Abstract
The use of Cre/loxP recombination in mammalian cells has expanded rapidly. We describe here that Cre expression in cultured mammalian cells may result in a markedly reduced proliferation and that this effect is dependent on the endonuclease activity of Cre. Chromosome analysis after Cre expression revealed numerous chromosomal aberrations and an increased number of sister chromatid exchanges. Titration experiments in mouse embryo fibroblasts with a ligand-regulatable Cre-ERT show that toxicity is dependent on the level of Cre activity. Prolonged, low levels of Cre activity permit recombination without concomitant toxicity. This urges for a careful titration of Cre activity in conditional gene modification in mammalian cells.
Keywords: genotoxicity, conditional knockout
The bacteriophage P1 Cre recombinase is a member of the phage λ integrase family of site-specific recombinases, which share a similar mechanism of action (1–3). The Cre DNA recombinase can catalyze the excision of a “floxed” DNA segment, i.e., a region flanked by 34-bp loxP elements, placed in direct orientation (4). For recombination, Cre does not depend on additional host factors (2, 5). Two Cre monomers bind to each recombination site and synapse the DNA through protein–protein interactions (6). During cleavage, Tyr324 becomes covalently linked to the 3′ phosphate (7, 8). The released 5′ hydroxyl groups attack the phosphotyrosines of the partner substrates, resulting in a Holliday intermediate. A second round of cleavage and strand exchange completes the recombination reaction (4, 7–10).
The dyad-symmetric loxP site is composed of two 13-bp inverted repeats, the Cre-binding sites, separated by an 8-bp spacer region (3, 11, 12). Cre requires as few as 8–10 matches in each 13-bp binding domain for efficient recombination (13, 14). Mutation analysis has revealed that for efficient recombination between loxP sites, homology of the spacer region is required (12). Recombination between a mutant loxP site, containing a deletion in the spacer region, and a wild-type loxP site in vitro with purified Cre leads to the formation of single- and double-strand breaks (15).
Cre-mediated, site-specific recombination has become an important tool for modifying DNA in mammalian systems. In conditional mouse models the Cre/loxP system can be used to inactivate genes in a tissue-specific and time-controlled fashion (reviewed in refs. 16–19). It is used mostly to generate small deletions (20) but also can be used to generate deletions up to several centimorgans (21) and to induce unequal sister chromatid exchanges in cultured cells and mice (21, 22). An important concern is the presence of secondary recombination sites. Earlier studies have shown that Cre recombinase is capable of catalyzing recombination between cryptic loxP sites naturally present in Escherichia coli and yeast (23–25). Cre expression in yeast has been shown to induce mitotic crossovers at these cryptic loxP sites (25). The recent discovery that pseudo-loxP sites are also present in the mammalian genome and that these sequences can serve as substrates for Cre recombinase in human cells may compromise the usefulness of Cre/loxP technology in mammalian cells (26). It has been shown that Cre expression in developing spermatids can lead to male sterility in mice and induces gross chromosomal rearrangements in spermatozoa, possibly because of Cre-induced recombination between cryptic loxP sites (27). These observations obligate a precise determination of the effects of Cre expression in mammalian cells. Here, we report on the effects of Cre recombinase in cultured mammalian cells in the absence of introduced loxP sites and describe a detailed analysis of the genomic damage induced by the Cre recombinase.
Materials and Methods
Cell Culture.
Heads and organs of embryonic day 13.5 embryos were removed, and the remaining fetal tissue was minced, rinsed in PBS, and incubated for 1 h in 0.5 ml of 0.1% trypsin/EDTA solution at 37°C. Subsequently, the cell aggregates were dissociated in DMEM supplemented with 10% FBS (GIBCO) and 1% penicillin-streptomycin (GIBCO). After removal of large clumps, the cells were plated in a 175-cm2 flask. After 2–3 days, confluent cultures were frozen down and considered as being passage 1. For growth curves, passage 1 cells were counted by using a hemocytometer. Cells (2 × 105) were plated into 10-cm2 plates. At various time points, cells were trypsinized and samples were counted by using a hemocytometer. All cell culture experiments were repeated at least three times, and representative curves are shown for each experiment.
Genotyping and Southern Blot Analysis.
Isolation of DNA from embryonic tissues and cultured cells was performed as described (28). Genotyping of mouse embryos by PCR, and Southern blot analysis of the floxed Brca2 allele were performed as described (29).
Retroviral Constructs.
cDNAs encoding Cre, Cre-ERT, or Cre(R173K)-ERT were cloned into the unique EcoRI site of a murine leukemia virus-based, replication-defective retroviral vector, containing an internal ribosomal entry site (IRES) and the enhanced green fluorescent protein (GFP) reporter gene (LZRS-IRES-GFP) (30). Ecotropic Phoenix cells were transfected with retroviral vectors by the calcium phosphate method to produce infectious virus. For infection, mouse embryo fibroblasts (MEFs) were incubated with the retroviral supernatant in the presence of 4 μg⋅ml−1 polybrene. After 5 h, the viral supernatant was replaced by complete medium. Percentages of transduced cells were determined on a FACScalibur flow cytometer (Becton Dickinson) by using cell quest software.
Western Blot Analysis.
For protein analysis, cells were lysed on ice in RIPA buffer (150 mM NaCl/1% Nonidet P-40/0.1% SDS/0.5% deoxycholate/50 mM Tris⋅HCl, pH 8.0/2 mM EDTA, pH 8.0/0.2 mM PMSF/0.5 mM DTT) containing protease inhibitors. Lysates were assayed for protein concentration. Equal amounts (30 μg) of protein were separated on 12.5% SDS/PAGE and transferred to nitrocellulose. Western blot analysis was performed by using a 1:10,000 diluted anti-Cre rabbit polyclonal antibody (Novagen) according to standard methods. Visualization was performed by using enhanced chemiluminescence (Amersham Pharmacia).
Propidium Iodide Stainings.
For measurement of DNA content, MEFs were seeded at a density of 2 × 105 cells per 10-cm2 plate and cultured for 48 h. Cells were collected, fixed, stained with propidium iodide, and analyzed by flow cytometry as described (31). Cell cycle profiles were analyzed by using modfit software.
Cytogenetics.
ROSA26 (R26)cre-ERT MEFs and wild-type (wt) MEFs were cultured in the presence or absence of 1 μM 4-hydroxytamoxifen (OHT) for 48 h. Cells were treated with colcemid (0.1 μg⋅ml−1) for 2 h, trypsinized, and pelleted at 1,000 rpm for 10 min. After hypotonic swelling in 0.075 mM KCl for 10 min at room temperature, cells were fixed in three changes of 3:1 methanol/glacial acetic acid. Cells resuspended in a drop of fixative were dropped onto humidified, clean, glass slides. Slides were stained in 3% Giemsa (BDH) in PBS. Individual metaphases were photographed. Photographs were scored double-blind for the presence of chromosomal aberrations.
Spectral Karyotype (SKY).
Metaphase preparations of R26cre-ERT and wt MEFs either untreated or treated with 1 μM OHT for 48 h were hybridized by using the mouse SkyPaint probe mixture (Applied Spectral Imaging, Migdal Ha'Emek, Israel) according to the manufacturer's instructions and mounted in 4′,6-diamidino-2-phenylindole/antifade solution. Using the SpectraCube 200 system and the skyview analysis software (Applied Spectral Imaging), 8–17 metaphases from each MEF culture were examined.
Sister Chromatid Exchanges (SCEs).
SCE analysis was performed in R26cre-ERT and wt MEFs either untreated or treated with 1 μM OHT for 48 h, according to standard procedures. At least 40 diploid metaphases per cell culture were analyzed for both the number of SCEs and chromosomal abnormalities.
Results
Cre Recombinase Inhibits Growth.
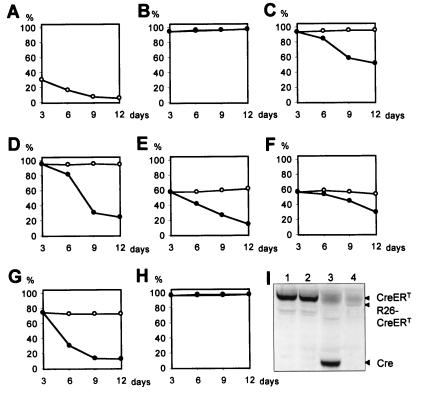
To express the bacteriophage P1 Cre recombinase in mammalian cells in culture, we constructed a bicistronic retroviral vector encoding Cre and a GFP. Expression of GFP allows the rapid recognition of Cre-expressing cells after retroviral infection. We infected primary MEFs with the virus and determined the percentage of GFP-positive cells. During continued passaging, we observed a reduction in the percentage of GFP-positive cells in the population (Fig. 1A). Southern blot analysis showed that this reduction was due to loss of retrovirally transduced cells in the population and, thus, to loss of Cre-expressing cells (not shown). A cell population infected with a retrovirus expressing GFP alone showed normal growth characteristics and no selection for uninfected cells, indicating that the reduced proliferation can be ascribed to Cre and not to other factors (Fig. 1B). Similar results were obtained when MEFs were infected with a bicistronic retroviral vector encoding GFP and Cre-ERT, a OHT-regulatable form of Cre (32, 33), and, subsequently, cultured in the presence of OHT (Fig. 1C). Expression of Cre-ERT in various established cell lines of human, monkey, and mouse origin resulted in an impaired growth upon culturing in the presence of OHT, indicating that the growth defect is not restricted to MEFs (Fig. 1 D–G). Our results are in agreement with the toxic effects observed by others in cells expressing a Cre-GFP fusion protein (34, 35).
Figure 1.
Repressive effects of Cre endonuclease activity on proliferation of mammalian cells. (A) Selective growth repression of MEFs infected with bicistronic retroviruses expressing Cre linked to GFP. Every 72 h, cells were enumerated before replating and analyzed by flow cytometry. (B) Growth rates of MEFs expressing GFP alone are comparable to wt cells. Primary MEFs (C), NIH 3T3 (D), COS-7 (E), HeLa (F), and U2OS (G) cells infected with bicistronic retroviruses expressing Cre-ERT and GFP show a reduced proliferation upon culturing in the presence of 1 μM OHT. No difference in proliferation is observed between wt cells and infected cells in the absence of OHT. (H) MEFs infected with retroviruses encoding an endonuclease-deficient Cre-ERT fusion protein do not suffer from a Cre-induced proliferation defect when cultured in the presence of OHT. The curves representing growth in the presence and absence of OHT are indistinguishable. For B–H, the data points are as follows: ○, cultured without OHT; ●, cultured in presence of 1 μM OHT. (I) Western blot analysis of expression levels of Cre and Cre fusion proteins in MEFs infected with viruses encoding Cre-ERT (lane 1), Cre(R173K)-ERT (lane 2), and Cre (lane 3), and in R26cre-ERT MEFs (lane 4). The R26cre-ERT fusion protein is slightly shorter than the retrovirally encoded Cre-ERT and Cre(R173K)-ERT fusion proteins because of a 21-aa deletion in domain D of ERT (29).
An Endonuclease-Deficient Cre Does Not Influence Proliferation.
To determine whether the growth-inhibiting effect of Cre expression was a result of Cre-mediated DNA modification, we tested a mutant version of Cre for its effect on proliferation. This Cre mutant contains a single amino acid substitution (R173K), which results in loss of the endonuclease activity but does not affect other properties of the Cre protein (36). We replaced a part of the Cre-ERT-encoding retrovirus with the mutation containing part of Cre(R173K). Expression of the Cre(R173K)-ERT protein in MEFs after retroviral infection was confirmed by Western blotting and found to be comparable to retroviral expression of Cre and Cre-ERT (Fig. 1I). MEFs infected with the Cre(R173K)-ERT retrovirus and cultured in the presence of 1 μM OHT did not show reduced proliferation (Fig. 1H). These results demonstrate that strand cleavage activity of Cre is required for the observed proliferation arrest, indicating that the Cre toxicity is inherent to its recombinase activity.
Low Level of Prolonged Cre Expression Induces Recombination Without Growth Inhibition.
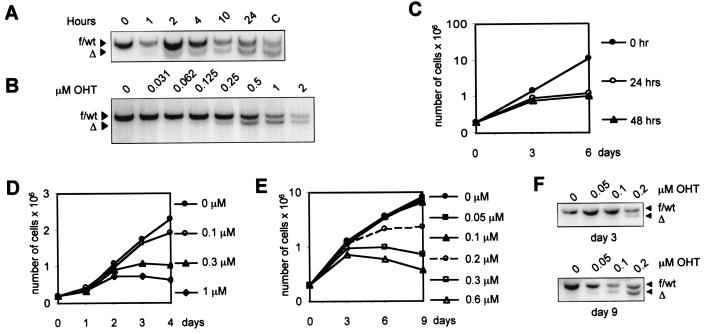
Because Western blot analysis revealed high protein levels of Cre in MEFs infected with Cre-encoding retroviral vectors, we decided to test the effects of moderate Cre expression on MEFs. We therefore isolated MEFs from embryos with a Cre-ERT knock-in allele under control of the endogenous ROSA26 promoter (R26cre-ERT) (29). Western blot analysis showed very low levels of Cre-ERT protein in the R26cre-ERT MEFs, compared with MEFs infected with the Cre-ERT retrovirus (Fig. 1I). R26cre-ERT MEFs cultured in the absence of OHT displayed growth rates comparable to MEFs isolated from wild-type littermate embryos. After growth in the presence of 1 μM OHT during 24 h and a subsequent period of 24 h to allow for clearance of the OHT-bound Cre-ERT protein, ≈90% of the MEFs had undergone recombination of a floxed target gene [in this case, a floxed exon 11 allele of Brca2 (29) (Fig. 2A)]. Lower OHT concentrations or culturing for a shorter time in the presence of 1 μM OHT resulted in a markedly reduced recombination efficiency (Fig. 2 A and B). However, culturing R26cre-ERT cells lacking exogenous loxP sites in the presence of 1 μM OHT for 24 or 48 h also resulted in severely reduced proliferation (Fig. 2C). A clear, inverse correlation could be found between the OHT concentration in the culturing medium and the proliferation rate (Fig. 2D). Treatment of a confluent population of contact-inhibited R26cre-ERT MEFs with 0.6 μM OHT did not affect proliferation after removal of OHT and subsequent passaging of the cells. Also, under these conditions, no Cre-mediated recombination of the floxed Brca2 allele occurred. This suggests that both Cre-mediated growth arrest and recombination can occur only in proliferating cells (data not shown).
Figure 2.
Characteristics of Cre-mediated recombination and growth inhibition in R26cre-ERT MEFs. (A and B) Time and dose dependence of Cre-ERT-mediated recombination. (A) Southern blot analysis of DNA from Brca2F11F/wt; R26cre-ERT MEFs that were treated with 1 μM OHT for the indicated periods of time. Control DNA from Brca2Δ/wt cells is included (lane C). (B) Southern blot analysis of DNA from Brca2F11F/wt; R26cre-ERT MEFs that were treated with various concentrations of OHT for 24 h. After OHT treatment, cells were cultured for another 24 h to allow for clearance of OHT-bound Cre-ERT. Hybridization signals indicate the floxed exon 11 or wt alleles (f/wt) or the deleted exon 11 allele (Δ) of Brca2. (C) Growth inhibition in R26cre-ERT MEFs after treatment with 1 μM OHT. Cells were plated at 2 × 105 per 10-cm2 dish and cultured in medium with 1 μM OHT for 0 h (●), 24 h (○), or 48 h (▴). On day 3 of the experiment, cells were counted, replated at initial densities, and cultured for another 3 days. The growth curves represent cumulative cell numbers. (D) Kinetic analysis for growth of R26 Cre-ERT MEFs in the presence of 0 μM (●), 0.1 μM (○), 0.3 μM (▴), or 1 μM (■) of OHT. (E and F) Prolonged, low-level expression of Cre results in detectable recombination without growth inhibition. (E) Growth curves of Brca2F11F/wt; R26cre-ERT MEFs cultured in the presence of 0 μM (●), 0.05 μM (■), 0.1 μM (▴), 0.2 μM (○), 0.3 μM (□), or 0.6 μM (▵) of OHT. Cells were plated at 2 × 1055 per 10-cm2 dish, and every 3 days cells were enumerated before replating. The growth curves represent cumulative cell numbers. (F) After 3 and 9 days of OHT treatment, DNA from the cell cultures described in E was used to detect deletion of the floxed Brca2 exon 11 by Southern blotting. OHT concentrations are indicated above the panes; recombination percentages, below.
Using various OHT concentrations and different OHT exposure times, we observed that the recombinase activity of R26cre-ERT is both time- and dose-dependent (Fig. 2 A and B). Conceivably, if the toxicity associated with Cre expression would be solely dose-dependent, prolonged culturing of conditional R26cre-ERT MEFs in the presence of low concentrations of OHT might result in recombination without toxicity. To test this, we measured proliferation of R26cre-ERT MEFs during prolonged culture in medium with various concentrations of OHT. Whereas R26cre-ERT MEFs rapidly arrested in the presence of 0.2–0.6 μM OHT, growth in medium with 0.05 μM or 0.1 μM OHT was comparable to growth in normal medium (Fig. 2E). Recombination of a floxed substrate was determined after culturing cells in the presence of OHT for 3 and 9 days, respectively (Fig. 2F). During this period, a marked increase in Cre-mediated gene deletion was observed in cells grown in the presence of low (0.05–0.1 μM) OHT concentrations, demonstrating that prolonged, low-level expression of Cre leads to detectable recombination without any apparent growth inhibition.
Cre Expression Induces Accumulation in G2/M and Aneuploidy.
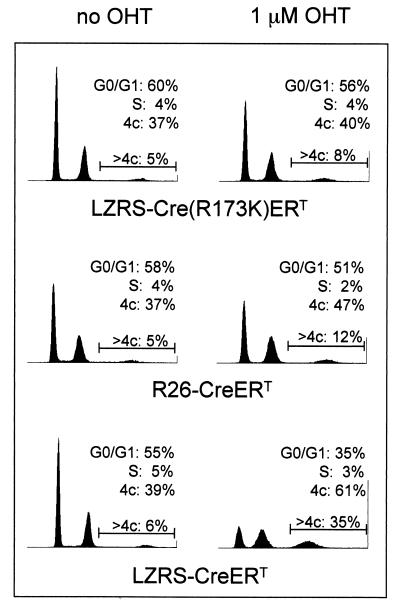
To further characterize the effect of Cre on proliferation, cell cycle profiles of R26cre-ERT MEFs and FACS-sorted MEFs infected with Cre-ERT virus or Cre(R173K)-ERT virus were determined after growth in medium with or without 1 μM OHT for 3 days. Cells were stained with propidium iodide, and cellular DNA content was analyzed by flow cytometry. Cells infected with the Cre(R173K)-ERT virus cultured in the presence of OHT showed a cell cycle profile comparable to untreated cells (Fig. 3). In contrast, Cre-ERT virus-infected MEFs and R26cre-ERT MEFs treated with OHT showed an increase in G2/M phase. Moreover, an increase was observed in the number of cells with polyploid nuclei of ≥4N DNA content, suggesting aberrations in chromosome segregation during mitosis. The observed accumulation of cells in the G2/M phase is suggestive of intolerable DNA damage (37). In addition, a slight increase in the percentage of cells with a sub-G1 DNA content also was observed, indicating that exposure of cells to Cre may result in increased apoptosis (not shown).
Figure 3.
Cre expression in MEFs induces accumulation in G2/M and aneuploidy. Cell cycle analysis of MEFs infected with viruses encoding Cre-ERT, Cre(R173K)-ERT, Cre (lane 3), and R26cre-ERT MEFs, either untreated or treated with 1 μM OHT, for 3 days. Cells were stained with propidium iodide for DNA content and analyzed by FACS. The numbers indicate the percentage of nonpolyploid cells in G0/G1, S, and G2/M phases of the cell cycle. In addition, a region marker is set to determine the percentage of polyploid cells in the total population.
Cre Expression Results in Aberrant Chromosomal Structures.
The aforementioned results are compatible with a model in which Cre expression in cultured cells induces DNA damage, resulting in proliferation arrest. To find evidence for DNA damage, we performed chromosome analysis on cells cultured in the presence of active Cre. Wild-type MEFs and R26cre-ERT MEFs were treated with 1 μM OHT for 48 h, and metaphase spreads were produced. Metaphases were scored by eye for the presence of abnormal structures. Control metaphases showed only low percentages of abnormalities (Fig. 4A). R26cre-ERT MEFs cultured in the presence of OHT displayed a high frequency of chromosomal aberrations, including chromatid breaks, dicentric chromosomes, exchange figures, and ring-shaped chromosomes. In addition, numerous micronuclei were detected in OHT-treated R26cre-ERT MEFs (not shown). R26cre-ERT MEFs, cultured in medium lacking OHT, showed a small increase in the number of abnormalities compared with wt MEFs, indicating that activity of the Cre-ERT fusion protein is not completely prevented in the absence of OHT. In support of this, R26cre-ERT expression in the absence of ligand already results in partial recombination of the very sensitive floxed Rb allele (29).
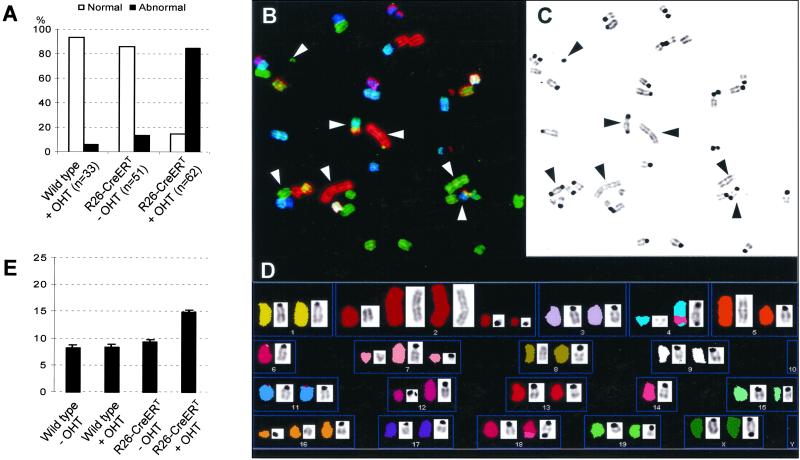
Figure 4.
Cre expression in MEFs induces chromosome abnormalities and increased frequencies of SCE. (A) Expression of active Cre induces chromosome aberrations in MEFs. Metaphase preparations of wt or R26cre-ERT MEFs either untreated or treated with 1 μM OHT for 48 h were examined by eye for the presence of chromosomal aberrations. n, number of metaphases examined. (B–D) SKY analysis of a typical metaphase from R26cre-ERT MEFs treated for 48 h with 1 μM OHT. (B) An individual metaphase after hybridization with fluorescent probes. The 4′,6-diamidino-2-phenylindole-banded image of the same metaphase is shown in C and in display colors by assignation of hybridization signals to specific spectral ranges (D). The arrowheads in B and C indicate examples of dicentric and acentric chromosome fusions and fragmented chromosomes. (D) Multiple fragmented chromosomes (2, 4, 7, 8, 12, and 16), two acentric fusions of chromosome 2 and one of chromosome 5, and dicentric fusions of chromosomes 4 and 14, chromosomes 14 and 18, and chromosomes X. (E) Expression of active Cre causes increased SCE in MEFs. Metaphase preparations of wt or R26cre-ERT MEFs either untreated or treated with 1 μM OHT for 48 h were labeled with BrdUrd before preparation of metaphase spreads. The number of SCEs per cell was determined for at least 40 diploid metaphases per culture.
Induction of Breaks and Fusion Products in OHT-Treated R26cre-ERT MEFs.
To determine the nature of the abnormalities found in the MEF metaphases after OHT-induced R26cre-ERT expression, we performed SKY analysis. Metaphases of wt cells, grown in the absence or presence of OHT, were normal. Metaphases of R26cre-ERT MEFs treated with OHT for 48 h showed severe aberrations. A representative metaphase shown in Fig. 4 B–D revealed acentric fragments of chromosomes 2, 4, 7, and 16. In addition, this metaphase contained two acentric fusions of chromosome 2 and one of chromosome 5 and dicentric fusions of chromosomes 4 and 14, 14 and 18, and X. Several chromosomes showed deletions. In the 17 analyzed metaphases of OHT-treated R26cre-ERT MEFs, a large number of double-strand breaks (DSBs) and 20 dicentric chromosomes were present, of which 8 consisted of fusions between chromosomes of different chromosome pairs. Interestingly, no reciprocal translocations were found. Furthermore, the metaphases with dicentric chromosomes composed of fragments belonging to different chromosome pairs did not contain reciprocal fusions of acentric fragments from the same chromosome pairs. This indicates that these dicentric fusions are probably the result of repair of centromere-containing fragments via nonhomologous end joining and not illegitimate, Cre-mediated recombination between these chromosomes.
Cre Expression Induces SCEs.
Postreplicative repair of double-strand breaks via homologous recombination leads to an increase of SCEs (38). We therefore compared the number of SCEs in wt and R26cre-ERT MEFs, either untreated or treated, with OHT. The level of SCEs found in wt cells, cultured in the presence of OHT, was similar to that observed in the absence of OHT (Fig. 4E). In R26cre-ERT cells treated with OHT, we observed a 50% increase in the number of SCEs. The number of SCEs in untreated R26cre-ERT MEFs was increased slightly. In line with our previous observations, a large number of chromosomal abnormalities was observed in metaphases from the OHT-treated R26cre-ERT MEFs.
Discussion
In this study we describe the growth-inhibitory and genotoxic effects of Cre expression in cultured cells. We have shown that Cre expression in primary MEFs and in established cell lines leads to a proliferation defect that depends on nuclease activity of the Cre recombinase. These results are comparable to the growth-inhibitory effects observed by others in cells expressing a Cre-GFP fusion protein (34, 35). In MEFs with a ligand-inducible Cre knock-in allele, we observed a strong correlation between Cre recombinase activity and toxicity after short-term exposure to ligand. At concentrations required for efficient switching of the floxed Brca2 locus, Cre expression also severely affected the proliferation rate of primary MEFs. We have analyzed extensively the Cre genotoxicity in primary MEFs, which is characterized by the occurrence of micronuclei, aneuploidy, karyotypic abnormalities, and an increased SCE frequency. The occurrence of Cre-induced DNA damage in MEFs is extremely rapid: already after 48 h of Cre induction, the vast majority of metaphases displays numerous karyotypic abnormalities, consisting predominantly of chromosome fragments and chromosome fusions. Moreover, the background activity of the moderately expressed R26cre-ERT in the absence of ligand resulted in detectable DNA damage, although proliferation was not affected. This background activity of R26cre-ERT has been shown to be sufficient for partial recombination of the very sensitive floxed Rb allele, but not for recombination of floxed p53, Brca2, or R26R alleles (29, 39).
Although the mechanism responsible for Cre-mediated induction of genomic damage is not clear, several reports have described promiscuity of the Cre recombinase. Cre can mediate recombination between cryptic loxP sites naturally present in E. coli, yeast, and mammalian genomes (23–26), and earlier studies already reported the induction of nicks and breaks after incubation of cryptic loxP sites with purified Cre recombinase in vitro (15). Given that mammalian genomes contain multiple cryptic loxP sites (26), the inadvertent effects of Cre presented here may be explained by aberrant Cre activity on these sites. This notion is supported by the recent observation that Cre expression in developing spermatids can induce gross chromosomal rearrangements (27). Our data are most compatible with a model in which illegitimate Cre nuclease activity induces DSBs or nicks that are converted to DSBs during DNA replication. The main part of the damage we observe in MEFs after Cre expression consists of chromosome fragments and chromosome fusions that may be the result of DSB repair via nonhomologous recombination. Although illegitimate recombination between cryptic loxP sites could result in chromosome fusions, it is not predicted to result in the accumulation of fragmented DNA. Moreover, the metaphases with dicentric fusions of chromosomes from different pairs do not contain reciprocal acentric fusions from the same chromosome pairs, indicating that these fusions are probably the result of repair via nonhomologous end joining and not illegitimate, Cre-mediated recombination of pseudo-loxP sites. In addition, the occurrence of micronuclei, the increased percentage of aneuploid cells, the increased SCE frequency, and the G2/M arrest in Cre-expressing MEFs are in line with a model in which Cre results in the accumulation of DSBs. Taken together, the growth-inhibitory and genotoxic effects of Cre expression in MEFs are comparable to the effects of the topoisomerase I ligase inhibitor camptothecin in mammalian cells (40, 41). Culturing cells in the presence of camptothecin results in the accumulation of DNA nicks, and, during S phase, this leads to induction of DSBs, an increase in SCEs, and a block in the G2 phase of the cell cycle, preventing entrance into mitosis (42).
The growth-inhibitory and genotoxic effects of Cre in primary MEFs and established cell lines raise the question of why no such effects have been described in several studies employing Cre/loxP technology in cultured cells and in only 1 of more than 70 published Cre transgenic mouse strains (http://www.mshri.on.ca/nagy/cre.htm; ref. 27). An explanation for this apparent discrepancy may be provided by our observation that the recombinase activity of R26-ERT is both time- and dose-dependent, whereas the growth-inhibitory effect appeared to be solely dose-dependent. Prolonged culturing of R26-ERT MEFs in the presence of low OHT concentrations (0.05 μM) resulted in detectable recombination of the floxed Brca2 allele without growth inhibition. These results indicate that sustained low-level expression of Cre causes a slow, cumulative increase in recombination without significant toxicity, possibly because of higher specificity of the Cre recombinase for the introduced lox sequences, to repair of Cre-mediated DNA damage or to removal of damaged cells from the population by proliferative disadvantage. Interestingly, in primary R26cre-ERT keratinocytes, this low dose of OHT is already sufficient for ≥90% recombination of the floxed Brca2 allele within 48 h (29). This result points at marked differences in recombination efficiency in different primary cell types and indicates that complete recombination can occur in the absence of a growth-inhibitory effect.
By analogy with our observations in primary R26cre-ERT MEFs, selection for low-level expression may occur during the generation of cre transgenic mouse strains and during transient or sustained Cre expression in cultured cells. In many experimental settings, such selection would remain unnoticed. For example, Cre toxicity after transgene microinjection might cause selection in utero against embryos with high Cre expression, thus resulting in a decreased yield of transgenic founders. Likewise, clonogenic outgrowth, drug-selection protocols, or prolonged culture of cells after transient transfection or retroviral transduction with Cre constructs may allow for unnoticed removal of cells with high Cre expression already before start of the actual experiment. Notwithstanding any selection against Cre toxicity, our results urge for a careful examination of the effects of Cre expression when used in mammalian conditional experimental settings. Apparently normal cre transgenic mice may reveal subtle phenotypes related to Cre toxicity; also, in cell culture experiments, the aforementioned phenotypic consequences of Cre expression may confound phenotypic analysis of the conditional mutation. Therefore, analysis of Cre-expressing mice or cells with no exogenous loxP sites should be included as controls.
The growth-inhibitory and genotoxic effects of Cre expression call for the development of Cre tools with regulatable recombinase activity to minimize toxicity. Our data show that the ligand-regulatable R26cre-ERT knock-in allele, which is ubiquitously expressed at low levels (29), may serve this purpose because prolonged culture of R26-ERT cells in the presence of low concentrations of ligand resulted in recombination without growth inhibition. An alternative solution has been developed by Silver and Livingston, who have generated a self-excising retroviral Cre expression vector (34). This self-deleting (hit and run) vector was found to efficiently excise itself and a target DNA sequence situated in trans, without causing detectable toxicity. In view of the results presented here, it can be expected that more self-titrating or regulatable Cre tools for applications in mouse models and in cultured cells will be developed in future years.
Acknowledgments
We thank Dr. G van Duyne for the Cre(R173K) mutant cDNA and Dr. P. Chambon for the Cre-ERT cDNA. LZRS-derived viral constructs and Phoenix packaging cell lines were kindly provided by Dr. G. I. Nolan. We also thank R. Beijersbergen, R. van Amerongen, and H. te Riele for critically reading the manuscript and E. Noteboom and A. Pfauth for help with the FACS analysis. This work was supported by the Dutch Cancer Society (NKB/KWF).
Abbreviations
- DSB
double-strand break
- MEF
mouse embryo fibroblast
- OHT
4-hydroxy tamoxifen
- R26
ROSA26
- SCE
sister chromatid exchange
- wt
wild type
- GFP
green fluorescent protein
References
- 1.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V, Pierson L S, 3rd, et al. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sternberg N, Hamilton D. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 3.Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Cold Spring Harbor Symp Quant Biol. 1981;45:297–309. doi: 10.1101/sqb.1981.045.01.042. [DOI] [PubMed] [Google Scholar]
- 4.Sauer B, Henderson N. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abremski K, Hoess R. J Biol Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 6.Mack A, Sauer B, Abremski K, Hoess R. Nucleic Acids Res. 1992;20:4451–4455. doi: 10.1093/nar/20.17.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gopaul D N, Guo F, Van Duyne G D. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo F, Gopaul D N, van Duyne G D. Nature (London) 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 9.Wierzbicki A, Kendall M, Abremski K, Hoess R. J Mol Biol. 1987;195:785–794. doi: 10.1016/0022-2836(87)90484-0. [DOI] [PubMed] [Google Scholar]
- 10.Hoess R, Abremski K, Irwin S, Kendall M, Mack A. J Mol Biol. 1990;216:873–882. doi: 10.1016/S0022-2836(99)80007-2. [DOI] [PubMed] [Google Scholar]
- 11.Hoess R, Ziese M, Sternberg N. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoess R H, Wierzbicki A, Abremski K. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abremski K, Hoess R. J Mol Biol. 1985;184:211–220. doi: 10.1016/0022-2836(85)90374-2. [DOI] [PubMed] [Google Scholar]
- 14.Abremski K, Frommer B, Wierzbicki A, Hoess R H. J Mol Biol. 1988;202:59–66. doi: 10.1016/0022-2836(88)90518-9. [DOI] [PubMed] [Google Scholar]
- 15.Abremski K, Wierzbicki A, Frommer B, Hoess R H. J Biol Chem. 1986;261:391–396. [PubMed] [Google Scholar]
- 16.Metzger D, Feil R. Curr Opin Biotechnol. 1999;10:470–476. doi: 10.1016/s0958-1669(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 17.Sauer B. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 18.Rajewsky K, Gu H, Kuhn R, Betz U A, Muller W, Roes J, Schwenk F. J Clin Invest. 1996;98:600–603. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossant J, McMahon A. Genes Dev. 1999;13:142–145. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 20.Gu H, Zou Y R, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 21.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 22.Herault Y, Rassoulzadegan M, Cuzin F, Duboule D. Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- 23.Sternberg N, Hamilton D, Hoess R. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- 24.Sauer B. J Mol Biol. 1992;223:911–928. doi: 10.1016/0022-2836(92)90252-f. [DOI] [PubMed] [Google Scholar]
- 25.Sauer B. Nucleic Acids Res. 1996;24:4608–4613. doi: 10.1093/nar/24.23.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thyagarajan B, Guimaraes M J, Groth A C, Calos M P. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt E E, Taylor D S, Prigge J R, Barnett S, Capecchi M R. Proc Natl Acad Sci USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. . (First Published November 21, 2000; 10.1073/pnas.240471297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vooijs M, Jonkers J, Berns A. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kinsella T M, Nolan G P. Hum Gene Ther. 1996;7:1405–1413. doi: 10.1089/hum.1996.7.12-1405. [DOI] [PubMed] [Google Scholar]
- 31.Rowan S, Ludwig R L, Haupt Y, S, B, Lu X, Oren M, Vousden K H. EMBO J. 1996;15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 32.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Proc Natl Acad Sci USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzger D, Clifford J, Chiba H, Chambon P. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silver, D. P. & Livingston, D. M. (2001) Mol. Cell, in press. [DOI] [PubMed]
- 35.Moreno de Alboran I, O'Hagan R C, Gärtner F, Malynn B, Davidson L, Rickert R, Rajewski K, DePinho R A, Alt F W. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 36.Abremski K E, Hoess R H. Protein Eng. 1992;5:87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- 37.Paulovich A G, Toczynski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 38.Dronkert M L G, Beverloo B H, Johnson R D, Hoeijmakers J H J, Jasin M, Kanaar R. Mol Cell Biol. 2000;20:3147–3156. doi: 10.1128/mcb.20.9.3147-3156.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 40.Horwitz M S, Horwitz S B. Biochem Biophys Res Commun. 1971;45:723–727. doi: 10.1016/0006-291x(71)90476-1. [DOI] [PubMed] [Google Scholar]
- 41.Hsiang Y H, Hertzberg R, Hecht S, Liu L F. J Biol Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 42.Li L H, Fraser T J, Olin E J, Bhuyan B K. Cancer Res. 1972;32:2643–2650. [PubMed] [Google Scholar]