Abstract
Aims
To develop and evaluate the performance of a panel of isothermal real-time recombinase polymerase amplification (RPA) assays for detection of common bacterial urinary tract infection (UTI) pathogens.
Methods and Results
The panel included RPAs for Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Pseudomonas aeruginosa, and Enterococcus faecalis. All five RPAs required reaction times of under 12 minutes to reach their lower limit of detection of 100 genomes per reaction or less, and did not cross-react with high concentrations of non-target bacterial genomic DNA. In a 50-sample retrospective clinical study, the five-RPA assay panel was found to have a specificity of 100% (95% CI, 78%-100%) and a sensitivity of 89% (95% CI, 75%-96%) for UTI detection.
Conclusions
The analytical and clinical validity of RPA for the rapid and sensitive detection of common UTI pathogens was established.
Significance and Impact
Rapid identification of the causative pathogens of urinary tract infections (UTIs) can be valuable in preventing serious complications by helping avoid the empirical treatment necessitated by traditional urine culture’s 48–72 hour turnaround time. The routine and widespread use of RPA to supplement or replace culture-based methods could profoundly impact UTI management and the emergence of multidrug-resistant pathogens.
Keywords: Detection, Rapid methods, Identification, Diagnosis, Urinary tract infection
INTRODUCTION
Urinary tract infections (UTIs) annually affect 150 million people worldwide, and accounted for an estimated 10.5 million office visits and 2–3 million emergency department visits in the United States in 2007.(Stamm and Norrby, 2001, Schappert and Rechtsteiner, 2011, Niska et al., 2010) UTIs, which can be symptomatic or asymptomatic, are a notable cause of morbidity in infant or elderly males, and females of all ages.(Foxman, 2010) Sequelae range in severity from frequent recurrences to renal scarring and life-threatening sepsis.(Foxman, 2002) For most UTIs, antibiotics are the most recommended and effective treatment option following diagnosis.(Foxman, 2010)
The diagnosis of UTIs is currently based on clinical symptoms, nonculture methods such as dipstick tests for nitrites or leukocyte esterase activity, and pathogen detection/identification by the gold standard urine culture method. Clinical symptoms overlap with other non-infectious conditions and therefore have poor specificity, while dipstick tests, which don’t identify pathogens, are known to have poor positive predictive values despite being useful to rule out infections.(Devillé et al., 2004, Semeniuk and Church, 1999, Zaman et al., 1998) Urine cultures, while mostly accurate, can take up to 72 hours to provide a result. Consequently, initial UTI treatments are mostly empirical and broad-spectrum in nature with the goal of reducing the duration of patient discomfort. This widespread, injudicious use of antibiotics has accelerated the emergence of multidrug-resistant UTI pathogens.(Chen et al., 2013) It is clear that methods for rapid UTI pathogen detection and identification at the point-of-care or in a clinical laboratory could help avoid imprecise treatments, and thereby have a positive impact on UTI management.
There is an increase interest in developing rapid UTI diagnostics to supplement or replace urine cultures. Various assays based on electrochemical biosensing, mass spectrometry, microcalorimetry, Raman spectroscopy, and real-time PCR have been recently reported.(Liao et al., 2006, Burillo et al., 2014, Wang et al., 2013, Bonkat et al., 2012, Kloß et al., 2013, Lehmann et al., 2011, Lehmann et al., 2010) Among these, real-time PCR and other nucleic acid amplification techniques (NAATs) are already well-established in clinical diagnostics, and have recently been developed into automated multiplex-capable platforms suitable for use in near-patient settings.(Tang et al., 1997, Raja et al., 2005, Poritz et al., 2011) Isothermal NAATs, which offer performance comparable to PCR and a much faster time-to-result in many cases, are more amenable to low complexity, small-footprint devices due to the lack of thermal cycling requirements.(Niemz et al., 2011, Kim and Easley, 2011, Craw and Balachandran, 2012, Asiello and Baeumner, 2011, Gill and Ghaemi, 2008) Of the DNA-specific isothermal NAATs, RPA stands out by virtue of being the least complex to design and optimize, and the fastest-to-result.
RPA uses T4 phage recombinases UvsX and UvsY to facilitate the site-specific strand invasion of 30–35 nt primers into a DNA template, following which the strand-displacing Sau polymerase (Staphylococcus aureus) mediates primer extension; the cyclic repetition of this process at a constant temperature of 37–42°C yields dsDNA amplicons exponentially as in PCR. In end-point RPA, agarose gel electrophoresis is used to visualize amplified DNA after 20–30 minutes. In real-time RPA, recombinases facilitate the hybridization of 46–52 nt long TwistAmp exo probes to either the positive or negative sense strand of the target amplicon. TwistAmp exo probes are oligonucleotides carrying a fluorophore and quencher separated by an abasic nucleotide analog (tetrahydrofuran, THF) located approximately 15 nt upstream of the 3’ end. Following target hybridization, the double-strand-specific Escherichia coli exonuclease III recognizes and cleaves the THF site, releasing the quencher and resulting in the amplification-dependent development of fluorescence.(Piepenburg et al., 2006)
In this paper, we describe the development of a panel of highly sensitive and specific exo probe-based real-time RPA assays for the detection of five bacterial pathogens that are among the most commonly implicated etiologic agents of UTIs. These pathogens – Escherichia coli, Proteus mirabilis, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Enterococcus spp. - were collectively detected in 84% and 85% of North American and European UTI samples respectively in the SENTRY Antimicrobial Surveillance Program;(Fluit et al., 2000, Jones et al., 1999) they could be more or less frequent in specific patient populations. All five real-time RPA assays were able to detect 100 genomes per reaction or less, and showed no cross-reactivity with high concentrations of non-target genomic DNA. In a pilot retrospective study for clinical evaluation, the assay panel was found to have a specificity of 100% (95% CI, 78%-100%) and a sensitivity of 89% (95% CI, 75%-96%) for the detection of a UTI.
MATERIALS AND METHODS
Quantitative genomic DNA standards
For analytical sensitivity and specificity determination, standards were generated by isolation and purification of genomic DNA (gDNA) from bacterial colonies grown on culture plates. 5% sheep blood agar plates (Beckton, Dickinson and Company, NJ) were streaked with American Type Culture Collection (ATCC; Manassas, VA) strains of E. coli (ATCC 35218; 4.64 Mbp genome), P. aeruginosa (ATCC 27853; 6.3 Mbp genome), K. pneumoniae (ATCC 700603; 5.3 Mbp genome), E. faecalis (ATCC 29212; 3.3 Mbp genome), and P. mirabilis (ATCC 25933; 4.1 Mbp genome), and incubated overnight at 37°C. An UltraClean® Microbial DNA Isolation Kit (Mo Bio Laboratories, Carlsbad, CA) was used, following the manufacturer’s protocol, to isolate pure gDNA of each pathogen. The absorbances of extracted gDNA samples at 230 nm, 260 nm, and 280 nm (A230, A260 and A280) were obtained using a Nanodrop 1000 instrument (NanoDrop Instruments, Wilmington, DE). Both A260/A280 and A260/A230 values were near or greater than 2.0 for all samples, indicating negligible contamination by proteins or organic solvents. The concentration, in genome copies/µL, of each reference strain’s pure gDNA stock was calculated using A260 values, the extinction coefficient of double stranded DNA (0.020 µg−1.mL.cm−1), the average molar mass of double stranded DNA (650 Daltons/base pair), the genome size of each reference strain, and the Avogadro constant. Appropriate dilutions of these stocks were made, using deionized water, and stored at −20°C as quantitative gDNA standards in 5 µL aliquots containing 107, 106, 105, 104, 103, 102 or 10 genomes each.
RPA primer and probe design
A combination of publicly-available bioinformatics tools, end-point RPA, confirmatory Sanger sequencing of amplicons, and real-time RPA was used for primer and TwistAmp exo probe design. Species-specific genes with sequence regions highly conserved between different strains were chosen for amplicon design. Publicly-available GenBank reference sequences of these genes (http://www.ncbi.nlm.nih.gov/genbank) were downloaded and aligned, if necessary, using SeaView software.(Gouy et al., 2010) The National Center for Biotechnology Information’s (NCBI) BLAST tool (http://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed September 21, 2015) was used to verify that the downloaded reference sequences were homologous to all publicly-available sequences of the gene. The target genes’ most highly conserved regions were used for primer design, either manually or using NCBI’s primer-BLAST tool (http://www.ncbi.nlm.nih.gov/tools/primer-blast/, last accessed September 21, 2015), while exo probes were designed manually.(Ye et al., 2012) Secondary structure prediction and free energy calculations were performed using UNAfold software, which is freely available through the OligoAnalyzer tool (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer, last accessed September 21, 2015).(Markham and Zuker, 2008, Owczarzy et al., 2008)
RPA primers were designed to be between 30 and 35 nt long (for optimal recombinase activity), with GC content between 30 and 70% and either a G or a C at the 3’ end to provide a more stably clamped target for the polymerase. Sequence elements likely to contribute to hairpin-like secondary structures or primer-primer interactions were avoided. For each target gene, two or three sets of forward and reverse primers were designed to overlap a 150–250 bp conserved region in a staggered manner. To lower the possibility of non-specifically amplifying human DNA or unrelated UTI pathogens, the sequence-specificity of all designed primers was evaluated by performing a BLAST search against publicly available nucleotide sequences of the following species: Homo sapiens, Escherichia coli, Proteus mirabilis, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterococcus faecalis, Enterococcus faecium, Citrobacter freundii, Enterobacter cloacae, Acinetobacter baumannii, Staphylococcus aureus, Providencia rettgeri, Providencia stuartii, Morganella morganii, and Streptococcus agalactiae. Primers that had less than 6 overall mismatches to a non-target sequences and/or less than 4 mismatches to a non-target sequence in the last 6 nucleotides at the 3’ end were rejected. Primers were purchased from Integrated DNA Technologies (Coralville, IA).
For verification of RPA product specificity, and to aid exo probe design, the outermost primers designed for each target gene were used in end-point RPA using purified gDNA as the template, followed by Sanger sequencing of the amplification product. End-point RPA was performed in a 50 µL volume using TwistAmp Basic kits (TwistDx, Cambridge, UK). Master mixes containing 480 nmol.l−1 RPA primers and TwistAmp rehydration buffer were prepared and distributed, in 42.5 µL volumes, into 0.2 mL reaction tubes containing dried enzyme pellets. The enzyme pellet in each tube was rapidly solubilized by pipetting the master mix up and down upon addition. Subsequently, 5 µL of template was added to the tubes. 2.5 µL of magnesium acetate (bringing the concentration of Mg2+ to 14 mmol.l−1) was then pipetted into the tube lids, centrifuged into the tubes using a mini centrifuge, and immediately placed into a PCR thermal cycler (Bio-Rad Laboratories, Hercules, CA) programmed to operate at 42oC for 30 minutes. RPA products were purified using a PCR purification kit (Qiagen, Valencia, CA) and bi-directionally Sanger-sequenced (SeqWright, Houston, Texas). Sanger sequencing results were used to confirm amplicon size and homology to GenBank sequences at a single-nucleotide level. Using these sequences, TwistAmp exo probes (46–52 nt) were designed to be homologous to the positive- or negative-sense strands of the target regions flanked by the primers, and not to overlap with the primers themselves. All probes (synthesized by LGC Biosearch Technologies) contained a dT-FAM (thymine labeled with 6-carboxyfluorescein), and a dT-BHQ1 (thymine labeled with Black Hole Quencher®-1, which quenches FAM in close proximity) separated by a THF cleavable linker; they also contained a three carbon spacer at the 3’ end to block polymerase-mediated extension. OligoAnalyzer was used to analyze secondary structures and their free energies.
Real-time RPA experiments and data analysis
Real-time RPA reactions were performed in 50 µL volumes using TwistAmp exo kits (TwistDx, Cambridge, UK). Master mixes containing 420 nmol.l−1 RPA primers, 120 nmol.l−1 RPA exo probes, and Twist Amp rehydration buffer were prepared and distributed, in 42.5 µL volumes, into reaction tubes supplied with dried enzyme pellets. 5 µL of template (quantitative gDNA standards or total DNA from clinical sample) was then added to the tubes. The 5 µL aliquot has been optimized for detecting between 102 to 106 CFU.ml−1 in urine. To start the reaction, 2.5 µL of magnesium acetate was added to each tube (to bring the total Mg2+ concentration to 14 mmol.l−1 ) by pipetting into tube lids and centrifuging immediately before placing the tubes in an Agilent MxPro 3005 real-time PCR machine (Agilent Technologies, Santa Clara, CA). The real-time PCR machine was programmed to run for 20 minutes at 42°C and collect fluorescence data at 10 s intervals.
Fluorescence data from real-time RPA assays using different exo probes was normalized (subtraction of baseline function from raw fluorescence) and thresholded individually, using MxPro software (Agilent Technologies, Santa Clara, CA). Baseline functions for each probe were calculated by fitting the raw fluorescence data obtained between 2 and 3 minutes from the beginning of the reaction to a line using a linear least mean squares algorithm. This baseline function was subtracted from the raw fluorescence (R) function to obtain baseline-corrected fluorescence (dR). A 9-point moving average was applied to smooth the amplification curve that plotted baseline-corrected fluorescence (dR) against time. Threshold times (analogous to threshold cycles in real-time PCR) represented the times, from the beginning of reactions, at which the baseline-corrected fluorescence exceeded a threshold value located in the exponential growth regions of amplification curves. Threshold values of dR were empirically set to be between 25 and 100 standard deviations above background fluorescence for each exo probe, and used consistently thereafter.
Determination of analytical sensitivity and specificity
The analytical sensitivities (limits of detection) of the panel of real-time RPA assays were estimated by performing real-time RPA on three to six replicates of different ten-fold serial dilutions of the quantitative gDNA standards, from 107 genome copies per reaction down to 10 genome copies per reaction. The threshold time was plotted against genome copies detected, and a semi-log regression was calculated. For analytical specificity determination, each assay on the panel was performed in triplicate using 106 genome copies of non-target gDNA, from each other pathogen on the panel individually, as the template.
Clinical evaluation of RPA panel
The panel of the 5-RPA assays was clinically evaluated using 50 randomly selected clinical urine samples (from 50 unique patients) from among those collected by Medical Center Laboratories in Houston, Texas, between August 5 and August 18, 2014. These samples were collected from symptomatic and asymptomatic in- and outpatients in the Greater Houston area. The median age of the patients was 80.5 years (range 13–104 years); 70% of the patients were female. “Gold standard” comparison results were obtained by urine culture. Briefly, urine specimens were first inoculated on 5% sheep blood agar plates (Becton, Dickinson and Company, New Jersey) using an InfoLab 1 µL calibrated loop. Following aerobic incubation at 37°C for 12, 24 or 48 hours, a VITEK® 2 system (bioMérieux, Marcy l’Etoile, France) was used according to the manufacturer’s instructions to establish pathogen identity in what were considered “true positive” samples - culture-positive plates with more than 10 colonies, indicating the presence of >104 CFU.mL−1 urine. If multiple pathogens were present, all pathogens were identified and reported.
Total DNA was extracted from clinical urine samples (within 12 hours of collection) using a simple, high-throughput method.(Lu et al., 2011) Briefly, 1 mL urine was pipetted into a 1.5 ml microcentrifuge tube and centrifuged at 12,000 x g for 5 min. The supernatant was discarded and 100 µL lysis buffer (1% Tween-20, 1% NP-40, 0.03% SDS, 5% Chelex 100 and 400 µg.mL−1 proteinase K) was added to the pellet and thoroughly mixed. The mixture was incubated at 56°C for 1 h, followed by 100°C for 10 min. The lysate was then centrifuged at 13,000 x g for 5 min, and 50 µL of the supernatant was aliquoted and stored at −20°C. The University of Houston’s Committee for the Protection of Human Subjects approved an application (No. 4228–14209) to exempt the RPA analysis of these samples from Institutional Review Board approval due to their de-identified nature. To test each clinical sample against the UTI RPA panel being evaluated, 5 µL of lysate supernatant was used as the template in five separate real-time RPA assays. Fluorescence data from the five tubes for each sample were normalized and thresholded as described above. A threshold time of less than 15 minutes in at least one reaction tube indicated a “positive” result, while no amplification above the threshold in all five tubes indicated a “negative” result. Sensitivity and specificity were calculated using the number of true positives (TP), false negatives (FN), false positives (FP), and false negatives (FN) as follows: Sensitivity = (TP)/(TP+FN); Specificity = (TN)/(TN+FP). Confidence intervals were calculated using the widely-used Wilson score method.(Wilson, 1927)
RESULTS
RPA primer and probe design
The following species-specific target genes were used for amplicon design: chuA for E. coli, khe for K. pneumoniae, lasB for P. aeruginosa, ureR for P. mirabilis, and rpoA for Enterococcus spp.(Clermont et al., 2000, Braun et al., 2014, Shi et al., 2012, Zhang et al., 2013, Naser et al., 2005) The most challenging primer and probe design was that for Enterococcus. Although twelve Enterococcus species have been identified as being pathogenic to humans, E. faecalis and E. faecium account for approximately 85–90% and 5–10% of clinical enterococcal isolates, respectively.(Gin and Zhanel, 1996, Gold, 2001, Murray, 1990, Murray, 2000, Zhanel et al., 2003) Several candidate genes including atpD, phoE, ddl, rpoB, tuf, and rpoA were initially evaluated for RPA amplicon-appropriate conserved regions between E. faecalis and E. faecium (preliminary probe/primer design for Enterococcus spp not shown). rpoA was chosen because it had the least evenly spread mismatches between the enterococcal species; it also resulted in the most sequence-specific primers. For each pathogen on the panel, between six and nine primer pairs were screened for optimal performance using real-time RPA with quantitative gDNA standards as the template. Primer pairs that resulted in the shortest threshold times for the lowest concentration of gDNA were selected. Final amplicon lengths ranged from 161 to 211 bp, as shown in Table 1. Primer and probe sequences are indicated in Table 2.
Table 1.
Final RPA amplicon design
Table 2.
RPA primers and probes.
| Pathogen | Sequence (5’-3’) |
|---|---|
|
| |
| E. coli | F – ATATGGCGGTGAGTATTATCGTCAGGAACAACATC |
| R – GAGATGACCATTTGTCGGCATCAACATCTTTGTAG | |
| P – AGCCCAAAACCGTACTCCTGAGTTTCGTTAG(FAM)H(BHQ)CCGGACGTAAGTTC | |
|
| |
| K. pneumoniae | F – TTATCCCGACAGCCCGGAGCGTTTTTCGATTGG |
| R – CAGCTTCCAGAGATAGCCGTTTATCCACACTTCCG | |
| P – CACGCGGAGAGCGATGAGGAAGAGT(FAM)CH(BHQ)CTACGTGCTGGAGGGC | |
|
| |
| P. aeruginosa | F – GAGAATGACAAAGTGGAACTGGTGATCCGCCTG |
| R – GCCAGGCCTTCCCACTGATCGAGCACTTCGCCG | |
| P – GAACAACATCGCCCAACTGGTCTACAACG(FAM)H(BHQ)CCTACCTGATTCCC | |
|
| |
| P. mirabilis | F – CAAAAACGCTCTATACTACACCATCAACATTAC |
| R – GTTTAAATGCGTCACAAAAATAAGCATTACTAC | |
| P – GTCGCCATTTAAGTAAAGAGGGCGTTTCG(FAM)H(BHQ)TGCCAATTACTGTT | |
|
| |
| E. faecalis | F – GGACCCGCTACCGTGACTGCCGGCGATATTATCG |
| R – GAATCAACTGGAAGTACACCGATTGGCATATC | |
| P – TCTGCTTGAACATAGCCACGACCAGGTTTCAC(FAM)H(BHQ)TAAGCGAGCATGGAA | |
F, forward primer; R, reverse primer; P – exo probe; (FAM), Fluorescein coupled to a thymine; H, terahydrofuran; (BHQ), Black Hole Quencher-1 coupled to a thymine.
RPA analytical sensitivity and specificity
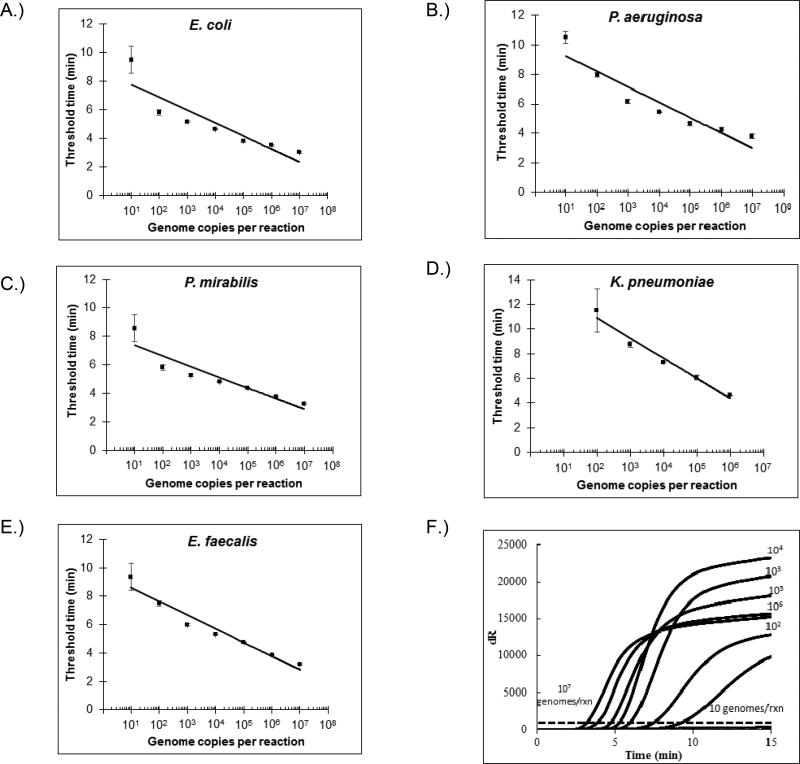
Quantitative gDNA standards from reference strains were used to test the analytical sensitivities of all five real-time RPA assays. Three replicates were tested for gDNA concentrations ≥1000 genome copies per reaction, while six replicates were tested for 10 and 100 genome copies per reaction. Threshold times were plotted against gDNA concentrations and fit to a semi-log regression line [Figure 1]. All replicates tested positive at gDNA concentrations ≥100 genome copies per reaction. Replicates for gDNA concentrations of 10 genomes copies per reaction for E. coli, E. faecalis, P. mirabilis, and P. aeruginosa were positive in 3/6, 5/6, 4/6, and 4/6 cases, respectively, putting the analytical sensitivity of those assays between 10 and 100 genome copies per reaction. All six replicates of the 10 genome copies per reaction sample were negative for the K. pneumoniae assay, putting its analytical sensitivity between 100 and 1000 genomes copies per reaction. Average threshold times for the lowest gDNA concentration detected by assays for E. coli, E. faecalis, P. mirabilis, P. aeruginosa, and K. pneumoniae were 9.5, 9.4, 8.6, 10.5, and 11.5 minutes, respectively.
FIGURE 1. Analytical performance of real-time RPAs.
(A) – (E): semi-log regression lines for detection of different dilutions of pathogens included on the UTI panel. Data points were derived from three replicates for 103 or more genome copies per reaction, and six replicates for 10 and 102 genome copies per reaction. (F) Representative amplification curves, averaged from three replicates, of different dilutions of E. faecalis gDNA.
The analytical specificity of all RPAs was determined by cross-testing each primer/probe set against 106 genome copies per reaction of each panel UTI pathogen separately, i.e. E. coli (ATCC 35218), P. aeruginosa (ATCC 27853), K. pneumoniae (ATCC 700603), E. faecalis (ATCC 29212), and P. mirabilis (ATCC 25933). Only specific detection was observed; amplification curves for all non-specific reactions did not exceed threshold dR values during the 20 minute reaction time.
Clinical evaluation of UTI RPA panel
In total, 50 urine samples were included in the study, with 36 true positives (72%) and 14 true negatives (28%) as determined by urine culture for pathogen detection and VITEK 2 for pathogen identification. Of the 36 culture-positive plates analyzed by VITEK 2, 26 had monomicrobial growth, 9 had bimicrobial growth, and 1 had trimicrobial growth, with the identities of all pathogens reported. Total DNA was extracted from 1 mL of all 50 samples by the method described in the Materials and Methods section, following which 5 µL of extracted DNA from each sample (equivalent to 50 µL of urine) was used as the amplification template for RPA reactions targeting each of the five pathogens. 4/36 culture-positive samples returned negative results from all five RPA reactions, and were considered false negatives. 1/4 false negatives was a M. morganii infection (not covered by the panel), while the others were samples containing E. coli, P. mirabilis, and K. pneumoniae. 32/36 culture-positive samples returned at least one positive RPA reaction. 14/14 culture-negative samples returned negative results from all five RPA reactions. Based on these results, the overall clinical sensitivity and specificity for the detection of a UTI by the RPA panel can be estimated at 89% (95% CI, 75%-96%) and 100% (95% CI, 78%-100%), respectively [Table 3].
Table 3.
Detection of infection in 50 samples.
| Microbiological culture (n = 50 samples) | ||||
|---|---|---|---|---|
| Positive UTI | Negative UTI | Total | ||
| RPA panel (n = 50 samples) | Positive UTI | 32 | 0 | 32 |
| Negative UTI | 4 | 14 | 18 | |
| Σ | 36 | 14 | 50 | |
| Sensitivity (%) | 89 | |||
| Specificity (%) | 100 | |||
Diagnostic accuracy analyses were also performed at a species specific level. Among the pathogens included in the RPA panel, Escherichia coli was the most commonly identified (n=14), followed by P. mirabilis (n=12), Enterococcus spp. and K. pneumoniae (n=5), and P. aeruginosa (n=2). Three pathogens not included in the RPA panel (M. morganii, S. aureus, and Group B Streptococcus) were identified by VITEK 2 in one monomicrobial sample and four polymicrobial samples. Table 4 shows the species-specific true- and false-positive and -negative results, sensitivities, and specificities for each pathogen covered by the panel.
Table 4.
Species-specific performance of real-time RPA assays.
| Pathogen | TP | FN | FP | TN | Sensitivity (CI) (%) | Specificity (CI) (%) |
|---|---|---|---|---|---|---|
| E. coli | 14 | 1 | 0 | 35 | 93 (70–99) | 100 (90–100) |
| K. pneumoniae | 5 | 1 | 0 | 44 | 83 (44–97) | 100 (91–100) |
| P. mirabilis | 12 | 1 | 0 | 37 | 92 (67–99) | 100 (91–100) |
| P. aeruginosa | 2 | 0 | 0 | 48 | 100 (34–100) | 100 (93–100) |
| E. faecalis | 5 | 1 | 0 | 44 | 83 (44–97) | 100 (91–100) |
TP – true positives, FP – false positives, TN – true negatives, FN – false negatives, CI – confidence interval.
DISCUSSION
Culture-based detection and identification of UTI-causing pathogens is responsible for up to 40% of the workload of clinical microbiology laboratories.(Wilson and Gaido, 2004) While it can be argued that cultures provide greater diagnostic reliability compared to dipstick methods, the 24- to 72-hour time span before the availability of culture results means they may have little impact on initial antibiotic therapy of 72 hour duration, which ends up being empirical and potentially inappropriate.(Burd and Kehl, 2011) Point-of-care testing panels for rapid identification of pathogens and/or antibiotic susceptibility – with assay targets determined by the setting (in-patient/out-patient), patient population (first-episode vs. recurrent infection, pediatric vs. adult, etc.), or severity (complicated vs. uncomplicated infection) – could therefore contribute significantly to lowering the clinical and economic burden of UTIs. In this study, we developed a panel of real-time recombinase polymerase amplification (RPA) assays for five of the most common bacterial UTI pathogens, and evaluated it in a small clinical study. We chose RPA over other isothermal NAATs such as loop-mediated isothermal amplification (LAMP) and helicase-dependent amplification (HDA) for the following reasons: faster enzyme kinetics for DNA amplification to a detectable level at low target concentrations – 10 minutes for RPA compared to 30–60 minutes for LAMP and HDA;(Hill et al., 2008, Kim et al., 2011) easier oligonucleotide design, with only three conserved regions required; easy availability of lyophilized reagents not requiring cold chain storage;(Crannell et al., 2014b) and ease of implementation in point-of-care-friendly devices and formats – examples include the ESEquant Tubescanner system, the lateral flow assay, and digital and centrifugal microfluidic platforms.(Euler et al., 2012, Crannell et al., 2015a, Kalsi et al., 2015, Kim et al., 2014, Lutz et al., 2010) These advantages have made RPA a popular choice for infectious disease diagnostics, including assays for Klebsiella pneumoniae, Category A bioterrorism agents, Group B streptococci, Chlamydia trachomatis, HIV, and Cryptosporidium spp.(Valiadi et al., 2016, Euler et al., 2013, Daher et al., 2014, Krolov et al., 2014, Crannell et al., 2014a, Rohrman and Richards-Kortum, 2012) To the best of our knowledge, this is the first report on the development a panel of RPA assays specifically targeting UTI pathogens.
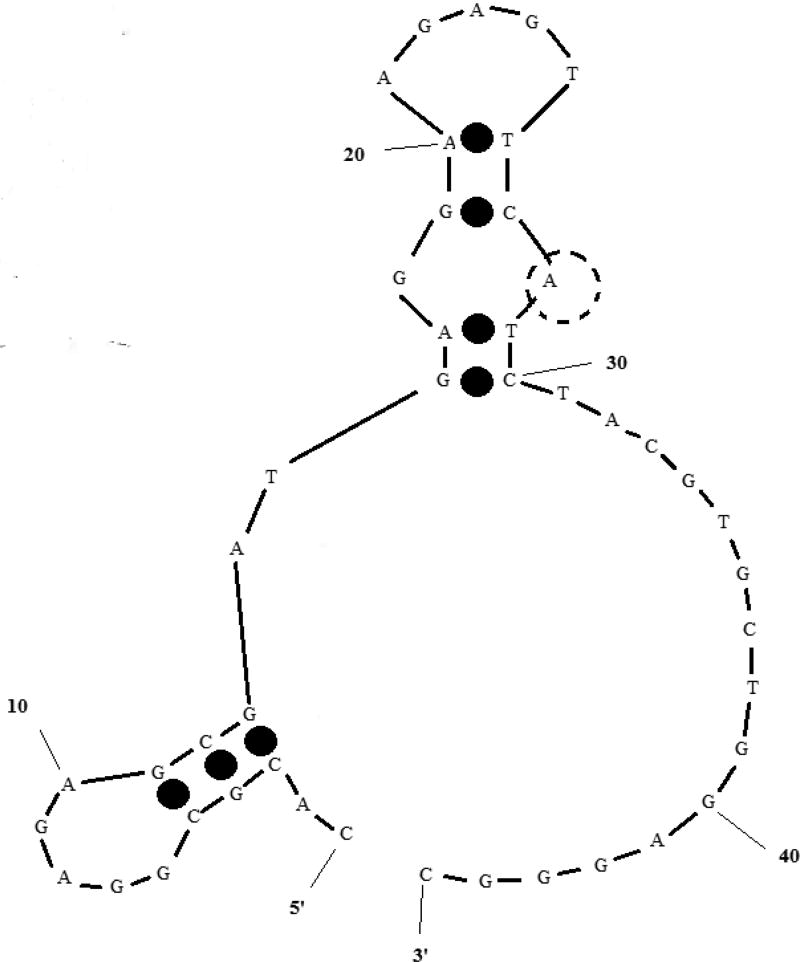
With regard to analytical sensitivity and specificity, the RPAs developed demonstrated limits of detection comparable to other RPAs for DNA amplification in the published literature (10–100 or 100–1000 molecules per reaction), and showed no cross-detection among the targets on the panel. The assays achieved their detection limits much faster than PCR or other isothermal NAATs, emphasizing the rapid reaction kinetics of the DNA polymerase and exonuclease enzymes used. The design of oligonucleotides for real-time RPA is as flexible as probe-based real-time PCR, with only three conserved regions required, as opposed to other isothermal NAATs like LAMP that require four to six primers. However, the long required length of the exo probe is a potential impediment to rapid design, especially for RPAs targeting multiple species with relatively low sequence homology. Longer oligonucleotides also make undesirable secondary structures with highly negative free energies much more likely, potentially affecting assay performance. The exo probe targeting K. pneumoniae, for example, was predicted to have a secondary structure (ΔG = −3.52 kcal mol−1) that creates a double-stranded neighborhood around the nucleotide to be replaced by exonuclease-cleavable THF [Figure 2]. This likely resulted in the assay for K. pneumoniae having a higher background fluorescence (data not shown) and worse analytical sensitivity compared to other assays on the panel.
FIGURE 2. Most stable secondary structure of exo probe targeting K. pneumoniae.
ΔG = −3.62 kcal mol−1; the transparent dotted circle (position 28) marks the nucleotide position replaced by THF and eventually cleaved by exonuclease III. RPA salt conditions − 14 mM Mg2+, 100 mM K+ – were used for secondary structure prediction.
In a retrospective clinical study using 50 urine samples of patients suspected of having UTIs, our panel of real-time RPA assays was determined to have a sensitivity and specificity of 89% (95% CI, 75%-96%) and 100% (95% CI, 78%-100%), respectively, for UTI detection. A number of the urine-derived samples tested by the RPA panel could be expected to have had a high concentration of human DNA (due to pyuria) and DNA from non-pathogenic microbes (skin flora). High background DNA has been shown to lower RPA sensitivity, and might have been responsible for the false negative results among pathogens included on the panel.(Rohrman and Richards-Kortum, 2015) Another potential cause of the false negatives is the inhibitory effect of detergents on RPA enzymes, which can be overcome by adapting a different approach for sample preparation; for example, heating urine at 90°C for 5 minutes has been shown to be effective in releasing pathogen DNA.(Krolov et al., 2014)
It should be noted that we used a cut-off of ≥10,000 CFU.mL−1 of urine, as determined by colony counting on a culture plate, to designate a urine sample as being UTI-positive(Wilson and Gaido, 2004). Our current sample preparation method, in which total DNA from 50 µL of urine was used as the template for each RPA reaction, was optimized for this cut-off. Lower and higher cut-offs in the range of 102 to 106 CFU.mL−1 are recommended for specific patient populations.(Stark and Maki, 1984, Coulthard et al., 2010, Hooton et al., 2010) To avoid potential false positives and false negatives caused by testing samples outside the cut-off range sample preparation protocol would need to be modified to ensure greater enrichment (for cutoffs <104 CFU.mL−1) or dilution (for cut-offs >104 CFU.mL−1) of total urine-derived DNA per reaction. Semi-quantitative RPA, based on threshold time cut-off (analogous to semi-quantitative PCR), or fully quantitative RPA, based on internal positive controls, could be used to supplement alternate sample preparation methods.(Hansen et al., 2013, Crannell et al., 2015b, Crannell et al., 2014c)
The accurate definition of an assay is imperative to ensure its clinical relevance. The panel of assays defined in this study covered a majority of the pathogens in the clinical samples tested; however, the samples included in the study were only representative of the predominantly geriatric patient population served by a single clinical microbiology laboratory in Houston. The etiology of UTIs can vary significantly by geography and patient age.(Kahlmeter, 2000, Langley et al., 2001) The healthcare setting from which the sample originates is also important – the higher propensity for complicated UTIs in in-patient settings like hospitals or assisted-living facilities such as nursing homes results in significantly different distributions of causative bacteria compared to ambulatory and out-patient settings.(Flores-Mireles et al., 2015, Laupland et al., 2007, Wilson and Gaido, 2004) Our choice of pathogens might therefore not be optimal for other patient populations.
In summary, we have demonstrated the analytical and clinical validity of a panel of very rapid and highly sensitive real-time RPAs for the isothermal detection of common UTI pathogens. The ease of developing RPA assays, along with the plethora of formats available for easy implementation, make it an excellent tool to develop highly tailored panel of assays that targeti UTI pathogens and/or common antibiotic susceptibility genes. The routine and widespread use of RPA to supplement or replace culture-based methods could profoundly impact UTI management and the emergence of multidrug-resistant pathogens by obviating empirical broad-spectrum therapies.
Acknowledgments
This work was supported by Welch Foundation (E-1264), NSF (CBET-1511789), and NIAID/NIH (U54 AI057156). The authors would like to thank the administrative staff members of Medical Center Laboratories for their assistance in this project.
Footnotes
CONFLICT OF INTEREST
No conflict of interest declared.
References
- Asiello PJ, Baeumner AJ. Miniaturized isothermal nucleic acid amplification, a review. Lab Chip. 2011;11:1420–1430. doi: 10.1039/c0lc00666a. [DOI] [PubMed] [Google Scholar]
- Bonkat G, Braissant O, Widmer AF, Frei R, Rieken M, Wyler S, Gasser TC, Wirz D, Daniels AU, Bachmann A. Rapid detection of urinary tract pathogens using microcalorimetry: principle, technique and first results. BJU Int. 2012;110:892–897. doi: 10.1111/j.1464-410X.2011.10902.x. [DOI] [PubMed] [Google Scholar]
- Braun SD, Monecke S, Thurmer A, Ruppelt A, Makarewicz O, Pletz M, Reibetaig A, Slickers P, Ehricht R. Rapid identification of carbapenemase genes in gram-negative bacteria with an oligonucleotide microarray-based assay. PLoS One. 2014;9:e102232. doi: 10.1371/journal.pone.0102232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd EM, Kehl KS. A Critical Appraisal of the Role of the Clinical Microbiology Laboratory in the Diagnosis of Urinary Tract Infections. Journal of Clinical Microbiology. 2011;49:S34–S38. [Google Scholar]
- Burillo A, RodríGuez-Sánchez B, Ramiro A, Cercenado E, RodríGuez-CréIxems M, Bouza E. Gram-Stain Plus MALDI-TOF MS (Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry) for a Rapid Diagnosis of Urinary Tract Infection. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Ko WC, Hsueh PR. Emerging resistance problems and future perspectives in pharmacotherapy for complicated urinary tract infections. Expert Opin Pharmacother. 2013;14:587–596. doi: 10.1517/14656566.2013.778827. [DOI] [PubMed] [Google Scholar]
- Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard MG, Kalra M, Lambert HJ, Nelson A, Smith T, Perry JD. Redefining urinary tract infections by bacterial colony counts. Pediatrics. 2010;125:335–341. doi: 10.1542/peds.2008-1455. [DOI] [PubMed] [Google Scholar]
- Crannell ZA, Cabada MM, Castellanos-Gonzalez A, Irani A, White AC, Richards-Kortum R. Recombinase polymerase amplification-based assay to diagnose Giardia in stool samples. Am J Trop Med Hyg. 2015a;92:583–587. doi: 10.4269/ajtmh.14-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crannell ZA, Castellanos-Gonzalez A, Irani A, Rohrman B, White AC, Richards-Kortum R. Nucleic Acid Test to Diagnose Cryptosporidiosis: Lab Assessment in Animal and Patient Specimens. Analytical Chemistry. 2014a;86:2565–2571. doi: 10.1021/ac403750z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crannell ZA, Rohrman B, Richards-Kortum R. Equipment-Free Incubation of Recombinase Polymerase Amplification Reactions Using Body Heat. PLoS ONE. 2014b;9:e112146. doi: 10.1371/journal.pone.0112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crannell ZA, Rohrman B, Richards-Kortum R. Quantification of HIV-1 DNA using real-time recombinase polymerase amplification. Anal Chem. 2014c;86:5615–5619. doi: 10.1021/ac5011298. [DOI] [PubMed] [Google Scholar]
- Crannell ZA, Rohrman B, Richards-Kortum R. Development of a quantitative recombinase polymerase amplification assay with an internal positive control. J Vis Exp. 2015b doi: 10.3791/52620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craw P, Balachandran W. Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab Chip. 2012;12:2469–2486. doi: 10.1039/c2lc40100b. [DOI] [PubMed] [Google Scholar]
- Daher RK, Stewart G, Boissinot M, Bergeron MG. Isothermal recombinase polymerase amplification assay applied to the detection of group B streptococci in vaginal/anal samples. Clin Chem. 2014;60:660–666. doi: 10.1373/clinchem.2013.213504. [DOI] [PubMed] [Google Scholar]
- Devillé W, Yzermans JC, Van Duijn NP, Bezemer PD, Van Der Windt Dë AWM, Bouter LM. The urine dipstick test useful to rule out infections. A meta-analysis of the accuracy. BMC Urol. 2004;4:4. doi: 10.1186/1471-2490-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler M, Wang Y, Heidenreich D, Patel P, Strohmeier O, Hakenberg S, Niedrig M, Hufert FT, Weidmann M. Development of a panel of recombinase polymerase amplification assays for detection of biothreat agents. J Clin Microbiol. 2013;51:1110–1117. doi: 10.1128/JCM.02704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euler M, Wang Y, Otto P, Tomaso H, Escudero R, Anda P, Hufert FT, Weidmann M. Recombinase polymerase amplification assay for rapid detection of Francisella tularensis. J Clin Microbiol. 2012;50:2234–2238. doi: 10.1128/JCM.06504-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluit AC, Jones ME, Schmitz FJ, Acar J, Gupta R, Verhoef J. Antimicrobial resistance among urinary tract infection (UTI) isolates in Europe: results from the SENTRY Antimicrobial Surveillance Program 1997. Antonie Van Leeuwenhoek. 2000;77:147–152. doi: 10.1023/a:1002003123629. [DOI] [PubMed] [Google Scholar]
- Foxman B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med. 2002;113(Suppl 1A):5s–13s. doi: 10.1016/s0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27:224–243. doi: 10.1080/15257770701845204. [DOI] [PubMed] [Google Scholar]
- Gin AS, Zhanel GG. Vancomycin-resistant enterococci. Ann Pharmacother. 1996;30:615–624. doi: 10.1177/106002809603000610. [DOI] [PubMed] [Google Scholar]
- Gold HS. Vancomycin-resistant enterococci: mechanisms and clinical observations. Clin Infect Dis. 2001;33:210–219. doi: 10.1086/321815. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Hansen WLJ, Van Der Donk CFM, Bruggeman CA, Stobberingh EE, Wolffs PFG. A Real-Time PCR-Based Semi-Quantitative Breakpoint to Aid in Molecular Identification of Urinary Tract Infections. PLoS ONE. 2013;8:e61439. doi: 10.1371/journal.pone.0061439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J, Beriwal S, Chandra I, Paul VK, Kapil A, Singh T, Wadowsky RM, Singh V, Goyal A, Jahnukainen T, Johnson JR, Tarr PI, Vats A. Loop-mediated isothermal amplification assay for rapid detection of common strains of Escherichia coli. J Clin Microbiol. 2008;46:2800–2804. doi: 10.1128/JCM.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton TM, Bradley SF, Cardenas DD, Colgan R, Geerlings SE, Rice JC, Saint S, Schaeffer AJ, Tambayh PA, Tenke P. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clinical Infectious Diseases. 2010;50:625–663. doi: 10.1086/650482. [DOI] [PubMed] [Google Scholar]
- Jones RN, Kugler KC, Pfaller MA, Winokur PL. Characteristics of pathogens causing urinary tract infections in hospitals in North America: results from the SENTRY Antimicrobial Surveillance Program, 1997. Diagn Microbiol Infect Dis. 1999;35:55–63. doi: 10.1016/s0732-8893(98)00158-8. [DOI] [PubMed] [Google Scholar]
- Kahlmeter G. The ECO*SENS Project: a prospective, multinational, multicentre epidemiological survey of the prevalence and antimicrobial susceptibility of urinary tract pathogens-interim report. J Antimicrob Chemother. 2000;46(Suppl A):15–22. [PubMed] [Google Scholar]
- Kalsi S, Valiadi M, Tsaloglou MN, Parry-Jones L, Jacobs A, Watson R, Turner C, Amos R, Hadwen B, Buse J, Brown C, Sutton M, Morgan H. Rapid and sensitive detection of antibiotic resistance on a programmable digital microfluidic platform. Lab Chip. 2015;15:3065–3075. doi: 10.1039/c5lc00462d. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Tong Y, Tang W, Quimson L, Cope VA, Pan X, Motre A, Kong R, Hong J, Kohn D, Miller NS, Poulter MD, Kong H, Tang YW, Yen-Lieberman B. A rapid and simple isothermal nucleic acid amplification test for detection of herpes simplex virus types 1 and 2. J Clin Virol. 2011;50:26–30. doi: 10.1016/j.jcv.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Easley CJ. Isothermal DNA amplification in bioanalysis: strategies and applications. Bioanalysis. 2011;3:227–239. doi: 10.4155/bio.10.172. [DOI] [PubMed] [Google Scholar]
- Kim T-H, Park J, Kim C-J, Cho Y-K. Fully Integrated Lab-on-a-Disc for Nucleic Acid Analysis of Food-Borne Pathogens. Analytical Chemistry. 2014;86:3841–3848. doi: 10.1021/ac403971h. [DOI] [PubMed] [Google Scholar]
- Kloß S, Kampe B, Sachse S, RöSch P, Straube E, Pfister W, Kiehntopf M, Popp J. Culture Independent Raman Spectroscopic Identification of Urinary Tract Infection Pathogens: A Proof of Principle Study. Analytical Chemistry. 2013;85:9610–9616. doi: 10.1021/ac401806f. [DOI] [PubMed] [Google Scholar]
- Krolov K, Frolova J, Tudoran O, Suhorutsenko J, Lehto T, Sibul H, Mager I, Laanpere M, Tulp I, Langel U. Sensitive and rapid detection of Chlamydia trachomatis by recombinase polymerase amplification directly from urine samples. J Mol Diagn. 2014;16:127–135. doi: 10.1016/j.jmoldx.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Langley JM, Hanakowski M, Leblanc JC. Unique epidemiology of nosocomial urinary tract infection in children. Am J Infect Control. 2001;29:94–98. doi: 10.1067/mic.2001.111537. [DOI] [PubMed] [Google Scholar]
- Laupland KB, Ross T, Pitout JD, Church DL, Gregson DB. Community-onset urinary tract infections: a population-based assessment. Infection. 2007;35:150–153. doi: 10.1007/s15010-007-6180-2. [DOI] [PubMed] [Google Scholar]
- Lehmann LE, Hauser S, Malinka T, Klaschik S, Stuber F, Book M. Real-time polymerase chain-reaction detection of pathogens is feasible to supplement the diagnostic sequence for urinary tract infections. BJU Int. 2010;106:114–120. doi: 10.1111/j.1464-410X.2009.09017.x. [DOI] [PubMed] [Google Scholar]
- Lehmann LE, Hauser S, Malinka T, Klaschik S, Weber SU, Schewe JC, Stuber F, Book M. Rapid qualitative urinary tract infection pathogen identification by SeptiFast real-time PCR. PLoS One. 2011;6:e17146. doi: 10.1371/journal.pone.0017146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JC, Mastali M, Gau V, Suchard MA, Moller AK, Bruckner DA, Babbitt JT, Li Y, Gornbein J, Landaw EM, Mccabe ER, Churchill BM, Haake DA. Use of electrochemical DNA biosensors for rapid molecular identification of uropathogens in clinical urine specimens. J Clin Microbiol. 2006;44:561–570. doi: 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Yu R, Yan Y, Zhang J, Ren X. Use of Pyromark Q96 ID pyrosequencing system in identifying bacterial pathogen directly with urine specimens for diagnosis of urinary tract infections. J Microbiol Methods. 2011;86:78–81. doi: 10.1016/j.mimet.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Muller C, Mark D, Roth G, Munday P, Armes N, Piepenburg O, Zengerle R, Von Stetten F. Microfluidic lab-on-a-foil for nucleic acid analysis based on isothermal recombinase polymerase amplification (RPA) Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- Murray BE. The life and times of the Enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BE. Vancomycin-resistant enterococcal infections. N Engl J Med. 2000;342:710–721. doi: 10.1056/NEJM200003093421007. [DOI] [PubMed] [Google Scholar]
- Naser SM, Thompson FL, Hoste B, Gevers D, Dawyndt P, Vancanneyt M, Swings J. Application of multilocus sequence analysis (MLSA) for rapid identification of Enterococcus species based on rpoA and pheS genes. Microbiology. 2005;151:2141–2150. doi: 10.1099/mic.0.27840-0. [DOI] [PubMed] [Google Scholar]
- Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niska R, Bhuiya F, Xu J. National Hospital Ambulatory Medical Care Survey: 2007 emergency department summary. Natl Health Stat Report. 2010:1–31. [PubMed] [Google Scholar]
- Owczarzy R, Tataurov AV, Wu Y, Manthey JA, Mcquisten KA, Almabrazi HG, Pedersen KF, Lin Y, Garretson J, Mcentaggart NO, Sailor CA, Dawson RB, Peek AS. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008;36:W163–W169. doi: 10.1093/nar/gkn198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DH, Ririe KM. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One. 2011;6:e26047. doi: 10.1371/journal.pone.0026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja S, Ching J, Xi L, Hughes SJ, Chang R, Wong W, Mcmillan W, Gooding WE, Mccarty KS, Jr, Chestney M, Luketich JD, Godfrey TE. Technology for automated, rapid, and quantitative PCR or reverse transcription-PCR clinical testing. Clin Chem. 2005;51:882–890. doi: 10.1373/clinchem.2004.046474. [DOI] [PubMed] [Google Scholar]
- Rohrman B, Richards-Kortum R. Inhibition of Recombinase Polymerase Amplification by Background DNA: A Lateral Flow-Based Method for Enriching Target DNA. Analytical Chemistry. 2015;87:1963–1967. doi: 10.1021/ac504365v. [DOI] [PubMed] [Google Scholar]
- Rohrman BA, Richards-Kortum RR. A paper and plastic device for performing recombinase polymerase amplification of HIV DNA. Lab Chip. 2012;12:3082–3088. doi: 10.1039/c2lc40423k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schappert SM, Rechtsteiner EA. Ambulatory medical care utilization estimates for 2007. Vital Health Stat. 2011;13:1–38. [PubMed] [Google Scholar]
- Semeniuk H, Church D. Evaluation of the leukocyte esterase and nitrite urine dipstick screening tests for detection of bacteriuria in women with suspected uncomplicated urinary tract infections. J Clin Microbiol. 1999;37:3051–3052. doi: 10.1128/jcm.37.9.3051-3052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Trinh Q, Xu W, Zhai B, Luo Y, Huang K. A universal primer multiplex PCR method for typing of toxinogenic Pseudomonas aeruginosa. Appl Microbiol Biotechnol. 2012;95:1579–1587. doi: 10.1007/s00253-012-4277-8. [DOI] [PubMed] [Google Scholar]
- Stamm WE, Norrby SR. Urinary tract infections: disease panorama and challenges. J Infect Dis. 2001;183(Suppl 1):S1–S4. doi: 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- Stark RP, Maki DG. Bacteriuria in the catheterized patient. What quantitative level of bacteriuria is relevant? N Engl J Med. 1984;311:560–564. doi: 10.1056/NEJM198408303110903. [DOI] [PubMed] [Google Scholar]
- Tang YW, Procop GW, Persing DH. Molecular diagnostics of infectious diseases. Clin Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- Valiadi M, Kalsi S, Jones IGF, Turner C, Sutton JM, Morgan H. Simple and rapid sample preparation system for the molecular detection of antibiotic resistant pathogens in human urine. Biomedical Microdevices. 2016;18(1):18. doi: 10.1007/s10544-016-0031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XH, Zhang G, Fan YY, Yang X, Sui WJ, Lu XX. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods. 2013;92:231–235. doi: 10.1016/j.mimet.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Wilson EB. Probable Inference, the Law of Succession, and Statistical Inference. Journal of the American Statistical Association. 1927;22:209–212. [Google Scholar]
- Wilson ML, Gaido L. Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis. 2004;38:1150–1158. doi: 10.1086/383029. [DOI] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman Z, Borremans A, Verhaegen J, Verbist L, Blanckaert N. Disappointing dipstick screening for urinary tract infection in hospital inpatients. Journal of Clinical Pathology. 1998;51:471–472. doi: 10.1136/jcp.51.6.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhanel GG, Laing NM, Nichol KA, Palatnick LP, Noreddin A, Hisanaga T, Johnson JL, Hoban DJ. Antibiotic activity against urinary tract infection (UTI) isolates of vancomycin-resistant enterococci (VRE): results from the 2002 North American Vancomycin Resistant Enterococci Susceptibility Study (NAVRESS) J Antimicrob Chemother. 2003;52:382–388. doi: 10.1093/jac/dkg352. [DOI] [PubMed] [Google Scholar]
- Zhang W, Niu Z, Yin K, Liu P, Chen L. Quick identification and quantification of Proteus mirabilis by polymerase chain reaction (PCR) assays. Annals of Microbiology. 2013;63:683–689. [Google Scholar]