Abstract
The triblock copolymer polyethylenimine-polycaprolactone-polyethylene glycol (PEI-PCL-PEG) has been shown to spontaneously assemble into nano-sized particulate carriers capable of complexing with nucleic acids for gene delivery. The objective of this study was to investigate micelleplex characteristics, their in vitro and in vivo fate following microfluidic preparation of siRNA nanoparticles compared to the routinely used batch reactor mixing technique. Herein, PEI-PCL-PEG nanoparticles were prepared with batch reactor or microfluidic mixing techniques and characterized by various biochemical assays and in cell culture. Microfluidic nanoparticles showed a reduction of overall particle size as well as a more uniform size distribution when compared to batch reactor pipette mixing. Confocal microscopy, flow cytometry and qRT-PCR displayed the subcellular delivery of the microfluidic formulation and confirmed the ability to achieve mRNA knockdown. Intratracheal instillation of microfluidic formulation resulted in a significantly more efficient (p < 0.05) knockdown of GAPDH compared to treatment with the batch reactor formulation. The use of microfluidic mixing techniques yields an overall smaller and more uniform PEG-PCL-PEI nanoparticle that is able to more efficiently deliver siRNA in vivo. This preparation method may prove to be useful when a scaled up production of well-defined polyplexes is required.
Keywords: microfluidic mixing, siRNA delivery, triblock copolymer, endosomal escape, nanoparticle formulation, pulmonary delivery
1. Introduction
One of the most promising strategies to address various untreatable diseases involves the use of RNA interference (RNAi) through the delivery of small-interfering RNA (siRNA). Ever since its discovery in 1998, siRNA has been proposed as a highly selective approach of disease treatment due to its ability to potentially knock down any gene of interest involved in disease development or progression [1]. Specifically, the administration of therapeutic siRNA has long been proposed as an alternative to conventional cancer therapy [2]. However, the transition of these potential therapies to clinical trials remains slow due to multiple barriers involved in the intracellular deposition of siRNA [3]. In order to overcome the hurdles associated with therapeutic siRNA, nanoparticles have long been the preferred method of condensation and protection with a large variety of formulations taking advantage of different delivery methods or carrier identity to achieve gene knockdown [4].
Recent studies from our lab have shown the ability of PEI-g-PCL-b-PEG triblock copolymers to self-assemble into micelleplexes capable of efficiently condensing and delivering siRNA in vitro to mediate the knockdown of toll-like receptor 4 (TLR4) [5]. However, due to the reaction vessel restraints and the undefined nature of the self-assembly process, it is well known that these polymeric systems tend to have broader size distributions, reflected in higher polydispersity indices (PDI > 0.3), than other well-defined nanoparticle systems developed to deliver siRNA (7). Due to the electrostatic interaction of the positively charged hyperbranched PEI (hy-PEI) with the negatively charged siRNA, the formation and rearrangement of the micelleplexes occurs in what is known as self-assembly (5). The ability of these particles to self-assemble when the polymer and siRNA come into contact makes them attractive candidates when considering costlier multi-step nanoparticle production that can limit the overall scale of particles generated (6). As such, self-assembling polymeric nanoparticle systems are often prepared using a batch reactor technique in which the two components (polymer and nucleic acids) are diluted to the proper concentration and are mixed in a single reaction vessel for an empirically derived amount of time (48). Often, it is unclear how the complexation of such particles takes place and the order in which components need to be added which ultimately results in highly disperse solutions of nanoparticles that could lead to aggregation and diminished transfection efficiency (49). Recently, microfluidic devices have been investigated as a potential strategy to overcome the issues associated with conventional batch reactor mixing techniques (50). It is proposed that the constant ratio at which the cationic polymer comes into contact with the anionic nucleic acid will serve to increase particle uniformity and allow for a tighter complexation, thereby increasing colloidal stability (51). We hypothesize that this increase in stability and complexation may lead to a difference in siRNA protection, nanoparticle uptake and potentially overall gene knockdown.
The purpose of this study was to investigate microfluidic mixing of our PEI-g-PCL-b-PEG triblock copolymers with siRNA and characterize the resulting micelleplexes that are assembled compared to those made via traditional batch reactor mixing. Specifically, the knockdown efficiency and intracellular trafficking of the two micelleplex formulations were studied.
2. Methods
2.1. Materials
Dicer substrate double-stranded siRNA (DsiRNA) targeting the enhanced green fluorescent protein gene (EGFP siRNA, 25/27), human (hGAPDH) and mouse GAPDH (msGAPDH), scrambled nonspecific control (siNegCon) DsiRNA as well as amine-modified and TYE™-563 labelled siRNA were purchased from Integrated DNA Technologies (IDT, Coralville, IA, USA). Fluorescein-conjugated wheat germ agglutinin (WGA) was purchased from Molecular Probes, Inc. (Eugene, OR, USA). Hyperbranched polyethylenimine (hyPEI, 25 kDa) was obtained from BASF (Ludwigshafen, Germany). Heterobifunctional polyethylene glycol (HO-PEGCOOH, 3.5 and 5 kDa), as well as monofunctional (CH3-PEG-COOH, 5 kDa) was purchased from JenKem Technologies (Plano, TX, USA). All other reagents for synthesis were obtained from Sigma-Aldrich (St. Louis, MO, USA) and used without further modification. Ham’s F-12K (Kaighn’s) Medium was purchased from Gibco (Carlsbad, CA, USA). Dulbecco’s phosphate buffered saline (PBS), heat-inactivated fetal bovine serum (FBS), D-(+)-glucose, sodium bicarbonate, sodium pyruvate, 2-mercaptoethanol, dimethyl sulfoxide (DMSO, ⩾ 99.7%), ethylenediaminetetraacetic acid (EDTA, 99.4%–100.06%) and trypan blue (0.4%, sterile filtered) were purchased from Sigma-Aldrich (St. Louis, MO, USA). SYBR Gold dye and Lipofectamine 2000 were obtained from Life Technologies (Carlsbad, CA, USA). The following antibodies were purchased from Abcam (Cambridge, UK): rabbit anti-caveolin-1, mouse anti-RAB7, rabbit anti-LAMP1, rabbit anti-Rab5 along with the Alexa Fluor® 488-conjugated IgG secondary antibodies goat anti-rabbit and goat anti-mouse.
2.2. Synthesis of triblock copolymer and characterization
Briefly, the triblock copolymer consisting of polyethylenimine-graft-polycaprolactone-block-polyethylene glycol (PEI-g-PCL-b-PEG) was synthesized by coupling the heterobifunctional diblock copolymer acrylated-PCL-b-PEG alkyne to hy-PEI (25 kDa), as previously described [1]. All compounds synthesized were characterized by 1HNMR and UV Spectroscopy [2].
2.3. Batch reactor preparation of PEI-g-PCL-b-PEG micelleplexes
The triblock copolymer was dissolved in water to yield a 0.5 mg ml−1 solution and was then filtered through a 0.22 μm filter for sterilization. This stock solution was then diluted to precalculated concentrations with a sterile 5% glucose solution to prepare micelleplexes at an N/P (the molar ratio of nitrogen in PEI to phosphate in siRNA) ratio of 6. To form micelleplexes via batch reactor mixing, 25 μl of the diluted polymer was mixed with an equal volume of 50 pmol siRNA by vigorous pipetting and incubated at room temperature for 20 min, as described in our previous study [2].
2.4. Microfluidic preparation of PEI-g-PCL-b-PEG micelleplexes
To form micelleplexes via a microfluidic mixing chip, 200 μl (minimum volume) of both the diluted PEI-g-PCL-b-PEG and siRNA solution (2 pmol μl−1) was transferred into two separate 3 ml sterile syringes (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). These fluid filled syringes were then loaded onto a KD Scientific 220 syringe pump (KD Scientific Inc., Holliston, MA, USA) and connected to a hydrophobic Dolomite micromixer chip (Dolomite Microfluidics, Charlestown, MA, USA) via 0.25 mm i.d. FEP tubing. Unless otherwise stated, the polymer and siRNA solution were then mixed at a constant flow rate of 0.5 ml min−1 and the formed micelleplexes were collected after the mixed solution reached steady state flow. Micelleplexes that were prepared via the micromixer chip did not require further incubation at room temperature and were used directly after collection.
2.5. Hydrodynamic diameter and zeta (ζ) potential measurements
Hydrodynamic diameter measurements of micelleplexes prepared with both mixing techniques were performed by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern Instruments Inc., Malvern, UK). Micelleplexes were formed via batch reactor or microfluidic chip as described above at N/P 6 complexing 2 pmol μl−1 of scrambled siRNA. A total volume of 50 μl of each sample was added into a disposal cuvette (Brand GMBH, Wertheim, Germany) and the 173° backscatter angle measurement was read in triplicates with each run consisting of 15 scans. Results are represented as average size (nm) ± standard deviation (SD). The samples were then diluted to 1 ml with filtered Nanopure water and transferred to a folded capillary cell (Malvern Instruments Inc., Malvern, UK), and ζ-potential measurements were taken. ζ-Potential measurements were read in triplicates by laser Doppler anemometry, with each run consisting of 30–50 scans. Results are shown in mV ± SD.
2.6. SYBR gold and heparin assays
A SYBR® gold assay was used to ensure the ability of micelleplexes formed by either mixing technique to successfully condense siRNA at N/P ratio 6. The SYBR® Gold fluorescent dye (Life Technologies, Carlsbad, CA, USA) undergoes a more than 1000-fold signal enhancement upon intercalation with uncomplexed double-stranded nucleic acids. However, this signal is reduced if the siRNA is condensed within micelleplexes, thereby blocking the dye from intercalating with the nucleic acids. To test the micelleplexes that were prepared using the microfluidic chip, samples were prepared as described above with the Dolomite micromixer chip and 100 μl of the mixed solution was transferred to each well of a white FluoroNunc 96-well plate (Thermo Fisher Scientific, Waltham, MA, USA). For the batch reactor samples, 50 μl of siRNA in 5% glucose (1 pmol μl−1) was distributed in each well of the white FluoroNunc 96-well plate followed by adding 50 μl of the diluted PEI-g-PCL-b-PEG solution to form micelleplexes at an N/P ratio of 6. Once mixed, the plate was incubated for 20 min at room temperature. Following incubation, 30 μl of a 4 × SYBR® gold solution was added to each well and incubated for 10 min in the dark at room temperature. The fluorescence of each sample was quantified using a Synergy 2 multi-mode microplate reader (BioTek Instrument, Winooski, VT, USA) at excitation wavelength of 485/20 nm and emission wavelength of 520/20 nm. Samples were normalized based on the following criteria. The fluorescence level of 100% free siRNA was calculated based on the fluorescence of uncondensed free siRNA incubated with SYBR® gold dye. The fluorescence of zero percent free siRNA was calculated with SYBR® gold dye in the absence of any siRNA. All measurements were performed in triplicates and the results are displayed as mean values and SD.
Similarly, heparin assays were used to determine the stability of the micelleplexes formed with the different mixing techniques and to compare the amount of siRNA within the particles generated by these two production methods. To test this, the release of siRNA from particles that were formed at a physiologically relevant pH (7.4), as well as at a lower pH (4.5, resembles the late endosome/lysosome) in the presence of the heparin was determined by a modified SYBR® gold assay [2]. Briefly, batch reactor or microfluidic micelleplexes measured at pH 7.4 were made in 5% glucose solution, while those measured at pH 4.5 were made in sodium acetate buffer. Samples were prepared as described above for the SYBR® gold assay with the exception of adding 10 μl of a 1.0 IU heparin solution and incubating for an additional 30 min at room temperature. Subsequently, 30 μl of a 4 × SYBR® gold solution was added to each well and incubated for 10 min in the dark at room temperature. The fluorescence of each sample was quantified using a Synergy 2 multi-mode microplate reader (BioTek Instrument, Winooski, VT, USA) at excitation wavelength of 485/20 nm and emission wavelength of 520/20 nm. Results of the heparin assays were analysed as described for the SYBR® gold assays.
2.7. Cell culture
A549 cells are a human adenocarcinoma alveolar based lung cancer cell line and were obtained from ATTC (LG Promochem, Wesel, Germany). A549 lung adenocarcinoma cells were cultured in Ham’s F-12K (Kaighn’s) medium and supplemented with 10% heat inactivated fetal bovine serum and 1% penicillin/streptomycin (Corning Incorporated, Corning, NY, USA). Cells were grown in 75 cm2 cell culture flasks (Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C and 5% CO2 and passaged every 2–3 days when they reached confluency.
2.8. In vitro cellular uptake
In 24-well plates, (Corning Incorporated, Corning, NY, USA) 50 000 A549 cells were seeded and incubated overnight at 37 °C and 5% CO2. Amine modified siRNA was labelled with succinimidyl ester (NHS) modified Alexa Fluor-488 (AF488) (Life Technologies) following the manufacturer’s protocol and purified by ethanol precipitation and spin column binding as described previously [3]. For each experiment, micelleplexes were freshly prepared by either mixing technique with AF488 labelled siRNA. Negative controls consisted of blank/untreated cells and cells treated with free siRNA. Positive control cells were transfected with Lipofectamine 2000 (Life Technologies) lipoplexes, which were prepared according to the manufacturer’s protocol. Briefly, every 10 pmol siRNA were formulated with 0.5 μl Lipofectamine (LF) solution. Unless otherwise stated, cells were transfected for 4 h in 37 °C and 5% CO2 with 50 μl of micelleplex solution containing 50 pmol siRNA within a total volume of 400 μl of serum containing cell culture media. Cells were then washed with PBS, trypsinised and spun down at 350 g for 10 min. After centrifugation, the supernatant was decanted, and the cells were washed three times and resuspended in 400 μl PBS/2 mM EDTA. Samples were analysed via flow cytometry (Applied Biosystems Attune® Acoustic Focusing Cytometer, Life Technologies), and the median fluorescence intensity (MFI) was measured using 488 excitation and 530/30 bandpass emission filter set. Samples were run in triplicates, with each sample gated by morphology based on forward/sideward scattering for a minimum of 10 000 viable cells. Analysis and presentation of the data was performed by GraphPad Prism 5.0 software calculating mean values and SD.
2.9. Route of cellular uptake in vitro
To study the route of micelleplex uptake, a modified in vitro cellular uptake was performed [4]. In short, 50 000 A549 cells were incubated with different types of uptake inhibitors such as chlorpromazine (10 μg ml−1), nystatin (10 μg ml−1), wortmannin (12 ng ml−1), and methyl-β-cyclodextrin (3 mg ml−1) for 1 h followed by incubation with PEI-g-PCL-b-PEG polyplexes containing AF488 siRNA for 4 h. Positive control cells were transfected with AF488 micelleplexes or LF lipoplexes for 4 h and untreated cells served as a blank control. After the incubation period, the cells were either trypsinised (unquenched samples) or treated with a 0.4% trypan blue solution for 5 min (quenched samples) before they were trypsinised. Subsequently, the cells were washed and subjected to flow cytometric detection of siRNA uptake as described above.
2.10. In vitro GAPDH gene knockdown
For gene silencing experiments, 50 000 A549 cells were seeded and incubated overnight at 37 °C and 5% CO2 in a 24 well plate. Micelleplexes were freshly prepared by either mixing technique with hGAPDH siRNA or scrambled siRNA at an N/P ratio of 6. Negative controls consisted of blank/untreated cells while positive control cells were transfected with Lipofectamine 2000 (Life Technologies) lipoplexes, which were prepared as discussed above. Cells were transfected for 24 h in 37 °C and 5% CO2 with 50 μl of micelleplex solution containing 50 pmol siRNA within a total volume of 400 μl of serum containing cell culture media. Cells were then washed with PBS, harvested and processed to isolate total RNA using the PureLink™ RNA mini kit (Life Technologies) according to the manufacturer’s protocol with DNase I digestion (Thermo Scientific). Synthesis of cDNA from total RNA and PCR amplification were performed with Brilliant III ultra-fast SYBR® green qRT-PCR master mix kit (Agilent Technologies, Santa Clara, CA, USA) using a Stratagene Mx3005P qPCR system (Agilent Technologies). Cycle threshold (Ct) values were determined by MxPro software (Agilent Technologies). A standard curve including 5 points was made from a 1:5 serial dilution of an untreated sample and assigned concentrations of each point (1, 0.2, 0.004, 0.0008, and 0.000 16) were plotted versus their corresponding Ct values. The gene expression of GAPDH was normalized by the expression of β-actin. Hs_GAPDH_2_SG primers for GAPDH and Hs_ACTB_2_SG primers for β-actin (Qiagen, Valencia, CA, USA) were used in the experiment.
2.11. Confocal scanning laser microscopy
For intracellular location and colocalization experiments, 50 000 A549 cells were seeded and incubated overnight at 37 °C and 5% CO2 on 12 mm diameter sterile coverslips (Fisher Scientific) which were placed in the bottom of each well of a 24 well plate. Micelleplexes were freshly prepared using either mixing technique with TYE-563 labelled siRNA at an N/P ratio of 6. The adherent cells on the coverslips were then transfected for either 4 or 24 h in 37 °C and 5% CO2 with 50 μl of micelleplex solution containing 50 pmol siRNA within a total volume of 400 μl of serum containing cell culture media. Following transfection, cells were washed with PBS and fixed with 4% paraformaldehyde solution in PBS (Affymetrix, Santa Clara, CA, USA) for 20 min at room temperature. For endosomal colocalization studies, cells were permeabilized with either 0.05% triton in PBS (PBST) for 1 h at room temperature or 0.03% (w/v) saponin in PBS for 15 min at room temperature. Next, the cells were incubated in a 1% BSA blocking buffer (PBS + 0.05% triton) for 1 h at room temperature. The various primary antibodies were diluted 1:250 in blocking solution and the cells were incubated overnight at 4 °C with gentle shaking. After washing with PBST, the cells were incubated with diluted Alexa Fluor®-488 secondary antibody (1:500) for 2 h at room temperature. Finally, the nuclei were stained with 5 μM of DRAQ5 (Thermo Fisher) for 5 min and rinsed with PBS. Slides were mounted with FluorSave™ mounting medium (EMD Millipore, Billerica, MA, USA) and were imaged by a Leica TCS SPE-II laser scanning confocal microscope (Leica, Wetzlar, Germany). The images were exported from the Leica Image Analysis Suite (Leica) and processed with the Fiji distribution of ImageJ [5].
2.12. In vivo GAPDH gene knockdown
In order to measure in vivo transfection efficiency, a murine GAPDH knockdown study was conducted using methods previously described with slight modifications [6]. All animal experiments were approved by a Wayne State University Institutional Animal Care and Use Committee. Balb/c mice were purchased from Charles River Laboratories (Boston, MA, USA). Briefly, mice were intratracheally instilled (under ketamine/xylazine anaesthesia) with 50 μl of freshly prepared batch reactor or microfluidic micelleplexes, prepared with 750 pmol msGAPDH siRNA or scrambled siRNA at N/P 6. Control mice were treated with the same volume of 5% glucose. After 24 h, the mice were sacrificed and lungs were flash frozen using liquid nitrogen and stored at −80 °C. To isolate whole RNA, the frozen tissue was placed into a mortar with a small amount of liquid nitrogen and grinded into a fine powder using the pestle. Following homogenization, the frozen lung powder was processed with the TRIzol™ Plus RNA Purification Kit (Thermo Scientific) according to the manufacturer’s protocol to yield purified RNA. Samples were stored at −80 °C and isolated mRNA was further quantified by qRT-PCR as described above. Mm_GAPDH_3_SG primers for GAPDH and Mm_ACTB_2_SG primers for β-actin (Qiagen, Valencia, CA, USA) were used in the experiment.
2.13. Statistics
All results are given as mean value ± SD. One-way ANOVA with Bonferroni posthoc post-test, two-way ANOVA and calculation of area under the curve (AUC) were performed in GraphPad Prism software (Graph Pad Software, La Jolla, CA).
3. Results and discussion
3.1. Microfluidic (MF) system to prepare PEI-g-PCL-b-PEG micelleplexes
A schematic illustration of our microfluidic mixing system utilizing the commercially available Dolomite Micromixer Chip is shown in figure 1. The Micromixer Chip is a static mixer that utilizes lamination of the three available input fluid flow streams in addition to a serpentine microchannel design [7]. The lamination of the fluid flow streams decreases the mixing time needed to achieve complete diffusion of triblock copolymer and siRNA while the serpentine microchannel design induces advection at each of the channel turns [8].
Figure 1.
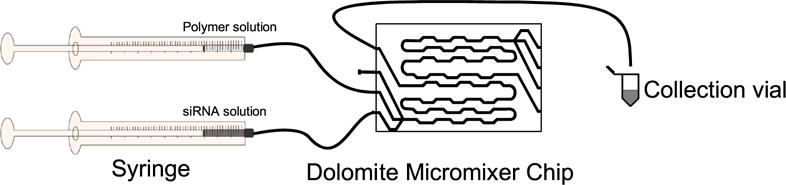
Schematic representation of microfluidic mixing system. Not shown is the syringe pump that is used to mix both the polymer and siRNA solution at a constant rate of 0.5 ml min−1.
3.2. Characterization of nanoparticles prepared with MF system
Two of the most important characteristics when investigating nanoparticle drug delivery are the hydrodynamic diameter and surface charge of the particle that is being developed. In order to be effective, carriers should form nanoparticles that have monodisperse hydrodynamic size profiles within the nanometer scale [9]. In addition, in order to help facilitate the binding and subsequent endocytosis of the particle, a slightly positive surface charge is desirable [10]. More specifically, numerous studies have reported that the exact parameters for these characteristics are highly dependent on the route of nanoparticle administration along with the particle’s destined organ/area of interest [11–13]. For example, intravenously injected nanoparticles greater than 150 nm have been shown to accumulate readily in lung capillaries [11]. Additionally, natural defence mechanisms such as macrophages are known to detect and clear particles with sizes greater than 260 nm [14]. Our group has previously reported batch reactor (BR) prepared PEI-g-PCL-b-PEG/siRNA micelleplexes at an N/P ratio of 6 with hydrodynamic diameters around 260 nm with polydispersity values (PDI) around 0.3 [2]. Here, micelleplex formulations with 50 pmol of siRNA at an NP 6 were made with the standard BR technique or with our MF system at various flow rates and were characterized by DLS. As seen in table 1, the hydrodynamic diameters of the particles made with the microfluidic mixing system were measurably smaller (<170 nm) than those that were made by batch reactor mixing (260 ± 11.9 nm). In addition, the flow rate of 0.5 ml min−1 produced particles that had the smallest size (122 ± 19.7 nm) and were the most uniform (PDI = 0.299). The smaller and more uniform distribution for nanoparticles assembled using microfluidic mixing shown here are a result of the laminar flow that is achieved within the microfluidic channel that leads to a distinct interfacial region and an increase in the diffusional mass-transfer between the fluids containing the polymer and nucleic acid. The decrease in particle size and distribution that we demonstrate agrees well with several other previous studies reported with other PEI-based polyplexes [15–17]. This suggests that our MF system is able to assemble micelleplexes into smaller, more uniform particles that will have favourable overall biodistribution and are able to evade immune system detection.
Table 1.
Micelleplex hydrodynamic diameter and zeta potential.
| Mixing technique | Flow rate (ml min−1) | Particle size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|---|
| Batch reactor | N/A | 260 ± 11.9 | 0.380 | 7.08 ± 1.7 |
| Microfluidic | 0.5 | 122 ± 19.7 | 0.299 | 4.46 ± 0.4 |
| 1 | 168 ± 15.7 | 0.423 | 5.6 ± 3.0 | |
| 2 | 150 ± 39.9 | 0.520 | 5.37 ± 1.0 |
The overall surface charge of the nanoparticle is another critical characteristic to be evaluated upon particle assembly. Higher surface charge has been found to increase the stability of nanoparticles in suspension due to limiting the aggregation of the particle [18]. In terms of delivery to cells, if the particle is negatively charged, it may lead to a slight repulsive effect when it comes into contact with the negatively charged cell membrane [19]. In addition, the positive charge on the particle will help to promote binding to the cell membrane which may in turn lead to higher, nonspecific endocytosis [20]. On the other hand, too strong of a positive charge on the particle’s surface may lead to toxicity that is associated with many polymeric based nanoparticles, in addition to increased detection and uptake by phagocytes [10]. For our study, table 1 shows that the corresponding zeta potential for micelleplexes assembled by both mixing techniques were positive and below +10 mV. These results corroborate our previous formulations in addition to other similar nanoparticle complexes [21]. Consequently, this slightly positive zeta potential suggests that both mixing techniques allow for proper charge neutralization and micelle arrangement between the strong cation PEI and the anionic siRNA with PEG chains on the surface, partially shielding the positive charges of PEI. Based on the size and zeta potential results, MF particles were made at an NP ratio of 6 using a flow rate of 0.5 ml min−1 continuously for all subsequent experiments.
3.3. Stability of batch reactor and microfluidic-assembled micelleplexes
As mentioned above, our PEI-g-PCL-b-PEG triblock copolymers utilize the polycation PEI to electrostatically condense the negatively charged siRNA and form micelleplexes. One of the biggest challenges to systemic administration of polymeric nanoparticles that electrostatically interact with siRNA is the presence of competing anions in serum that are able to out compete the negative charges found on the siRNA molecules [22]. In order to mimic the stability of the micelleplexes when in the presence of serum at physiologically relevant conditions, we performed a heparin assay [23]. This assay utilized SYBR® gold fluorescent dye to assess the relative amount of free, uncondensed siRNA after micelleplexes were treated with neutral (pH 7.4) or acidic (pH 4.5) buffers in the presence of the polyanion heparin. Figure 2 shows that both the BR and MF micelleplexes only released ∼15% of the total siRNA at the high concentration of heparin tested here. Conversely, we needed to verify that the particles would be able to release the siRNA when in a low pH environment similar to what is present in endocytic vesicles (pH 4.5). To this end, we treated the BR and MF micelleplexes with 1.0 IU of heparin and then added sodium acetate buffer to mimic the environment of the endocytic vesicles. Again, figure 2 demonstrates that both the BR and MF micelleplexes are able to successfully release over 85% of their total siRNA in the presence of low pH and the polyanion. In addition, both the BR and MF samples show similar levels of SYBR® gold fluorescent dye in each buffer system tested. These data suggest that PEI-g-PCL-b-PEG/siRNA particles maintain the same concentration of siRNA following microfluidic assembly.
Figure 2.
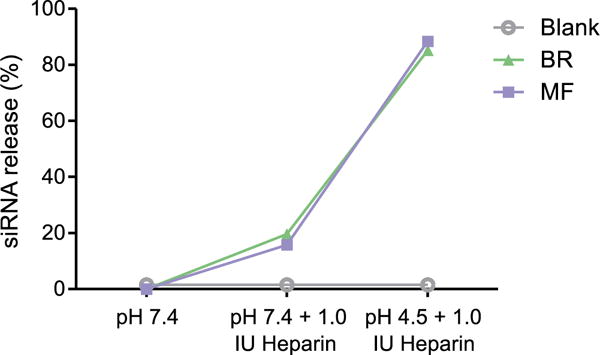
Micelleplexes were assembled via BR (batch reactor) and MF (microfluidic mixing) at an N/P ratio of 6. Both formulations were tested at N/P 6 and incubated in the presence of heparin for 30 min at pH 7.4 or 4.5. This heparin assay at pH 7.4 mimics the pH during in vivo circulation while pH 4.5 mimics the late endosome acidic environment. Free siRNA represents 100% siRNA release.
3.4. In vitro cellular uptake of micelleplexes
After ensuring that the MF system was able to assemble micelleplexes that are of the appropriate size, zeta potential and siRNA retention/release profiles, ability of the MF particles to be delivered to cells needed to be investigated. Therefore, Alexa Fluor-488 labelled siRNA was used to form micelleplexes with the PEI-g-PCL-b-PEG triblock copolymers via batch reactor or microfluidic mixing and lung adenocarcinoma A549 cells were treated for 24 h. Following transfection, cells were submitted to flow cytometry analysis and the level of fluorescence was quantified. Unmodified hy-PEI polyplexes and Lipofectamine 2000 (LF) served as positive controls while cells treated with free uncondensed siRNA or 5% glucose were included as negative controls. Consistent with our previously published results, figure 3 shows that BR and MF micelleplexes exhibited similar levels of cellular uptake as hy-PEI but were not taken up as readily as LF after a 24 h transfection. By delivering Alexa Fluor-488 labelled siRNA into the cell after microfluidic-assisted assembly, we are able to conclude that the microfluidic process of forming micelleplexes generates particles that have similar uptake profiles to those that are made by the conventional batch reactor process. The slightly but non-significantly lower uptake of MF particles in cell culture can potentially be explained by the slower sedimentation of smaller particles as obtained by MF mixing [24].
Figure 3.

Median fluorescence intensity (MFI) was determined by flow cytometry to evaluate the cellular uptake in lung adenocarcinoma cells (A549) of micelleplexes loaded via BR (batch reactor) and MF (microfluidic mixing) at an N/P ratio of 6 with 50 pmol Alexa Fluor-488 labelled siRNA. Lipofectamine 2000 (LF) polyplexes/lipoplexes were prepared according to manufacturer’s protocol with 50 pmol Alexa Fluor-488 labelled siRNA. Data points indicate mean ± SD (n = 3). One-way ANOVA.
3.5. In vitro transfection efficiency of micelleplexes
While we have shown that the microfluidic assembled particles are able to be internalized by A549 cells, we next wanted to investigate the ability of the microfluidic micelleplexes to efficiently mediate gene knockdown. Hence, A549 cells were transfected with BR and MF micelleplexes that contained hGAPDH siRNA or scrambled siRNA for 24 h. Negative controls consisted of blank/untreated cells while positive control cells were transfected with Lipofectamine 2000 and hy-PEI (N/P 10). GAPDH gene expression was quantified via real time PCR (qRT-PCR) and was normalized to β-actin gene expression5. As seen in figure 4, A549 cells that were treated with BR and MF micelleplexes show analogous levels of reduced GAPDH expression. Furthermore, GAPDH expression in these samples was reduced approximately 2 fold in comparison to cells that were treated with scrambled siRNA nanoparticles. The comparable levels of gene knockdown achieved by both BR and MF micelleplexes indicates that our microfluidic mixing generates PEI-g-PCL-b-PEG particles that maintain the ability to deliver siRNA that elicits robust gene knockdown in vitro.
Figure 4.
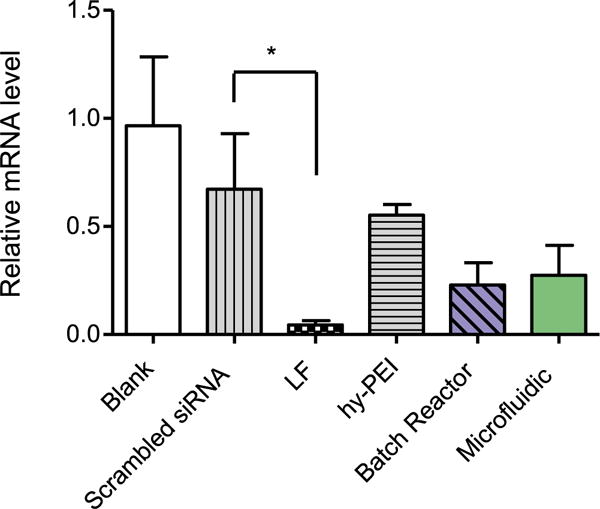
GAPDH gene knockdown efficiency was validated in lung adenocarcinoma cells (A549) after 24 h treatment with micelleplexes assembled via batch reactor and microfluidic mixing at an N/P ratio of 6 with 50 pmol hGAPDH siRNA. GAPDH expression was normalized with β-actin expression and quantified by real time PCR. Data points indicate mean ± SD (n = 3). One-way ANOVA, *P < 0.01.
3.6. Route of cellular uptake of micelleplexes in vitro
To further characterize the microfluidic assembled micelleplexes, we next investigated their specific route of uptake through the various endocytosis pathways. More specifically, we evaluated whether the MF nanoparticles were taken up in a manner similar to that of the BR nanoparticles. Accordingly, we conducted flow cytometry analysis and compared the measured MFI of A549 cells treated with AF488 siRNA micelleplexes in the presence of different endocytosis inhibitors is shown in figure 5. Nystatin is known to inhibit the internalization of caveolae and lipid rafts through depletion of the cholesterol in the cell membrane [25]. For the cells transfected with BR micelleplexes, nystatin treatment significantly (P < 0.001) reduced the cellular uptake as shown by the increase of MFI for those samples that were not quenched with trypan blue (figure 5(A)). In those samples that were quenched with trypan blue, the observed MFI decreases, indicating that nystatin treatment caused significant accumulation of micelleplexes on the cell surface. This trend in cellular uptake can also be observed in those cells treated with MF micelleplexes, albeit at a lower overall scale (figure 5(B)). Along with inhibiting receptor mediated endocytosis, wortmannin has been shown to block macropinocytosis, an endocytic pathway that has long been thought to be a possible internalization pathway of cationic polymer/nucleic acid complexes [26, 27]. In cells that were transfected with BR micelleplexes, wortmannin treatment also significantly (P < 0.001) reduced the cellular uptake of both quenched and unquenched populations. Since nystatin and wortmannin both had the largest impact on cellular uptake of BR micelleplexes, we hypothesize that these particles are internalized mainly via macropinocytosis and to a lesser extent, caveolae-meditated endocytosis. Conversely, treatment with the various endocytic inhibitors did not significantly reduce the uptake of MF micelleplexes (figure 5(B)). We hypothesize that the apparent absence of uptake inhibition for these particles may have to do with multiple pathways of endocytosis that are available for the smaller micelleplexes assembled by microfluidic mixing. Numerous studies have reported the key role that particle size plays on the route of cellular uptake which in turn affects the overall biodistribution of the particle. [10, 28, 29] Specifically, slightly positive charged polymeric nanoparticles with a size range of 100–200 nm have been shown to achieve the best cellular uptake due to an accumulation of favourable properties that allow them to be taken up via multiple different endocytic pathways [19]. As such, when we treated the A549 cells with the different specific uptake inhibitors, the MF particles were still taken up through other methods of endocytosis and were not blocked by the inhibitor treatment.
Figure 5.
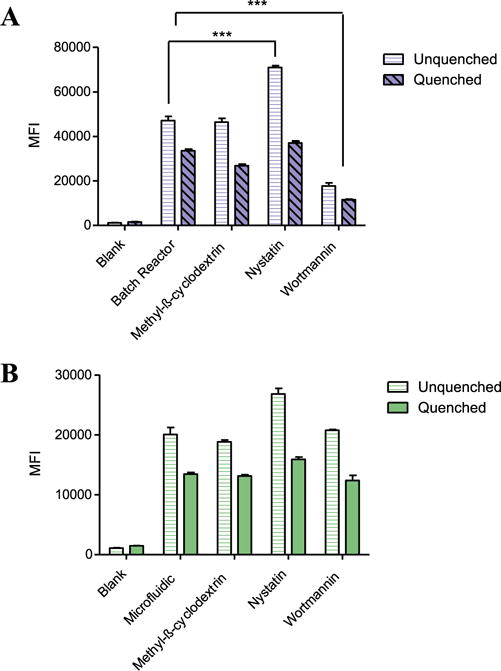
Route of micelleplex cellular uptake was investigated in human lung adenocarcinoma epithelial cells (A549) following a 1 h treatment with the various endocytosis inhibitors nystatin (10 μg ml−1), wortmannin (12 ng ml−1), and methyl-β-cyclodextrin (3 mg ml−1). Median fluorescence intensity was measured via flow cytometry after 4 h transfection with micelleplexes assembled via batch reactor (A) or microfluidic mixing (B) at an N/P ratio of 6 with 50 pmol AF488 siRNA. Data points indicate mean ± SD (n = 3). One-way ANOVA ***P < 0.001.
3.7. Investigation of endosomal escape via confocal microscopy
In order to further investigate the micelleplexes’ cellular uptake profile and method of endosomal escape, we performed a series of confocal microscopy experiments utilizing TYE™-563 fluorescently labelled siRNA and immunostaining. Following a 24 h transfection with the TYE™-563 siRNA micelleplexes, A549 cells were incubated with a variety of primary antibodies specific for the endocytic pathway and the intracellular colocalization of the nanoparticles within endocytic membranes was observed with a Leica TCS SPE-II laser scanning confocal microscope after incubation with an Alexa Fluor-488 conjugated secondary antibody. Figure 6 confirms the intracellular deposition of the siRNA after transfection with micelleplexes assembled with both mixing techniques. To begin our colocalization studies, we probed the cells treated with either BR or MF micelleplexes with Rab7, a well-known marker of late endosome maturation and lysosome fusion [30]. Figure 6(A) shows that both methods of nanoparticle preparation yielded no overlap of the fluorescent siRNA within the late endosomes. After staining for LAMP-1, a protein associated with the lysosome, a modest level of colocalization can be observed for the BR particles which is somewhat further reduced for the MF particles (figure 6(B)). Caveolin-1 (CAV), is a scaffolding protein that plays a critical role in the formation of caveolae mediated endocytosis [31]. Reinforcing our endocytosis inhibitor uptake study results, figure 6(C) shows a strong coinciding signal in the cells that were treated with the BR micelleplexes and stained for the CAV protein. However, the cells that were treated with the MF particles show no visible colocalization with the caveolae marker at all. Finally, WGA is a widely used lectin in the field of fluorescence microscopy for its ability to stain lipids in the plasma membrane. When the membrane is permeabilised, WGA is also able internalise into cells and stain the lipid rich vesicles of the trans-Golgi apparatus [32]. Interestingly, those cells that were stained with WGA showed an extremely selective and strong colocalization of signal from both the WGA stain and the TYE-563 siRNA delivered via BR and MF micelleplexes. Taken together, these images lend support to our previous cellular uptake experiments and give us insight into the possible mechanisms of intracellular delivery. We hypothesized from the inhibitor and cell uptake study that the BR particles might be taken up through macropinocytosis and/or caveolae mediated endocytosis. Our confocal microscopy images show that the micelleplexes that were assembled via the batch reactor mixing technique had overlap with three markers known to play a direct role in those pathways. We hypothesize that the BR particles are able to be taken up by caveolae and then are brought straight to the trans-Golgi apparatus largely avoiding degradation in the lysosome [33]. In conjunction, we infer that the A549 cell is also using macropinocytosis to ‘gulp’ the larger BR micelleplexes in a non-specific manner [34]. After doing so, the particles may then facilitate the generation of macropinosomes which may or may not ultimately fuse with the lysosome [27]. In addition, macropinosomes are thought to be ‘leaky’ which would also help the particle to reach the cytoplasm and achieve gene knockdown. On the other hand, the particles that were generated via microfluidic mixing show a strong colocalization within the trans-Golgi apparatus when stained with WGA along with a minimal overlap with the lysosomal protein LAMP-1. Given the smaller size of the MF micelleplexes in comparison to the BR micelleplexes, we postulate that the MF particles are not taken up through caveola-mediated endocytosis but rather move quickly through the endocytic pathway, avoid lysosome degradation and associate with the trans-Golgi apparatus until further processed. The rather low extent of colocalization of both BR and MF micelleplexes with the endo-lysosome confirms their endosomal escape and emphasizes the repeatedly reported value of PEI-PCL-PEG as siRNA nanocarriers for efficient gene silencing [2, 3, 35, 36].
Figure 6.
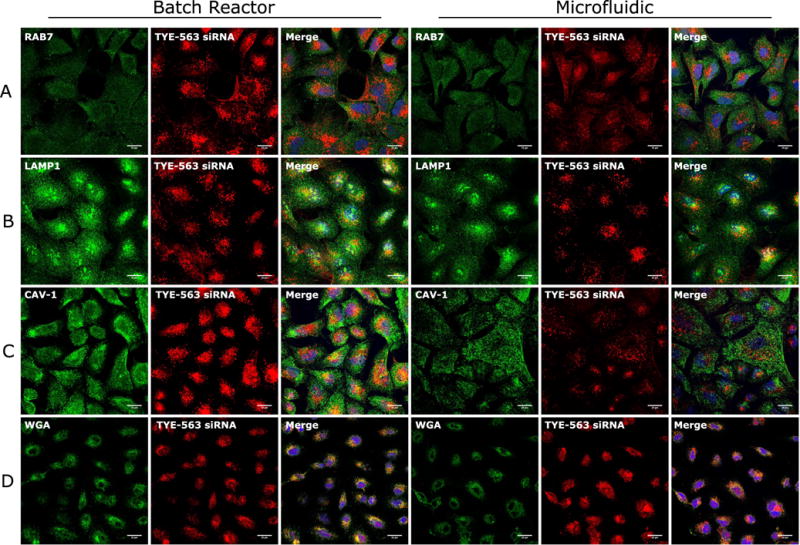
Intracellular delivery of TYE™-563 fluorescently labelled siRNA was evaluated via confocal scanning laser microscopy following 24 h transfection of micelleplexes. Immunostaining was performed on cell samples in order to investigate colocalization of siRNA within the following subcellular locations: (A) late endosomes (RAB7), (B) lysosome (LAMP-1), (C) caveolae (CAV-1) or (D) trans-Golgi (WGA).
3.8. In vivo delivery of micelleplexes
To further elucidate the impact that microfluidic mixing has on in vivo transfection efficiency of PEI-g-PCL-b-PEG micelleplexes, a murine model for pulmonary delivery was used [6]. Using local siRNA delivery, the first pass effect in the liver can be largely circumvented [37]. Mice were intratracheally administered BR or MF micelleplexes that were made with either Mm_GAPDH or scrambled control siRNA. Untreated mice were instilled with the same volume of 5% glucose solution. The following day, the animals were sacrificed and the lungs were collected for total RNA isolation. Following qRT-PCR analysis, GAPDH mRNA was normalized by the expression of β-actin (figure 7). The mice that were treated with the microfluidic micelleplexes had a significantly lower expression of GAPDH than all of the other treatment groups evaluated. Of most interest, there was a significant difference in the GADPH expression level between those animals treated with BR particles versus those treated with MF particles. Previous studies have reported on this phenomenon with varying explanations for its existence [15, 38]. Wagner et al observed that in vitro PEI/DNA particles that are formed by simple pipetting at low N/P ratios tend to aggregate and increase in overall size which, in turn, leads to more efficient gene knockdown of the aggregated particles in vitro due to differences in sedimentation rates [24]. However, in vivo sedimentation plays a secondary role, and larger particles are phagocytosed and cleared by macrophages more rapidly, while smaller particles more efficiently diffuse through mucus and surfactant in the lung and are more efficiently endocytosed by the epithelium [39]. Additionally, another possible explanation for the increase in gene efficiency lies in the possibilty for our particles to undergo transcytosis. Recently, there have been multiple reports that describe the ability for cationic nanoparticles with a diameter of 100–200 nm to undergo transcytosis when administered to lung epithelial cells [39–41]. This mechanism of uptake allows for the endocytosed particle on the apical plasma membrane to penetrate through the multiple cell layers of the lung to the lateral plasma membrane and furthermore be released into the blood stream [40]. One of the key organelles involved in the sorting and recycling process that allows for the nanoparticles to pass through in this manner is the trans-Golgi apparatus [42]. Not only do our microfluidic-assembled micelleplexes demonstrate size profiles smaller than <200 nm, our confocal microscopy demonstrated a strong association of the microfluidic micelleplexes with the trans-Golgi apparatus (figure 6(D)). Therefore, we hypothesize that these particles are able to extravasate thorough the lung tissue due to their uniform size distribution around 100 nm and strong intracellular delivery to the Golgi apparatus, which then repackages the particles to be sent onward into the lateral plasma membrane.
Figure 7.
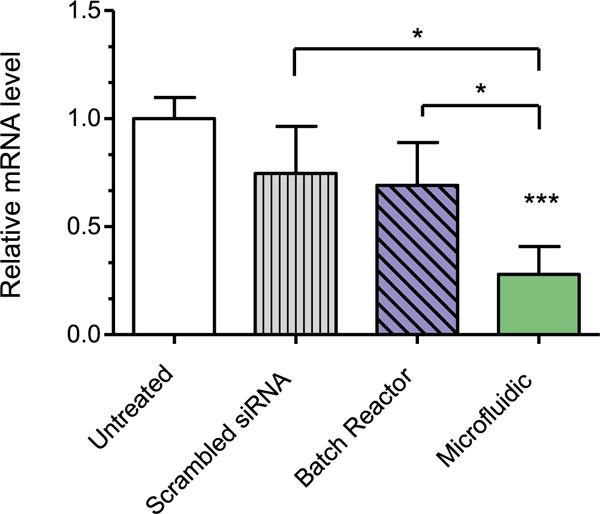
In vivo GAPDH gene knockdown efficiency was validated in whole lung RNA isolated from normal BALB/c mice that were intratracheally instilled with micelleplexes assembled via batch reactor and microfluidic mixing at an N/P ratio of 6 with 750 pmol msGAPDH siRNA. GAPDH expression was normalized with β-actin expression and quantified by real time PCR. Data points indicate mean ± SD (n = 4). One-way ANOVA, *P < 0.01, ***P < 0.001.
4. Conclusion
Herein we present an application of microfluidic devices utilizing a Dolomite micromixer chip for the assembly of a well-established micelleplex delivery system consisting of the triblock copolymer PEI-g-PCL-b-PEG. These microfluidic-assembled micelleplexes were then characterized through various physical techniques and compared to those particles assembled via conventional batch reactor pipetting. Our present work demonstrates that the use of microfluidics was able to assemble particles that had a reduced hydrodynamic diameter and an increase in the overall particle uniformity, a parameter that is difficult for self-assembled polymeric nanoparticles to achieve. Although we did not observe an increase in in vitro uptake or knockdown efficiency, endocytic inhibitor studies along with confocal microscopy colocalization studies suggest a difference in cellular uptake mechanisms and intracellular trafficking for the particles made by either method. Lastly, we show that while the preparation method did not affect in vitro knockdown efficiency, pulmonary delivery of the microfluidic micelleplexes in a murine model showed a significantly stronger decrease of GAPDH expression than conventionally prepared particles. While previous studies describe benefits regarding transfection efficiency for these microfluidic particles, this study is one of the first to demonstrate their potential in vivo. Along with the increase of in vivo knockdown efficiency, the use of such devices increases the scalability of well-defined nanoparticle production, a factor which is crucial for any clinical trial consideration. Along these lines, further in vivo targeting is underway with targeted triblock copolymer in addition to long term storage and stability studies of particles assembled with this technique.
Acknowledgments
This work was supported by the Wayne State Start-Up and ERC-2014-StG—637830 to Olivia Merkel as well as the National Institutes of Health (NIDA) R01 grant DA034783 supporting Anna Moszczynska. The NIH Center grant P30CA22453 supporting the Wayne State Microscopy, Imaging and Cytometry Resources (MICR) is gratefully acknowledged as well as the Wayne State School of Medicine for GRA support of Daniel Feldmann.
Footnotes
(Some figures may appear in colour only in the online journal)
A slight difference in β-actin expression towards lower expression levels may cause a normalized value of more than 1.0.
References
- 1.Liu L, Zheng M, Librizzi D, Renette T, Merkel OM, Kissel T. Efficient and tumor targeted siRNA delivery by polyethylenimine-graft-polycaprolactone-block-poly (ethylene glycol)-folate (PEI–PCL–PEG–Fol) Mol Pharm. 2015;13:134–43. doi: 10.1021/acs.molpharmaceut.5b00575. [DOI] [PubMed] [Google Scholar]
- 2.Jones SK, Lizzio V, Merkel OM. Folate receptor targeted delivery of siRNA and paclitaxel to ovarian cancer cells via folate conjugated triblock copolymer to overcome TLR4 driven chemotherapy resistance. Biomacromolecules. 2015;17:76–87. doi: 10.1021/acs.biomac.5b01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Béhé M, Kissel T. In vivo SPECT and real-time gamma camera imaging of biodistribution and pharmacokinetics of siRNA delivery using an optimized radiolabeling and purification procedure. Bioconjugate Chem. 2008;20:174–82. doi: 10.1021/bc800408g. [DOI] [PubMed] [Google Scholar]
- 4.Nadithe V, Liu R, Killinger BA, Movassaghian S, Kim NH, Moszczynska AB, Masters KS, Gellman SH, Merkel OM. Screening nylon-3 polymers, a new class of cationic amphiphiles, for siRNA delivery. Mol Pharm. 2014;12:362–74. doi: 10.1021/mp5004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Kim NH, Nadithe V, Schalk D, Thakur A, Kılıç A, Lum LG, Bassett DJ, Merkel OM. Targeted delivery of siRNA to activated T cells via transferrin-polyethylenimine (Tf-PEI) as a potential therapy of asthma. J Control Release. 2016;229:120–9. doi: 10.1016/j.jconrel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolomite Microfluidics Product Datasheet for Micromixer Chip 3200401. 2017 (Accessed: 1 December 2016) ( http://dolomite-microfluidics.com/images/stories/PDFs/datasheets/micromixer_chip_product_datasheet)
- 8.Zahn JD. Methods in Bioengineering: Biomicrofabrication and Biomicrofluidics. Boston, MA: Artech House; 2009. [Google Scholar]
- 9.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, Bawendi MG, Frangioni JV. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–70. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He C, Hu Y, Yin L, Tang C, Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–66. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 11.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33:941–51. doi: 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pathak Y, Benita S. Antibody-mediated Drug Delivery Systems: Concepts, Technology, and Applications. New York: Wiley; 2012. ( https://doi.org/10.1002/9781118229019) [Google Scholar]
- 13.Jiang W, Kim BY, Rutka JT, Chan WC. Nanoparticle-mediated cellular response is size-dependent. Nat Nanotechnol. 2008;3:145–50. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 14.Lauweryns JM, Baert JH. Alveolar clearance and the role of the pulmonary lymphatics 1–3. Am Rev Respiratory Dis. 1977;115:625–83. doi: 10.1164/arrd.1977.115.4.625. [DOI] [PubMed] [Google Scholar]
- 15.Endres T, Zheng M, Beck-Broichsitter M, Samsonova O, Debus H, Kissel T. Optimising the self-assembly of siRNA loaded PEG-PCL-lPEI nano-carriers employing different preparation techniques. J Control Release. 2012;160:583–91. doi: 10.1016/j.jconrel.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 16.Kasper JC, Schaffert D, Ogris M, Wagner E, Friess W. The establishment of an up-scaled micro-mixer method allows the standardized and reproducible preparation of well-defined plasmid/LPEI polyplexes. Eur J Pharm Biopharm. 2011;77:182–5. doi: 10.1016/j.ejpb.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Koh CG, Kang X, Xie Y, Fei Z, Guan J, Yu B, Zhang X, Lee LJ. Delivery of polyethylenimine/DNA complexes assembled in a microfluidics device. Mol Pharm. 2009;6:1333–42. doi: 10.1021/mp900016q. [DOI] [PubMed] [Google Scholar]
- 18.Lee WH, Loo CY, Young PM, Traini D, Mason RS, Rohanizadeh R. Recent advances in curcumin nanoformulation for cancer therapy. Expert Opin Drug Deliv. 2014;11:1183–201. doi: 10.1517/17425247.2014.916686. [DOI] [PubMed] [Google Scholar]
- 19.Win KY, Feng SS. Effects of particle size and surface coating on cellular uptake of polymeric nanoparticles for oral delivery of anticancer drugs. Biomaterials. 2005;26:2713–22. doi: 10.1016/j.biomaterials.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 20.Gary DJ, Puri N, Won YY. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Yu B, Tang C, Yin C. Enhanced antitumor efficacy of folate modified amphiphilic nanoparticles through co-delivery of chemotherapeutic drugs and genes. Biomaterials. 2014;35:6369–78. doi: 10.1016/j.biomaterials.2014.04.095. [DOI] [PubMed] [Google Scholar]
- 22.Merkel OM, Librizzi D, Pfestroff A, Schurrat T, Buyens K, Sanders NN, De Smedt SC, Béhé M, Kissel T. Stability of siRNA polyplexes from poly (ethylenimine) and poly (ethylenimine)-g-poly (ethylene glycol) under in vivo conditions: effects on pharmacokinetics and biodistribution measured by fluorescence fluctuation spectroscopy and single photon emission computed tomography (SPECT) imaging. J Control Release. 2009;138:148–59. doi: 10.1016/j.jconrel.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Elsayed M, Corrand V, Kolhatkar V, Xie Y, Kim NH, Kolhatkar R, Merkel OM. Influence of oligospermines architecture on their suitability for siRNA delivery. Biomacromolecules. 2014;15:1299–310. doi: 10.1021/bm401849d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogris M, Steinlein P, Kursa M, Mechtler K, Kircheis R, Wagner E. The size of DNA/transferrin-PEI complexes is an important factor for gene expression in cultured cells. Gene Ther. 1998;5:1425–33. doi: 10.1038/sj.gt.3300745. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Exocytosis Endocytosis. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- 26.Labat-Moleur F, Steffan AM, Brisson C, Perron H, Feugeas O, Furstenberger P, Oberling F, Brambilla E, Behr JP. An electron microscopy study into the mechanism of gene transfer with lipopolyamines. Gene Ther. 1996;3:1010–7. [PubMed] [Google Scholar]
- 27.Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 28.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem J. 2004;377:159–69. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurmi BD, Kayat J, Gajbhiye V, Tekade RK, Jain NK. Micro-and nanocarrier-mediated lung targeting. Expert Opin Drug Deliv. 2010;7:781–94. doi: 10.1517/17425247.2010.492212. [DOI] [PubMed] [Google Scholar]
- 30.Humphries IVWH, Szymanski CJ, Payne CK. Endolysosomal vesicles positive for Rab7 and LAMP1 are terminal vesicles for the transport of dextran. PloS One. 2011;6:e26626. doi: 10.1371/journal.pone.0026626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu P, Rudick M, Anderson RG. Multiple functions of caveolin-1. J Biol Chem. 2002;277:41295–8. doi: 10.1074/jbc.R200020200. [DOI] [PubMed] [Google Scholar]
- 32.Kanazawa T, Takematsu H, Yamamoto A, Yamamoto H, Kozutsumi Y. Wheat germ agglutinin stains dispersed post-golgi vesicles after treatment with the cytokinesis inhibitor psychosine. J Cell Physiol. 2008;215:517–25. doi: 10.1002/jcp.21328. [DOI] [PubMed] [Google Scholar]
- 33.Kou L, Sun J, Zhai Y, He Z. The endocytosis and intracellular fate of nanomedicines: implication for rational design. Asian J Pharm Sci. 2013;8:1–10. [Google Scholar]
- 34.García-Pérez BE, Hernández-González JC, García-Nieto S, Luna-Herrera J. Internalization of a non-pathogenic mycobacteria by macropinocytosis in human alveolar epithelial A549 cells. Microbial Pathogenesis. 2008;45:1–6. doi: 10.1016/j.micpath.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Endres T, Zheng M, Beck-Broichsitter M, Kissel T. Lyophilised ready-to-use formulations of PEG-PCL-PEI nano-carriers for siRNA delivery. Int J Pharm. 2012;428:121–4. doi: 10.1016/j.ijpharm.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Endres T, Zheng M, Kılıç A, Turowska A, Beck-Broichsitter M, Renz H, Merkel OM, Kissel T. Amphiphilic biodegradable PEG-PCL-PEI triblock copolymers for FRET-capable in vitro and in vivo delivery of siRNA and quantum dots. Mol Pharm. 2014;11:1273–81. doi: 10.1021/mp400744a. [DOI] [PubMed] [Google Scholar]
- 37.Feldmann DP, Merkel OM. The advantages of pulmonary delivery of therapeutic siRNA. Therapeutic Deliv. 2015;6:407–9. doi: 10.4155/tde.15.8. [DOI] [PubMed] [Google Scholar]
- 38.Thorley AJ, Ruenraroengsak P, Potter TE, Tetley TD. Critical determinants of uptake and translocation of nanoparticles by the human pulmonary alveolar epithelium. ACS Nano. 2014;8:11778–89. doi: 10.1021/nn505399e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachler G, Losert S, Umehara Y, von Goetz N, Rodriguez-Lorenzo L, Petri-Fink A, Rothen-Rutishauser B, Hungerbuehler K. Translocation of gold nanoparticles across the lung epithelial tissue barrier: combining in vitro and in silico methods to substitute in vivo experiments. Part Fibre Toxicol. 2015;12:18. doi: 10.1186/s12989-015-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harush-Frenkel O, Rozentur E, Benita S, Altschuler Y. Surface charge of nanoparticles determines their endocytic and transcytotic pathway in polarized MDCK cells. Biomacromolecules. 2008;9:435–43. doi: 10.1021/bm700535p. [DOI] [PubMed] [Google Scholar]
- 41.Voigt J, Christensen J, Shastri VP. Differential uptake of nanoparticles by endothelial cells through polyelectrolytes with affinity for caveolae. Proc Natl Acad Sci. 2014;111:2942–7. doi: 10.1073/pnas.1322356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maier O, Hoekstra D. Trans-golgi network and subapical compartment of HepG2 cells display different properties in sorting and exiting of sphingolipids. J Biol Chem. 2003;278:164–73. doi: 10.1074/jbc.M208259200. [DOI] [PubMed] [Google Scholar]


