Abstract
Cancer and stem cells appear to share a common metabolic profile that is characterized by high utilization of glucose through aerobic glycolysis. In the presence of sufficient nutrients, this metabolic strategy provides sufficient cellular ATP while additionally providing important metabolites necessary for the biosynthetic demands of continuous cell proliferation. Recent studies indicate that this metabolic profile is dependent on genes that regulate the fusion and fission of mitochondria. High levels of mitochondrial fission activity are associated with high proliferation and invasiveness in some cancer cells and with self-renewal and resistance to differentiation in some stem cells. These observations reveal new ways in which mitochondria regulate cell physiology, through their effects on metabolism and cell signaling.
Introduction
Depending on the cell type, a mammalian cell can have several to thousands of mitochondria. In metabolically active cells, such as hepatocytes and cardiomyocytes, the mitochondria make up 20–30% of the cell volume. These mitochondria do not operate as isolated organelles; rather, they function as a collective whose activity is orchestrated by mitochondrial dynamics. The identities of individual mitochondria are constantly altered by continuous cycles of membrane fusion and fission, which serve to mix the contents of the mitochondrial population, promote homogeneity of the organelles, control the morphology of mitochondria, and maintain their high functionality. The term "mitochondrial dynamics" has come to encompass additional behaviors of mitochondria, such as their transport along the cytoskeleton, their selective degradation by the mitophagy pathway, and their interactions with other organelles, including the endoplasmic reticulum. This review focuses on the role of fusion/fission dynamics in the biology of cancer and stem cells.
Mitochondria play central roles in determining the physiology, and particularly the metabolism, of most eukaryotic cells. Cell physiology, in turn, is critically important in regulating the proliferative and developmental potential of cells. Given this, it is perhaps not surprising that bidirectional links have been uncovered between mitochondrial dynamics and cell replication. We introduce the fundamentals of mitochondrial fusion and fission, and then discuss the effect of mitochondrial dynamics in cell cycle control and how interruption of this control can contribute to tumorigenesis. We then describe emerging evidence that the distinct metabolism of tumor cells is regulated by mitochondrial dynamics, along with the striking parallels in regenerating stem cells. Apoptotic pathways also play important roles in cancer biology and are regulated by mitochondrial dynamics, but are beyond the scope of this review and have been extensively discussed elsewhere (Sheridan and Martin, 2010; Suen et al., 2008).
Mitochondrial fusion and fission
Fusion and fission are opposing processes that must be balanced to enable efficient content exchange between mitochondria while maintaining the proper mitochondrial morphology (Chan, 2012; Labbe et al., 2014). Although the machineries for fusion and fission are distinct, there is evidence that fission events are temporally coordinated with fusion events (Twig et al., 2008), a collaboration that helps to keep the two processes balanced. Severe defects in either mitochondrial fusion or fission lead to mitochondrial dysfunction. In part, this dysfunction occurs because the processes become unbalanced, and cell physiology can be restored by manipulating mitochondrial dynamics to obtain a new, re-balanced setpoint (Bleazard et al., 1999; Chen et al., 2015; Sesaki and Jensen, 1999).
Because mitochondria are double-membraned organelles, mitochondrial fusion involves outer membrane fusion followed by inner membrane fusion (Chan, 2012; Labbe et al., 2014). The net result is the coordinated merger of four lipid bilayers and the formation of a single, fused mitochondrial matrix (the compartment encased by the inner membrane). These two membrane fusion events are normally coupled in vivo but can be uncoupled in vitro, because they have distinct requirements for co-factors and metabolism. Outer membrane fusion is mediated by mitofusins, large GTPases of the dynamin family that are embedded on the mitochondrial outer membrane. Mammals have two mitofusins termed MFN1 (Mitofusin 1) and MFN2 (Mitofusin 2). In the absence of MFN1 and MFN2, mitochondria fail to fuse their outer membranes, either in live cells or in biochemically isolated organelles. Following outer membrane fusion, OPA1 (Optic Atrophy 1), another dynamin family member, mediates inner membrane fusion. Cells lacking OPA1 show outer membrane fusion intermediates that cannot progress to full fusion and that ultimately resolve by fission.
Disruption of mitochondrial fusion genes lead to neurodegenerative disease (Carelli and Chan, 2014). Charcot-Marie-Tooth type 2A, an axonopathy of long peripheral nerves, is caused by autosomal dominant mutations in MFN2. Similarly, autosomal dominant mutations in OPA1 are the predominant cause of dominant optic atrophy, a disease of the optic nerve that results in reduced visual acuity or blindness. In both these diseases, the heterozygous mutations result in mild reductions in mitochondrial fusion that are compatible with development into adulthood but lead to defects in specific neurons. In contrast, when homozygous null alleles are constructed in mouse models, they result in severe embryonic and neuromuscular phenotypes (Chen et al., 2003; Chen et al., 2007; Chen et al., 2010).
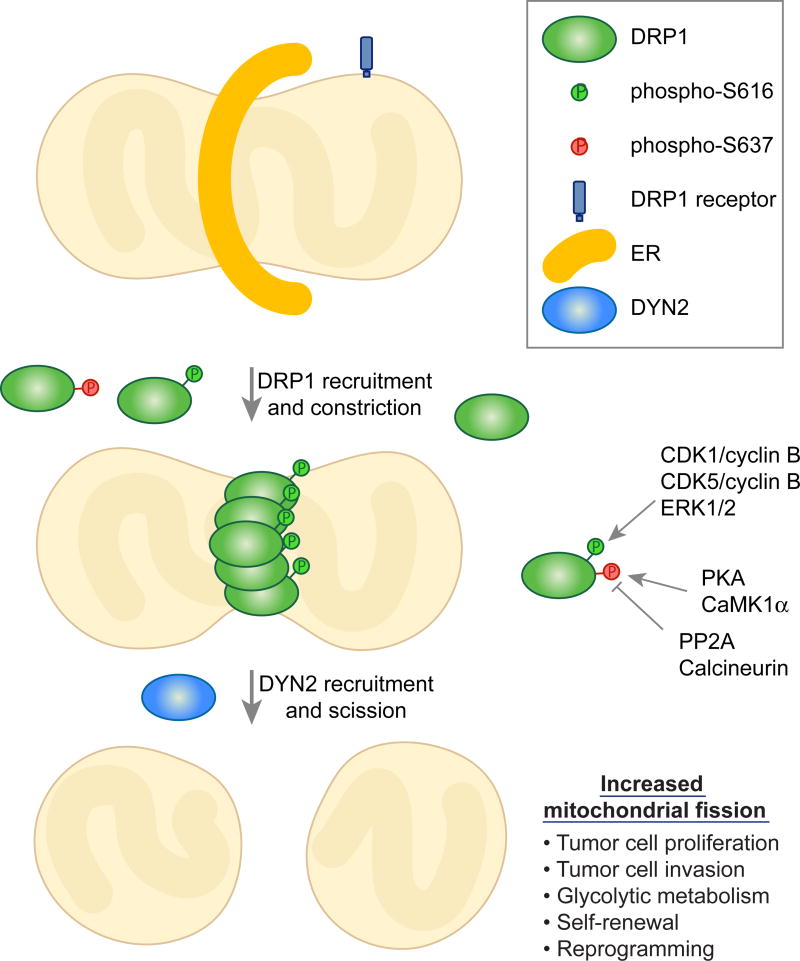
The central player in mitochondrial fission is DRP1 (Dynamin-related protein 1), a large GTPase (Chan, 2012; Labbe et al., 2014) (Figure 1). A pool of DRP1 resides in the cytosol and must be recruited to the mitochondrial surface to mediate fission. Once on the mitochondrial surface, DRP1 functions as a mechano-chemical enzyme that forms helical assemblies that constrict the mitochondrial tubule. Its mechanism of action is conceptually analogous to the role of classical dynamins in constricting the necks of endocytic vesicles at the cell surface (Ferguson and De Camilli, 2012; Schmid and Frolov, 2011). Recent results, however, suggest that the mechanical properties of Dp1 may be insufficient to complete the scission process (Lee et al., 2016). When DYN2 (Dynamin 2) is depleted, mitochondrial tubules with narrow constrictions accumulate. The accumulation of these apparent fission intermediates suggests that whereas DRP1 constricts mitochondrial tubules, this compression is not sufficient to complete fission, and that DYN2 instead may be additionally required at the final step of scission.
Figure 1. Regulation of mitochondrial fission and its role in cancer and stem cells.
The fission of mitochondria begins with an interaction with the endoplasmic reticulum, which causes initial constriction of the mitochondrial tubule. DRP1, recruited by one of several DRP1 receptors, assembles on the mitochondrial surface and causes further constriction. The final stage of membrane scission requires DYN2. The activity of DRP1 is regulated by molecules that control the phosphorylation of DRP1 at two serine residues, which have opposing effects. The level of mitochondrial fission has significant effects on tumor and stem cell phenotypes.
Mitochondrial fission begins with interaction of mitochondria with the endoplasmic reticulum, which causes initial constriction of the mitochondrial tubule (Figure 1) (Friedman et al., 2011). DRP1 is subsequently recruited to the mitochondrial surface through association with several DRP1 receptors embedded in the outer membrane. One important DRP1 receptor is MFF (mitochondrial fission factor), whose depletion leads to dramatic mitochondrial elongation (Gandre-Babbe and van der Bliek, 2008) and reduction of DRP1 on mitochondria (Loson et al., 2013; Otera et al., 2010). Two other proteins, MID49 (mitochondrial dynamics protein of 49 kDa) and MID51 (mitochondrial dynamics protein of 51 kDa), also play significant roles in recruiting DRP1 (Loson et al., 2013; Otera et al., 2016; Palmer et al., 2013). FIS1 (Fission 1) was initially identified as the first DRP1 receptor, based on its clear involvement in mitochondrial fission in budding yeast. However, FIS1 appears to have little, if any, role in mitochondrial fission in mammalian cells (Loson et al., 2013; Otera et al., 2010). More recent work has implicated FIS1 in the degradation of dysfunctional mitochondria (Rojansky et al., 2016; Shen et al., 2014; Yamano et al., 2014).
As with mitochondrial fusion, loss of mitochondrial fission results in human disease. Mutations in DRP1 result in clinical phenotypes with a strong neurological component, ranging from microcephaly with multi-organ failure and neonatal lethality to intractable epilepsy in childhood (Fahrner et al., 2016; Nasca et al., 2016; Sheffer et al., 2016; Vanstone et al., 2016; Waterham et al., 2007; Yoon et al., 2016). Homozygous mutations in MFF result in severe neuromuscular disease (Koch et al., 2016; Shamseldin et al., 2012). The severe phenotypes of mouse DRP1 or MFF knockouts also point to the critical role of mitochondrial fission in tissue physiology (Chen et al., 2015; Ishihara et al., 2009; Wakabayashi et al., 2009). Interestingly, loss of either fusion or fission can lead to mitochondrial DNA (mtDNA) instability, which may exacerbate pathological phenotypes (Amati-Bonneau et al., 2008; Chen et al., 2007; Chen et al., 2010; Ishihara et al., 2009; Parone et al., 2008).
Mitochondrial dynamics during the cell cycle
It has been known for many decades that mitochondria are symmetrically partitioned to daughter cells during a typical cell division (Christiansen, 1949). Detailed imaging studies of cultured cells indicate that mitochondrial morphology undergoes stereotyped changes during progression through the cell cycle (Margineantu et al., 2002; Mitra et al., 2009; Taguchi et al., 2007). The two most apparent features are tubulation of the mitochondria network at the G1/S transition and extensive fragmentation during mitosis (Figure 2). The elongated, "hyperfused" network at G1/S is associated with higher ATP production and affects entry into S phase by controlling the levels of cyclin E (Mitra et al., 2009). Extensive fragmentation during mitosis likely increases the likelihood of equitable inheritance of mitochondria to daughter cells, because partitioning is thought to occur passively during cytokinesis. Nevertheless, cells lacking DRP1 are capable of undergoing cell division, even though they maintain filamentous mitochondria throughout the cell cycle (Ishihara et al., 2009; Taguchi et al., 2007). Daughter cells arising from DRP1-deficient cells do inherit mitochondria, suggesting that the cytokinesis machinery is capable of dividing mitochondria in a DRP1-independent manner; however, the partitioning of mitochondria to daughter cells appears less equitable (Ishihara et al., 2009). Although mitochondrial inheritance during cell division in most animal cells has been assumed to be passive, it should be noted that budding yeast has active mechanisms to transport a subset of mitochondria into the daughter cell and retain others in the mother cell (Vevea et al., 2014; Westermann, 2014). In addition, stem-like cells from the mammary gland display DRP1-dependent, asymmetric inheritance of mitochondria, suggesting that, even in mammalian cells, not all mitochondrial segregation is passive (Katajisto et al., 2015).
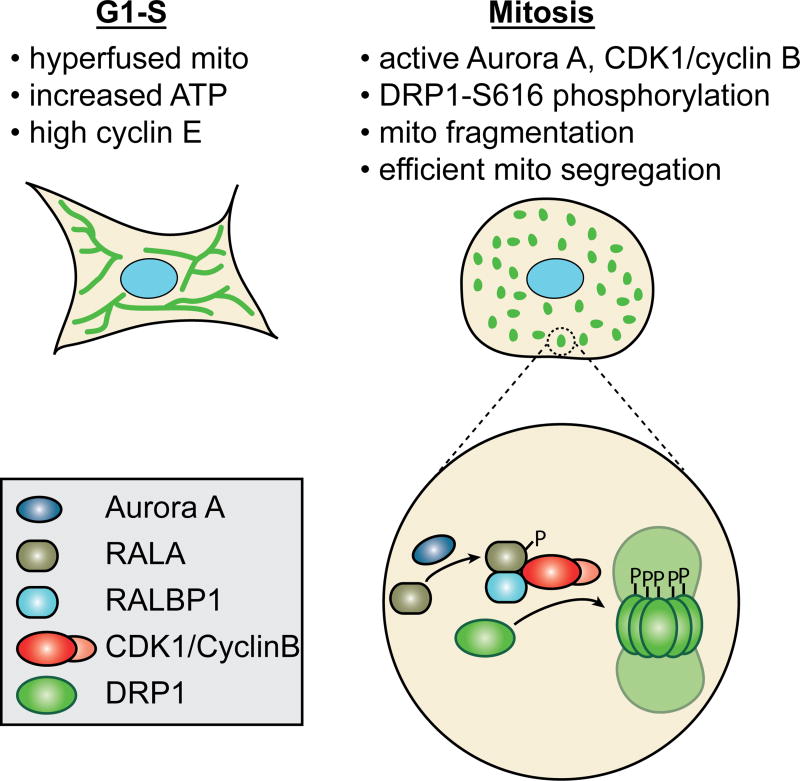
Figure 2. Mitochondrial dynamics during the cell cycle.
The two signature features of mitochondrial structure during the cell cycle are the hyperfused network at G1-S and the extensively fragmented state at mitosis. The hyperfused network at G1-S is associated with increased ATP and high cyclin E levels. The fragmented state during mitosis facilitates equitable distribution of mitochondria to daughter cells. Fragmentation is driven by phosphorylation of DRP1 by CDK1/cyclin B, causing the activation of DRP1 and mitochondrial fission. Aurora A works upstream to activate CDK1/cyclin B.
DRP1 plays a crucial role in controlling cell-cycle-associated changes in mitochondrial morphology (Figure 2). Phosphorylation of a conserved serine (S616 in human DRP1 variant 1) by the mitotic kinase CDK1/cyclin B causes fragmentation of the mitochondrial network early in M phase (Taguchi et al., 2007). This phosphorylation is accompanied by an MFF-dependent rise in DRP1 levels on the mitochondrial surface (Kashatus et al., 2011). Aurora A, a mitotic kinase, works upstream to direct the activity of CDK1/cyclin B onto DRP1. Aurora A phosphorylates RALA, a RAS-like GTPase. Phosphorylated RALA is released from the plasma membrane and concentrates, along with its effector protein RALBP1, at the mitochondrial surface. RALBP1 associates with CDK1/cyclin B and activates its kinase activity, leading to DRP1 phosphorylation at position 616 (Kashatus et al., 2011). Loss of either RALA or RALBP1 disrupts mitotic mitochondrial fission and results in decreased cell proliferation. Underscoring the importance of DRP1 in mitotic progression, inhibition of DRP1 results in delayed S-phase entry (Mitra et al., 2009) and partial G2/M arrest (Qian et al., 2012).
Another level of control for DRP1 during the cell cycle resides in its alternative splicing of three exons (Strack et al., 2013). Splice variants which contain the third alternative exon without the second one bind to microtubules, thus sequestering DRP1 away from mitochondria. Interestingly, this association is only found with highly bundled microtubules, as depolymerization by nocodazole removes DRP1 from α-tubulin and increases DRP1-mitochondria interactions. Phosphorylation of DRP1-S616 by CDK1 promotes dissociation of the DRP1 from microtubules, increases DRP1 levels on mitochondria, and thereby augments mitochondrial fragmentation.
DRP1 also demonstrates strong interactions with another cell cycle protein, cyclin E. Although it is unclear how this interaction regulates normal cell cycle progression, it is evident that inhibition of DRP1 leads to increased levels of cyclin E throughout the cell cycle, and especially during the G2/M transition (Mitra et al., 2009; Parker et al., 2015; Qian et al., 2012). This cyclin E buildup causes replication stress in the form of premature entry into a prolonged S-phase, chromosomal instability, and subsequent aneuploidy. Some stem cells demonstrate a similar profile of enhanced cyclin E, short G1, and long S-phase, and indeed, progenitor cell markers are upregulated in DRP1-inhibited cells (Parker et al., 2015). Interestingly, concurrent knockdown of the mitochondrial fusion protein, OPA1, with DRP1 abrogates the elevation of cyclin E levels (Qian et al., 2012), suggesting it may be the hyperfused mitochondrial morphology of DRP1-inhibited cells that causes increased cyclin E levels. It is as yet unclear how mitochondrial morphology controls cyclin E, but intriguingly, a mitochondrial pool of cyclin E, which is not phosphorylated and degraded as the nuclear pool is, was discovered recently (Parker et al., 2015).
DRP1 is also regulated by phosphorylation at S637. A number of kinases and phosphatases act on this site, including protein kinase A (PKA), calcium/calmodulin-dependent protein kinase 1α (CaMK1α), calcineurin, and protein phosphatase 2A (PP2A) (Chang and Blackstone, 2010). Most studies have found phosphorylation at S637 to inhibit the fission activity of DRP1. Although there is no evidence demonstrating involvement of S637 phosphorylation during normal cell cycle progression, several cancer studies suggest that increased tumorigenesis correlates with decreased S637 phosphorylation, as described below (Rehman et al., 2012; Xie et al., 2015).
Enhanced mitochondrial fission in cancer cells
Cancer progression consists of tumorigenesis, during which cells multiply in an unrestrained manner, and subsequent cell invasion, which allows metastatic spread of tumor cells to other tissues and organs. Biochemical pathways present in normal cells are often co-opted in tumor cells to promote their growth. Cell cycle pathways are prime targets, and given the prominent effect of DRP1 on cell cycle, it is perhaps unsurprising that DRP1 seems to be activated in several cancer types.
In a survey of several lung cancer cell lines, the mitochondria were found to be highly fragmented, when compared to unrelated primary cells obtained from the lung (Rehman et al., 2012). In multiple cultured lung adenocarcinoma cell lines and human patient samples, mitochondrial fragmentation is correlated with reduced levels of MFN2 and higher levels of DRP1. In addition, there is more abundant phosphorylation of the activating S616 site and less phosphorylation of the inactivating S637 site. When MFN2 is overexpressed or DRP1 is inhibited, tumor growth is reduced in vitro and in vivo. This growth reduction involves both reduced proliferation and increased apoptosis. Therefore, as in normal cell cycle, increased fission/decreased fusion seems to push these cancer cells into mitosis, thus increasing cell replication.
Similarly, a specific pathway for increasing DRP1-mediated mitochondrial fission has been uncovered in RAS-induced tumors (Kashatus et al., 2015; Serasinghe et al., 2015) (Figure 1). In cultured cells transformed by the oncogenic RASG12V mutant, mitochondria are fragmented due to increased activity of DRP1. DRP1 is activated by phosphorylation of S616 by the MAP kinases ERK1 and ERK2. Importantly, inhibition of this pathway reduces the proliferation of RASG12V transformed cells and their ability to generate tumors in a xenograft model. These results indicate that the MAP kinase pathway can regulate mitochondrial fission to remodel mitochondria during cell transformation. Whereas DRP1 activation by CDK1 phosphorylation is specific to mitosis, in cancer cells, ERK1/2 activation of DRP1 leads to fragmented mitochondria throughout the cell cycle.
There is increasing evidence that tumors often contain a subpopulation of cells with enhanced capacity to initiate new tumors when transplanted to a novel site. Such tumor-initiating cells have been termed cancer stem cells, and they share some parallels with normal stem cells that function during development, in that they can undergo both self-renewal and differentiation (Pattabiraman and Weinberg, 2014; Zhou et al., 2009). A recent study suggests that brain tumor initiating cells (BTICs) have a distinct mitochondrial profile, compared to non-initiating tumor cells, that is driven by mitochondrial fission (Xie et al., 2015). In surgical samples of glioblastomas, the initiating cell subpopulation (isolated by cell surface markers) has elevated levels of DRP1-S616 phosphorylation due to the activity of CDK5, and downregulated levels of inhibitory DRP1-S637 phosphorylation. Inhibition of DRP1 activity hinders growth of the initiating cells in culture and in xenografts. Importantly, the level of DRP1-S616 phosphorylation negatively correlates with glioblastoma patient survival.
Of note, the opposing force of mitochondrial fusion seems to slow growth of several cancer lines. As described above, MFN2 overexpression can reduce lung cancer growth (Rehman et al., 2012; Xie et al., 2015). In another example, mouse medulloblastoma cells and tumors express lower levels of MFN1/2 as compared to non-transformed cells (Malhotra et al., 2016). Medulloblastoma is the most common malignant brain tumor found in children. Upon mitofusin overexpression, cell proliferation, as measured by BrdU labeling and cyclin D2 levels, is decreased.
Cancer cell migration
During progression to a metastatic phenotype, cancer cells undergo an epithelial-to-mesenchymal transition and acquire greater mobility and invasiveness. These qualities are required for cancer cells to invade surrounding tissue, enter the bloodstream, migrate throughout the body and seed additional organs. Once again, mitochondrial fragmentation has been found to increase in malignant cells and to promote tumor cell invasion. As compared to non-metastatic breast tumors, invasive breast carcinomas and metastases express higher levels of DRP1 and lower levels of MFN1 (Zhao et al., 2013). Using in vitro transwell cell invasion assays with metastatic and non-metastatic breast cancer cell lines, inhibition of DRP1 activity or overexpression of MFN1/2 greatly reduces cell migration and invasion, whereas silencing of mitofusins increases invasive activity. Enhanced migration is accompanied by more lamellipodia formation and mitochondrial accumulation in the lamellipodia periphery. Mitochondrial respiration is required for this process, presumably due to the high energy demands of F-actin polymerization, a central step in formation of lamellipodia. Similar patterns were found in malignant, oncoctyic thyroid carcinomas (Ferreira-da-Silva et al., 2015). A sampling of human tumors found increased DRP1 expression in malignant cells. Furthermore, either mdivi-1, a chemical inhibitor of DRP1 (Cassidy-Stone et al., 2008), or dominant-negative DRP1 can inhibit migration and invasion of thyroid cancer cell lines in scratch-wound assays and transwell experiments.
Mechanistically, NF-κB-inducing kinase (NIK) seems to regulate DRP1 involvement in cell migration in multiple cancer types, including glioma, breast cancer, and pancreatic cancer (Jung et al., 2016). Loss of NIK greatly reduces invasive activity and causes congregation of mitochondria around the nucleus as opposed to their normal, predominantly anterograde movement toward the leading edge of migrating cancer cells. NIK is localized to the mitochondria, recruits DRP1, and promotes (perhaps indirectly) phosphorylation of DRP1-S616 and dephosphorylation of DRP1-S637, thus increasing mitochondrial fission. DRP1 seems to act downstream of NIK as deletion of DRP1 abrogates NIK-dependent invasiveness.
As for cell growth, mitochondrial fusion seems to oppose fission and inhibits invasiveness of both breast and lung cancers (Xu et al., 2017). Low expression of MFN2 for patients of both diseases correlates with poor prognosis. In vitro transwell assays and in vivo xenograft metastasis experiments demonstrate that MFN2 loss in cancer cell lines stimulate cell migration. Biochemically, MFN2 binds directly to Rictor in the mammalian target of rapamycin complex 2 (mTORC2) and suppresses AKT signaling.
Reactive oxygen species (ROS) in cancer
While a comprehensive examination of the role of ROS in cancer is beyond the scope of this review, it must be mentioned that cancer cells often contain increased levels of ROS, which are mostly generated by the mitochondria (Panieri and Santoro, 2016; Sullivan and Chandel, 2014). ROS promote tumor growth by altering cellular metabolism and acting as signaling molecules in growth factor pathways. Interestingly, ROS can also affect expression and post-translational modifications of mitochondrial dynamic proteins, which may further stimulate tumorigenesis (Willems et al., 2015). Heteroplasmic mtDNA mutation levels correlate with ROS production, tumorigenicity, and metastastic potential (Sullivan and Chandel, 2014). Given the importance of mitochondrial dynamics in maintaining mtDNA stability and ROS levels (Amati-Bonneau et al., 2008; Chen et al., 2007; Chen et al., 2010; Chen et al., 2015; Ishihara et al., 2009; Parone et al., 2008), it is likely that mitochondrial fusion and fission impinge on cancer dynamics at least partially through ROS activity.
Parallels between the mitochondrial and metabolic profiles of cancer cells and stem cells
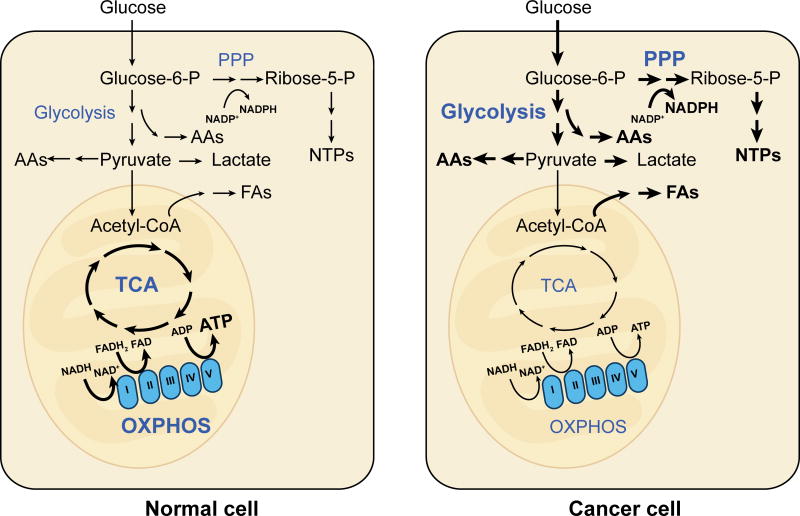
Cancer cells are known to have distinct metabolic profiles compared to their normal counterparts. Many cancer cells display the Warburg effect, in which cells predominantly rely on glycolysis over oxidative phosphorylation (OXPHOS), even when oxygen is plentiful (Pavlova and Thompson, 2016; Vander Heiden et al., 2009). In the clinical setting, this prominent property has been exploited in the use of positron emission tomography to image some tumors based on their high uptake of positron-emitting glucose analogs. Cancer cells displaying the Warburg effect use predominantly glycolysis over OXPHOS, even though they generally have functional mitochondria (Figure 3). Given that glycolysis is far less efficient than OXPHOS at extracting energy from glucose, generating only 2 ATP instead of 36 ATP, why would rapidly reproducing cells undergo metabolic reprogramming to preferentially utilize glycolysis? The prevailing hypothesis is that in the presence of ample nutrients, high glycolytic flux generates sufficient cellular energy while providing the building blocks for cell growth, such as acetyl-CoA for fatty acids, glycolytic intermediates for amino acids, and ribose for nucleotides (Figure 3). In addition, there is evidence that lactate produced by glycolytic tumor cells is not merely a waste product but can be used to “feed” adjacent oxidative cancer cells and/or to signal vascular endothelial cells to initiate angiogenesis (Doherty and Cleveland, 2013; Sonveaux et al., 2008). The latter point emphasizes that cancer cells are notoriously heterogeneous, and that the Warburg effect is not a feature of all types of cancers, or even of all cancer cells in a given tumor.
Figure 3. Metabolic rewiring in normal versus cancer cells.
Differentiated cells typically rely heavily on the OXPHOS activity of mitochondria. In contrast, many cancer cells show the Warburg effect, characterized by reliance on aerobic glycolysis and reduced emphasis on OXPHOS. Glycolysis, though less energy efficient than OXPHOS, generates metabolic intermediates that provide building blocks for synthesis of amino acids (AAs), fatty acids (FAs), and nucleotides (NTPs).
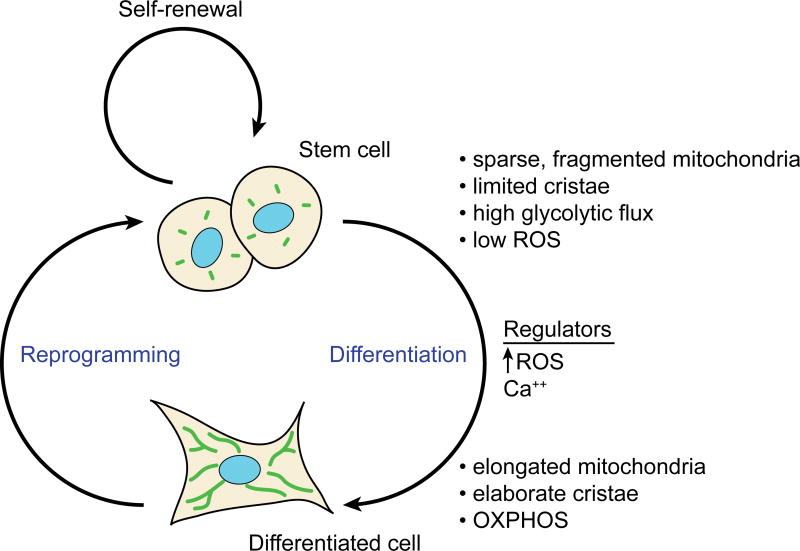
Given the similarities between stem cells and aggressive cancer cells in terms of replicative needs and non-differentiated status, it is quite interesting that recent studies reveal that stem cells have a metabolic profile reminiscent of that of cancer cells. Stem cells rely primarily on “aerobic glycolysis” to generate energy. This is defined as preferential utilization of glycolysis over OXPHOS even in an environment rich with oxygen. Pluripotent embryonic stem cells (ESCs) and induced pluripotent cells (iPSCs)--which are generated by reprogramming of adult, differentiated cells by expression of the Yamanaka factors (OCT4, SOX2, KLF4 and c-MYC)--have high glycolytic flux and relatively low mitochondrial respiration (Kondoh et al., 2007; Prigione et al., 2010; Zhang et al., 2011). Mitochondria in these cells are generally characterized as sparse and “immature,” containing less mtDNA and demonstrating underdeveloped cristae structure. Upon differentiation to terminal cell types, mitochondrial content and utilization of OXPHOS are typically increased (Figure 4). For example, human ESC lines have sparse mitochondria, but during differentiation into cardiomyocytes, they accumulate mitochondria and turn from glycolysis to fatty acid oxidation as their primary source of energy (St John et al., 2005). It should be noted that these differences in mitochondrial mass may be exaggerated due to the small size of pluripotent cells. One study found that the differences in mitochondrial mass between human iPSCs and ESCs compared to fibroblasts are minimal when normalized to total protein content (Zhang et al., 2011).
Figure 4. Mitochondrial and metabolic profiles of stem cells versus differentiated cells.
ESCs have less reliance on mitochondrial metabolism, and this is reflected in the ultrastructure of their mitochondria. Similar differences exist when somatic cells are reprogrammed into iPSCs. As noted in the main text, these generalizations for mitochondrial structure in stem cells do not apply to neural stem cells. ROS and calcium are two regulators of differentiation that are regulated by mitochondrial function.
Several reasons have been proposed for why it may be beneficial for ESCs versus differentiated cells to favor a higher glycolysis:OXPHOS ratio for energy production (Zhang et al., 2012). First, in the presence of ample glucose and other nutrients, glycolysis provides sufficient cellular ATP while reducing the production of reactive oxygen species (ROS) that are a normal byproduct of cellular respiration. Stem cells are a long-term source for cellular regeneration, and therefore longevity and genome maintenance are priorities. Containment of ROS may be important for longevity by minimizing ROS-induced telomere shortening and for genome integrity by reducing DNA and protein damage. Second, as for cancer cells, glycolytic metabolism can be regulated to yield important chemical building blocks that are critical for synthesis of amino acids, lipids, and nucleic acids, which are important for cells that continually replicate and divide (Figure 3). For example, shunting of glycolytic intermediates into the pentose phosphate pathway generates ribose-5-phosphate, a precursor for nucleotide synthesis, and NADPH, which provides reducing power to drive nucleotide and lipid biosynthesis. Moreover, under oxidative stress conditions, mouse ESCs additionally depend on NADPH from the pentose phosphate pathway for detoxification of reactive oxygen species (Filosa et al., 2003). Finally, cytosolic acetyl-CoA, derived from glycolytically produced pyruvate, is critical to maintain histone acetylation and thus, pluripotency (Moussaieff et al., 2015). Therefore, stem cells and differentiated cells have distinct metabolic profiles that are tailored to their divergent cellular needs and functions. It should be noted that whereas cancer cells and stem cells both favor glycolysis over OXPHOS, the usage of downstream effectors, such as lactate and acetyl-CoA, may differ greatly (Shyh-Chang and Daley, 2015).
The likely functional importance of these metabolic profiles is highlighted by the observation that reprogramming of somatic cells to iPSCs is associated with downregulation of respiratory chain activity and increased glycolysis (Folmes et al., 2011; Zhu et al., 2010). Whereas mouse embryonic fibroblasts have organized mitochondrial networks displaying ample cristae, the iPSCs derived from them have few and "regressed" mitochondria with sparse cristae, morphological features suggesting reduced mitochondrial activity (Zhu et al., 2010). Glucose utilization increases, along with lactate production and its efflux from the cell. Drugs that inhibit glycolysis, such as deoxyglucose, substantially reduce the efficiency of reprogramming, whereas glucose supplementation or stimulation of glycolysis has the opposite effect.
Moreover, there is evidence that mitochondrial changes occur prior to the acquisition of pluripotential markers. In mouse embryonic fibroblasts (MEFs) that are undergoing reprogramming, mitochondrial membrane potential and expression of glycolytic gene expression are increased well before the pluripotential state is achieved (Folmes et al., 2011). Consistent with this, during differentiation of human pluripotent stem cells, a metabolic shift from glycolysis towards oxidative phosphorylation occurs prior to the loss of pluripotency markers, such as OCT4 (Zhang et al., 2011). These temporal correlations suggest that remodeling of mitochondrial activity may be important in determining cell state. Given the striking changes in mitochondrial architecture that occur when stem cells differentiate and when somatic cells are reprogrammed, it is reasonable to ask what role mitochondrial dynamics might play. Furthermore, the parallels between cancer cells and stem cells in terms of metabolism and rapid division argue that common mechanisms may govern the biochemical changes in stem cells and cancer cells.
Mitochondrial dynamics in stem cells
Mitochondrial fission driven by DRP1 seems to play a crucial role in developing and maintaining pluripotency. Mdivi-1, a chemical inhibitor of DRP1, inhibits the transition of MEFs into iPSC colonies when applied early in reprogramming (Vazquez-Martin et al., 2012). This compound is now known to have off-target effects (Bordt et al., 2017), including inhibition of respiratory Complex I, so such experiments need to be re-evaluated. Nevertheless, downregulation of DRP1 function by other means--RNAi and overexpression of a dominant negative DRP1--greatly diminishes the re-programming efficiency of fibroblasts, as measured by the formation of colonies positive for alkaline phosphatase, a marker of pluripotency (Prieto et al., 2016b). During the reprogramming process, DRP1 is activated by phosphorylation of S616 by ERK1/2. This phosphorylation is maintained partially by downregulation of DUSP6, a MAPK phosphatase. However, the changes in mitochondrial dynamics during reprogramming are complex, with mitochondria undergoing severe fragmentation during the early phase of reprogramming, followed by restoration to shorter tubules when pluripotent colonies are formed (Prieto et al., 2016b).
During MEF reprogramming, mitochondrial fission appears to depend primarily on DRP1, MID51 and GDAP1 (Prieto et al., 2016a). MID51 is one of several mitochondrial outer membrane receptors for DRP1. GDAP1 is a mitochondrial outer membrane protein that is involved in some forms of Charcot-Marie-Tooth disease and regulates mitochondrial fission, but its mechanism of action remains unclear (Niemann et al., 2005). In addition, there is evidence for a moderate role for MFF, but no role for FIS1 and MID49. GDAP1 knockout MEFs form only 25% of the number of alkaline-phosphatase-positive colonies upon reprogramming when compared to control MEFs. Interestingly, a G2/M block seems to occur in GDAP1-null cells (Prieto et al., 2016a), similar to that found during DRP1 inhibition (Qian et al., 2012).
Even in naturally pluripotent human ESCs (hESCs), maintenance of pluripotency is dependent on DRP1 activity (Son et al., 2013). REX1, a transcriptional activator of cyclins B1 and B2, promotes DRP1-S616 phosphorylation, mitochondrial fragmentation, and increased glycolytic activity. In stable knockdowns of REX1, hESC cells lose proliferative capacity and partially arrest at the G2/M transition of the cell cycle. The REX1-deficient ESCs differentiate at a higher rate and have defects in embryoid body and teratoma formation. Knockdown of DRP1 also promotes hESC differentiation, whereas overexpression of a constitutively active DRP1-S616D mutant dramatically increases pluripotency markers, such as TRA-1–60, OCT4, and REX1 itself.
As in cancer cells, mitochondrial fusion seems to work in opposition to fission by driving differentiation and decreasing cellular proliferation. For example, the differentiation of human iPSCs into functional neurons requires MFN2 function (Fang et al., 2016). Knockdown of MFN2 reduces OXPHOS activity and inhibits dendrite and synapse formation, whereas overexpression of MFN2 enhances dendritic length, synaptophysin expression, and ATP production. Depending on the cellular context, loss of MFN2 (Mourier et al., 2015), of both MFN1 and MFN2, or of OPA1 (Chen et al., 2005) results in severe respiratory chain function, which is likely to be critical for cell differentiation.
Despite the plethora of evidence demonstrating the importance of DRP1 and mitochondrial fission and fragmentation in establishing and maintaining pluripotency, cell type differences exist and suggest that the relationship of mitochondrial dynamics to metabolism and cell identity is not straightforward. Genetic studies in the mouse brain suggest that mitochondrial fusion, rather than fission, may play a role in promoting self-renewal of neural stem cells (NSCs) of the cortex (Khacho et al., 2016). In contrast to embryonic and hematopoietic stem cells, which contain fragmented and immature mitochondria, neural stem cells have relatively abundant tubular mitochondria. These cells progress to form committed progenitors, which have fragmented mitochondria. The committed progenitors then differentiate into post-mitotic neurons, which have elongated mitochondria. The NSCs therefore go through at least two structural transitions on their way to forming terminally differentiated neurons. When the mitochondrial fusion proteins MFN1 and MFN2 are removed from NSCs, they show mitochondrial fragmentation, increased tendency to differentiate, and reduced capacity for self-renewal (Khacho et al., 2016). Similar results were obtained for depletion of OPA1. Maintenance of elongated mitochondria therefore seems important for self-renewal of these neural stem cells. Interestingly, despite the differences in mitochondrial morphology, the metabolic requirements for neural stem cell pluripotency remained consistent with that of ESCs. Namely, the NSCs rely on aerobic glycolysis and progressively shift towards cellular respiration during cell fate commitment and differentiation. NSCs express high levels of UCP2 (Uncoupling protein 2) (Khacho et al., 2016), consistent with the high expression of UCP2 in human iPSCs and ESCs, where it inhibits mitochondrial glucose oxidation (Zhang et al., 2011). Biochemical analysis of recombinant UCP2 indicates that it functions as a metabolite transporter to export TCA cycle intermediates from the mitochondria (Vozza et al., 2014), and thus may suppress mitochondrial oxidation of glucose even in the presence of oxygen and functional mitochondrial respiration complexes. Therefore, in neural stem cells, aerobic glycolysis is still critical for maintaining pluripotency and replicative function, but mitochondria are maintained in elongated form as opposed to the fragmented form found in ESCs. Based on the divergent results from NSCs versus ESCs, it seems that the type of mitochondrial profile associated with stem cells is highly context dependent, as is the biological effect of inhibiting either mitochondrial fusion or fission. Clearly, there are still large gaps in our understanding of how mitochondrial dynamics and morphology intersect with metabolism and cell identity. Nevertheless, it is becoming clear that mitochondrial dynamics plays an important role in controlling stem cell metabolism, which is critical in determining the ability of these cells to self-renew or differentiate.
An interesting parallel to the NSCs may exist in the Drosophila egg chamber (Mitra et al., 2012), where mitochondrial dynamics has been shown to control the differentiation of follicle cells. Expression of dominant-negative DRP1 in follicle cells causes their hyper-proliferation, instead of differentiation. Conversely, inhibition of MARF-1 (a mitofusin ortholog) induced premature induction of differentiation markers. These effects correlate with the levels of cyclin E, suggesting that mitochondrial dynamics may affect cell differentiation by regulating cell cycle exit.
The observations with NSCs noted above suggest that different states of stem cells can have distinct mitochondrial and metabolic profiles. It is worth noting that pluripotent stem cells may also belong to two distinct states of pluripotency. Naive pluripotent stem cells are derived from the inner cell mass of preimplantation embryos, whereas primed pluripotent stem cells are derived from post-implantation embryos and represent a more mature state (Nichols and Smith, 2009; Weinberger et al., 2016). Among their differences in cellular properties, there is evidence that naive versus primed pluripotent stem cells are metabolically distinct (Zhou et al., 2012), pointing towards the complexity in understanding the metabolism of stem cells.
Adding to the complexity, one group found that knockdown of DRP1 did not affect the reprogramming of MEFs into iPSCs (Wang et al., 2014). Surprisingly, even though fibroblasts depleted of DRP1 have very elongated mitochondria, the derived iPSCs have fragmented mitochondrial typical of iPSCs, raising the possibility that DRP1 is not involved in structural remodeling of mitochondria during this process. The DRP1-deficient iPSCs also showed reduced tendency to differentiate into neuroepithelium, consistent with the situation in neural stem cells (Khacho et al., 2016). Further testing will be required to understand the discrepancies among the studies regarding reprogramming of MEFs to iPSCs.
The role of mitochondrial dynamics in signaling differentiation
During the transition of neural stem cells to committed progenitor cells, the appearance of fragmented mitochondria is associated with increased ROS (Khacho et al., 2016). Experimental depletion of MFN1 and MFN2 or of OPA1 results in an increase in ROS that correlates with reduced NSC self-renewal and their increased differentiation to progenitor cells. The authors propose that this increase in ROS serves as an internal signal to drive cell differentiation. It is difficult to rule out that these effects are not due to mitochondrial dysfunction caused by depletion of mitochondrial fusion. However, the study showed that no gross reduction in respiratory chain function occurs.
A role for ROS in cell differentiation has been supported by studies in other systems. A particularly compelling case occurs in keratinocyte differentiation in the mouse epidermis (Hamanaka et al., 2013). Deletion of mitochondrial transcription factor A (TFAM) from the epidermis results in excessive basal layer proliferation and defective keratinocyte differentiation and hair development. Because TFAM is essential for maintenance of mitochondrial DNA, such cells cannot assemble functional respiratory chain complexes and have a severe bioenergetic defect. They also lack the ROS that is normally generated from endogenous respiration. Importantly, application of exogenous hydrogen peroxide caused restoration of differentiation markers to primary keratinocytes lacking TFAM, arguing that it is ROS, and not cellular ATP generation, that is essential for differentiation. A signaling function for ROS has been implicated to several other cell differentiation systems, including adipocytes, hematopoietic cells, neurons, and glial cells (Owusu-Ansah and Banerjee, 2009; Smith et al., 2000; Tormos et al., 2011; Tsatmali et al., 2005).
In addition to ROS, calcium signaling may also be affected by mitochondrial dynamics during cell differentiation. Mitochondrial fusion has been shown to be important for differentiation of mouse cardiomyocytes (Kasahara et al., 2013). Mice with MFN1 and MFN2 deletions in cardiomyocytes have hypoplastic hearts during embryonic development, and gene expression analysis shows reduced levels of markers for cardiac differentiation and proliferation. The differentiation of mouse ESCs into cardiomyocytes was found to require MFN2 and OPA1. In their absence, calcium entry, Notch and calcineurin signaling, known to inhibit cardiomyocyte differentiation, were hyperactive (Kasahara et al., 2013). A role of MFN2 in regulating intracellular calcium has also been reported for hematopoietic stems cells of the lymphoid lineage (Luchsinger et al., 2016).
Perspectives
Recent studies provide persuasive evidence that mitochondrial dynamics plays an important role in regulating the unique metabolism of cancer cells and their ability to rapidly proliferate. Moreover, there are striking parallels between the metabolism of cancer cells and pluripotential stem cells. Mitochondrial dynamics appears to regulate the glycolytic metabolism of stem cells and their tendency towards self-renewal versus differentiation.
Looking ahead, there are several issues that need clarification. First, most of the cancer cell studies have focused on how mitochondrial dynamics affects cell proliferation, and it will be important to test whether the metabolic insights from stem cell studies also apply to tumor cells. Related to this issue, it is unclear whether the effects of mitochondrial fission on tumor cell proliferation arise from a change in metabolism or another cellular process. Second, there is not a simple relationship between mitochondrial morphology and metabolism. There is often an assumption that glycolytic cells have fragmented mitochondria, and that OXPHOS-intensive cells have elongated mitochondria. However, the discrepancies in mitochondrial profiles between glycolytic ESCs and neuronal stem cells suggest that this idea is an oversimplification. We will need additional studies to better understand the molecular mechanisms linking mitochondrial dynamics to cell metabolism. Third, it will be important to understand whether the effects of mitochondrial dynamics are cell-type-specific. Because changes in mitochondrial dynamics can directly or indirectly impact multiple processes--including metabolism, ROS signaling, and calcium signaling--the outcome may be critically dependent on cell type. Finally, once the interconnections between mitochondrial dynamics, cell metabolism, and proliferation, and differentiation are better understood, we will hopefully be able to harness this information to devise therapies for tumorigenesis and to modulate stem cell function.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amati-Bonneau P, Valentino ML, Reynier P, Gallardo ME, Bornstein B, Boissiere A, Campos Y, Rivera H, de la Aleja JG, Carroccia R, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy 'plus' phenotypes. Brain. 2008;131:338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordt EA, Clerc P, Roelofs BA, Saladino AJ, Tretter L, Adam-Vizi V, Cherok E, Khalil A, Yadava N, Ge SX, et al. The Putative Drp1 Inhibitor mdivi-1 Is a Reversible Mitochondrial Complex I Inhibitor that Modulates Reactive Oxygen Species. Developmental cell. 2017;40:583–594.e586. doi: 10.1016/j.devcel.2017.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carelli V, Chan DC. Mitochondrial DNA: impacting central and peripheral nervous systems. Neuron. 2014;84:1126–1142. doi: 10.1016/j.neuron.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, et al. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Developmental cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annual review of genetics. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Annals of the New York Academy of Sciences. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–26192. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- Chen H, McCaffery JM, Chan DC. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell. 2007;130:548–562. doi: 10.1016/j.cell.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ren S, Clish C, Jain M, Mootha V, McCaffery JM, Chan DC. Titration of mitochondrial fusion rescues Mff-deficient cardiomyopathy. J Cell Biol. 2015;211:795–805. doi: 10.1083/jcb.201507035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen EG. Orientation of the mitochondria during mitosis. Nature. 1949;163:361. doi: 10.1038/163361a0. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrner JA, Liu R, Perry MS, Klein J, Chan DC. A novel de novo dominant negative mutation in DNM1L impairs mitochondrial fission and presents as childhood epileptic encephalopathy. Am J Med Genet A. 2016;170:2002–2011. doi: 10.1002/ajmg.a.37721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Yan S, Yu Q, Chen D, Yan SS. Mfn2 is Required for Mitochondrial Development and Synapse Formation in Human Induced Pluripotent Stem Cells/hiPSC Derived Cortical Neurons. Scientific reports. 2016;6:31462. doi: 10.1038/srep31462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol. 2012;13:75–88. doi: 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-da-Silva A, Valacca C, Rios E, Populo H, Soares P, Sobrinho-Simoes M, Scorrano L, Maximo V, Campello S. Mitochondrial dynamics protein Drp1 is overexpressed in oncocytic thyroid tumors and regulates cancer cell migration. PloS one. 2015;10:e0122308. doi: 10.1371/journal.pone.0122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filosa S, Fico A, Paglialunga F, Balestrieri M, Crooke A, Verde P, Abrescia P, Bautista JM, Martini G. Failure to increase glucose consumption through the pentose-phosphate pathway results in the death of glucose-6-phosphate dehydrogenase gene-deleted mouse embryonic stem cells subjected to oxidative stress. The Biochemical journal. 2003;370:935–943. doi: 10.1042/BJ20021614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Glasauer A, Hoover P, Yang S, Blatt H, Mullen AR, Getsios S, Gottardi CJ, DeBerardinis RJ, Lavker RM, et al. Mitochondrial reactive oxygen species promote epidermal differentiation and hair follicle development. Sci Signal. 2013;6:ra8. doi: 10.1126/scisignal.2003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, et al. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- Jung JU, Ravi S, Lee DW, McFadden K, Kamradt ML, Toussaint LG, Sitcheran R. NIK/MAP3K14 Regulates Mitochondrial Dynamics and Trafficking to Promote Cell Invasion. Current biology : CB. 2016;26:3288–3302. doi: 10.1016/j.cub.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara A, Cipolat S, Chen Y, Dorn GW, 2nd, Scorrano L. Mitochondrial fusion directs cardiomyocyte differentiation via calcineurin and Notch signaling. Science. 2013;342:734–737. doi: 10.1126/science.1241359. [DOI] [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, Kashatus DF. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Molecular cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katajisto P, Dohla J, Chaffer CL, Pentinmikko N, Marjanovic N, Iqbal S, Zoncu R, Chen W, Weinberg RA, Sabatini DM. Stem cells. Asymmetric apportioning of aged mitochondria between daughter cells is required for stemness. Science. 2015;348:340–343. doi: 10.1126/science.1260384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khacho M, Clark A, Svoboda DS, Azzi J, MacLaurin JG, Meghaizel C, Sesaki H, Lagace DC, Germain M, Harper ME, et al. Mitochondrial Dynamics Impacts Stem Cell Identity and Fate Decisions by Regulating a Nuclear Transcriptional Program. Cell Stem Cell. 2016;19:232–247. doi: 10.1016/j.stem.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Koch J, Feichtinger RG, Freisinger P, Pies M, Schrodl F, Iuso A, Sperl W, Mayr JA, Prokisch H, Haack TB. Disturbed mitochondrial and peroxisomal dynamics due to loss of MFF causes Leigh-like encephalopathy, optic atrophy and peripheral neuropathy. J Med Genet. 2016;53:270–278. doi: 10.1136/jmedgenet-2015-103500. [DOI] [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annual review of cell and developmental biology. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- Lee JE, Westrate LM, Wu H, Page C, Voeltz GK. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529:528–531. doi: 10.1038/nature16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Dey A, Prasad N, Kenney AM. Sonic Hedgehog Signaling Drives Mitochondrial Fragmentation by Suppressing Mitofusins in Cerebellar Granule Neuron Precursors and Medulloblastoma. Mol Cancer Res. 2016;14:114–124. doi: 10.1158/1541-7786.MCR-15-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margineantu DH, Gregory Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, Rikhy R, Lilly M, Lippincott-Schwartz J. DRP1-dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis. J Cell Biol. 2012;197:487–497. doi: 10.1083/jcb.201110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourier A, Motori E, Brandt T, Lagouge M, Atanassov I, Galinier A, Rappl G, Brodesser S, Hultenby K, Dieterich C, et al. Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J Cell Biol. 2015;208:429–442. doi: 10.1083/jcb.201411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, et al. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Nasca A, Legati A, Baruffini E, Nolli C, Moroni I, Ardissone A, Goffrini P, Ghezzi D. Biallelic Mutations in DNM1L are Associated with a Slowly Progressive Infantile Encephalopathy. Human mutation. 2016;37:898–903. doi: 10.1002/humu.23033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Miyata N, Kuge O, Mihara K. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J Cell Biol. 2016;212:531–544. doi: 10.1083/jcb.201508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem. 2013;288:27584–27593. doi: 10.1074/jbc.M113.479873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panieri E, Santoro MM. ROS homeostasis and metabolism: a dangerous liason in cancer cells. Cell Death Dis. 2016;7:e2253. doi: 10.1038/cddis.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker DJ, Iyer A, Shah S, Moran A, Hjelmeland AB, Basu MK, Liu R, Mitra K. A new mitochondrial pool of cyclin E, regulated by Drp1, is linked to cell-density-dependent cell proliferation. Journal of cell science. 2015;128:4171–4182. doi: 10.1242/jcs.172429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PloS one. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J, Leon M, Ponsoda X, Garcia-Garcia F, Bort R, Serna E, Barneo-Munoz M, Palau F, Dopazo J, Lopez-Garcia C, et al. Dysfunctional mitochondrial fission impairs cell reprogramming. Cell Cycle. 2016a;15:3240–3250. doi: 10.1080/15384101.2016.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J, Leon M, Ponsoda X, Sendra R, Bort R, Ferrer-Lorente R, Raya A, Lopez-Garcia C, Torres J. Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. Nat Commun. 2016b;7:11124. doi: 10.1038/ncomms11124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Qian W, Choi S, Gibson GA, Watkins SC, Bakkenist CJ, Van Houten B. Mitochondrial hyperfusion induced by loss of the fission protein Drp1 causes ATM-dependent G2/M arrest and aneuploidy through DNA replication stress. Journal of cell science. 2012;125:5745–5757. doi: 10.1242/jcs.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, Archer SL. Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J. 2012;26:2175–2186. doi: 10.1096/fj.11-196543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojansky R, Cha MY, Chan DC. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. eLife. 2016;5 doi: 10.7554/eLife.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: functional design of a membrane fission catalyst. Annual review of cell and developmental biology. 2011;27:79–105. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Serasinghe MN, Wieder SY, Renault TT, Elkholi R, Asciolla JJ, Yao JL, Jabado O, Hoehn K, Kageyama Y, Sesaki H, et al. Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Molecular cell. 2015;57:521–536. doi: 10.1016/j.molcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamseldin HE, Alshammari M, Al-Sheddi T, Salih MA, Alkhalidi H, Kentab A, Repetto GM, Hashem M, Alkuraya FS. Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J Med Genet. 2012;49:234–241. doi: 10.1136/jmedgenet-2012-100836. [DOI] [PubMed] [Google Scholar]
- Sheffer R, Douiev L, Edvardson S, Shaag A, Tamimi K, Soiferman D, Meiner V, Saada A. Postnatal microcephaly and pain insensitivity due to a de novo heterozygous DNM1L mutation causing impaired mitochondrial fission and function. Am J Med Genet A. 2016;170:1603–1607. doi: 10.1002/ajmg.a.37624. [DOI] [PubMed] [Google Scholar]
- Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, Martin SJ. Mitochondrial fission/fusion dynamics and apoptosis. Mitochondrion. 2010;10:640–648. doi: 10.1016/j.mito.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ. Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab. 2015;21:349–350. doi: 10.1016/j.cmet.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Smith J, Ladi E, Mayer-Proschel M, Noble M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10032–10037. doi: 10.1073/pnas.170209797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MY, Choi H, Han YM, Cho YS. Unveiling the critical role of REX1 in the regulation of human stem cell pluripotency. Stem Cells. 2013;31:2374–2387. doi: 10.1002/stem.1509. [DOI] [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008;118:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, Simerly CR, Schatten GP. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–153. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- Strack S, Wilson TJ, Cribbs JT. Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J Cell Biol. 2013;201:1037–1051. doi: 10.1083/jcb.201210045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Chandel NS. Mitochondrial reactive oxygen species and cancer. Cancer Metab. 2014;2:17. doi: 10.1186/2049-3002-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, Chandel NS. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsatmali M, Walcott EC, Crossin KL. Newborn neurons acquire high levels of reactive oxygen species and increased mitochondrial proteins upon differentiation from progenitors. Brain research. 2005;1040:137–150. doi: 10.1016/j.brainres.2005.01.087. [DOI] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. The EMBO journal. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstone JR, Smith AM, McBride S, Naas T, Holcik M, Antoun G, Harper ME, Michaud J, Sell E, Chakraborty P, et al. DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur J Hum Genet. 2016;24:1084–1088. doi: 10.1038/ejhg.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Martin A, Cufi S, Corominas-Faja B, Oliveras-Ferraros C, Vellon L, Menendez JA. Mitochondrial fusion by pharmacological manipulation impedes somatic cell reprogramming to pluripotency: new insight into the role of mitophagy in cell stemness. Aging (Albany NY) 2012;4:393–401. doi: 10.18632/aging.100465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vevea JD, Swayne TC, Boldogh IR, Pon LA. Inheritance of the fittest mitochondria in yeast. Trends in cell biology. 2014;24:53–60. doi: 10.1016/j.tcb.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozza A, Parisi G, De Leonardis F, Lasorsa FM, Castegna A, Amorese D, Marmo R, Calcagnile VM, Palmieri L, Ricquier D, et al. UCP2 transports C4 metabolites out of mitochondria, regulating glucose and glutamine oxidation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:960–965. doi: 10.1073/pnas.1317400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ye X, Zhao Q, Zhou Z, Dan J, Zhu Y, Chen Q, Liu L. Drp1 is dispensable for mitochondria biogenesis in induction to pluripotency but required for differentiation of embryonic stem cells. Stem cells and development. 2014;23:2422–2434. doi: 10.1089/scd.2014.0059. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Westermann B. Mitochondrial inheritance in yeast. Biochimica et biophysica acta. 2014;1837:1039–1046. doi: 10.1016/j.bbabio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ. Redox Homeostasis and Mitochondrial Dynamics. Cell Metab. 2015;22:207–218. doi: 10.1016/j.cmet.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Xie Q, Wu Q, Horbinski CM, Flavahan WA, Yang K, Zhou W, Dombrowski SM, Huang Z, Fang X, Shi Y, et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci. 2015;18:501–510. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Chen G, Li X, Wu X, Chang Z, Xu J, Zhu Y, Yin P, Liang X, Dong L. MFN2 suppresses cancer progression through inhibition of mTORC2/Akt signaling. Scientific reports. 2017;7:41718. doi: 10.1038/srep41718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon G, Malam Z, Paton T, Marshall CR, Hyatt E, Ivakine Z, Scherer SW, Lee KS, Hawkins C, Cohn RD. Lethal Disorder of Mitochondrial Fission Caused by Mutations in DNM1L. The Journal of pediatrics. 2016;171:313–316. e311–312. doi: 10.1016/j.jpeds.2015.12.060. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A, et al. UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. The EMBO journal. 2011;30:4860–4873. doi: 10.1038/emboj.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, Tu Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene. 2013;32:4814–4824. doi: 10.1038/onc.2012.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–823. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C, et al. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. The EMBO journal. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]