanking the SB transposon are shown in bold uppercase.
For further evidence of transposition events taking place at the molecular level, flanking regions at the novel integration sites were isolated by means of ligation-mediated PCR (15), and their sequences were determined together with the corresponding wild-type locus (Fig. 4e). Mice 1-8 and 1-9 were used because the absence of the multiple copies of transposons at the original integration site (Fig. 4 b and c) would avoid amplification of the vector sequences and result in specific amplification of the novel integration sites. Although there was a possibility that the potential mosaicism mentioned earlier could interfere with the PCR amplification, flanking sequences could be obtained easily. One flanking region from mouse 1-8 and two from mouse 1-9 were PCR amplified, which corresponded to the number of the discrete novel bands seen in Fig. 4 b and c, and one flanking region for each was characterized as seen in Fig. 4e. The TA dinucleotides were identified at both ends of the transposon sequence, and one copy of these dinucleotides was observed at the corresponding wild-type locus. This is the typical pattern of integration mediated by SB transposase (10, 11) and demonstrates that the transposition in fact did occur in the mice. We also determined the flanking sequences in 15 mice, and none of the sequences were identical (data not shown), indicating that the transposition happened at different chromosomal loci.
Chromosomal Localization of the Transposition Sites by FISH Analysis.
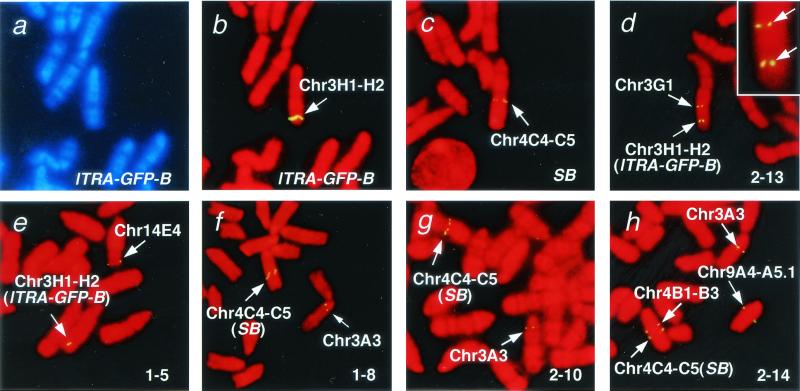
Transposition in the lTRA-GFP-B line was examined by means of FISH (Fig. 5), using the 4.1-kb SalI–Asp-718 region of the pTransCX-GFP:Neo (Fig. 1b) as a probe. The parental integration site of the transposon was mapped at chromosome 3H1-H2 (Fig. 5 a and b). The SB transgene was also detected at chromosome 4C4-C5 (Fig. 5c) because the probe contained the CAG promoter present in pCX-SB (Fig. 1b). We included the CAG promoter region in the probe to maintain the signal required for FISH analysis. The results of the five progeny from the lTRA-GFP-B:SB doubly positive mouse are shown in Fig. 5 d–h. Transmission of either the parental integration site (lTRA-GFP-B, Fig. 5 d and e) or the SB transgene (SB, Fig. 5 f–h) from the lTRA-GFP-B:SB doubly positive mouse was seen in all of the mice, as well as other novel signals representing transpositions that had occurred in these mice. Various patterns of transposition were observed: near the parental integration site in mouse 2-13 (Fig. 5d), at a site distant from the parental site in mice 1-5 (Fig. 5e), 1-8 (Fig. 5f), and 2-10 (Fig. 5g), and on three separate chromosomes in mouse 2-14 (Fig. 5h). The parental integration site was not observed in mouse 2-14 (Fig. 5h), although it was detected with Southern blot analysis (Fig. 4b). We speculate that more than one copy of the transposons was transposed at a time together with the vector backbone sequences, thus generating the 4-kb AseI–KpnI fragment from the novel integration site seen in Southern blot analysis (Fig. 4b). The number of novel signals in mice 2-13 (Fig. 5d), 1-5 (Fig. 5e), and 2-14 (Fig. 5h) was less than that of the novel bands detected with Southern blot analysis (Fig. 4b), probably because some of the novel integration sites could not be detected under the conditions for FISH analysis. In fact, no novel signals could be found in three other mice (data not shown). Another explanation is that the integration occurred near the parental integration site and that the novel signal could not be identified, owing to the limited resolution of FISH analysis.
Figure 5.
Chromosomal localization of the parental integration site (a and b), the SB transposase gene (c), and the novel integration sites in the progeny from the lTRA-GFP-B:SB doubly transgenic mouse (d–h) by FISH analysis. The names of the mice examined are shown at the right bottom corner of each panel, and the chromosomal localizations of the signals are presented by arrows. In d–h, the parental integration site and the SB transposase gene transmitted from their parent are indicated by lTRA-GFP-B and SB in parentheses, respectively. In d, the signals are shown at high magnification at the upper right corner. G-banded and R-banded patterns are demonstrated in a and b–h, respectively.
Results from FISH analysis clearly show that the transposon jumps to various locations of the chromosome and, when combined with the result of Southern blot analysis (Fig. 4), demonstrate the efficient chromosomal transposition by the SB transposon system in mice.
Discussion
Luo et al. tested SB transposition efficiency in ES cells (11). They produced ES cell clones in which the transposon is inserted between the PGK promoter and the puromycin resistance gene to inhibit puromycin expression. Excision of the transposon reconstituted the functional puromycin expression unit, and scoring of the frequency of transposon excision was based on the number of puromycin-resistant cells. They examined both transient and permanent expression of SB transposase and found that excision frequency was at most 3.5 × 10−5 events/cell per generation (11). We conducted an independent test of the transposon in the ES cells. We created an ES cell clone containing a single copy of the transposon and used PCR to measure the excision frequency after transient expression of the SB transposase (data not shown). Excision was detectable only by nested PCR, suggesting that the excision frequency was extremely low, so that our result was in agreement with that reported by Luo et al. (11).
In this study, we found that SB transposition in mice was much more frequent than that reported in cultured cells. Because our transgenic mice contained ≈20 copies of transposons at the original integration site, SB transposase should have a better chance to recognize and transpose them. However, the frequency observed in vivo was much more than the simple sum of that found in the cultured cells multiplied by the number of transposon copies, so that the multiple array of transposons might have a synergistic effect, thus strongly increasing the transposition frequency. Alternatively, there may be specific stage(s) during mouse development advantageous for transposition. The more frequent transposition found in the progeny from the male lTRA-GFP-B:SB doubly positive mouse than in that from the female mouse (Table 1) is consistent with this explanation. We have started to address this issue by using a tetracycline-inducible system that makes it possible for SB transposase to be expressed at different time points in mouse development (19).
Although we obtained GFP-active mice from the mating of lTRA-GFP:SB doubly positive mice and wild-type mice (Fig. 3), GFP expression was not detected in the peripheral blood or tails of lTRA-GFP:SB doubly positive mice despite the high frequency of excision (Fig. 2d). We consider that the following mechanisms may account for this finding. One mechanism involves epigenetic repression of the GFP gene at the novel integration sites. GFP genes at the original integration site are inactive, owing to the epigenetic mechanism(s), such as methylation, conferred by the flanking sequences of the transposon. The transposed GFP gene without the original flanking sequences remains inactive because the repression status is maintained even at the new chromosomal location. However, this condition is eliminated in germ cells, and the transposed GFP gene in the progeny from lTRA-GFP:SB doubly positive mice becomes active. GFP expression may also be affected epigenetically by the genetic background of the mice, in which case GFP expression may not reflect the transposition event. However, this is unlikely for the following reason. Both lTRA-GFP mice and SB mice were generated from BCF1 × BCF1 fertilized eggs, and each line was established by crossing the founder mice with C57BL/6 mice so that the genetic background is a mixture of C57BL/6 and C3H. To minimize the effect of the genetic background on GFP expression, we mated the male lTRA-GFP-A:SB and lTRA-GFP-B:SB doubly positive mice with female BCF1 mice. We obtained GFP-active mice (data not shown), suggesting that the GFP expression may not be affected by the genetic background.
Another possible explanation of the lack of GFP signals in lTRA-GFP:SB doubly positive mice is that extensive mosaicism of the newly integrated transposons occurs among tissues. Mosaicism can be seen when novel DNA sequences are introduced into multicellular organisms. For example, extensive mosaicism has been reported when ecotropic MuLV proviruses were acquired in a mouse germ line (20). In our case, mosaicism of the newly integrated transposons carrying the GFP gene could differ substantially among tissues if transposition efficiency varies from tissue to tissue. Transposition efficiency could be very high in the germ line as indicated by the high frequency of GFP-active mice in the progeny of the lTRA-GFP:SB doubly transgenic mice, but it could be very low in somatic cells. This seems inconsistent with the high frequency of excision in peripheral blood and tail (Fig. 2d) and the finding by Luo et al. (11) that more than half of the excised transposons were shown to have been integrated into the genome in ES cells. However, the possibility remains that the integration in mouse somatic cells is extremely inefficient. This would be similar to the transposase activity recently found in the RAG1 and RAG2 proteins (21, 22). The RAG proteins were previously known to catalyze the V(D)J recombination to generate the diversification of T cell receptors and immunoglobulins. Recent studies have demonstrated that the RAG proteins can excise a DNA segment from a donor DNA and integrate it into a target DNA in the test tube (21, 22). The V(D)J recombination during lymphocyte development represents the excision step of the transpositional reaction, whereas the integration step is rare in vivo if it occurs at all, indicating that the integration step is inefficient in vivo. Future investigations need to determine whether a similar regulation exists in the SB transposon system.
We examined the reactivation of a novel integration site in a GFP-positive mouse bearing a single copy of the transposon and the SB transposase gene. Thirty-one progeny were examined, but no transposition was detected (data not shown). This may reflect low copy number of the transposon at the donor site. However, conditions at the donor site, such as chromatin structure or methylation status, would be quite different from those of the GFP-negative mice, and this may be the cause for the different transposition efficiency values.
Future Direction
Recently, chemical mutagenesis has been used for the large-scale generation of mutant animals (2, 3). The mutation rate obtained with chemical mutagenesis is orders of magnitude higher than the rate of 1.25 transpositions per genome per animal with the transposon system. This high-powered chemical mutagenicity can cover a large number of genes in a short time. Germ-line mutagenesis by infection of replication-competent retroviruses has also been reported (20) with an efficiency of integration similar to that for the transposon system. However, the transposon system has unique features that complement other approaches. First, the transposon system can introduce exogenous sequences as a tag of the mutated genes. This makes it possible to use gene trap schemes, such as the promoter trap (23–25) to study the expression pattern of the mutated genes and the polyA trap (26) to identify the insertion events within the endogenous genes. Second, the SB transposon has been reported to transpose preferentially into linked loci (11). This feature will be useful to introduce mutations with high frequency at specific loci. Mice with chromosomal deletions at various loci have been generated by means of the Cre/loxP system (27) or radiation (28). Transposon-tagged mutagenesis at the wild-type loci allelic to the deleted regions will create null alleles efficiently and facilitate genetic screens for recessive mutations.
The transposon system could be used to induce insertional mutagenesis for cancer genetics if transposition occurs with high frequency in somatic tissues. The frequency of transposon insertion in specific organs would be estimated by comparing the number of cells with and without a new transposon insertion site determined by the ligation-mediated PCR method (15). In fact, we analyzed more than 100 integration sites in testicular cells obtained from doubly transgenic mice and calculated the approximate efficiency of the transposition (K.H., K. Yusa, J. Odajima, and J.T., unpublished observation). The same protocol may be used for examining the rate of transposition in somatic tissues to assess the usefulness of the transposon system for cancer genetics.
In view of this evidence, the transposon system can be expected to open a new avenue for functional genomics in mice.
Acknowledgments
We thank P. B. Hackett for providing pSB10 and pT/neo plasmids, E. Meyer for comments on the manuscript, and Y. Nakano, H. Koike, T. Nakanishi, and S. Takagi for technical assistance.
Abbreviations
- SB
Sleeping Beauty
- ES
embryonic stem
- EGFP
enhanced green fluorescent protein
- IR/DR
inverted repeats/direct repeats
- FISH
fluorescence in situ hybridization
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Capecchi M R. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 2.Nolan P M, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray I C, Vizor L, Brooker D, Whitenhill E, et al. Nat Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 3.de Angelis M H, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, et al. Nat Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 4.Bellen H J, O'Kane C J, Wilson C, Grossniklaus U, Pearson R K, Gehring W J. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 5.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plasterk R H. Curr Top Microbiol Immunol. 1996;204:125–143. doi: 10.1007/978-3-642-79795-8_6. [DOI] [PubMed] [Google Scholar]
- 7.Osborne B I, Baker B. Curr Opin Cell Biol. 1995;7:406–413. doi: 10.1016/0955-0674(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 8.Vos J C, De Baere I, Plasterk R H. Genes Dev. 1996;10:755–761. doi: 10.1101/gad.10.6.755. [DOI] [PubMed] [Google Scholar]
- 9.Lampe D J, Churchill M E, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 10.Ivics Z, Hackett P B, Plasterk R H, Izsvak Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 11.Luo G, Ivics Z, Izsvak Z, Bradley A. Proc Natl Acad Sci USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 13.Kawagoe K, Takeda J, Endo Y, Kinoshita T. Genomics. 1994;23:566–574. doi: 10.1006/geno.1994.1544. [DOI] [PubMed] [Google Scholar]
- 14.Nakano Y, Kondoh G, Kudo T, Imaizumi K, Kato M, Miyazaki J I, Tohyama M, Takeda J, Takeda M. Eur J Neurosci. 1999;11:2577–2581. doi: 10.1046/j.1460-9568.1999.00698.x. [DOI] [PubMed] [Google Scholar]
- 15.Devon R S, Porteous D J, Brookes A J. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi E, Hori T, O'Connell P, Leppert M, White R. Hum Genet. 1990;86:14–16. doi: 10.1007/BF00205165. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda Y, Chapman V M. Electrophoresis. 1995;16:261–272. doi: 10.1002/elps.1150160142. [DOI] [PubMed] [Google Scholar]
- 18.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 19.Shin M K, Levorse J M, Ingram R S, Tilghman S M. Nature (London) 1999;402:496–501. doi: 10.1038/990040. [DOI] [PubMed] [Google Scholar]
- 20.Jenkins N A, Copeland N G. Cell. 1985;43:811–819. doi: 10.1016/0092-8674(85)90254-5. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal A, Eastman Q M, Schatz D G. Nature (London) 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 22.Hiom K, Melek M, Gellert M. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 23.Gossler A, Joyner A L, Rossant J, Skarnes W C. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 24.Skarnes W C, Auerbach B A, Joyner A L. Genes Dev. 1992;6:903–918. doi: 10.1101/gad.6.6.903. [DOI] [PubMed] [Google Scholar]
- 25.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 26.Zambrowicz B P, Friedrich G A, Buxton E C, Lilleberg S L, Person C, Sands A T. Nature (London) 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 27.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 28.You Y, Bergstrom R, Klemm M, Lederman B, Nelson H, Ticknor C, Jaenisch R, Schimenti J. Nat Genet. 1997;15:285–288. doi: 10.1038/ng0397-285. [DOI] [PubMed] [Google Scholar]