Abstract
Th17 cells play a role as an inflammation mediator in a variety of autoimmune disorders, including inflammatory bowel disease (IBD) and thus are widely considered to be pathogenic. However, Th17 cells are present in the normal intestine and show a homeostatic phenotype, i.e., they participate in the maintenance of intestinal homeostasis rather than inducing inflammation. We observed an enlarged Th17 population in the small intestine of C57BL/6.IgA−/− mice compared to wild-type mice, which was further amplified with cholera toxin (CT) immunization without causing intestinal inflammation. The increased Th17 induction and the correspondingly 10-fold higher CTB-specific serum IgG response in C57BL/6.IgA−/− mice after CT immunization was microbiota dependent and was associated with increased segmented filamentous bacteria (SFB) in the small intestine of C57BL/6.IgA−/− mice. Oral administration of vancomycin greatly dampened both CT immunogenicity and adjuvanticity, and the differential CT responses in IgA−/− and wild-type mice disappeared after intestinal microbiota equalization. Using gnotobiotic mouse models, we found that CT induction of homeostatic intestinal Th17 responses was supported not only by SFB but also by other commensal bacteria. Furthermore, transcriptome analysis using IL-17AhCD2 reporter mice revealed a similar gene expression profile in CT-induced intestinal Th17 cells and endogenous intestinal Th17 cells at homeostasis, with upregulated expression of a panel of immune regulatory genes, which was distinctly different from the gene expression profile of pathogenic Th17 cells. Taken together, we identified a non-pathogenic signature of intestinal homeostatic Th17 cells, which are actively regulated by the commensal microbiota and can be selectively stimulated by CT.
INTRODUCTION
There are trillions of microbes residing in the normal mammalian intestine1. These microbes include bacteria, archaea, fungi and viruses, of which bacteria are the most abundantly studied. The role of intestinal microbiota in the induction and modulation of the host immune system has been increasingly recognized in recent years2,3. At steady state, the mammalian intestine is the biggest reservoir of activated effector T cells, which provide protection against potential intestinal pathogens together with other immune cell subsets. Th17 cells are among the most abundant effector CD4+ T cells in the intestinal lamina propria (LP)4. They are characterized by the expression of master transcription factor Rorγt and the production of cytokines including IL-17A, IL-17F, IL-21 and IL-225. Th17 cells show heterogeneity in their gene expression profile in different research models and conditions. For instance, they are involved in the pathogenesis of an array of autoimmune diseases, including rheumatoid arthritis, multiple sclerosis, experimental autoimmune encephalomyelitis, psoriasis and inflammatory bowel diseases (IBD). Such pathogenic Th17 cells frequently co-express IFNγ and TNFα6. In other studies, Th17 cells have been shown to be indispensable in fighting against respiratory fungal infections and intestinal bacterial pathogens via the production of IL-17A and IL-227.
Segmented filamentous bacteria (SFB) are a group of Gram-positive, spore forming commensal bacteria that form long filaments, and preferentially adhere tightly to the epithelium and Peyer’s patches (PPs) of the terminal ileum. Colonization with SFB strongly induces endogenous Th17 cells as well as a broad range of other pro-inflammatory cytokines in the murine small intestine8,9. Such induction is considered as a beneficial stimulation of the immune system, and shown to provide mice with better protection against Citrobacter rodentium infection. However, SFB may be only one among a large pool of microbes that are likely to elicit Th17 cells10, and other bacterial species that could do so still await identification.
Cholera toxin (CT), which is a potent mucosal immunogen and adjuvant, also has the capacity to induce mucosal Th17 cell differentiation after intranasal administration11. Mucosal delivery of CT induces strong mucosal and systemic humoral immune responses to itself and co-administered protein antigens12, but does not cause intestinal inflammation. CT does so by activating Gsα and the nucleotide-binding oligomerization domain containing 2 (Nod2) expressed in CD11c+ cells13,14. Th17 cells can convert into T follicular helper (Tfh) cells in the PPs, thus enhancing high-affinity intestinal IgA production15, suggesting an important role of Th17 cells in the induction of antigen-specific IgA in the gut, which in turn is involved in the protection against microbial antigens at the mucosal surface16.
IgA deficiency is the most commonly seen primary immunodeficiency17, and has been shown to relate to IBD pathogenesis on genome wide association studies18. Interestingly, in mice that are deficient of IgA, we observed increased Th17 cells in their normal small intestine, which can be further augmented with mucosal CT immunization without inducing any inflammation. This increased Th17 and antibody response to CT in IgA−/− mice was dependent on intestinal microbiota. Experiments with gnotobiotic mice indicate that not only SFB but also other commensal bacteria participate in CT induction of the Th17 response. Moreover, we performed mRNA sequencing analysis and found that CT-induced intestinal Th17 cells share a similar expression profile with endogenous intestinal Th17 cells at homeostasis, and both of them are distinctly different from pathogenic Th17 cells.
MATERIALS AND METHODS
Mice
C57BL/6 (B6) mice and IL-6−/− mice were purchased from The Jackson Laboratory. IgA−/− mice were kindly provided by Dr. I. N. Mbawuike from Baylor College of Medicine, Houston, TX. 10BiT.Foxp3gfp mice were kindly provided by Dr. Craig Maynard, UAB. IgA−/−.10BiT.Foxp3gfp mice were generated by crossing 10BiT.Foxp3gfp mice with IgA−/− mice. IL-17AhCD2 mice were generated and kindly provided by Dr. Stacey Harbour and Dr. Casey Weaver, UAB. IgA−/−.IL-17AhCD2 mice were generated by crossing IL-17AhCD2 mice with IgA−/− mice, using the latter as the dam. All mice were on C57BL/6 background and were maintained in the Animal Facility at the University of Alabama at Birmingham (UAB) under specific pathogen free conditions. Germ free mice derivation and maintenance was as described before19. For inoculation of GF/ASF mice with SFB, fecal pellets were purchased from CGIBD Gnotobiotic Core of UNC (collected from SFB-monocolonized mice using sterilized test tubes in the vinyl-isolator) and were preserved frozen under dry ice/−80°C until immediately before oral administration. SFB colonization was performed by oral gavage with 300–400μl of suspension obtained by homogenizing the fecal pellets from SFB-monocolonized mice in water. Fecal pellets of the recipients were collected 2 weeks after gavage and SFB colonization was confirmed with PCR (primers: forward, 5′-ACG CTA CAT CGT CTT ATC TTC CCG C-3′; reverse, 5′-TCC CCC AAG ACC AAG TTC ACG A-3′). Vancomycin treatment was performed with indicated schedules with 0.5mg/day in the drinking water. For intragastric immunization, 10μg CT and 1mg OVA in 500μl 0.2mM NaHCO3 was gavaged on days 0, 7, and 14. Mice were sacrificed on day 28 for examination. Intracolonic immunization was performed with 10μg CT and 1mg OVA in 100μl PBS, with the same immunization schedule. SCFA repletion was performed with 100mM butyrate or 150mM propionate in the drinking water from day 0 of CT immunization till mice were sacrificed. 8–12 week-old mice were used in these experiments unless specified otherwise. All experiments were reviewed and approved by the Institutional Animal Care and Use Committee of UAB.
Reagents and materials
Reagents and materials were purchased from the following sources: anti–mouse CD4 (RM4-5), CD25 (PC61) antibodies were purchased from BD Biosciences; anti-mouse IL-17A (TC11-18H10.1), IFN-γ (XMG1.2), CD90.1 (OX7) antibodies were purchased from BioLegend; anti-mouse Foxp3 (FJK-16s) and IL-22 (1H8PWSR) antibodies as well as Foxp3 staining buffer set were purchased from eBioscience; anti-mouse CD90.1 (19E.2) and anti-human CD2 (OKT11) were purchased from the hybridoma lab of Epitope Recognition and Immunoreagent Core at UAB; live/dead staining kit was purchased from Invitrogen; mouse lamina propria dissociation kit and regulatory T cell isolation kit were purchased from Miltenyi Biotec; mouse LCN-2 ELISA kit was purchased from R&D Systems; fecal DNA extraction kit was purchased from Zymo Research.
Isolation and in vitro culturing of LP cells
Small and large intestines were removed, sliced, and washed with ice-cold PBS to remove fecal content. After removing epithelium by gentle shaking in Ca2+/Mg2+-free HBSS supplemented with 5mM EDTA, 1mM DTT and 2% FBS for 40 min at 37°C, the tissue was washed and resuspended in digestion medium following Miltenyi protocol and then incubated for 30 min at 37°C by gentle shaking. Cells were passed through a mesh, centrifuged, and the pellet was resuspended in 40% Percoll and carefully overlaid onto 70% Percoll. The interface containing the LP leukocytes was collected and washed with PBS supplemented with 10% FBS. 106 LPLs were co-cultured with 5x104 APCs isolated from naïve C57BL/6 mouse spleen (CD4− fraction) with the addition of either SFB, CTB or OVA T cell epitope peptide (SFB 568-580, DVQFSGAVPNKTD; CTB 89-100, NNKTPHAIAAIS; OVA 323-339, ISQAVHAAHAEINEAGR)20–22 for 5 days. Culture supernatant was then collected and measured for cytokine production.
Intracellular staining for cytokine production
As described previously23, isolated cells were stimulated for 4–5h with phorbol 12-myristate 13-acetate (PMA, 50ng/ml, Sigma) and ionomycin (750ng/ml, Sigma) in the presence of Golgi stop (10μg/ml, BD Biosciences). After staining of cell surface antigens, cells were fixed and permeabilized using Foxp3 staining buffer set (eBioscience), followed with intracellular cytokine staining. Dead cells were excluded by surface staining with Live/Dead staining kit. Flow cytometry was performed on BD LSR II and analyzed with FlowJo software (V9.3.3, TreeStar)
ELISA and Multi-plex assay
96-well ELISA plates (Thermo Fisher Scientific MaxiSorp) were coated with capture antibody in PBS or coating buffer at 4°C overnight. Plates were washed and blocked, then samples and standards were added and incubated at room temperature for 2 hours. After washing, biotin labeled detection antibodies and streptavidin-HRP were added and incubated for another hour. Followed by final washes, plates were developed with TMB solutions for 15mins, and read at 450nm. Sample concentrations were calculated according to the standard curve. Milliplex mouse Th17 magnetic kit was purchased from Millipore. Multi-plex assay was conducted following manufacturer’s instructions. 9 cytokines including IFNγ, IL-4, IL-5, IL-13, IL-6, IL-17A, IL-21, IL-22, and IL-10 were measured simultaneously.
Histopathologic assessment
Naïve IgA−/− mice at 9-month-old and CT immunized mice at 12-week-old (immunization started at 8-week-old with the schedule described above) were sacrificed for histopathologic assessment. At necropsy, the small intestine, cecum, and colon were separated and cut open longitudinally. Tissues were fixed in 10% buffered formalin and paraffin embedded. The sections (5 μm) were stained with hematoxylin and eosin. All slides were scored by an experienced veterinary pathologist (T. Schoeb, UAB) without knowledge of their origin.
Microbiome sequencing and data analysis
Bacterial genomic DNA from mouse ileal and cecal contents was extracted with ZR Fecal DNA MiniPrep kit following manufacture’s protocol. The V4 region of 16S rRNA gene was targeted for PCR amplification using modified F515/R806 primer24. Duplicates were run for all PCR reactions. The 150 paired-end sequencing reaction was performed using the MiSeq platform (Illumina) at the Gut Microbiome and Large Animal Biosecurity Laboratories, Department of Animal Science, University of Manitoba, Winnipeg, MB, Canada. The FLASH assembler was used to merge overlapping paired-end Illumina fastq files25. The output fastq file was then further analyzed by downstream computational pipelines of the open source software package QIIME26. Taxonomies were assigned to the representative sequence of each OTU using RDP classifier, and were aligned with the Greengenes Core reference database using PyNAST algorithms. Within community diversity (α-diversity) was calculated using QIIME. An Alpha rarefaction curve was generated using Chao 1 estimator of species richness with ten sampling repetitions at each sampling depth. An even depth of approximately 13,100 sequences per sample was used for calculation of richness and diversity indices. To compare microbial composition between samples, β-diversity was measured by calculating the weighted and unweighted Unifrac distances27 using QIIME default scripts to generate principal coordinate analysis (PCoA) plot and generated nonmetric multidimensional scaling (NMDS) plots using Bray-Curtis distance in R with package “picante”. Permutational multivariate analysis of variance (PERMANOVA) was used to calculate P-values and test for significant differences of β-diversity among treatment groups.
In vitro generation of pathogenic Th17 cells and gene expression analysis
CD4+CD25− cells were purified from spleen with Treg isolation kit (Miltenyi Biotech) and polarized to pathogenic Th17 cells with plate-bound anti-CD3 (5μg/ml; 145-2C11; BD Biosciences) and anti-CD28 (2μg/ml; 37.51; BD Biosciences) in the presence of recombinant human TGFβ1 (5ng/ml; R&D Systems), recombinant mouse IL-6 (20ng/ml; R&D Systems), recombinant mouse IL-23 (20ng/ml; R&D Systems) and recombinant mouse IL-1β (20ng/ml; R&D Systems). Cells were cultured for 5 days and CD4+hCD2+ cells were surface stained with fluorescent antibodies and sorted with BD FACSAria and stored in lysis buffer. mRNA was isolated using Dynabeads mRNA DIRECT purification kit (Life technologies) following manufacturer’s protocol. RNA-Seq and data analysis were performed at the Broad Institute of MIT and Harvard. In brief, cDNA libraries were generated using the Smart-seq2 protocol, modified for 500+ cells of input, and sequenced on a MiSeq (Illumina, San Diego, CA) per manufacturer’s instructions. Sequences were aligned against the Mus musculus genome (mm10) using TopHat + Bowtie28. HTSeq-count29 was used to count the transcripts associated with each gene, and a counts matrix containing the number of counts for each gene across different samples and stimulations was obtained. The counts were normalized using the TMM method30. To analyze differential expression across different stimulations, the counts matrix was analyzed with a generalized linear model, using the edgeR package31 with a threshold of q-value ≤ 0.05.
Statistical analysis
Unpaired Student t test was used for the determination of difference between groups unless other methods were stated. Data were presented as mean ± SEM of replicate experiments. The differences were considered statistically significant at P < 0.05 (*<0.05, **<0.01, ***<0.001, ****<0.0001).
RESULTS
1. IgA−/− mice have increased endogenous intestinal Th17 cells and stronger responses to CT than WT mice, but no intestinal inflammation
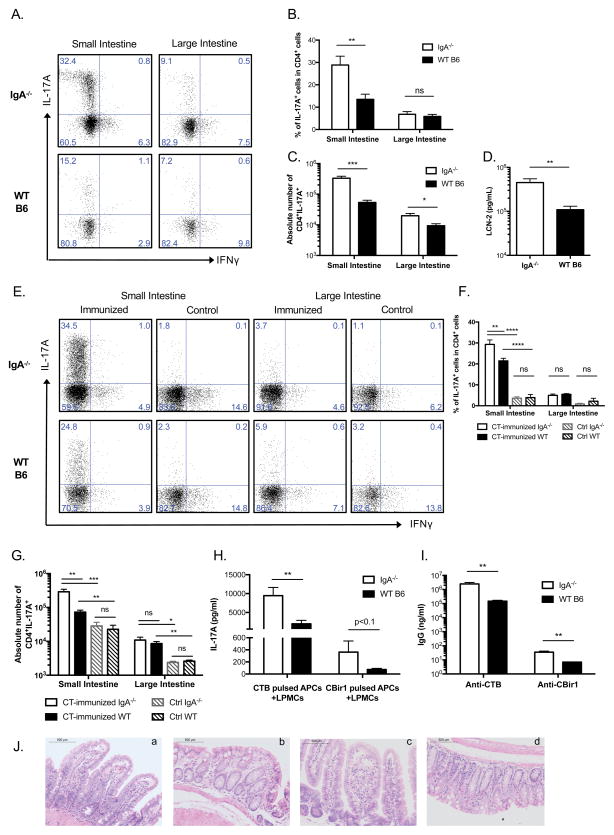
Intestinal lamina propria lymphocytes (LPLs) isolated from 9-month-old naïve C57BL/6.IgA−/− mice were compared to sex- and age-matched naïve C57BL/6 wild-type (WT) mice, and found to have a significantly enlarged Th17 population in the small intestine than WT mice (Figure 1A to C), a difference not observed in 8-week-old naïve mice (Figure 1E to G, control group). Correspondingly, there was a significantly higher serum lipocalin-2 (LCN-2) level in elderly IgA−/− mice compared to matched WT mice (Figure 1D), which is an antimicrobial protein that is downstream of IL-17A stimulation and plays important roles in epithelial homeostasis32–34.
Figure 1. IgA−/− mice have increased endogenous intestinal Th17 cells and stronger responses to CT than WT mice, but no intestinal inflammation.
(A–C) Small and large intestinal lamina propria lymphocytes (siLPL and liLPL) were isolated from naïve 9-month-old IgA−/− and WT mice. Representative plots from repeated experiments in (A) show IL-17A and IFNγ expression in CD4+ LPLs, and combined data (n=5 per group) of the percent of IL-17A expression in CD4+ LPLs (B) and absolute numbers are shown in (C). (D) Serum Lipocalin-2 (LCN-2) level was measured with ELISA (n=19 for IgA−/−; n=16 for WT B6). (E–G) 8–12-week-old mice were immunized intragastrically with 10μg CT and 100μg CBir1 flagellin in 500μl 0.2M sodium bicarbonate on days 0, 7, and 14, LPLs were isolated when mice were sacrificed on day 28 and from control naïve mice. Representative plots in (E) show IL-17A and IFNγ expression in CD4+ LPLs, and combined data (n=4 per group; two-way ANOVA) of the percent of IL-17A expression in CD4+ LPLs (F) and absolute numbers are shown in (G). (H) APCs were isolated from naïve C57BL/6 mouse spleen and pulsed with 50μg CTB or CBir1 flagellin, respectively, for 4 hours. Then pulsed APCs were cultured with siLPLs isolated from immunized mice for 5 days. IL-17A level in the culture supernatants was measured with ELISA (n=6 per group). (I) CTB- and CBir1- specific IgG responses in the sera were measured with ELISA (n=4 per group). (J) Histopathological assessment of the small intestine and colon of 9-month-old naïve IgA−/− mice (a, b) and CT immunized 12-week-old IgA−/− mice (c, d). All of the results are representative of at least two independent experiments.
Next, 8–12-week-old IgA−/− and WT mice were immunized with CT and CBir1 flagellin (a microbiota antigen) intragastrically. Th17 cells in small intestinal LPLs (siLPLs) of both strains increased considerably compared to control mice, and this increase was significantly higher in IgA−/− mice than in WT mice. The induction of Th17 cells in the large intestine was much less profound than that in the small intestine (Figure 1E to G). Different from a previous report investigating cellular response in the peripheral lymphoid organs after intranasal and intravenous CT immunization13, we did not observe significant changes of Th1, Th2, or regulatory T (Treg) cells either in the mucosal or in the peripheral sites (data not shown). siLPLs isolated from IgA−/− mice secreted almost 10-fold more IL-17A than those from WT mice upon antigen restimulation in vitro (Figure 1H). Also, IgA−/− mice developed 10-fold higher serum IgG responses against CTB (Figure 1I). Although Th17 cells have been shown to contribute to pathogenicity in various colitis models, this high percentage of endogenous or CT-induced Th17 cells in the small intestine did not cause spontaneous inflammation in IgA−/− mice in that all sections of intestinal tissue from duodenum to distal colon were normal on histologic evaluation by an experienced pathologist (T.R.S) (Figure 1J and data not shown), consistent with a homeostatic role of gut Th17 cells at steady state.
2. Intestinal microbiota contribute to the immune differences between IgA−/− and WT mice
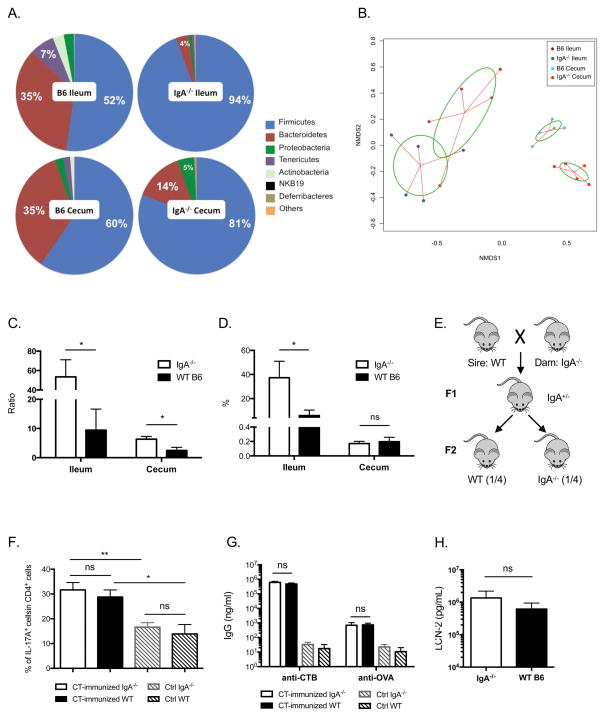
It has been shown that long-term isolated breeding of mutant and WT mice can cause marked compositional differences in their gut microbiota35, which plays a critical role in immune system development. Thus we asked whether the gut microbiota or the lack of IgA accounted for the enlarged endogenous and CT-induced Th17 responses in IgA−/− mice. First, we tested if the gut microbiota in IgA−/− mice differed from WT mice by comparing the bacterial composition in the ileum and cecum of age- and sex-matched adult mice. Pie charts of the phyla show that IgA−/− mice had higher proportions of Firmicutes than WT mice in both ileum and cecum (Figure 2A). In contrast, WT mice had more Bacteroidetes, Tenericutes and Actinobacteria. Principal coordinate analysis shows that bacteria in IgA−/− mice cluster separately from WT mice in both ileal and cecal samples (Figure 2B), indicating that the composition of the intestinal microbiota of IgA−/− mice differs from that of WT mice. Because Firmicutes and Bacteroidetes are the two major phyla of intestinal commensals, and are closely related to intestinal immune homeostasis, we compared the ratio of Firmicutes over Bacteroidetes in IgA−/− and WT mice. In both ileum and cecum, this ratio was significantly higher in IgA−/− mice (Figure 2C). Further analysis showed that a large part of Firmicutes in the ileum of IgA−/− mice was SFB, comprising about 40% of the total detected bacteria, much higher than that in WT mice (Figure 2D). Because SFB is a potent inducer of endogenous Th17 cells in the small intestine, we asked whether differences in microbiota composition, in particular elevated SFB in IgA−/− mice, could promote stronger CT-induced Th17 and antibody responses. To test this hypothesis, we equalized the intestinal microbiota of these two strains by crossing female IgA−/− mice to male WT mice, and then interbreeding of the heterozygous first generation (F1) mice to obtain littermate IgA−/− and IgA+/+ mice in the second generation (Figure 2E). The intestinal microbiota of all offspring were transmitted from maternal IgA−/− mice and remained equivalent in adulthood (Supplementary Fig. 1). Homozygous IgA−/− and WT littermates in the F2 generation were immunized with CT and ovalbumin (OVA) intragastrically. Surprisingly, the previously seen differences in intestinal Th17 induction and antigen-specific serum IgG responses did not occur (Figure 2F and 2G), indicating it was the microbiota composition in IgA−/− mice that led to the elevated host Th17 responses. Furthermore, naïve littermate IgA−/− and WT mice had comparable levels of serum LCN-2 as well (Figure 2H). Interestingly, because offspring WT mice were colonized with intestinal microbiota from the original IgA−/− mothers, their immune responses upon CT immunization resembled that of IgA−/− mice, but not the parent WT strain, providing strong evidence that IgA deficient mice are not intrinsically colonized by higher levels of Firmicutes, including SFB, different from a previous report36.
Figure 2. Intestinal microbiota contribute to the immune differences between IgA−/− and WT mice.
(A–D) Ileal and cecal contents were collected from 5 adult IgA−/− and WT mice of parent strains, respectively. Bacterial genomic DNA was extracted and used for 16s rDNA microbiome sequencing. (A) Pie charts show phyla as percent of total bacteria. (B) Principle coordinate analysis shows ileal and cecal bacterial composition of IgA−/− and WT mice of the parent strains. Each dot represents an individual mouse. The ratios of Firmicutes over Bacteroidetes in the ileum and cecum of parent strain IgA−/− and WT mice are shown in (C). (D) SFB prevalence in the ileum and cecum of parent strain IgA−/− and WT mice. (E) Breeding strategy for intestinal microbiota equalization between IgA−/− and WT mice. Female IgA−/− mice were crossed with male WT mice (which were removed from breeding cages before pups were born), to obtain F1 heterozygous mice. F1 mice were crossed to obtain homozygous IgA−/− and WT littermates in the F2 generation. Number in parentheses indicates expected frequency of homozygotes in the F2 generation. (F) Littermate IgA−/− and WT mice were immunized intragastrically with 10μg CT and 1mg Ovalbumin (OVA) in 500μl 0.2M sodium bicarbonate on days 0, 7, and 14, IL-17A expression in CD4+ siLPLs of immunized mice and naïve control mice is shown (n=4 per group). (G) CTB- and OVA- specific IgG responses (n=4 per group) and (H) LCN-2 level in the sera of littermate IgA−/− and WT mice (n=6 per group) were measured with ELISA. All of the results are representative of at least two independent experiments.
3. Selective ablation of Gram-positive bacteria impairs the Th17 response to CT immunization
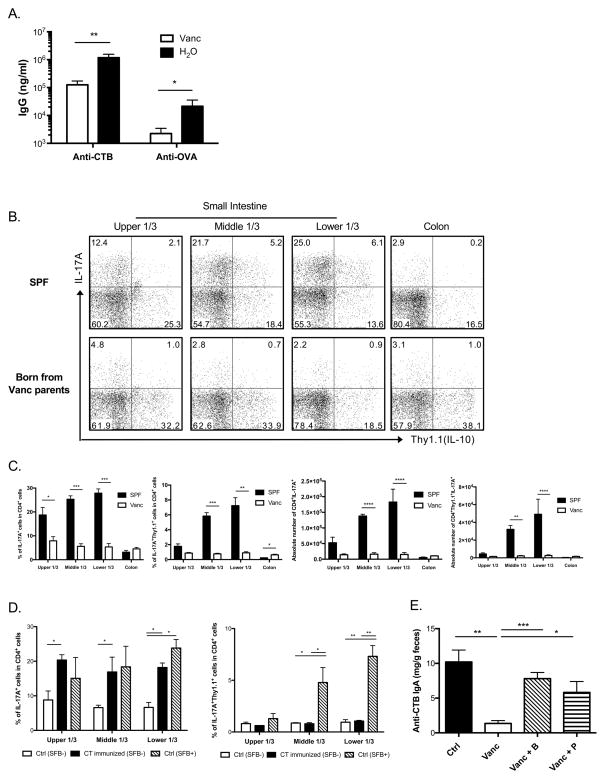
To investigate how intestinal microbiota modulates immune responses to CT immunization, IgA−/−.10BiT.Foxp3gfp mice, which were derived from IgA−/− mice and express a Thy1.1 (CD90.1) reporter for IL-10 expression and a GFP reporter for Foxp3 expression37, were treated with antibiotics in their drinking water for 2 weeks to ablate Gram-positive, spore forming bacteria, including SFB. Ablation of SFB was confirmed with PCR using DNA extracted from their fecal pellets (data not shown). IgA−/−.10BiT.Foxp3gfp mice receiving vancomycin or regular water were immunized intragastrically with CT and OVA as described before. On day 28 of immunization, mice receiving vancomycin developed one-log less anti-CTB and anti-OVA IgG responses in their sera compared to mice kept on regular water (Figure 3A), indicating that Gram-positive intestinal bacteria are actively regulating systemic immune responses to CT/OVA immunization.
Figure 3. Selective ablation of Gram-positive bacteria impairs the Th17 response to CT immunization.
(A) IgA−/−.10BiT.Foxp3gfp mice were treated with vancomycin (Vanc) in the drinking water for 2 weeks before intragastric CT/OVA immunization; antigen specific IgG responses in the sera on Day28 of immunization were measured with ELISA (n=6 per group). (B) IgA−/−.10BiT.Foxp3gfp parents were treated with vancomycin in the drinking water for 1 month prior to breeding and continuously during gestation, IL-17A and IL-10 expression in CD4+ LPLs in 6-week-old offspring and sex- and age-matched SPF IgA−/−.10BiT.Foxp3gfp mice are shown in representative flow cytometric plots, whereas combined data (n=3 per group, each sample was a pool of 2–3 mice) of the percent of IL-17A+ and IL-17A+Thy1.1+ in CD4+ LPLs and absolute numbers are shown in (C). (D) Same as described in (B), IgA−/−.10BiT.Foxp3gfp mice were born from vancomycin treated parents and maintained separately from other mice in the same facility. SFB presence in the offspring was measured twice, 4 weeks apart, using PCR. 8-week-old SFB-absent IgA−/−.10BiT.Foxp3gfp mice were immunized with 10μg CT in 500μl 0.2M sodium bicarbonate i.g. on days 0, 7, and 14, naïve IgA−/−.10BiT.Foxp3gfp mice from the same litter with and without the presence of SFB were used as controls. IL-17A and IL-17A+IL-10+ expressions in CD4+ siLPLs are shown (n=6 per group). (E) C57BL/6 mice were pre-treated with vancomycin in the drinking water for 10 days, and then received intracolonic CT immunization on days 0, 7, and 14. Indicated short-chain fatty acids (B=butyrate; P=propionate) were added to the drinking water together with vancomycin from the beginning of immunization till day 28. Fecal anti-CTB IgA level on day 28 was measured with ELISA (n=4 per group; one-way ANOVA). All of the results are representative of at least two independent experiments.
Two-week-treatment with vancomycin did not reduce intestinal Th17 on its own (data not shown), however when we treated parent IgA−/−.10BiT.Foxp3gfp mice with vancomycin for one month prior to breeding and during gestation, offspring had greatly decreased intestinal Th17 cells, as well as IL-17A and IL-10 double expressing cells in all parts of their small intestine compared to sex- and age-matched specific-pathogen-free (SPF) IgA−/−.10BiT.Foxp3gfp mice (Figure 3B and C). Th17 cells in the colonic LP or other T cell subsets in the intestine were not significantly affected, indicating that Gram-positive bacteria ablated by vancomycin are mainly present in the small intestine and locally modulate siLPLs, consistent with our microbiome sequencing data showing SFB outgrowth in the terminal ileum of parent IgA−/− mice (Figure 2D).
To test whether SFB is playing a role in modulating siLPLs in response to intragastric CT immunization, we immunized 8-week-old IgA−/−.10BiT.Foxp3gfp mice born from vancomycin treated parents lacking SFB, and divided the small intestine equally into three parts when examined. Using naïve littermate controls that either had SFB or not, we found that SFB alone was able to induce endogenous Th17 cells as well as IL-17A and IL-10 double producing cells in the small intestine, but this induction was the most prominent in the middle and lower thirds. In contrast, CT immunization increased Th17 cells in all parts of the small intestine, but did not induce IL-17A and IL-10 double producing cells (Figure 3D).
Consistent with previous reports38,39 that intracolonic CT immunization induces systemic humoral immune responses, our data show that intracolonic CT induced intestinal Th17 cells as well. The antibody response to intracolonic CT was also microbiota-dependent, because vancomycin treated C57BL/6 mice had impaired anti-CTB fecal IgA as well as total fecal IgA after immunization (Figure 3E and data not shown). Addition of the bacterial fermentation products butyrate or propionate to the drinking water40–42 partially rescued the production of anti-CTB fecal IgA as well as total fecal IgA (Figure 3E), indicating that decreased immune responses to CT after ablation of Gram-positive bacteria was partially due to deficiency of bacterial metabolic products in the intestine. Collectively, these findings indicate that Gram-positive bacterial ablation with vancomycin impairs the host intestinal Th17 response and its related antibody responses to CT immunization. SFB specifically induces an IL-17A+IL-10+ population in the distal small intestine, indicating that the intestinal microbiota can modulate the host Th17 response in some ways that are different from cholera toxin.
4. Bacterial colonization enhances host Th17 and antibody responses to CT
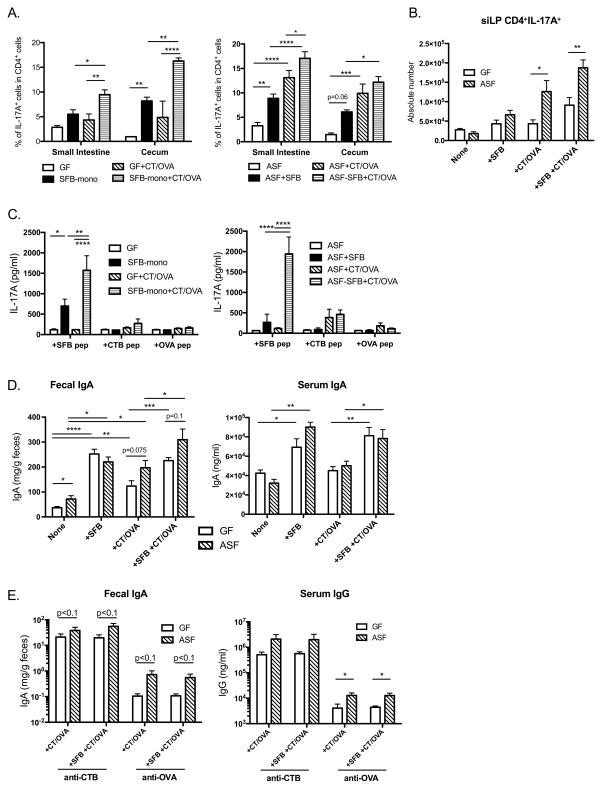
To better define the effect of SFB versus other microbes on the ability of CT to induce Th17 cells, gnotobiotic mice were used. Germ free (GF) and altered Schaedler flora (ASF) colonized C57BL/6 mice43 were selectively colonized with SFB to further test whether SFB alone is sufficient to modulate immune responses against CT or whether other commensal bacteria also contribute to this regulation. In both GF and ASF mice, SFB colonization alone induced Th17 cell differentiation in the small intestine and cecum (Figure 4A). However, the increase of Th17 cells in the small intestine in SFB mono-colonized mice versus GF mice was not statistically significant (Figure 4A and B). ASF colonization did not induce Th17 cells by itself, and largely suppressed Th1 cell differentiation in both small intestine and cecum (data not shown). In the absence of SFB, CT/OVA immunization significantly induced Th17 cells in the small intestine and cecum of ASF mice, but only slightly in GF mice, indicating that single or multiple bacteria belonging to ASF also have the potential to regulate Th17 response upon CT immunization (Figure 4A and 4B). SFB predominantly enhanced Th17 cell induction in response to CT immunization in mono-colonized mice, while not significantly in ASF mice, which already had a higher level (Figure 4A). Cytokine production (including Th1, Th2 and Th17 related cytokines) was measured by co-culturing of isolated siLPLs with naïve antigen presenting cells (APCs) and either SFB, CTB, or OVA CD4 T cell epitope peptide (see Methods). Intragastric CT immunization specifically induced lamina propria IL-17A response (Figure 4C) and slightly suppressed IFNγ response, without a significant effect on IL-4, IL-5, IL-13 response (data not shown). Although peptide restimulation resulted in increased IL-6 production in CT-immunized groups, IL-6 appeared not to be the sole pathway that contributed to Th17 differentiation in vivo, because there was no difference in Th17 cell induction or antibody responses with CT immunization in IL-6−/− mice (Supplementary Fig. 2). Upon SFB colonization, both GF and ASF mice receiving CT/OVA immunization significantly increased IL-17A production specific to SFB peptide stimulation, to a much greater extent than either CTB or OVA peptide, indicating that CT serves as an adjuvant for the proliferation of Th17 cells that are predominantly reactive to SFB. Collectively, these data provided clear evidence that CT induction of intestinal Th17 response is dependent on the gut microbiota. Indeed, siLPLs isolated from CT immunized SPF WT mice responded significantly more strongly than those of any group of gnotobiotic mice (Supplementary Fig. 3A and 3B), indicating that a more diverse intestinal microbiota provides optimal support for the development of host Th17 response to CT immunization. Although CT elicits strong B cell and T cell responses to itself, tetramer staining showed that most of the induced Th17 cells are not CTB-specific, indicating that CT induces Th17 response in a polyclonal fashion (Supplementary Fig. 3C and 3D).
Figure 4. Bacterial colonization enhances host Th17 and antibody responses to CT.
(A–E) GF and ASF mice were colonized with SFB by oral gavage of fecal pellets collected from SFB mono-associated mice. Colonization was confirmed by PCR with recipients’ fecal pellets. Intragastric CT immunization was given 2 weeks after colonization on days 0, 7, and 14. Mice were sacrificed on day 28, when intestinal LPLs were isolated and sera as well as fecal samples were collected. (A) Combined data of IL-17A expression in CD4+ LPLs in GF and ASF mice (n=4 per group; two-way ANOVA), respectively, while absolute numbers of CD4+IL-17A+ cells in the LPLs are shown in (B). (C) Isolated LPLs were cultured with indicated peptides and naïve APCs for 5 days, and IL-17A production in the culture supernatants was measured with multi-plex assay (n=4 per group; Two-way ANOVA). (D) Total IgA level in the feces and sera, (E) CTB- and OVA- specific fecal IgA and serum IgG levels were measured with ELISA (n=5 per group; Student t test and one-way ANOVA).
SFB colonization is a strong inducer of both fecal and serum IgA in gnotobiotic mice, whether naïve or immunized (Figure 4D). In contrast, ASF colonization enhanced fecal secretory IgA levels but not serum IgA, indicating that different commensals have distinct mechanisms to regulate host IgA production. Surprisingly, in contrast to our vancomycin experiment data, SFB colonization did not increase host antibody responses to CT or OVA in either GF mice or ASF mice, i.e., SFB did not act as an adjuvant for the response to these antigens. However, regardless of the presence of SFB, ASF colonized mice developed greater CTB and OVA specific antibody responses upon immunization than GF mice both mucosally and systemically (Figure 4E), indicating that the decreased antibody responses upon CT immunization in vancomycin treated mice were due to vancomycin sensitive commensal bacteria other than SFB. These experiments suggested that in addition to SFB, which is a known potent inducer for endogenous Th17 cells in the intestine, other commensal bacteria are also involved in the regulation of host Th17 and antibody responses upon stimulation.
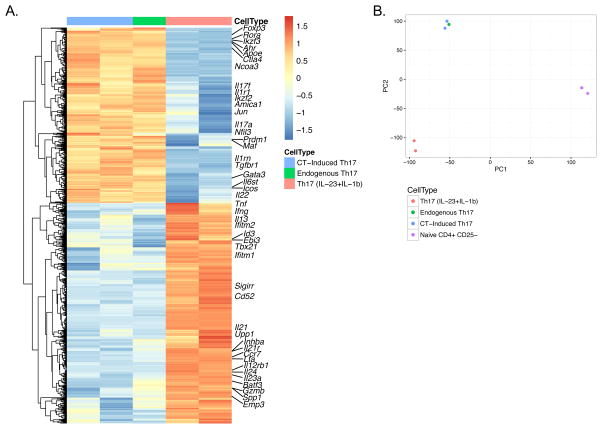
5. CT-induced intestinal Th17 cells share the same transcriptome profile with endogenous homeostatic Th17 cells, and both differ from pathogenic Th17 cells
To test the hypothesis that intestinal Th17 cells under steady state conditions are different from pathogenic Th17 cells, we used IgA−/−.IL-17AhCD2 reporter mice, in which the IL-17A promoter directs the transcription of a bicistronic mRNA such that IL-17A producing cells are marked by surface membrane expression of the human CD2 molecule (Supplementary Fig. 4A and manuscript in preparation), allowing us to recover IL-17A producing cells for RNA-Seq. Of note, almost none of the IL-17A producing cells co-express Foxp3 (Supplementary Fig. 4B). We isolated CD4+hCD2+ (Th17) and CD4+hCD2− (non-Th17) cells from the small intestine of either naïve or CT immunized IgA−/−.IL-17AhCD2 mice, and used splenic CD4+CD25− cells from naïve mice as negative controls for gene expression profiling. Comparing CD4+ cells isolated from naïve and CT immunized IgA−/−.IL-17AhCD2 intestines, we found that Th17 cells induced by CT only have one gene differently expressed compared to endogenous LP Th17 cells; similarly, non-Th17 cells isolated from CT immunized mice only had two genes differently expressed compared to non-Th17 cells isolated from naïve mice. Thus, although mucosal CT immunization enlarges the Th17 population in the intestinal LP, CT-induced Th17 cells share the same expression profile as endogenous intestinal Th17 cells. The gene expression of Th17 cells isolated from the healthy intestine or CT immunized intestine is very different from pathogenic Th17 cells induced with IL-6+IL-23+IL-1β44 as well as Th17 cells isolated from the LP of colitic tissues (Figure 5A and B, and data not shown). In particular, a series of genes related to Th17 pathogenicity, including Tnf, Ifng, Il23a, Il24, Il12rb1, and Gzmb, were significantly less expressed in endogenous and CT-induced intestinal Th17 cells compared to pathogenic Th17 cells, whereas genes involved in anti-inflammatory activities such as Ctla4, Icos, Il22, Ahr, Maf, and Ikzf3, showed at least 4 fold higher expression in homeostatic intestinal Th17 cells, consistent with previously reported Th17 signatures44–47. Additionally, functional analysis reveals a stronger immune regulatory profile of homeostatic intestinal Th17 cells, whereas pathogenic Th17 cells highlight pathways that are involved in various inflammatory diseases. In summary, comparison of gene expression levels shows great similarity between CT-induced Th17 cells and endogenous Th17 cells isolated from the healthy small intestine, and both of them are distinct from the signature of pathogenic Th17 cells.
Figure 5. Gene expression profile of endogenous, CT-induced, and pathogenic Th17 cells.
RNA-Seq was performed with CD4+hCD2+ cells isolated from the small intestine of naïve and CT-immunized IgA−/−.IL-17AhCD2 mice (a pool of at least 6 mice per sample) and these were compared to the transcriptome of pathogenic Th17 cells induced with IL-23 and IL-1β44. (A) Heatmap shows transcripts differentially expressed in Th17 cells from different origins and (B) shows the principle component analysis on their gene expression profiles, where each point represents an individual sample.
DISCUSSION
In this study, we showed that endogenous Th17 cells present in the normal intestine have a homeostatic phenotype. The increase of this homeostatic population and the elevated Th17 response upon CT immunization in IgA−/− mice are dependent on intestinal microbiota. Using gnotobiotic mouse models, we found that SFB, ASF and other commensal bacteria are actively involved in the regulation of CT immunogenicity and adjuvanticity. Furthermore, our RNA-Seq data indicate that CT-induced intestinal Th17 cells share a similar gene expression profile with endogenous Th17 cells in the intestine, both of which are quite different from the signature of pathogenic Th17 cells.
The age related increase of intestinal Th17 cells in IgA−/− mice suggested an environmental effect of Th17 cell development, in this case most likely the intestinal microbiota. Indeed, with activation-induced cytidine deaminase knockout (AID−/−) mice, Suzuki et al concluded that the lack of hypermutated IgA resulted in the excessive overgrowth of anaerobic bacteria in the intestine, the most predominant being SFB36. Although conflicting data has been reported with Igh-J−/− mice48, neither of them were perfect models for the study of IgA deficiency. Targeted deletion of the IgA constant region in our IgA−/− mice leads to specific deficiency of IgA, thus providing a more appropriate model of the effects of IgA deficiency on microbiota49. Our study shows that increased Firmicutes, including SFB, led to the exaggerated intestinal Th17 responses and humoral antibody induction upon CT immunization in IgA−/− mice. However, this microbiota difference is not intrinsic to IgA deficiency, in that crossing the WT with IgA−/− mice equalized the intestinal microbiota in the offspring, indicating a familial transmission rather than IgA deficiency caused microbiota shift35,36,50.
CT has been known as a potent mucosal immunogen and adjuvant for several decades; we confirm and extend a recent report linking these CT properties with the intestinal microbiota14. With SFB as a known stimulator of endogenous Th17 cells in the intestine, we found that in addition, other commensal bacteria such as those belonging to the altered Schaedler flora, can also participate in the regulation of intestinal Th17 and antibody responses to CT mucosal immunization. When the CTB subunit binds to the GM1 ganglioside present on cell membranes, CT induces strong activation of STAT3 signaling and massive production of IL-611,51,52, which is known to promote the differentiation of Th17 cells in vitro. However, in agreement with previously published in vitro studies53,54, IL-6−/− mice did not show impaired intestinal Th17 cell induction upon CT immunization compared to littermate WT mice (Supplementary Fig. 2), implying the involvement of other possible cytokine pathways such as IL-21 or IL-1β. Although both SFB and CT are strong inducers of intestinal Th17 cells, the induction tends to reach a saturated level at about 35–40 percent of CD4+ T cells in siLPLs, even in IgA−/− mice. In addition, CT had little effect on other CD4+ T cells in the intestine, including Th1, Th2, and Treg cells. This difference from the conclusions of a previous report13 was probably due to different microbiota in mice maintained in isolated facilities.
In this study, we show that both intragastric and intracolonic CT immunization induces profound Th17 cell increase in the intestinal LP, without causing any intestinal inflammation. Data from a collection of studies suggest that Th17 cells play a major role in the pathogenesis of IBD and experimental models of colitis, via secretion of various cytokines, particularly the co-expression of IL-17A and IFNγ23,55–57. However, IgA−/− mice, which lack the Treg-IgA homeostatic pathway58, and have an expanded Th17 population in their intestine, do not develop spontaneous intestinal inflammation under SPF conditions throughout life. Our data showed that a subset of endogenous Th17 cells induced by SFB colonization co-express IL-17A and IL-10. IL-10 is an anti-inflammatory cytokine that restrains the activities of effector T cells. Although CT immunization did not show induction of IL-17A/IL-10 co-producing Th17 cells on the cytokine level, RNA-Seq data indicated an immune regulatory transcriptome of CT-induced Th17 cells as well as endogenous Th17 cells, explaining why CT does not cause intestinal inflammation despite strongly expanding Th17 cells. For example, in keeping with a recent report59, we saw upregulated Foxp3 gene expression in endogenous intestinal Th17 cells, as well as in CT-induced intestinal Th17 cells. Ctla4 and Ikzf2 (encoding Helios), which are normally highly expressed in Treg cells, and the products of which contribute to the suppressive function of Treg cells60,61, were more highly expressed in homeostatic intestinal Th17 cells as well. Also, these homeostatic intestinal Th17 cells have an upregulated immune gene network that promotes IL-10 production, including Icos, Maf, Prdm1, and Ahr62–66. Single cell RNA-Seq is needed in future studies to fully reveal the nature of Foxp3 expressing Th17 cells, although we saw very little co-expression of Foxp3 and IL-17A on the protein level (Supplementary Fig. 4B). Interestingly, pathogenic Th17 cells had lower expression of both Il17a and Il17f but higher expression of Ifng compared to endogenous and CT-induced Th17 cells, consistent with what has been shown by Harbour et al., that Th17 convert into Th1 cells and contribute to the pathogenesis of colitis67. Most recently, Gaublomme and Wang et al. published their studies of Th17 cell pathogenicity using the single-cell RNA-Seq technique, in which they found an intestinal Th17 cell signature similar to ours, although we did not notice any differential expression of Cd5l in our RNA-Seq data47,68. The ratio of homeostatic Th17 cells to pathogenic Th17 cells might be critical in overt inflammation; it is an important topic of future research to understand whether these are separate lineages, and if not, what triggers the conversion from homeostatic Th17 cells to pathogenic Th17 cells and/or Th1 cells, and what determines the conversion from Th17 to Treg cells. This will provide us with new insights into the pathogenesis, prevention, and treatment of diseases of dysregulated immunity and inflammation.
Supplementary Material
Acknowledgments
We thank Dr. I. N. Mbawuike and Dr. C. L. Maynard for generously providing the IgA−/− and 10BiT.Foxp3gfp mice, respectively, M. L. Spell for cell sorting, and Dr. D. Botta for helping with the Multi-plex assay.
Funding: This work was supported by NIH P01 DK071176, NIH P30 AI27667, NIH P30 AR048311, G20RR022807-01, and NIH RR-20136.
Abbreviations used in this article
- IBD
inflammatory bowel disease
- CT
cholera toxin
- SFB
segmented filamentous bacteria
- LP
lamina propria
- LCN-2
lipocalin-2
- WT
wild type
- SPF
specific pathogen free
- GF
germ free
- ASF
altered Schaedler flora
Footnotes
Data deposition: The RNA-Seq data presented in this article have been submitted to the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE97571.
References
- 1.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 4.Niess JH, Leithauser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- 5.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 6.Gaffen SL, Jain R, Garg AV, Cua DJ. The IL-23-IL-17 immune axis: from mechanisms to therapeutic testing. Nat Rev Immunol. 2014;14:585–600. doi: 10.1038/nri3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008;28:454–467. doi: 10.1016/j.immuni.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 10.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta SK, Sabet M, Nguyen KP, Valdez PA, Gonzalez-Navajas JM, Islam S, Mihajlov I, Fierer J, Insel PA, Webster NJ, Guiney DG, Raz E. Mucosal adjuvant activity of cholera toxin requires Th17 cells and protects against inhalation anthrax. Proc Natl Acad Sci U S A. 2010;107:10638–10643. doi: 10.1073/pnas.1002348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elson CO, Ealding W. Generalized systemic and mucosal immunity in mice after mucosal stimulation with cholera toxin. J Immunol. 1984;132:2736–2741. [PubMed] [Google Scholar]
- 13.Mattsson J, Schon K, Ekman L, Fahlen-Yrlid L, Yrlid U, Lycke NY. Cholera toxin adjuvant promotes a balanced Th1/Th2/Th17 response independently of IL-12 and IL-17 by acting on Gsalpha in CD11b(+) DCs. Mucosal Immunol. 2015;8:815–827. doi: 10.1038/mi.2014.111. [DOI] [PubMed] [Google Scholar]
- 14.Kim D, Kim YG, Seo SU, Kim DJ, Kamada N, Prescott D, Philpott DJ, Rosenstiel P, Inohara N, Nunez G. Nod2-mediated recognition of the microbiota is critical for mucosal adjuvant activity of cholera toxin. Nat Med. 2016;22:524–530. doi: 10.1038/nm.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantis NJ, Rol N, Corthesy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yel L. Selective IgA deficiency. J Clin Immunol. 2010;30:10–16. doi: 10.1007/s10875-009-9357-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng T, Wang L, Schoeb TR, Elson CO, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J Exp Med. 2010;207:1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sette A, Buus S, Colon S, Smith JA, Miles C, Grey HM. Structural characteristics of an antigen required for its interaction with Ia and recognition by T cells. Nature. 1987;328:395–399. doi: 10.1038/328395a0. [DOI] [PubMed] [Google Scholar]
- 21.Cong Y, Bowdon HR, Elson CO. Identification of an immunodominant T cell epitope on cholera toxin. Eur J Immunol. 1996;26:2587–2594. doi: 10.1002/eji.1830261108. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y, Torchinsky MB, Gobert M, Xiong H, Xu M, Linehan JL, Alonzo F, Ng C, Chen A, Lin X, Sczesnak A, Liao JJ, Torres VJ, Jenkins MK, Lafaille JJ, Littman DR. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature. 2014;510:152–156. doi: 10.1038/nature13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elson CO, Cong Y, Weaver CT, Schoeb TR, McClanahan TK, Fick RB, Kastelein RA. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–2370. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 24.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012;13:31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amatya N, Garg AV, Gaffen SL. IL-17 Signaling: The Yin and the Yang. Trends Immunol. 2017 doi: 10.1016/j.it.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlsen JR, Borregaard N, Cowland JB. Induction of neutrophil gelatinase-associated lipocalin expression by co-stimulation with interleukin-17 and tumor necrosis factor-alpha is controlled by IkappaB-zeta but neither by C/EBP-beta nor C/EBP-delta. J Biol Chem. 2010;285:14088–14100. doi: 10.1074/jbc.M109.017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, Pamer EG. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maynard CL, Harrington LE, Janowski KM, Oliver JR, Zindl CL, Rudensky AY, Weaver CT. Regulatory T cells expressing interleukin 10 develop from Foxp3+ and Foxp3-precursor cells in the absence of interleukin 10. Nat Immunol. 2007;8:931–941. doi: 10.1038/ni1504. [DOI] [PubMed] [Google Scholar]
- 38.Haneberg B, Kendall D, Amerongen HM, Apter FM, Kraehenbuhl JP, Neutra MR. Induction of specific immunoglobulin A in the small intestine, colon-rectum, and vagina measured by a new method for collection of secretions from local mucosal surfaces. Infect Immun. 1994;62:15–23. doi: 10.1128/iai.62.1.15-23.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen X, Lagergard T, Yang Y, Lindblad M, Fredriksson M, Holmgren J. Systemic and mucosal immune responses in mice after mucosal immunization with group B streptococcus type III capsular polysaccharide-cholera toxin B subunit conjugate vaccine. Infect Immun. 2000;68:5749–5755. doi: 10.1128/iai.68.10.5749-5755.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, Rudensky AY. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 42.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dewhirst FE, Chien CC, Paster BJ, Ericson RL, Orcutt RP, Schauer DB, Fox JG. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl Environ Microbiol. 1999;65:3287–3292. doi: 10.1128/aem.65.8.3287-3292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, Shakya A, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK, Regev A. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–468. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gagliani N, Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limon P, Paiva RS, Ching T, Weaver C, Zi X, Pan X, Fan R, Garmire LX, Cotton MJ, Drier Y, Bernstein B, Geginat J, Stockinger B, Esplugues E, Huber S, Flavell RA. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 2015;523:221–225. doi: 10.1038/nature14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, Kuchroo VK, Park H, Regev A. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, Fritzen R, Eberl G, McCoy KD, Macpherson AJ, Reynaud CA, Cerf-Bensussan N, Gaboriau-Routhiau V. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 49.Harriman GR, Bogue M, Rogers P, Finegold M, Pacheco S, Bradley A, Zhang Y, Mbawuike IN. Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J Immunol. 1999;162:2521–2529. [PubMed] [Google Scholar]
- 50.Rogier EW, Frantz AL, Bruno ME, Wedlund L, Cohen DA, Stromberg AJ, Kaetzel CS. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A. 2014;111:3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGee DW, Elson CO, McGhee JR. Enhancing effect of cholera toxin on interleukin-6 secretion by IEC-6 intestinal epithelial cells: mode of action and augmenting effect of inflammatory cytokines. Infect Immun. 1993;61:4637–4644. doi: 10.1128/iai.61.11.4637-4644.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sjoblom-Hallen A, Marklund U, Nerstedt A, Schon K, Ekman L, Bergqvist P, Lowenadler B, Lycke NY. Gene expression profiling identifies STAT3 as a novel pathway for immunomodulation by cholera toxin adjuvant. Mucosal Immunol. 2010;3:374–386. doi: 10.1038/mi.2010.16. [DOI] [PubMed] [Google Scholar]
- 53.Kimura A, Naka T, Kishimoto T. IL-6-dependent and -independent pathways in the development of interleukin 17-producing T helper cells. Proc Natl Acad Sci U S A. 2007;104:12099–12104. doi: 10.1073/pnas.0705268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leppkes M, Becker C, Ivanov, Hirth S, Wirtz S, Neufert C, Pouly S, Murphy AJ, Valenzuela DM, Yancopoulos GD, Becher B, Littman DR, Neurath MF. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–267. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 2010;33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cong Y, Feng T, Fujihashi K, Schoeb TR, Elson CO. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci U S A. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, Ghosh S, Earl A, Snapper SB, Jupp R, Kasper D, Mathis D, Benoist C. MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science. 2015;349:993–997. doi: 10.1126/science.aaa9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 61.Thornton AM, Korty PE, Tran DQ, Wohlfert EA, Murray PE, Belkaid Y, Shevach EM. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184:3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu J, Yang Y, Qiu G, Lal G, Wu Z, Levy DE, Ochando JC, Bromberg JS, Ding Y. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–6236. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, Kuchroo VK. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. 2010;11:854–861. doi: 10.1038/ni.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jankovic D, Kugler DG, Sher A. IL-10 production by CD4+ effector T cells: a mechanism for self-regulation. Mucosal Immunol. 2010;3:239–246. doi: 10.1038/mi.2010.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinemann C, Heink S, Petermann F, Vasanthakumar A, Rothhammer V, Doorduijn E, Mitsdoerffer M, Sie C, Prazeres da Costa O, Buch T, Hemmer B, Oukka M, Kallies A, Korn T. IL-27 and IL-12 oppose pro-inflammatory IL-23 in CD4+ T cells by inducing Blimp1. Nat Commun. 2014;5:3770. doi: 10.1038/ncomms4770. [DOI] [PubMed] [Google Scholar]
- 66.Neumann C, Heinrich F, Neumann K, Junghans V, Mashreghi MF, Ahlers J, Janke M, Rudolph C, Mockel-Tenbrinck N, Kuhl AA, Heimesaat MM, Esser C, Im SH, Radbruch A, Rutz S, Scheffold A. Role of Blimp-1 in programing Th effector cells into IL-10 producers. J Exp Med. 2014;211:1807–1819. doi: 10.1084/jem.20131548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci U S A. 2015;112:7061–7066. doi: 10.1073/pnas.1415675112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang C, Yosef N, Gaublomme J, Wu C, Lee Y, Clish CB, Kaminski J, Xiao S, Meyer Zu Horste G, Pawlak M, Kishi Y, Joller N, Karwacz K, Zhu C, Ordovas-Montanes M, Madi A, Wortman I, Miyazaki T, Sobel RA, Park H, Regev A, Kuchroo VK. CD5L/AIM Regulates Lipid Biosynthesis and Restrains Th17 Cell Pathogenicity. Cell. 2015;163:1413–1427. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.