ABSTRACT
The quality of recombinant proteins such as monoclonal antibodies produced using Chinese hamster ovary cell-based mammalian systems is dependent on many factors, including cell line, process and cell culture media. Due to these factors, the generated product is heterogeneous and may have chemically-induced modifications or post-translational modifications that affect antibody stability, functionality and, in some cases, patient safety. This study demonstrates that S-sulfocysteine, a cysteine derivative, can increase the antibody specific productivity in different cell lines cultivated with different processes while minimizing trisulfide linkages in generated mAbs, mainly between heavy and light chain. The supplementation of a cell culture feed with S-sulfocysteine also proved to be useful to reduce the percentage of antibody fragments generated from the monoclonal antibody. Overall, this new component used in the upstream process allows a reduction of product heterogeneity.
KEYWORDS: Critical quality attributes, Fed-batch, fragments, monoclonal antibodies, productivity, S-sulfocysteine, trisulfide
Introduction
The production of recombinant proteins like monoclonal antibodies (mAbs) relies mainly on cell culture methods that involve cultivating mammalian cells, engineered to produce the protein of interest, in either fed-batch or continuous mode. During the cultivation period, factors such as cell culture media, process parameters and environmental conditions, may have effects on the final recombinant protein, leading to a heterogeneous product. Such heterogeneity can result from chemically-induced modifications such as oxidation, deamidation, and glycation, as well as post-translational modifications such as proteolytic maturation, glycosylation, phosphorylation, and disulfide bond formation. The presence of aggregates and fragments is also a key critical quality attribute impacting antibody safety.
The formation of trisulfide linkages is a molecular heterogeneity that has gained interest lately. Trisulfides are a post-translational modification formed by insertion of a sulfur atom into a disulfide bond. Several reports described the presence of trisulfide bonds in small proteins such as human growth hormone,1 a truncated IL-62 and a Cu, Zn-superoxide dismutase.3 Trisulfides were also described as a common modification in natural and recombinant antibodies of all IgG subtypes.4,5 Trisulfides were detected only in inter-chain linkages, particularly in light–heavy chain bonds. Several factors affecting trisulfide formation have already been described. First, several cell culture operating parameters such as cell density, feed strategies and time of harvest have been correlated with the amount of trisulfide bonds in the produced protein.4 Secondly, the level of hydrogen sulfide (H2S), produced during cell culture from the catabolism of cysteine, has been directly linked to the amount of trisulfide modifications.4,6
The biologic function of trisulfides is still largely unknown. Reports indicated no effect on EC504 or biologic activity of the protein.4 In vivo, rapid conversion of trisulfides to disulfides4 was observed, thus limiting the potential effect of this post-translational modification on the bioactivity of the drug.7 However, the heterogeneity introduced by variations in trisulfide levels has a direct impact on the formation of antibody-drug conjugates (ADCs). Indeed, an inverse correlation between trisulfide levels and the average drug to antibody ratio (DAR) values for a given amount of reductant was described.8 Thus, in process development for ADCs, monitoring and control of trisulfide levels is key to achieve a constant average DAR value, one of the most important quality attribute for ADCs.
Several methods aiming at removing or reducing trisulfide linkages have already been described for purified antibodies, such as buffer exchange, pH control, and mild reduction using sodium sulfide, tris(2-carboxyethyl)phosphine (TCEP) or cysteine.6,9,10 During cell culture, the use of inhibitors of cysteine degradation like antioxidants (e.g., glutathione) or keto-acids (e.g., pyruvate)9 was described. The level of trisulfides has also been controlled during the cell culture process by adjusting the cysteine feeding strategy, by addition of metal salts or by stripping the cell culture fluid with a gas such as nitrogen or argon.10,11 Cellular systems like the glutathione reductase enzyme12 were also effective at reducing the amount of trisulfide bonds in proteins, probably explaining the rapid conversion of trisulfides in disulfides in vivo.
Here, we show that S-sulfocysteine (SSC), a cysteine derivative obtained from the condensation of sulfuric acid and cysteine, affects the performance of cell culture processes by increasing culture duration and cell specific productivity (Qp). The increase in Qp was observed for three cell lines using different processes. Additionally, the use of SSC allowed a reduction of trisulfide bond levels in the produced IgGs, leading to an overall reduced heterogeneity of the mAb. Finally, a reduced amount of fragmentation was detected in antibodies produced using the SSC-containing process. Altogether, our data indicate that the use of SSC in cell culture feeds can improve the overall process performance, as well as reduce product heterogeneity.
Results
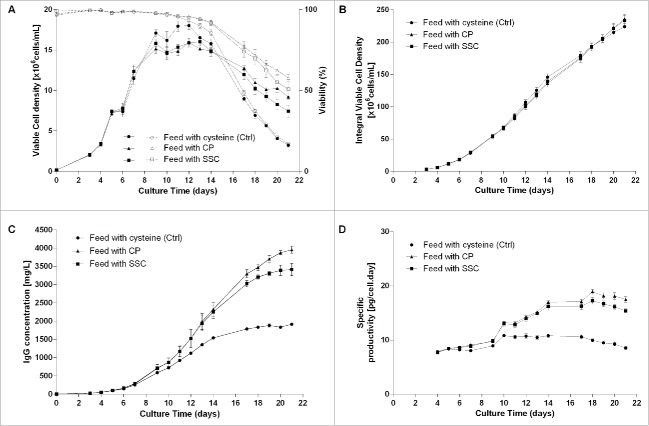
In the first part of this study, a suspension Chinese hamster ovary (CHO)-K1 cell line engineered to produce a human monoclonal IgG1 antibody was cultivated using three different fed-batch processes. In the control condition, cysteine was added throughout the process using a cysteine-containing feed. In the second condition, cysteine was allowed to interact with pyruvate in the feed, allowing formation of the thiazolidine molecule 2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid (CP).13,14 This condensation product has demonstrated its efficacy in stabilizing cell culture media formulations containing cysteine,15,16 and has been described as a tool to reduce trisulfide bond formation in IgG.9 It was used in this study as positive control. In the third condition, the cysteine present in the feed was replaced by SSC. This molecule has been described previously as a potent cysteine derivative having an anti-oxidative effect in CHO-based fed-batch processes.17 Results presented in Fig. 1A show the viable cell density of CHO cells cultivated using the three processes. Through feeding of CP and SSC, the maximum viable cell density was reduced by ∼10%, whereas the culture duration was prolonged after day 14, leading to an equivalent integral viable cell density (Fig. 1B). The antibody concentration at the end of the process (Fig. 1C) was significantly increased with both processes (titer of 3.4 and 4.0 g/L for the SSC and CP conditions on day 21) compared with the control condition (titer of 1.9 g/L on day 21). The Qp was also increased, with a specific productivity being nearly doubled in the CP and SSC conditions at day 18 compared with the control (Fig. 1D). The potential mechanisms explaining this increase in productivity were described elsewhere.16,17
Figure 1.
1.2 L bioreactor fed-batch experiment using a feed containing either cysteine (control, n = 2), cysteine stabilized with pyruvate (CP) or SSC (n = 5 each). Feed was added at 3% (v/v) at day 3 and 6% (v/v) at days 5, 7, 9 and 14. (A) Viable cell density (plain lines) and viability (dotted lines), (B) Integral viable cell density, (C) IgG concentration in the supernatant measured by a turbidometric method, (D) Specific productivity.
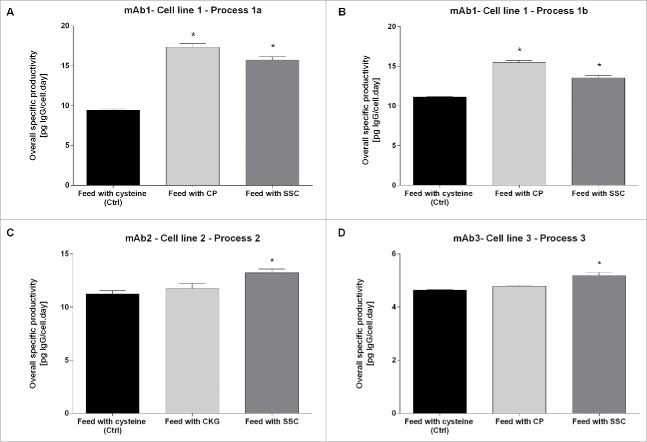
To demonstrate that the observed effect is not specific to the clone, the process or the cell culture media platform chosen for this study, similar experiments were performed using different processes, cell lines producing other mAbs and fed-batch platforms. The overall specific productivity obtained is presented in Fig. 2. Whereas the Qp was only significantly increased for CP with cell line 1 (process 1a and 1b), the difference compared with the control was significant for SSC for the three cell lines and three different mAbs according to the Mann-Whitney test. This indicates that SSC can robustly increase Qp in different CHO cell lines or processes.
Figure 2.
Overall cell specific productivity for mAb1 to 3 in cysteine, CP/CKG and SSC-containing processes. The overall specific productivity was determined by calculating the slope from the linear regression between titer and corrected integral VCD. Values are mean ± SEM obtained from 4 independent replicates. A p-value of less than 0.05 (*) was considered significant (Mann-Whitney test).
Independently of the effect of the molecules on Qp, the addition of pyruvate to cysteine-containing feed was described as a method to reduce the amount of trisulfide linkages in mAbs. 9 Thus, the following experiments were designed to evaluate whether the SSC supplementation in the feed may, similarly to CP, decrease the trisulfide content in mAb1. An overlay of extracted ion chromatograms is presented in Fig. 3A showing the ratio between the light chain (cysteine 213) and heavy chain (cysteine 224) linkage with a disulfide and a trisulfide bond. This linkage is the most frequent site of trisulfide formation in IgG.4 Fig. 3B and C display the corresponding mass spectra with information on the mass accuracy as well as the fragment ion spectrum, confirming the identity of the detected trisulfide linked light chain (LC)-heavy chain (HC) peptides.
Figure 3.
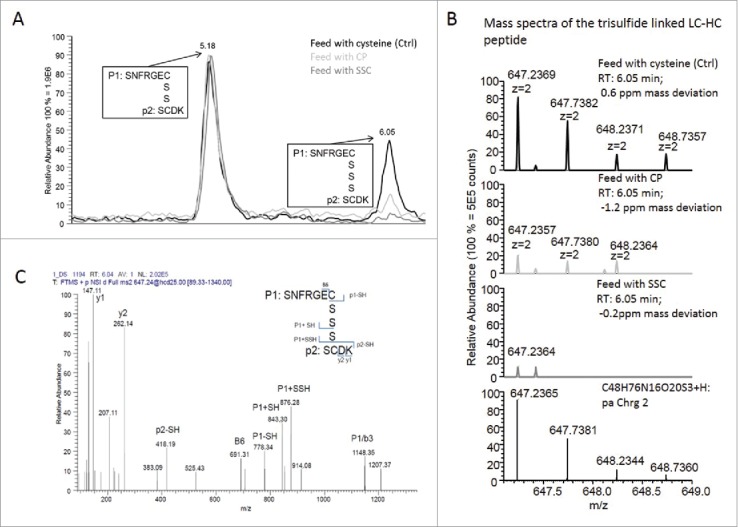
(A) Overlay of extracted ion chromatograms showing the relation between the LC-HC linkage with a disulfide bridge (elution at 5.18 min) and the LC-HC link with a trisulfide bridge (elution at 6.05 min). The black line corresponds to the control condition using cysteine-containing feed, light gray to the CP-containing feed and dark gray to the SSC-containing feed. (B) Mass spectra of the 3 samples showing the isotopic pattern compared with the theory and the mass accuracy. (C) Example of fragment ion spectrum of the trisulfide linked LC-HC peptide with annotation of the main peaks.
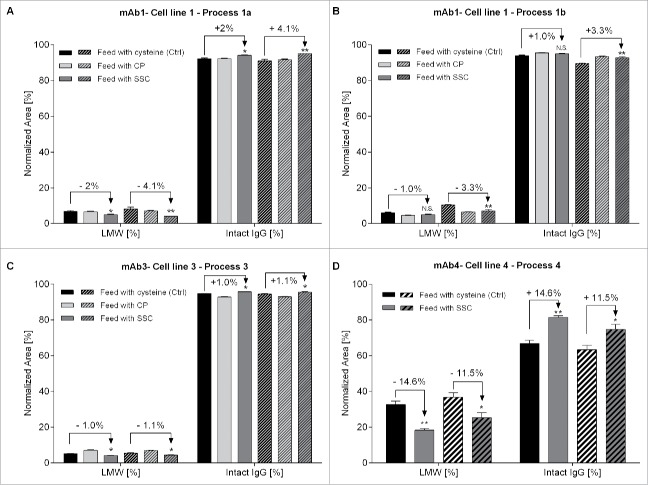
In Fig. 4, the relative percentage of trisulfide linked peptides was calculated by dividing the area of trisulfide linked peptides by the sum of areas of trisulfide and disulfide linked peptides. Results for mAb1 obtained with process 1a indicate that the highest amount of trisulfide linkages was found for LC-HC linked peptides in control samples produced using the cysteine feed with mean trisulfide bond levels of 46 and 42% on day 13 and day 18, respectively (Fig. 4A). The variability between biologic replicates resulting from different reactors was high, indicating an influence of several non-controlled factors on this post-translational modification, leading to product heterogeneity. This high value of trisulfide content is in the range of several reports already published.4,18 When using CP in process 1a, the relative amount of trisulfide linkages was drastically reduced to 9.6 and 7.2% on both days, confirming the results of Kshirsagar.9 Results obtained by using the SSC-containing process indicate a residual mean relative trisulfide content of 3.0 and 3.1% on day 13 and day 18, respectively, pointing out to the capacity of SSC to reduce trisulfide bond formation in mAb1. Additionally, another occurrence of trisulfide bond was detected at very low levels in an intramolecular disulfide bridge of the heavy chain (cysteine 259–319, Fig. S1). Although the relative amount was very low, a trend indicating a reduced level following feeding with CP and SSC was observed. Regarding the LC-HC peptide, the decrease in the amount of trisulfide linkages was observed for mAb1 produced using process 1b after CP and SSC supplementation (Fig. 4B), although the trisulfide reduction was higher for CP compared with SSC.
Figure 4.
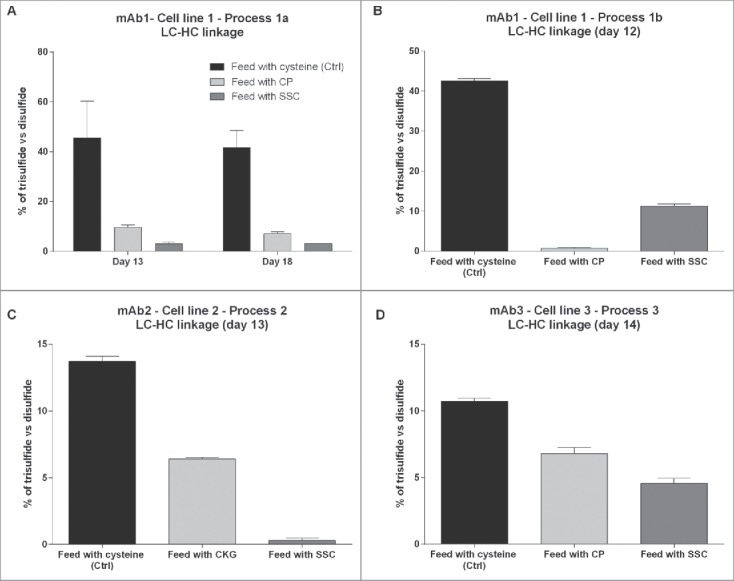
Relative quantification of the amount of trisulfide linkages between light chain and heavy chain of the IgG compared with the disulfide linkages quantified in 4 different monoclonal antibodies or processes. (A) Trisulfide content in mAb1 at day 13 and day 18 of the fed-batch process 1a. (B) Trisulfide content in mAb1 at day 12 of the fed-batch process 1b. (C) Trisulfide content in mAb2 at day 13. (D) Trisulfide content in mAb3 at day 14.
The trisulfide quantification was further performed on mAb2 and mAb3 produced in other CHO cells lines and using different processes or cell culture media platforms (Table 1). For mAb2, the trisulfide bond level was of 13.7% in the control, and a reduction to 6.4% was observed in the process using the thiazolidine CKG ((2-carboxyethyl)-1,3-thiazolidine-2,4-dicarboxylic acid), a component similar to CP as already described elsewhere16 (Fig. 4C). In the SSC-containing process, the trisulfide bond levels were drastically reduced to 0.3%. Finally, for mAb3, the trisulfide content between light chain and heavy chain had a mean value of 10.7% in the control process whereas the replacement of cysteine with either CP or SSC was able to decrease the occurrence of trisulfide bonds to values of 6.8% and 4.5%, respectively (Fig. 4D). The reasons for the differences in trisulfide content obtained for several mAbs are unknown, but may be related to the cell line, the process or the cell culture media and feed platform used. Altogether, the data indicate that the use of SSC in the feed is a robust method to reduce trisulfide bond formation in IgG. The main advantage of SSC compared with CP is its stability in highly concentrated feeds at neutral pH as described elsewhere.17
Table 1.
Overview of the scale, culture conditions and processes of cell culture fed-batch experiments.
| mAb | Cell line | Isotype | Process | FB platform | Scale | Culture conditions | Seeding density | Feeding Process |
|---|---|---|---|---|---|---|---|---|
| mAb1 | CHO-K1 | IgG1 | Process 1a | Cellvento™ CHO-220 | 1.2 L bioreactors | 37 °C (day 0 - day 5), 33°C (day 5–21), pH 6.95 ± 0.15, 50 % dissolved oxygen, agitation 140 rpm | 2.105 cells/mL | Main feed: 3% (day 3), 6% (days 5, 7, 9, 14)Glucose adjusted on demand to 6 g/L |
| mAb1 | CHO-K1 | IgG1 | Process 1b | Cellvento™ CHO-220 | 1.2 L bioreactors | 37 °C, pH 7.0 ± 0.02, 50 % dissolved oxygen, agitation 140 rpm | 2.105 cells/mL | Main feed: 3% (day 3, 5), 6% (day 7), 3% (day 10, 12))Glucose adjusted on demand to 6 g/L |
| mAb2 | CHO-K1 | IgG1 | Process 2 | Cellvento™ CHO-220 | 50 mL spin tubes | 37 °C, 5% CO2, 320 rpm, 80% humidity | 2.105 cells/mL | Main feed: 3% (day 3), 6% (days 5, 7, 10, 14)Glucose adjusted on demand to 6 g/L |
| mAb3 | CHO-DG44 | IgG1 | Process 3 | Cellvento™ CHO-210 | 50 mL spin tubes | 37 °C, 5% CO2, 320 rpm, 80% humidity | 3.105 cells/mL | Main feed: 6% (day 3, 5, 7, 10, 12)Glucose adjusted on demand to 6 g/L |
| mAb4 | CHOZN® | IgG1 | Process 4 | Excell® Advanced | 50 mL spin tubes | 37 °C, 5% CO2, 200 rpm, 80% humidity | 3.105 cells/mL | Main feed: 5% (day 3), 10% (day 6), 5% (day 8, 10)Glucose adjusted on demand to 6 g/L |
Since trisulfides are very reactive molecules, the trisulfide content was investigated in the raw materials used in the feed. For both cysteine and SSC, trisulfide signals were detected using LC-MS (data not shown) indicating that the differences between the processes might be correlated with the raw materials or their reactivity. Additional studies will be designed to investigate the mechanisms behind the trisulfide reduction.
In the final part of this work, we sought to study the effects of the CP and SSC supplementation on mAb fragmentation using non-reduced capillary gel electrophoresis. This method requires the formation of SDS-protein complexes through heating to provide similar charge densities, allowing an electrophoretic separation depending on protein size only. Since results are known to be highly dependent on the sample preparation, preliminary experiments were performed with mAb1 to define the experimental conditions leading to a minimum of thermally induced fragments due to disulfide reduction or shuffling19,20 (data not shown; final protocol detailed in the Material & Methods section below). Using our optimized method, we demonstrated that the use of a SSC-containing feed can significantly decrease the occurrence of low molecular weight (LMW) fragments with a concomitant increase in the main intact IgG form compared with the process using a cysteine-containing feed (Fig. 5). This effect was seen for mAb1 in process 1a (Fig. 5A) with a significant reduction of LMW fragments of 2.0% at day 13 (n = 6, p = 0.015) and 4.1% at day 18 (n = 6, p = 0.0068), as well as process 1b (Fig. 5B) with a reduction of LMW fragments of 1.0% at day 5 (n = 8, p = 0.066) and 3.3% at day 9 (n = 8, p = 0.007). The effect was also detected for mAb3, although the extent of the reduction was much lower (1.0% and 1.1% at day 14 and 18 with p-value of 0.0472 and 0.0324, respectively, Fig. 5C).
Figure 5.
Relative quantification of intact IgG and total low molecular weight forms using capillary gel electrophoresis. Fragments were separated according to their size under non-reduced conditions using CE-SDS. (A) Quantification of mAb1 at day 13 (n = 6, plain bars) and day 18 (n = 6, hatched bars) of the fed-batch process 1a. (B) Quantification of mAb1 at day 5 (n = 8, plain bars) and day 9 (n = 8, hatched bars) of the fed-batch process 1b. (C) Quantification of mAb3 at day 14 (n = 8, plain bars) and day 18 (n = 8, hatched bars). (D) Quantification of mAb3 at day 8 (n = 6, plain bars) and day 12 (n = 6, hatched bars). Statistical differences were assessed by Kruskal-Wallis and Dunn's multiple comparison tests. P-values of less than 0.05 (*) and 0.01 (**) were considered significant. P-values higher than 0.05 are marked as non-significant (N.S.).
To confirm the impact of SSC on the level of fragmentation, the analysis was performed on mAb4, an IgG1 known to generate LMW fragments. As shown in Fig. 5D, SSC supplementation allowed the reduction of LMW fragments by 14.6% on day 8 (n = 6, p = 0.0043) and 11.5% on day 12 (n = 6, p = 0.0173), respectively. However, the decrease in fragmentation cannot be correlated with the decrease in trisulfide bond formation since the effect on LMW fragments was only seen for SSC supplementation, and not for CP supplementation. The mechanism explaining the observed effect for SSC is still unclear and will require more investigations. Detailed results for the individual fragments such as LC, HC, heavy-light chain (HL), heavy-heavy chain (HH), heavy-heavy-light chain (HHL) fragments are presented in Table S1. Additionally, the feeding with SSC had no impact on the amount of aggregates for mAb1, process 1a (data not shown). However, to evaluate the effect of SSC on aggregate occurrence, other mAb isotypes or fusion proteins known to significantly aggregate need to be studied, since the IgG1s used in this work present very low levels of aggregates (levels below 2%).
Discussion
The quality of monoclonal antibodies or recombinant proteins produced using CHO-based mammalian systems is dependent on many factors, including cell line, process and cell culture media. This study shows how the use of SSC, a cysteine derivative, can help increase antibody productivity while minimizing antibody fragments and trisulfide bonds. Overall, this new strategy allows a reduction of product heterogeneity.
In standard fed-batch processes, cysteine is commonly added in both media and feeds and is essential for the survival and productivity of CHO cells.21 However, the addition of cysteine also introduces some variability in the process. Indeed, due to the reactivity of its thiol-group, cysteine is a highly reactive component in the media, reacting with trace metals like copper or iron22 and also oxidizing to form cystine. In particular, this oxidation is dependent on environmental factors like the partial pressure of oxygen, the aeration strategy and the overall media composition. Cysteine is also prone to degradation leading to the generation of hydrogen sulfide,23 which has been directly correlated with the amount of trisulfide linkages in mAbs.6 As expected, our results confirm the effect of cysteine on the trisulfide content, with a detection of 10–45% of trisulfide bonds between the light chain and heavy chain of mAbs depending on the molecule, the process and the cell culture media formulation. The standard deviation observed in the cysteine condition revealed a high heterogeneity, which may be attributed to factors described above and which may be detrimental in particular when the antibody has to be linked to a drug to form ADCs.8 Thus, our study sought to develop a method allowing to drastically reduce the amount of trisulfide bonds in the final product through modification of the cell culture media formulation. With the use of SSC, the amount of trisulfides detected between heavy chain and light chain was significantly decreased for mAb1. The final amount of trisulfide bonds obtained using the developed process was between 2.3 and 3.8%, indicating also a decreased heterogeneity between biologic replicates. This effect was confirmed in another process for mAb1 and for two other IgG1 molecules.
The use of SSC-containing feeds was also correlated with a lower occurrence of low molecular weight fragments (e.g., LC, HHL) compared with the cysteine-containing feed. From an efficacy standpoint, the decrease in the amount of fragments is beneficial since fragmentation can have adverse effects, including reduced biologic activity, shorter half-life and increased immunogenicity reactions, and hence provoke patient safety issues.24 This effect was not found to be correlated with the occurrence of trisulfide bonds since no effect on the level of fragmentation was detected for the positive control CP. However, a potential link may be hypothesized based on the work of Cumnock et al.,8 who demonstrated that, when disulfide and trisulfide bonds are present as mixtures, the addition of a reducing agent like TCEP induces a preferential reduction of trisulfide bonds. This implies that trisulfide bonds are, in general, more likely to be reduced, thus generating fragments. Further investigations are needed to understand differences in the effect observed for different IgG, different time points as well as to elucidate the mechanism of reduced fragmentation in the SSC-containing process.
Finally, the use of SSC in our fed-batch process was correlated with an increase in specific productivity. This may be linked to the protein processing mechanisms and the oxidative environment needed for proper folding of the protein in the endoplasmic reticulum (ER). Indeed, several studies indicated that the productivity of CHO cells may be limited by protein folding in the ER.25,26 Since SSC has been described as a potent inducer of the glutathione pool,17 this effect may be linked to a more favorable redox environment in the ER, allowing the correct folding of mAbs.
Altogether, our results indicate that SSC can be used in cell culture processes to reduce mAb heterogeneity, in particular to decrease the amount of trisulfide bonds and fragments. This cysteine derivative may also be used to increase cell specific productivity while reducing the overall IgG heterogeneity.
Materials and methods
Reagents
All chemicals were purchased from Merck, Germany unless stated otherwise. SSC sodium salt was synthesized according to published protocols27 or was purchased from Bachem (Bachem AG, Bubendorf, Switzerland).
Cell culture fed-batch process
Four CHO cell lines producing four different IgG were used in this study. Cell lines 1 and 2 are recombinant CHO-K1 clones expressing mAb1 and mAb2, respectively. Cell line 3 is a CHO-DG44 clone expressing mAb3, whereas cell line 4 is a CHOZN® clone expressing mAb4. All media and feeds used in this study are fully chemically defined formulations. mAb1 and mAb2 were produced in the Cellvento™ CHO-220 fed-batch platform whereas mAb3 was produced in the Cellvento™ CHO-210 fed-batch platform. Both media contain 1.5 mM cysteine, whereas feeds contain mainly vitamins, trace elements and common amino acids. Feeds were supplemented with either 15 mM cysteine alone or 15 mM cysteine mixed with 50 mM pyruvate or 15 mM of SSC. Previously performed dose response studies with SSC indicated that the 1:1 replacement of cysteine in the feed provided optimal results (data not shown). mAb4 was produced in the EX-CELL® Advanced fed-batch platform. SSC was added to the EX-CELL® Advanced feed at a concentration of 5 mM. Cell culture conditions, processes as well as scales are summarized in Table 1.
Offline analysis, IVCD and specific productivity
Viable cell density (VCD) and viability were monitored using Beckman Coulter ViCell® (Beckman Coulter, Fullerton, CA). IgG concentration was determined after centrifugation of the cell culture supernatant 5 min at 1500 rpm using an automatic turbidometric method (Cedex Bio HT, Roche). The productivity per cell per day was calculated daily by dividing the titer by the corrected integral VCD to take into account the dilution resulting from feeding. Overall Qp was determined by calculating the slope from the linear regression between titer and corrected integral VCD.
MAb purification
IgG were purified using protein A binding (Phytips, PhyNexus) and were eluted in either 200 mM NaH2PO4, 140 mM NaCl, 96 mM Tris HCl or 25 mM citrate, 56 mM tris and quantified again.
Fragments analysis using capillary gel electrophoresis
Fragments were separated according to their size in non-reduced conditions using capillary gel electrophoresis with SDS (CE-SDS) and the Beckman Coulter IgG Purity/Heterogeneity assay. Briefly, 100 µg of IgG were resuspended in a total of 45 µL phosphate-buffered saline and mixed with a 10 kDa internal standard, 12.5 mM iodoacetamide (IAM) and 50 µL of SDS-MW sample buffer. A non-reduced IgG control standard was prepared daily by adding 5 µL of 250 mM IAM and 2 µL of internal standard to a 95 µL aliquot provided in the kit. All samples were denatured for 10 min at 70°C, 500 rpm agitation using a thermomixer followed by a cooling step at RT in a water bath.
Relative amounts of intact IgG and fragments were determined using the Pharmaceutical Analysis System CESI 8000 (Sciex) with a photodiode array detector set at 220 ± 10 nm. The separation was performed in a 50 µm (i.d.) fused silica capillary with a total length of 30.2 cm and an effective length of 20 cm. After an initial conditioning step performed daily, the capillary surface was rinsed with 0.1 N NaOH for 3 min at 70 psi. Surface silanol groups were neutralized with 0.1 N HCl for 1 min applying 70 psi. To remove acidic residues, a water rinsing step of 1 min at 70 psi was included. The capillary was finally filled with SDS-MW Gel Buffer (Beckman-Coulter) for 10 min at 70 psi. After 2 additional water dipping steps to clean capillary tips, samples were injected electrokinetically for 20 sec using 5 kV. The actual separation run was performed for 33 min at 15 kV. Peaks were identified according to their individual migration time (determined previously using PNGase F treatment or gradual reduction using β-mercaptoethanol) and integrated according to the following parameters: peak width 0.4, threshold 400 and shoulder sensitivity 100.
Trisulfide characterization and quantification
First, free cysteine residues were alkylated by addition of 10 mM N-ethylmaleimide for 30 min in the dark. For digestion with Lys-C, 55 μL digestion buffer (8 M urea, 0.1 M Tris, pH 7) and 15 μL Lys-C (3 µg) enzyme (Promega) were added and samples were incubated for 4 hours at 37 °C. Subsequently, samples were diluted to 1 M urea by addition of 468 μL digestion buffer and were digested by addition of 12 μL (3 µg) trypsin overnight at 37 °C. The final antibody concentration was 50 mg/L. For the analysis of disulfide and trisulfide bonds, samples were treated with 10% formic acid to stop the digestion and analyzed by liquid chromatography–mass spectrometry (LC-MS)/MS. The peptide mixture obtained was injected and separated using reversed phase HPLC (RSLC3000 nanoLC, Thermo Scientific Dionex). A nanoLC column (Acquity UPLC M-class HSS T3, 1.8 μm, 75 μm x 150 mm, Waters) with a cartridge (PM100, C18, 5 μm, 0.3 × 5 mm, Thermo Scientific Dionex) was used for pre-concentration and separation of samples. An optimized 26 minutes linear gradient with varying slopes was applied at 50 °C as follows (minute / % B): 0/2, 0.25/2, 8/16, 20/29, 26.25/45, 27.25/99, 30.25/99 31.25/20 32.25/20, 33.25/99, 34.25/99, 35.25/2, 38/2. The injection amount was 0.35 μg. Samples were loaded at a flow rate of 120 μL/min onto the cartridge for 0.25 minutes using an eluent composed of 97.95% water, 2% acetonitrile and 0.05% trifluoroacetic acid. After switching the valve, samples were transferred to the analytical column and separated using a flow rate of 0.6 μL/min. The HPLC eluate was directly infused into a Q Exactive Plus mass spectrometer (Thermo scientific). The mass spectrometer operated in positive ion mode, the spray voltage was 1.9 kV, the capillary temperature was 275 °C and the S-Lens RF voltage was 55 V. For MS/MS product ion scan, the activation type was collision-induced dissociation, the default charge state was 2. The 4-scan-event Q Exactive method applied consists of a full MS survey scan at m/z 200–2000 and resolution power of 70 000 followed by 3 cycles of data-dependent MS/MS scans on the top 3 most intense ions. The dynamic exclusion function was enabled and parameters were as follows: dynamic exclusion of 10 sec, isolation window of 2 m/z, resolution of 17 500, AGC target 3e6, maximum inject time of 250 msec, normalized collision energy of 25, intensity threshold of 1.2e4. Unassigned charge states and charge states ≥ 8 were rejected for MS/MS triggering and a reject mass list containing polysiloxane ions was enabled. Replicate runs were performed with and without charge state +1 exclusion. For data processing, the Mascot 2.3 error tolerant search (Matrix Science), based on an in-house protein database containing sequences of the light and heavy chain of the analyzed antibody, respectively, was used. MS/MS spectra were charge deconvoluted using MS2 spectrum processor before database search and Mascot settings were adapted to allow identification of small peptides: the shortest peptide tested in the search and evidenced in the report was set to 4 amino acids. Mascot search parameters were: enzyme was trypsin, 2 missed cleavages allowed, variable modification N-ethylmaleimide, peptide tolerance: 6 ppm, MS/MS tolerance 0.05 Da, peptide charge +2, +3, +4, error tolerant search activated, minimum ion score 15.
Statistics
Data are expressed as means ± SEMs. Statistical differences were assessed using non-parametric tests, the Kruskal-Wallis and Dunn's post test for multiple comparisons and the Mann-Whitney test for single comparisons. P-values of less than 0.05 (*) and 0.01(**) were considered significant. Statistical and graphic analyses were performed with Prism 6 software (GraphPad Software Inc.).
Contributors
R. Seibel performed the bioreactor experiments for mAb1 process 1 and 2 as well as the spin tube experiments for mAb3. Sandra Maier performed the quantification of disulfide and trisulfide bonds in all experiments. A. Schnellbaecher performed the small scale spin tube experiment for mAb2. S. Bohl and -M. Wehsling performed the CE-SDS method development as well as the analysis of fragments in all experiments. A. Zeck supervised the mass spectrometric part of the study. A. Zimmer designed, supervised the study and wrote the manuscript. All authors have approved the final article.
Supplementary Material
Disclosure of potential conflicts of interest
Ronja Seibel, Alisa Schnellbaecher, Susanne Bohl, Maria Wehsling and Aline Zimmer are employees of Merck, Darmstadt, Germany.
Acknowledgments
The authors would like to thank Jonathan Kaiser for the samples of the mAb4 produced using cell line 4 and Sascha Pering for insightful scientific discussions and critical review of the manuscript.
Funding
This study was entirely funded by Merck, Darmstadt, Germany.
References
- 1.Jespersen AM, Christensen T, Klausen NK, Nielsen PF, Sorensen HH. Characterisation of a trisulphide derivative of biosynthetic human growth hormone produced in Escherichia coli. Eur J Biochem 1994; 219:365-73; PMID:8307002; https://doi.org/ 10.1111/j.1432-1033.1994.tb19948.x [DOI] [PubMed] [Google Scholar]
- 2.Breton J, Avanzi N, Valsasina B, Sgarella L, La Fiura A, Breme U, Orsini G, Wenisch E, Righetti PG. Detection of traces of a trisulphide derivative in the preparation of a recombinant truncated interleukin-6 mutein. J Chromatogr A 1995; 709:135-46; PMID:7581842; https://doi.org/ 10.1016/0021-9673(95)00108-Y [DOI] [PubMed] [Google Scholar]
- 3.Okado-Matsumoto A, Guan Z, Fridovich I. Modification of Cysteine 111 in human Cu,Zn-superoxide dismutase. Free Radic Biol Med 2006; 41:1837-46; PMID:17157186; https://doi.org/ 10.1016/j.freeradbiomed.2006.09.011 [DOI] [PubMed] [Google Scholar]
- 4.Gu S, Wen D, Weinreb PH, Sun Y, Zhang L, Foley SF, Kshirsagar R, Evans D, Mi S, Meier W, et al.. Characterization of trisulfide modification in antibodies. Anal Biochem 2010; 400:89-98; PMID:20085742; https://doi.org/ 10.1016/j.ab.2010.01.019 [DOI] [PubMed] [Google Scholar]
- 5.Pristatsky P, Cohen SL, Krantz D, Acevedo J, Ionescu R, Vlasak J. Evidence for trisulfide bonds in a recombinant variant of a human IgG2 monoclonal antibody. Anal Chem 2009; 81:6148-55; PMID:19591437; https://doi.org/ 10.1021/ac9006254 [DOI] [PubMed] [Google Scholar]
- 6.Kshirsagar R, McElearney K, Gilbert A, Sinacore M, Ryll T. Controlling trisulfide modification in recombinant monoclonal antibody produced in fed-batch cell culture. Biotechnol Bioeng 2012; 109:2523-32; PMID:22473825; https://doi.org/ 10.1002/bit.24511 [DOI] [PubMed] [Google Scholar]
- 7.Thomsen MK, Hansen BS, Nilsson P, Nowak J, Johansen PB, Thomsen PD, Christiansen J. Pharmacological Characterization of a Biosynthetic Trisulfide-Containing Hydrophobic Derivative of Human Growth Hormone: Comparison with Standard 22 K Growth Hormone. Pharmacol Toxicol 1994; 74:351-8; PMID:7937569; https://doi.org/ 10.1111/j.1600-0773.1994.tb01372.x [DOI] [PubMed] [Google Scholar]
- 8.Cumnock K, Tully T, Cornell C, Hutchinson M, Gorrell J, Skidmore K, Chen Y, Jacobson F. Trisulfide modification impacts the reduction step in antibody-drug conjugation process. Bioconjug Chem 2013; 24:1154-60; PMID:23713462; https://doi.org/ 10.1021/bc4000299 [DOI] [PubMed] [Google Scholar]
- 9.Kshirsagar R, Gilbert A. Methods of preventing and removing trisulfide bonds. WO 2012/158551 A1. [Google Scholar]
- 10.Evans D, Pepinsky RB, Wen D, Kshirsagar RR, Lucas K. Methods of preventing and removing trisulfide bonds. WO 2011/041721 A1 [Google Scholar]
- 11.Hemmendorff B, Castan A, Persson A. Method for the Production of Recombinant Peptides with a Low Amount of Trisulfides. US 2007/0232792 A1 [Google Scholar]
- 12.Moutiez M, Aumercier M, Teissier E, Parmentier B, Tartar A, Sergheraert C. Reduction of a trisulfide derivative of glutathione by glutathione reductase. Biochem Biophys Res Commun 1994; 202:1380-6; PMID:8060317; https://doi.org/ 10.1006/bbrc.1994.2083 [DOI] [PubMed] [Google Scholar]
- 13.Wlodek L. The reaction of sulfhydryl groups with carbonyl compounds. Acta Biochim Pol 1988; 35:307-17; PMID:3247807 [PubMed] [Google Scholar]
- 14.Cavallini D. The coupled oxidation of pyruvate with glutathione and cysteine. Biochem J 1951; 49:1-5; PMID:14848019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy RE, Costa NA, Morris AE. Factors that determine stability of highly concentrated chemically defined production media. Biotechnol Prog 2015; 31:493-502; PMID:25641710; https://doi.org/ 10.1002/btpr.2047 [DOI] [PubMed] [Google Scholar]
- 16.Kuschelewski J, Schnellbaecher A, Pering S, Wehsling M, Zimmer A. Antioxidant effect of thiazolidine molecules in cell culture media improves stability and performance. Biotechnol Prog 2017; in press. PMID:28268250; https://doi.org/ 10.1002/btpr.2458 [DOI] [PubMed] [Google Scholar]
- 17.Hecklau C, Pering S, Seibel R, Schnellbaecher A, Wehsling M, Eichhorn T, Jv Hagen, Zimmer A. S-Sulfocysteine simplifies fed-batch processes and increases the CHO specific productivity via anti-oxidant activity. J Biotechnol 2016; 218:53-63; PMID:26654938; https://doi.org/ 10.1016/j.jbiotec.2015.11.022 [DOI] [PubMed] [Google Scholar]
- 18.Cornell C, Karanjit A, Chen Y, Jacobson F. A high-throughput hydrophilic interaction liquid chromatography coupled with a charged aerosol detector method to assess trisulfides in IgG1 monoclonal antibodies using tris(2-carboxyethyl)phosphine reaction products: Tris(2-carboxyethyl)phosphine-oxide and tris(2-carboxyethyl)phosphine-sulfide. J Chromatogr A 2016; 1457:107-15; PMID:27345209; https://doi.org/ 10.1016/j.chroma.2016.06.037 [DOI] [PubMed] [Google Scholar]
- 19.Salas-Solano O, Tomlinson B, Du S, Parker M, Strahan A, Ma S. Optimization and Validation of a Quantitative Capillary Electrophoresis Sodium Dodecyl Sulfate Method for Quality Control and Stability Monitoring of Monoclonal Antibodies. Anal Chem 2006; 78:6583-94; PMID:16970337; https://doi.org/ 10.1021/ac060828p [DOI] [PubMed] [Google Scholar]
- 20.Hunt G, Nashabeh W. Capillary Electrophoresis Sodium Dodecyl Sulfate Nongel Sieving Analysis of a Therapeutic Recombinant Monoclonal Antibody: A Biotechnology Perspective. Anal Chem 1999; 71:2390-7; PMID:10405607; https://doi.org/ 10.1021/ac981209m [DOI] [PubMed] [Google Scholar]
- 21.Stipanuk MH, Dominy JE Jr., Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr 2006; 136:1652S-9S; PMID:16702335 [DOI] [PubMed] [Google Scholar]
- 22.Rigo A, Corazza A, di Paolo ML, Rossetto M, Ugolini R, Scarpa M. Interaction of copper with cysteine: stability of cuprous complexes and catalytic role of cupric ions in anaerobic thiol oxidation. J Inorg Biochem 2004; 98:1495-501; PMID:15337601; https://doi.org/ 10.1016/j.jinorgbio.2004.06.008 [DOI] [PubMed] [Google Scholar]
- 23.Kimura H. Hydrogen sulfide: its production, release and functions. Amino Acids 2011; 41:113-21; PMID:20191298; https://doi.org/ 10.1007/s00726-010-0510-x [DOI] [PubMed] [Google Scholar]
- 24.Eon-Duval A, Broly H, Gleixner R. Quality attributes of recombinant therapeutic proteins: an assessment of impact on safety and efficacy as part of a quality by design development approach. Biotechnol Prog 2012; 28:608-22; PMID:22473974; https://doi.org/ 10.1002/btpr.1548 [DOI] [PubMed] [Google Scholar]
- 25.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. J Mol Biol 2011; 409:238-49; PMID:21435343; https://doi.org/ 10.1016/j.jmb.2011.03.024 [DOI] [PubMed] [Google Scholar]
- 26.Mohan C, Lee GM. Effect of inducible co-overexpression of protein disulfide isomerase and endoplasmic reticulum oxidoreductase on the specific antibody productivity of recombinant Chinese hamster ovary cells. Biotechnol Bioeng 2010; 107:337-46; PMID:20506311; https://doi.org/ 10.1002/bit.22781 [DOI] [PubMed] [Google Scholar]
- 27.Segel IH, Johnson MJ. Synthesis and characterization of sodium cysteine-S-sulfate monohydrate. Anal Biochem 1963; 5:330-7; PMID:13987685; https://doi.org/ 10.1016/0003-2697(63)90085-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.