Abstract
Oxidative metabolites of estrogens have been implicated in the development of breast cancer, yet relatively little is known about the metabolism of estrogens in the normal breast. We developed an experimental in vitro model of mammary estrogen metabolism in which we combined purified, recombinant phase I enzymes CYP1A1 and CYP1B1 with the phase II enzymes COMT and GSTP1 to determine how 17β-estradiol (E2) is metabolized. We employed both gas and liquid chromatography with mass spectrometry to measure the parent hormone E2 as well as eight metabolites, that is, the catechol estrogens, methoxyestrogens, and estrogen–GSH conjugates. We used these experimental data to develop an in silico model, which allowed the kinetic simulation of converting E2 into eight metabolites. The simulations showed excellent agreement with experimental results and provided a quantitative assessment of the metabolic interactions. Using rate constants of genetic variants of CYP1A1, CYP1B1, and COMT, the model further allowed examination of the kinetic impact of enzyme polymorphisms on the entire metabolic pathway, including the identification of those haplotypes producing the largest amounts of catechols and quinones. Application of the model to a breast cancer case-control population defined the estrogen quinone E2-3,4-Q as a potential risk factor and identified a subset of women with an increased risk of breast cancer based on their enzyme haplotypes and consequent E2-3,4-Q production. Our in silico model integrates diverse types of data and offers the exciting opportunity for researchers to combine metabolic and genetic data in assessing estrogenic exposure in relation to breast cancer risk.
Keywords: estrogen, metabolism, breast cancer, risk, genotype, model
Introduction
Estrogens have long been recognized as the prime risk factor for the development of breast cancer,1,2 but their assessment has not progressed beyond traditional exposure data, such as age, age at menarche, and age at first live birth. Although valuable in risk calculation, current models of breast cancer risk prediction based on cumulative estrogen exposure do not reflect observations of and data on mammary estrogen metabolism.3,4 Here we present a novel approach that is based on the molecular analysis of mammary estrogen metabolism.
Carcinogenesis is usually viewed as a stepwise process beginning with genotoxic effects (initiation) followed by enhanced cell proliferation (promotion). In the breast the main estrogen, 17β-estradiol (E2), is both a substrate for the phase I enzymes cytochrome P50 (CYP) 1A1 and 1B1 and a ligand for the estrogen receptor (ER). In its dual role of substrate and ligand, E2 has been implicated in the development of breast cancer by the way it simultaneously causes DNA damage via its oxidation products, the 2-OH and 4-OH catechol estrogens, and by how it stimulates cell proliferation and gene expression via the ER. Thus, E2 and its oxidative metabolites are unique carcinogens that affect both tumor initiation and promotion.5-8
Experiments on estrogen metabolism, formation of DNA adducts, mutagenicity, cell transformation, and carcinogenicity have implicated that certain estrogen metabolites, especially the catechol estrogen 4-hydroxyestradiol (4-OHE2), react with DNA via its quinone, causing mutations and initiating carcinogenesis.8-18 It is important to note that the concentration of the 4-OHE2 metabolite in human breast tissue is actually higher than that of the parent hormone, E2, evidence that oxidative estrogen metabolism may be a critical factor in the development of human breast cancer.14,19 Estrogen–DNA adducts have been detected in normal and malignant human breast tissues,20,21 and we have recently provided direct experimental proof that oxidative metabolism of E2 leads to the formation of 4-OHE2 and deoxyribonucleoside adducts.22 The collective evidence points to the importance of including mammary estrogen metabolism data into risk calculations, which is what we aim to accomplish with our new model.23
Models of Mammary Estrogen Metabolism
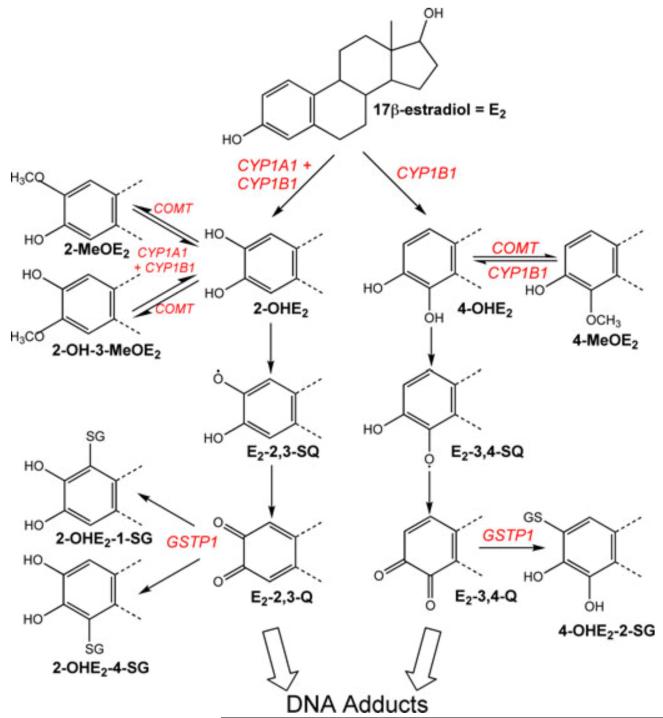
Several investigators have proposed a qualitative model of mammary estrogen metabolism regulated by oxidizing phase I and conjugating phase II enzymes.9,24 As shown in Figure 1, E2 is oxidized to catechol estrogens by CYP1A1 and 1B1. These enzymes further oxidize the catechol estrogens to semiquinones and quinones. The highly reactive estrogen quinones form Michael addition products with deoxyribonucleosides.25-27 Thus, estrogen quinones share a common feature of many chemical carcinogens, that is, the ability to covalently modify DNA.12,28,29 Furthermore, estrogen semiquinones and quinones undergo redox-cycling, which results in the production of reactive oxygen species that can cause oxidative DNA damage.30-32 It is postulated that the genotoxicity of the oxidative estrogen metabolism pathway is mitigated by alternate reactions of the metabolites with phase II enzymes. Specifically, COMT catalyzes the methylation of catechol estrogens to methoxy estrogens, which lowers the amount of catechol estrogens available for conversion to estrogen quinones.33,34 In turn, the estrogen quinones undergo conjugation with glutathione (GSH) via the catalytic action of GSTP1.35 The formation of GSH–estrogen conjugates would reduce the level of-estrogen quinones and thereby lower the potential for DNA damage.
Figure 1.
Oxidative estrogen metabolism causes DNA adduct formation. The estrogen metabolism pathway is regulated by oxidizing phase I and conjugating phase II enzymes. CYP1A1 and CYP1B1 catalyze the oxidation of E2 to catechol estrogens 2-OHE2 and 4-OHE2. The catechol estrogens are either methylated by COMT to methoxyestrogens (2-MeOE2, 2-OH-3-MeOE2, 4-MeOE2) or further oxidized by CYPs to semiquinones (E2-2,3-SQ, E2-3,4-SQ) and quinones (E2-2,3-Q, E2-3,4-Q). The estrogen quinones are conjugated by GSTP1 to GSH-conjugates (2-OHE2-1-SG, 2-OHE2-4-SG, 4-OHE2-2-SG). Alternatively, the quinones can form quinone-DNA adducts (e.g., 4-OHE2-N7-guanine, 2-OHE2-N2-deoxyguanosine) or cause oxidative adducts (e.g., 8-OH-deoxyguanosine) via reactive oxygen species resulting from redox-cycling between semiquinones and quinones. The three adducts and their estrone (E1) and adenine counterparts have been identified in human breast tissues.20,21 Recently, we demonstrated experimentally that CYP1B1-mediated oxidation of E2 in the presence of deoxyguanosine caused the formation of the 4-OHE2-N7-guanine adduct.22 Our results provide direct evidence that metabolism of the parent hormone can initiate DNA damage.
The current model of mammary estrogen metabolism has several limitations. First, only single enzymes, for example, CYP1B1 and COMT, have been analyzed to date with simple substrate-product kinetics, which clearly generates an incomplete picture of the metabolic pathway. Second, while the model incorporates the functional roles of the phase I and II enzymes, it does so only qualitatively, and it remains uncertain how the enzymes interact quantitatively. Third, each of the phase I and II enzymes contains genetic polymorphisms.33,36-38 Studies from several laboratories, including our own, have examined the functional implications of the polymorphisms on estrogen metabolism, again focusing on single enzymes.33,34,38-40 Thus, the multitude of potential kinetic reactions resulting from the complex genetic variations of the phase I and II enzymes is completely outside the scope of the current model of estrogen metabolism. In contrast to the relatively small number of functional studies of estrogen metabolism multiple epidemiological studies have investigated breast cancer risk in relation to genetic variation in the critical enzymes involved in estrogen metabolism with inconsistent findings.41,42 Such studies are limited by their ability to consider only one or two of the enzymes in the estrogen metabolic pathway. Furthermore, those studies that examined all of the component enzymes were not able to assess underlying metabolic interactions in the pathway.43,44 The drawback of any purely genetic assessment is the lack of information about functional interactions inherent in complex metabolic pathways such as the estrogen metabolism pathway. Moreover, DNA analysis identifies the variant alleles but does not quantify the variation in the dynamics of the pathway. In contrast, the functional protein analysis will provide a quantitative assessment with each variation in protein structure likely to have a different effect. Thus, a pathway-based functional and quantitative approach is needed to overcome the current limitation in genotype assessment.
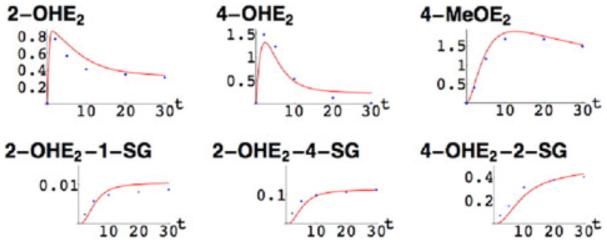
We recently developed an experimental in vitro model of mammary estrogen metabolism, in which we combined purified, recombinant phase I enzymes CYP1A1 and CYP1B1 with the phase II enzymes COMT and GSTP1 to determine how E2 is metabolized.45 We employed both gas and liquid chromatography with mass spectrometry (GC–MS and LC-MS) to measure the parent hormone E2 as well as eight metabolites, i.e., the catechol estrogens, methoxyestrogens, and estrogen–GSH conjugates. We then used these experimental data to develop an in silico model of the metabolic pathway.23 The in silico model allowed the kinetic simulation of converting E2 into eight metabolites. The simulations showed excellent agreement with experimental results and provided a quantitative assessment of the metabolic interactions (Fig. 2). Using rate constants of genetic variants of CYP1A1, CYP1B1, and COMT, the model further allowed examination of the kinetic impact of enzyme polymorphisms on the entire estrogen metabolic pathway, including the identification of those haplotypes producing the largest amounts of catechols and quinones (Fig. 3).
Figure 2.
Comparison of mathematical model with experimental data. The red curves are plots of the solutions to the nonlinear system of differential equations and the blue dots are experimental data.45 As shown, the model allowed simulations of all reactions in the pathway, which agreed well with the experimentally determined results.23
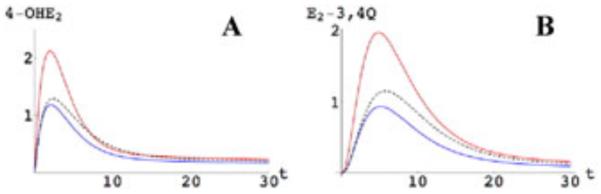
Figure 3.
Kinetic–genomic modeling of catechol estrogen (A) 4-OHE2 and estrogen quinone (B) E2-3,4-Q using rate constants for wild-type and variant CYP1A1, CYP1B1, and COMT. The Area Under the Curve = AUC represents the metabolite production over time. Only the highest, lowest, and wild-type (dotted line) AUCs are shown.23
Our in silico model is pertinent to the numerous epidemiological studies that have examined the association of genetic variants of enzymes involved in estrogen metabolism with breast cancer risk.41,42 These studies were handicapped because they investigated only one or two enzymes, but even those examining all enzymes have been fundamentally limited by not having the means to assess the underlying metabolic interactions.43,44 Our model attempts to fill this gap, and we applied it to a hospital-based case-control population that was analyzed previously with respect to CYP1A1, CYP1B1, and COMT genotypes.43,46,47 Here we went beyond genotypes and used the model to determine for each woman the effect of her composite CYP1A1, CYP1B1, and COMT haplotypes on estrogen metabolite production. Inherited variations in enzyme genotype persist throughout life and can therefore be regarded as constants for each individual.
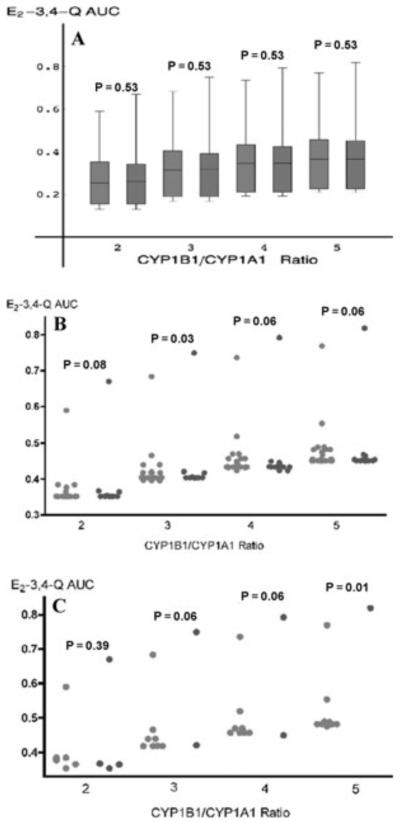
Application of the model to a breast cancer case-control population (221 invasive breast cancer cases, 217 controls) defined the estrogen quinone E2-3,4-Q as a potential risk factor. In this exploratory study, the model identified a subset of women with an increased risk of breast cancer based on their enzyme haplotype and consequent E2-3,4-Q production (Fig. 4).23 Whether the E2-3,4-Q is an independent risk factor, as suggested by our exploratory study, will need to be confirmed by a larger separate study.
Figure 4.
Correlation of E2-3,4-Q AUC with CYP1B1/CYP1A1 ratio for cases and controls.23 (A) Box and whisker graph of E2-3,4-Q AUCs for entire population of 221 cases (red) and 217 controls (blue). Each box includes 84% of the respective group while the whiskers represent the top and bottom 8 percentiles. As indicated by the medians (center line in each box), the AUCs for cases and controls rise with increasing CYP ratio. However, there are no significant differences between case and control medians at any CYP ratio tested (see P-values). (B) Column scatter graph of E2-3,4-Q AUCs for top 8 percentile (35 subjects) of entire study population. Each dot represents an individual case (red) or control (blue). Subjects with the same composite CYP1A1-CYP1B1-COMT enzyme haplotype have the same E2-3,4-Q AUC. As the CYP ratio increases, their E2-3,4-Q AUC changes in the same manner. However, subjects with different composite enzyme haplotypes may yield different E2-3,4-Q AUC values, resulting in a change in their ranking with increasing CYP ratio. (C) Column scatter graph of E2-3,4-Q AUCs for top 2 percentile (10 subjects) of entire study population. There are significantly more cases (red) than controls (blue) (P = 0.01 at CYP1B1/CYP1A1 = 5).
Summary
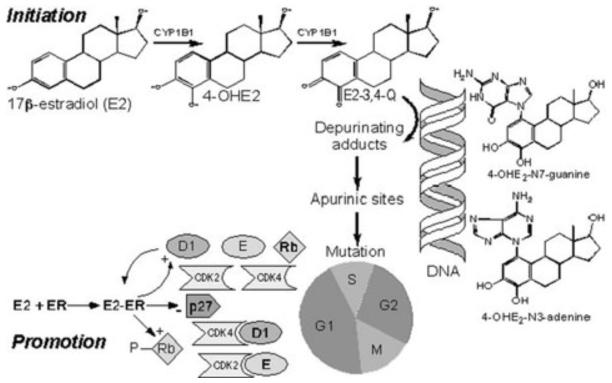
Carcinogenesis is usually viewed as a stepwise process that begins with genotoxic effects (initiation) followed by enhanced cell proliferation (promotion). The main estrogen, E2, is a substrate for oxidizing enzymes such as CYP1B1 and a ligand for the ER. In its dual role of substrate and ligand, E2 has been implicated in the development of breast cancer by its simultaneously causing DNA damage via its oxidation products, the catechol estrogens, and estrogen quinones and by stimulating cell proliferation and gene expression via the ER. Thus, E2 and its oxidative metabolites are unique carcinogens that affect both tumor initiation and promotion (Fig. 5).5-8
Figure 5.
Overview of estrogen carcinogenesis as a two-step process beginning with genotoxic effects (initiation) followed by enhanced cell proliferation (promotion). In the breast, E2 is a substrate for CYP1B1 and a ligand for the estrogen receptor. The main enzyme, CYP1B1, oxidizes E2 to catechol estrogens and further to quinones, such as 4-OHE2 and E2-3,4-Q. The highly reactive E2-3,4-Q forms Michael addition products with deoxyribonucleosides. The resulting depurinating adducts, such as 4-OHE2-N7-guanine and 4-OHE2-N3-adenine, leave apurinic sites in the DNA. Unless repaired during G1 of the cell cycle, DNA replication during the S phase may lead to mutations that can be propagated into daughter cells during the M phase. Studies of ER-positive breast cancer cell lines demonstrated that E2 increased the rate of cell proliferation by two mechanisms, i.e., by recruiting noncycling cells from a quiescent G0 state into the cell cycle and by shortening the overall cell cycle time due predominantly to a reduction in length of the G1 phase.48 Binding of E2 to ER stimulates progression of G1 by increasing the concentration of cyclin D1, which, in turn, enhances ER-mediated transcription. E2 also causes a decrease in the concentration of CDK inhibitor p27 and a rise in cyclin D1-CDK4 and cyclin E-CDK2 activities accompanied by hyperphosphorylation of Rb.
Basing further research on our kinetic-genomic model, it should be possible for investigators to develop more refined risk prediction models that integrate known reproductive and lifestyle factors with predicted exposure to E2-3,4-Q as determined by inherited variation in critical genes involved in the estrogen metabolic pathway. Being based upon a validated laboratory model and high quality epidemiologic data, this research has the potential to significantly enhance our ability to predict breast cancer risk in individual women—a potential with direct implications for breast cancer screening and earlier detection of disease.
Acknowledgments
Supported by NIH (1R01CA/ES83752, U54CA113007) and the Vanderbilt Integrative Cancer Biology Center.
Financial Support
This work was partially supported under grant U54CA113007
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.MacMahon B, Feinleib M. Breast cancer in relation to nursing and menopausal history. J. Natl. Cancer Inst. 1960;24:733–753. doi: 10.1093/jnci/24.3.733. [DOI] [PubMed] [Google Scholar]
- 2.Pike MC, et al. “Hormonal” risk factors, “breast tissue age” and the age-incidence of breast cancer. Nature. 1983;303:767–770. doi: 10.1038/303767a0. [DOI] [PubMed] [Google Scholar]
- 3.Gail MH, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl. Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 4.Rockhill B, et al. Validation of the Gail et al. model of breast cancer risk prediction and implications for chemoprevention. J. Natl. Cancer Inst. 2001;93:358–366. doi: 10.1093/jnci/93.5.358. [DOI] [PubMed] [Google Scholar]
- 5.Gaikwad NW, et al. The molecular etiology of breast cancer: Evidence from biomarkers of risk. Int. J. Cancer. 2008;122:1949–1957. doi: 10.1002/ijc.23329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parl FF. Estrogens, Estrogen Receptor and Breast Cancer. IOS Press; Amsterdam: 2000. [Google Scholar]
- 7.Yager JD, Davidson NE. Estrogen carcinogenesis in breast cancer. New Engl. J. Med. 2006;354:270–282. doi: 10.1056/NEJMra050776. [DOI] [PubMed] [Google Scholar]
- 8.Yue W, et al. Genotoxic metabolites of estradiol in breast: Potential mechanism of estradiol induced carcinogenesis. J. Steroid Biochem. Mol. Biol. 2003;86:477–486. doi: 10.1016/s0960-0760(03)00377-7. [DOI] [PubMed] [Google Scholar]
- 9.Cavalieri EL, et al. Molecular origin of cancer: Catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc. Natl. Acad. Sci. USA. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devanesan P, et al. Catechol estrogen metabolites and conjugates in mammary tumors and hyperplastic tissue from estrogen receptor-α knockout (ERKO)/Wnt-1 mice: Implications for initiation of mammary tumors. Carcinogenesis. 2001;22:1573–1576. doi: 10.1093/carcin/22.9.1573. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez SV, Russo IH, Russo J. Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int. J. Cancer. 2006;118:1862–1868. doi: 10.1002/ijc.21590. [DOI] [PubMed] [Google Scholar]
- 12.Li KM, et al. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 13.Newbold RR, Liehr JG. Induction of uterine adenocarcinoma in CD-1 mice by catechol estrogens. Cancer Res. 2000;60:235–237. [PubMed] [Google Scholar]
- 14.Rogan EG, et al. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: Potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 15.Russo J, et al. 17-beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J. 2006;20:1622–1634. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 16.Russo J, et al. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J. Steroid Biochem. Mol. Biol. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 17.Saeed M, et al. Formation of depurinating N3Adenine and N7Guanine adducts by MCF-10F cells cultured in the presence of 4-hydroxyestradiol. Int. J. Cancer. 2007;120:1821–1824. doi: 10.1002/ijc.22399. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Z, et al. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB Rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem. Res. Toxicol. 2006;19:475–479. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 19.Castagnetta LAM, et al. Tissue content of hydroxyestrogens in relation to survival of breast cancer patients. Clin. Cancer Res. 2002;8:3146–3155. [PubMed] [Google Scholar]
- 20.Embrechts J, et al. Detection of estrogen DNA-adducts in human breast tumor tissue and healthy tissue by combined nano LC-nano ES tandem mass spectrometry. J. Am. Soc. Mass. Spectrom. 2003;14:482–491. doi: 10.1016/S1044-0305(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 21.Markushin Y, et al. Spectral characterization of catechol estrogen quinone (CEQ)-derived DNA adducts and their identification in human breast tissue extract. Chem. Res. Toxicol. 2003;16:1107–1117. doi: 10.1021/tx0340854. [DOI] [PubMed] [Google Scholar]
- 22.Belous AR, et al. Cytochrome P450 1B1-mediated estrogen metabolism results in estrogen-deoxyribonucleoside adduct formation. Cancer Res. 2007;67:812–817. doi: 10.1158/0008-5472.CAN-06-2133. [DOI] [PubMed] [Google Scholar]
- 23.Crooke PS, et al. Estrogens, enzyme variants, and breast cancer: A risk model. Cancer Epidemiol. Biomarkers Prev. 2006;15:1620–1629. doi: 10.1158/1055-9965.EPI-06-0198. [DOI] [PubMed] [Google Scholar]
- 24.Yager JD, Liehr JG. Molecular mechanisms of estrogen carcinogenesis. Annu. Rev. Pharmacol. Toxicol. 1996;36:203–232. doi: 10.1146/annurev.pa.36.040196.001223. [DOI] [PubMed] [Google Scholar]
- 25.Akanni A, Abul-Hajj YJ. Estrogen-nucleic acid adducts: Reaction of 3,4-estrone o-quinone radical anion with deoxyribonucleosides. Chem. Res. Toxicol. 1997;10:760–766. doi: 10.1021/tx970026c. [DOI] [PubMed] [Google Scholar]
- 26.Bolton JL, et al. Role of quinoids in estrogen carcinogenesis. Chem. Res. Toxicol. 1998;11:1113–1127. doi: 10.1021/tx9801007. [DOI] [PubMed] [Google Scholar]
- 27.Stack DE, et al. Molecular characteristics of catechol estrogen quinones in reactions with deoxyribonucleosides. Chem. Res. Toxicol. 1996;9:851–859. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- 28.Bolton JL, et al. Role of quinones in toxicology. Chem. Res. Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 29.Cavalieri EL, et al. Catechol ortho-quinones: The electrophilic compounds that form depurinating DNA adducts and could initiate cancer and other diseases. Carcinogenesis. 2002;23:1071–1077. doi: 10.1093/carcin/23.6.1071. [DOI] [PubMed] [Google Scholar]
- 30.Han X, Liehr JG. Microsome-mediated 8-hydroxylation of guanine bases of DNA by steroid estrogens: Correlation of DNA damage by free radicals with metabolic activation to quinones. Carcinogenesis. 1995;16:2571–2574. doi: 10.1093/carcin/16.10.2571. [DOI] [PubMed] [Google Scholar]
- 31.Liehr JG, Roy D. Free radical generation by redox cycling of estrogens. Free Radical Biol. Med. 1990;8:415–423. doi: 10.1016/0891-5849(90)90108-u. [DOI] [PubMed] [Google Scholar]
- 32.Nutter LM, et al. An o-quinone form of estrogen produces free radicals in human breast cancer cells: Correlation with DNA damage. Chem. Res. Toxicol. 1994;7:23–28. doi: 10.1021/tx00037a004. [DOI] [PubMed] [Google Scholar]
- 33.Dawling S, et al. Catechol-O-methyltransferase (COMT)-mediated metabolism of catechol estrogens: Comparison of wild-type and variant COMT isoforms. Cancer Res. 2001;61:6716–6722. [PubMed] [Google Scholar]
- 34.Goodman JE, et al. Characterization of human soluble high and low activity catechol-O-methyltransferase catalyzed catechol estrogen methylation. Pharmacogenetics. 2002;12:517–528. doi: 10.1097/00008571-200210000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Hachey DL, et al. Sequential action of phase I and II enzymes cytochrome P450 1B1 and glutathione S-transferase P1 in mammary estrogen metabolism. Cancer Res. 2003;63:8492–8499. [PubMed] [Google Scholar]
- 36.Ali-Osman F, et al. Molecular cloning, characterization, and expression in Escherichia coli of full-length cDNAs of three human glutathione S-transferase Pi gene variants. J. Biol. Chem. 1997;15:10004–10012. doi: 10.1074/jbc.272.15.10004. [DOI] [PubMed] [Google Scholar]
- 37.Cascorbi I, Brockmoller J, Roots I. A C4887A polymorphism in Exon 7 of Human CYP1A1: Population frequency, mutation linkages, and impact on lung cancer susceptibility. Cancer Res. 1996;56:4965–4969. [PubMed] [Google Scholar]
- 38.Hanna IH, et al. Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: Association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res. 2000;60:3440–3444. [PubMed] [Google Scholar]
- 39.Li DN, et al. Polymorphisms in P450 CYP1B1 affect the conversion of estradiol to the potentially carcinogenic metabolite 4-hydroxyestradiol. Pharmacogenetics. 2000;10:343–353. doi: 10.1097/00008571-200006000-00008. [DOI] [PubMed] [Google Scholar]
- 40.Shimada T, et al. Specificity of 17β-oestradiol and benzo[a]pyrene oxidation by polymorphic human cytochrome P4501B1 variants substituted at residues 48, 119 and 432. Xenobiotica. 2001;31:163–176. doi: 10.1080/00498250110043490. [DOI] [PubMed] [Google Scholar]
- 41.Dunning AM, et al. A systematic review of genetic polymorphisms and breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 1999;8:843–854. [PubMed] [Google Scholar]
- 42.Mitrunen K, Hirvonen A. Molecular epidemiology of sporadic breast cancer. The role of polymorphic genes involved in estrogen biosynthesis and metabolism. Mutat. Res. 2003;544:9–41. doi: 10.1016/s1383-5742(03)00016-4. [DOI] [PubMed] [Google Scholar]
- 43.Ritchie MD, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas DC. The need for a systematic approach to complex pathways in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 2005;14:557–559. doi: 10.1158/1055-9965.EPI-14-3-EDB. [DOI] [PubMed] [Google Scholar]
- 45.Dawling S, et al. In vitro model of mammary estrogen metabolism: Structural and kinetic differences in mammary metabolism of catechol estrogens 2- and 4-hydroxyestradiol. Chem. Res. Toxicol. 2004;17:1258–1264. doi: 10.1021/tx0498657. [DOI] [PubMed] [Google Scholar]
- 46.Bailey LR, et al. Association of cytochrome P450 1B1 (CYP1B1) polymorphism with steroid receptor status in breast cancer [Erratum: Cancer Res 1999; 59: 1388] Cancer Res. 1998;58:5038–5041. [PubMed] [Google Scholar]
- 47.Bailey LR, et al. Breast cancer and CYP1A1, GSTM1, and GSTT1 polymorphisms: Evidence of a lack of association in Caucasians and African Americans. Cancer Res. 1998;58:65–70. [PubMed] [Google Scholar]
- 48.Foster JS, et al. Estrogens and cell-cycle regulation in breast cancer. Trends Endocrinol. Metab. 2001;12:320–327. doi: 10.1016/s1043-2760(01)00436-2. [DOI] [PubMed] [Google Scholar]