Abstract
Background:
One of the gene expression regulatory mechanisms is mediated by small noncoding RNAs called microRNA (miRNA). They interact with a recognition sequence located mostly in 3’-untranslated regions (3’-UTRs) of mRNAs. Polymorphisms in miRNAs recognition sequences could affect gene expression which in turn may alter disease susceptibility. SET8, a member of the SET domain-containing methyltransferase, acts in a variety of biological processes such as genomic stability. Here, we report correlation of rs16917496 polymorphism, located in the recognition sequence of miR-502 within 3’-UTR of SET8, with colorectal cancer (CRC) in Iranians.
Materials and Methods:
One hundred and seventy CRC patients and 170 noncancer counterparts were recruited in this case–control study. Genotyping of rs16917496 was performed using polymerase chain reaction-restriction fragment length polymorphism method.
Results:
There was no significant association of rs16917496 with CRC in population under study (P value for genotype and allele distribution were >0.05). However, stratification analysis based on smoking status revealed that TT+TC genotypes of SET8 rs16917496 are strongly associated with increased risk of CRC (odds ratio: 5.8, 95% confidence interval: 1.37–24.34, P - 0.005) in smoker subgroup.
Conclusion:
Correlation of rs16917496 T allele with CRC in smokers is emphasizing the importance of individuals’ genotype in the recruitment of adverse health hazards of smoking more profoundly for certain people compared to others.
Keywords: Colorectal cancer, microRNA, SET8 gene, single-nucleotide polymorphism
Introduction
Colorectal cancer (CRC) is the second most common cancer in women and the third prevalent one in men in developed countries.[1,2,3] According to the National Institute of Cancer Statistics as of 2013, 45 out of 100,000 people diagnosed with CRC in the world.[4,5] Although the incidence of CRC is lower in Asian than in Western countries, recent studies have shown increasing rates of CRC in developing countries including Iran.[4,6,7]
Genetic syndromes such as lynch and familial adenomatous polyposis, together with positive family history constituting up to 30% of CRC susceptibility; however, common genetic variants present in individuals’ genome considered as the main genetic risk of CRC.[8] Recent genome-wide association studies determined multiple CRC-related single nucleotide polymorphisms (SNPs).[9,10,11,12,13]
MicroRNAs (miRNAs) constitute a novel discovered a class of small noncoding RNAs (about 22 nucleotides long) that play important roles in posttranscriptional gene regulation by binding to the 3’-untranslated regions (3’-UTRs) of specific mRNAs resulting in mRNA cleavage or translational repression.[4,14,15,16] SNPs located in miRNAs-binding sites contribute to the disruption of miRNA recognition elements (MREs) or create new sites which eventually can lead to gene expression dysregulation and disease susceptibility.[2,17,18]
The previous studies have suggested that the SNP rs16917496 (CC genotype), which is located within the miR-502 binding site of SET8 (also known as SETD8 and PR-SET7) 3’UTR, is correlated with reduced SET8 protein levels.[19,20,21] SET8 is known as a histone H4-Lys-20 specific methyltransferase and has been recently reported that modulates p53 activity by particularly monomethylation p53 at lysine 382 (p53K382me1).[22,23] SET8 is involved in vital cellular processes such as transcriptional regulation, genomic stability, heterochromatin formation, DNA replication, and cell cycle arrest.[24,25,26] Therefore, it is proposed that deregulation of SET8 may affect individual's cancer risk. Considering regulatory effects of SET8 on p53 activity, in conditions such as DNA damage, with decreased expression of SET8, the proapoptotic and checkpoint activation functions of p53 would be enhanced consequently. However, CC genotype related to SNP rs16917496 could disrupt this regulation.[22,27,28]
According to the available reports, this polymorphism is associated with breast, ovarian, lymphoblastic leukemia and cervical cancer susceptibility, and clinical outcome of hepatocellular carcinoma (HCC).[19,20,27,28,29,30] However, CRC susceptibility in relation to SNP rs16917496 has not worked out yet.
Here, we report the results of our case–control study on the correlation of SNP rs16917496 with sporadic CRC in the Iranian population. In addition, other factors such as gender, age, body mass index (BMI), physical activity, smoking status, and nonsteroidal anti-inflammatory drugs (NSAIDs) consumption were also analyzed in the present study.
Materials and Methods
Study population
For this case–control study, a total of 340 individuals were selected among patients referred to the colonoscopy centers of the Isfahan University of Medical Sciences Hospitals, from mid-2013 to mid-2015. One hundred and seventy CRC cases were selected on the basis of diagnosis by colonoscopy and then followed their pathology report for the ultimate confirmation of their colonoscopy-based CRC diagnosis. Equal numbers of individuals who underwent colonoscopy procedure but diagnosed with no malignancy or other adverse colonic diseases were constituted our normal control group. All participants filled and signed an informed consent form according to the Isfahan University of Medical Sciences Ethical Committee instructions. To eliminate any established genetic risk factor for CRC, patients were individually interviewed to select cases with sporadic CRC without positive history of familial cancers. In addition, all participants were asked to fill up a structured questionnaire to register the parameters known to influence the CRC susceptibility risk including gender, age, BMI, physical activity, smoking status, and NSAIDs consumption (for NSAIDs consumption any person who use aspirin or nonaspirin NSAIDs occasional or daily for long term [>5 years] were considered as an NSAIDs consumer or regular consumer).
Single nucleotide polymorphism selection
Figure 1 summarizes the course of SNP selection in SET8 gene. As it is evident, SNPs located at 3’UTR of the target gene were considered for this study. Minor allele frequency higher than 0.05 and being located at MREs of miRNAs were the other two criteria considered for SNP selection. SNP rs16917496 was well fitted into our criteria and hence selected for this study.
Figure 1.
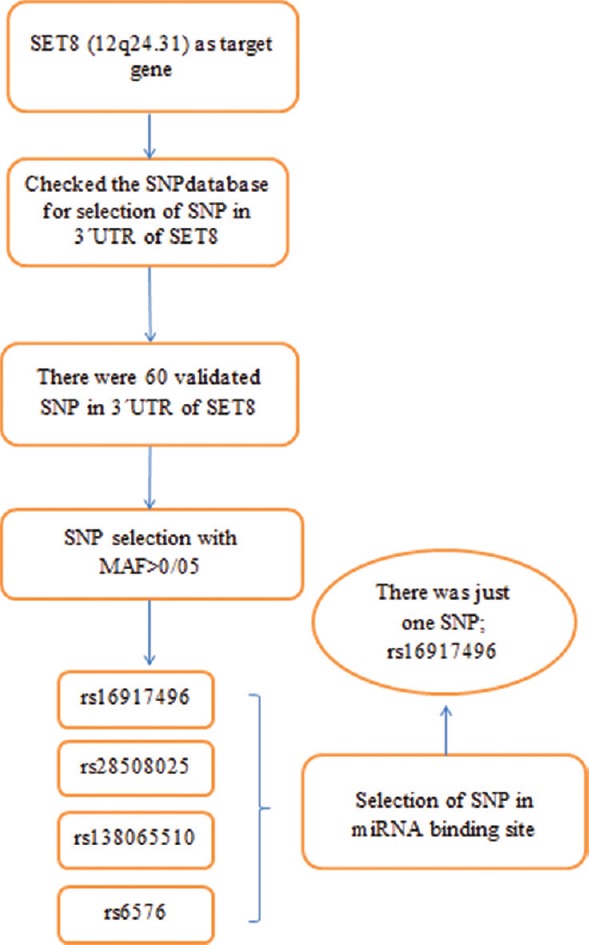
Single nucleotide polymorphism selection algorithm
Single nucleotide polymorphism genotyping
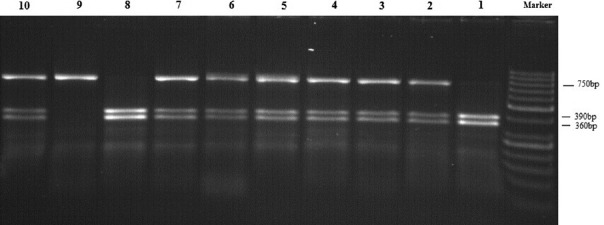
Genomic DNA was extracted from peripheral blood using PrimePrep Genomic DNA Isolation Kit (Genet Bio, Korea). The quality and quantity of the extracted DNA were assessed by agarose gel electrophoresis and spectrophotometry. SNP genotyping performed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method. PCR product size, restriction enzyme used, fragments subsequent to digestion, and primer sequences used are given in Table 1. PCR was performed in a total volume of 25 μl, including 2.5 μl of 10 × PCR buffer, 1 mM MgCl2, 200 μM each dNTPs, 0.2 μM each primer, 100 ng of genomic DNA, and 1.5 U of Taq DNA polymerase (provided from CinnaGen company, Iran). Thermal cycling condition included an initial denaturation of 94°C for 5 min and then 34 cycles of 94°C, 62°C, and 72°C, all for 30 s and a final extension at 72°C for 5 min. RFLP was carried out using SwaI restriction endonuclease enzyme (provided from Thermo Fisher Scientific Company, USA). Subsequently, digested products were differentiated on agarose gel to determine the genotype of each sample. Fragment sizes of 390 and 360 bp were indicative of TT homozygous genotype; a single 750 bp fragment displayed the presence of CC homozygous and three fragments of 390, 360, and 750 bp indicated TC genotype [Figure 2]. Confirmation of the RFLP-based genotyping was done by direct sequencing of randomly selected samples with different genotypes (10% of samples).
Table 1.
Primer sequences and characteristic of polymerase chain reaction product and digestion reaction
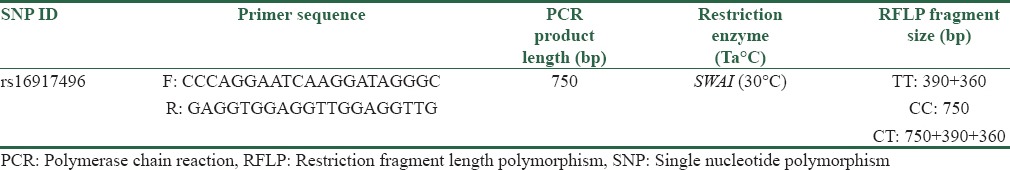
Figure 2.
Agarose gel electrophoresis analysis of restriction fragment length polymorphism-polymerase chain reaction products. Lines 1 and 8 are TT genotype, 2–7 and 10 are TC genotype, and 9 is CC genotype
Statistics
All statistical analyses were performed using SPSS version 22 (SPSS, Chicago, IL, USA). Genotype frequencies in cases and controls were tested for Hardy–Weinberg equilibrium using Chi-square test. Associations between SET8 (rs16917496) polymorphism with susceptibility to sporadic CRC were examined by logistic regression analysis. Age and sex were used as confounders. Correlation between this polymorphism and CRC was evaluated using odds ratios (ORs) and 95% confidence intervals (CIs). The significance level was set at P < 0.05. The difference in demographic and lifestyle characteristics distribution such as age, gender, smoking status, BMI, and NSAIDs consumption assessed by Pearson Chi-square test for categorical variables and t-test for continuous variables. Mann–Whitney test were used to compare physical activity between CRC and control groups.
Results
Demographic and lifestyle characteristics
Our study was intended to evaluate the relationship between rs16917496 SNP located at 3’UTR of SET8 gene and CRC susceptibility in a subset of the Iranian population. In this case–control study, 170 confirmed CRC patients (90 males and 80 females) and 170 (66 males and 104 females) unaffected individuals were selected among those referred to colonoscopy units of our University hospitals. Mean age in case and control groups was 57.72 ± 12.29 and 49.05 ± 13.79 years, respectively. Adjusted data (by age and gender) demonstrated that there was a considerable difference between case and control groups regarding physical activity (P < 0.001). Moreover, we found that healthy subjects in control group had more NSAIDs consumption compared with CRC patients group (P - 0.004). However, there were no statistically significant differences between patients and controls in terms of BMI and smoking status (P - 0.19 and P - 0.23, respectively). Characteristics of the study population are summarized in Table 2.
Table 2.
Baseline characteristics of colorectal cancer patients and controls in study

Genotype and allele distribution
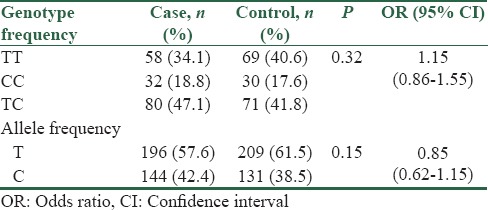
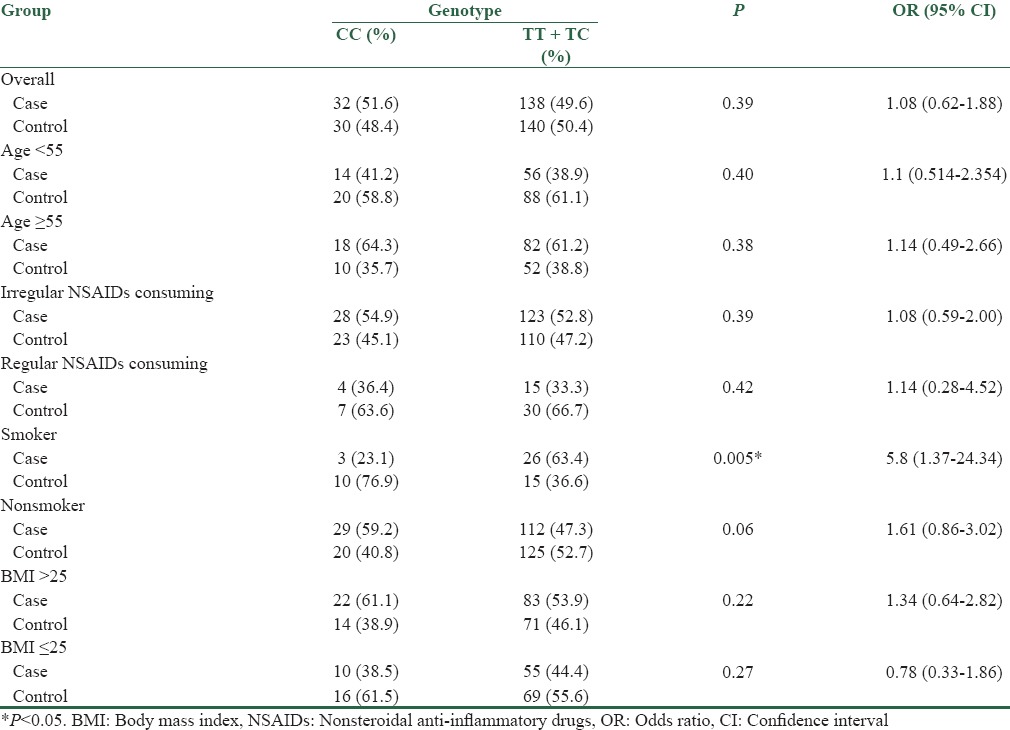
The genotype distribution of rs16917496 polymorphism in cases and controls was in agreement with Hardy–Weinberg equilibrium. The frequency of TT, CC, and TC genotype in control group was 40.6%, 17.6%, and 41.8%, respectively and this frequency in case group was 34.1%, 18.8%, and 47.1%, respectively. In addition, the frequencies of T and C alleles were 61.5% and 38.5% in controls, and 57.6% and 42.4% in cases, respectively. Statistical analysis revealed that no significant correlation exists between SNP rs16917496 and risk of CRC in Iranian population (P value for genotype and allele distribution, respectively: 0.32 and 0.15) [Table 3]. Because C is the ancestral allele for this SNP, we compared other two genotypes against CC genotype. It is determined that TT + TC genotype (variant genotypes) has no either pathologic or protective effect on sporadic CRC (OR: 1.08, 95% CI: 0.62–1.88). In addition, association between these genotypes (TT+TC) compared to CC with CRC risk was assessed in subgroups of participants stratified by age (under 55 and over 55) and BMI (under 25 and over 25 kg/m2). Our data indicated that there was no significant difference in two age groups (OR<55: 1.1, 95% CI: 0.51–2.35 and OR≥55: 1.14, 95% CI: 0.49–2.66) with genotype distribution. In addition, data for BMI stratification demonstrated that there was no correlation between BMI and the target genotypes (OR≤25: 0.78, 95% CI: 0.33–1.86 and OR>25:1.34, 95% CI: 0.64–2.82).
Table 3.
Association between genotypes and allele frequency with colorectal cancer risk
Further analyses were carried out to evaluate the association between TT+TC compared to CC and CRC stratified by NSAIDs consuming and smoking status. Stratification based on irregular and regular NSAID subgroups displayed no correlation between these subgroups and SNP genotypes regarding to CRC risk (ORirregular: 1.08, 95% CI: 0.59–2.00 and ORregular: 1.14, 95% CI: 0.28–4.52). TT+TC genotypes were determined to be associated with increased risk of CRC (OR: 5.8, 95% CI: 1.37–24.34) in smoker subgroup. While in nonsmoker subgroup, no significant correlation was determined (OR: 1.61, 95% CI: 0.86–3.02) [Table 4].
Table 4.
Stratification analysis of rs16917496 genotype frequency in sporadic colorectal cancer and control groups
Discussion
SNPs are the most frequent type of variation in the human genome.[31] SNPs in the miRNA-binding sites at 3’UTR region can modulate miRNA-binding by disrupting existing recognition elements (MREs) or creating new recognition sites.[17,18] Altered mRNA-miRNA interaction would affect the amount of particular gene translation products available in the cell at a given time point. However, the outcome of such deregulation would manifest as disease susceptibility such as malignancies.[14,32,33]
The previous studies demonstrated that SNP rs16917496 (CC genotype and C allele) which is located within the SET8 miR-502 binding site in its 3’UTR, is correlated with reduced SET8 protein levels.[27,28] Here, we report an association between SNP rs16917496 genotypes and sporadic CRC risk in Iranian population. SET8 encodes a histone H4 methyltransferase that is essential for various biological processes, such as transcriptional regulation, heterochromatin formation, DNA replication, cell-cycle arrest, and maintenance of genome integrity.[23,34] SET8 methyltransferase activity modifies p53 protein function by monomethylation of p53 at lysine 382.[22] In cellular stresses including DNA damage, p53-dependent cell cycle arrest or apoptosis activated for which depletion of cellular SET8 is an essential prerequisite as seen that p53K382me1 levels decrease with DNA damage.[22,27] Given, the importance of SET8-P53 interaction in cell-cycle and apoptosis control, deregulation of SET8 under influence of SNP rs16917496 T>C within the miR-502 binding site in the 3’-UTR of the SET8 gene would adversely affect this vital axis in the cell.[20]
Our data are indicative of no significant association between allele and genotype frequency of this polymorphism and sporadic CRC risk in Iranian population. Reviewing previous studies regarding rs16917496 T>C shows contradictory results. In Chinese population, CC and CC+CT genotypes are reported to be associated with longer survival of HCC, nonsmall-cell lung carcinoma (NSCLC) (CC genotype), and SCLC (CC+CT genotype) patients.[20,27,35] In addition, the other study in china indicated that CC genotype is associated with decreased risk of epithelial ovarian cancer and TT genotype is associated with increased risk of NSCLC.[29,36] On the other hand, a few studies revealed that CC genotype was associated with increased risk of breast cancer among the premenopausal women in the Chinese population, and CT+TT genotype is associated with decreased risk of acute lymphoblastic leukemia in comparison with CC genotype in the Iranian population.[19,28] Despite these inconsistent results, all analysis on SET8 expression indicated that C allele is associated with reduced SET8 protein level.[20,27]
Our results revealed no correlation of this polymorphism with sporadic CRC in Iranian population. In stratification analysis, neither CC genotype nor combined TT and TC genotypes, did not alter the risk of CRC among individuals under and over 55 years (P - 0.40 and P - 0.38, respectively), irregular and regular NSAIDs consumers (P - 0.39 and P - 0.42, respectively), and BMI under 25 and over 25 kg/m2 (P - 0.27 and P - 0.22, respectively). However, in the subgroup of smokers, combined TT+TC genotype was associated with increased risk of sporadic CRC. However, there may be other players taking part in SET8 regulatory pathway. For instance, we can mention SCF (β-TRCP) which is marking SET8 for ubiquitination and degradation in a casein kinase I-mediated manner.[37]
Our finding regarding rs16917496 T allele correlation with CRC in smokers compared to nonsmokers, would be an additional proof of cigarette health hazard. However, our findings regarding genotype-smoking interaction highlight this important point that certain genotypes are more readily accommodate adverse health effect of smoking compared to the other genotypes. Finally, it is the first evaluation of SNP rs16917496 located within the SET8 miR-502 binding site in relation with CRC. However, the reasons for the contradictory results may be owing to differences in ethnic background, and the different forms of CRC entities (familial vs. sporadic) and low-sample size that needs to more investigation in other population and more sample size.
Financial support and sponsorship
The study was supported by Isfahan University of Medical Sciences.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We would like to appreciate financial support provided by Isfahan University of Medical Sciences.
References
- 1.Huang SH, Chao Y, Wu YY, Luo JC, Kao CH, Yen SH, et al. Concurrence of UGT1A polymorphism and end-stage renal disease leads to severe toxicities of irinotecan in a patient with metastatic colon cancer. Tumori. 2011;97:243–7. doi: 10.1177/030089161109700221. [DOI] [PubMed] [Google Scholar]
- 2.Chae YS, Kim JG, Lee SJ, Kang BW, Lee YJ, Park JY, et al. A miR-146a polymorphism (rs2910164) predicts risk of and survival from colorectal cancer. Anticancer Res. 2013;33:3233–9. [PubMed] [Google Scholar]
- 3.Saito R, Suzuki H, Yamada T, Endo S, Moriwaki T, Ueno T, et al. Predicting skin toxicity according to EGFR polymorphisms in patients with colorectal cancer receiving antibody against EGFR. Anticancer Res. 2013;33:4995–8. [PubMed] [Google Scholar]
- 4.Bhaumik P, Gopalakrishnan C, Kamaraj B, Purohit R. Single nucleotide polymorphisms in microRNA binding sites: Implications in colorectal cancer. ScientificWorldJournal. 2014;2014:547154. doi: 10.1155/2014/547154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feik E, Baierl A, Hieger B, Führlinger G, Pentz A, Stättner S, et al. Association of IGF1 and IGFBP3 polymorphisms with colorectal polyps and colorectal cancer risk. Cancer Causes Control. 2010;21:91–7. doi: 10.1007/s10552-009-9438-4. [DOI] [PubMed] [Google Scholar]
- 6.Kolahdoozan S, Sadjadi A, Radmard AR, Khademi H. Five common cancers in Iran. Arch Iran Med. 2010;13:143–6. [PubMed] [Google Scholar]
- 7.Hosseini SV, Izadpanah A, Yarmohammadi H. Epidemiological changes in colorectal cancer in Shiraz, Iran: 1980-2000. ANZ J Surg. 2004;74:547–9. doi: 10.1111/j.1445-2197.2004.03064.x. [DOI] [PubMed] [Google Scholar]
- 8.Chang WS, Yueh TC, Tsai CW, Ji HX, Wu CN, Wang SC, et al. Contribution of DNA repair xeroderma pigmentosum group D Genotypes to colorectal cancer risk in Taiwan. Anticancer Res. 2016;36:1657–63. [PubMed] [Google Scholar]
- 9.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 10.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Civan J, Mukherjee S, Patel F, Yang H. Genetic variations in colorectal cancer risk and clinical outcome. World J Gastroenterol. 2014;20:4167–77. doi: 10.3748/wjg.v20.i15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daraei A, Salehi R, Salehi M, Emami MH, Janghorbani M, Mohamadhashem F, et al. Effect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadic colorectal cancer in Iranian population. Med Oncol. 2012;29:1044–9. doi: 10.1007/s12032-011-9980-2. [DOI] [PubMed] [Google Scholar]
- 13.Wu CZ, Ni XJ, Zheng SL, Yang YR, Xia P, Zeng YJ, et al. A fast SSP-PCR method for genotyping the ATP-binding cassette subfamily B member 1 gene C3435T and G2677T polymorphisms in Chinese transplant recipients. Tumori. 2009;95:338–42. doi: 10.1177/030089160909500311. [DOI] [PubMed] [Google Scholar]
- 14.Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, et al. Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis. 2008;29:579–84. doi: 10.1093/carcin/bgm304. [DOI] [PubMed] [Google Scholar]
- 15.Manikandan M, Munirajan AK. Single nucleotide polymorphisms in microRNA binding sites of oncogenes: Implications in cancer and pharmacogenomics. OMICS. 2014;18:142–54. doi: 10.1089/omi.2013.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahangari F, Salehi R, Salehi M, Khanahmad H. A miRNA-binding site single nucleotide polymorphism in the 3’-UTR region of the NOD2 gene is associated with colorectal cancer. Med Oncol. 2014;31:173. doi: 10.1007/s12032-014-0173-7. [DOI] [PubMed] [Google Scholar]
- 17.Vaishnavi V, Manikandan M, Munirajan AK. Mining the 3’UTR of autism-implicated genes for SNPs perturbing microRNA regulation. Genomics Proteomics Bioinformatics. 2014;12:92–104. doi: 10.1016/j.gpb.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Z, Wei S, Ma H, Zhao M, Myers JN, Weber RS, et al. A functional variant at the miR-184 binding site in TNFAIP2 and risk of squamous cell carcinoma of the head and neck. Carcinogenesis. 2011;32:1668–74. doi: 10.1093/carcin/bgr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashemi M, Sheybani-Nasab M, Naderi M, Roodbari F, Taheri M. Association of functional polymorphism at the miR-502-binding site in the 3’ untranslated region of the SETD8 gene with risk of childhood acute lymphoblastic leukemia, a preliminary report. Tumour Biol. 2014;35:10375–9. doi: 10.1007/s13277-014-2359-1. [DOI] [PubMed] [Google Scholar]
- 20.Guo Z, Wu C, Wang X, Wang C, Zhang R, Shan B. A polymorphism at the miR-502 binding site in the 3’-untranslated region of the histone methyltransferase SET8 is associated with hepatocellular carcinoma outcome. Int J Cancer. 2012;131:1318–22. doi: 10.1002/ijc.27352. [DOI] [PubMed] [Google Scholar]
- 21.Diao L, Su H, Wei G, Li T, Gao Y, Zhao G, et al. Prognostic value of microRNA 502 binding site SNP in the 3’-untranslated region of the SET8 gene in patients with non-Hodgkin's lymphoma. Tumori. 2014;100:553–8. doi: 10.1700/1660.18180. [DOI] [PubMed] [Google Scholar]
- 22.Shi X, Kachirskaia I, Yamaguchi H, West LE, Wen H, Wang EW, et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell. 2007;27:636–46. doi: 10.1016/j.molcel.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–99. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 24.Julien E, Herr W. A switch in mitotic histone H4 lysine 20 methylation status is linked to M phase defects upon loss of HCF-1. Mol Cell. 2004;14:713–25. doi: 10.1016/j.molcel.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–93. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 26.Jørgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–45. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu J, Yin Z, Gao W, Liu L, Yin Y, Liu P, et al. Genetic variation in a microRNA-502 minding site in SET8 gene confers clinical outcome of non-small cell lung cancer in a Chinese population. PLoS One. 2013;8:e77024. doi: 10.1371/journal.pone.0077024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song F, Zheng H, Liu B, Wei S, Dai H, Zhang L, et al. An miR-502-binding site single-nucleotide polymorphism in the 3’-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;15:6292–300. doi: 10.1158/1078-0432.CCR-09-0826. [DOI] [PubMed] [Google Scholar]
- 29.Yang S, Guo H, Wei B, Zhu S, Cai Y, Jiang P, et al. Association of miR-502-binding site single nucleotide polymorphism in the 3’-untranslated region of SET8 and TP53 codon 72 polymorphism with non-small cell lung cancer in Chinese population. Acta Biochim Biophys Sin (Shanghai) 2014;46:149–54. doi: 10.1093/abbs/gmt138. [DOI] [PubMed] [Google Scholar]
- 30.Yang S, Jiang P, Tang J. Association of miR-502-binding site single nucleotide polymorphism of SET8 gene with non-small cell lung cancer risk in Chinese people. J Bionanoscience. 2013;7:585–9. [Google Scholar]
- 31.Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res. 2007;35:4535–41. doi: 10.1093/nar/gkm480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi Y, Wang L, Zong L, Pei M, Lu Q, Huang P. Genetic variants in microRNA target sites of 37 selected cancer-related genes and the risk of cervical cancer. PLoS One. 2014;9:e86061. doi: 10.1371/journal.pone.0086061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naccarati A, Pardini B, Stefano L, Landi D, Slyskova J, Novotny J, et al. Polymorphisms in miRNA-binding sites of nucleotide excision repair genes and colorectal cancer risk. Carcinogenesis. 2012;33:1346–51. doi: 10.1093/carcin/bgs172. [DOI] [PubMed] [Google Scholar]
- 34.Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–13. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 35.Ding C, Li R, Peng J, Li S, Guo Z. A polymorphism at the miR-502 binding site in the 3’ untranslated region of the SET8 gene is associated with the outcome of small-cell lung cancer. Exp Ther Med. 2012;3:689–92. doi: 10.3892/etm.2012.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, Guo Z, Wu C, Li Y, Kang S. A polymorphism at the miR-502 binding site in the 3’ untranslated region of the SET8 gene is associated with the risk of epithelial ovarian cancer. Cancer Genet. 2012;205:373–6. doi: 10.1016/j.cancergen.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Dai X, Zhong J, Inuzuka H, Wan L, Li X, et al. SCF (ß-TRCP) promotes cell growth by targeting PR-Set7/Set8 for degradation. Nat Commun. 2015;6:10185. doi: 10.1038/ncomms10185. [DOI] [PMC free article] [PubMed] [Google Scholar]