Abstract
The human pathogen Helicobacter pylori requires nickel for colonization of the acidic environment of the stomach. HypA, a Ni metallochaperone that is typically associated with hydrogenase maturation, is also required for urease maturation and acid survival of H. pylori. There are two proposed Ni site structures for HypA; one is a paramagnetic 6-coordinate site characterized by X-ray absorption spectroscopy (XAS) in unmodified HypA while another is a diamagnetic four-coordinate planar site characterized by solution NMR in an N-terminally modified HypA construct. To determine the role of the N-terminal amine in Ni binding of HypA, an N-terminal extension variant, L2*-HypA, where a leucine residue was inserted into the second position of the amino acid sequence in the proposed Ni-binding motif, was characterized in vitro and in vivo. Structural characterization of the Ni site using x-ray absorption spectroscopy (XAS) showed a coordination change from 6-coordinate in WT-HypA to 5-coordinate pyramidal in L2*-HypA, which was accompanied by the loss of two N/O-donor protein ligands and the addition of an exogenous bromide ligand from the buffer. The magnetic properties of the Ni-sites in WT- compared to L2*-HypA confirmed that a spin-state change from high- to low-spin accompanied this change in structure. The L2*-HypA H. pylori strain was shown to be acid sensitive and deficient in urease activity in vivo. In vitro characterization showed that L2*-HypA did not disrupt the HypA-UreE interaction essential for urease maturation, but was at least 20-fold weaker in Ni-binding as compared to WT. Characterization of the L2*-HypA variant clearly demonstrates that the N-terminal amine of HypA is involved in proper Ni coordination and is necessary for urease activity and acid survival.
Keywords: Helicobacter pylori, Nickel protein, acid resistance, urease, HypA, nickel binding, XAS, Metallochaperone
TOC image
Helicobacter pylori is a Gram-negative bacterium that colonizes the human stomach and is a known cause of gastric and duodenal ulcers.1, 2 Despite recent declines, prevalence of infection is still greater than 50% in most countries.3 Although most infected individuals appear to be asymptomatic, chronic infection has been shown to cause gastric carcinomas and lymphomas; H. pylori is estimated to be responsible for 6.2% of the global burden of gastric cancers.4 One of the major adaptations that allow H. pylori to thrive in the acidic environment of the human stomach is the utilization of the Ni-dependent enzyme urease.5, 6 Urease catalyzes the hydrolysis of urea to carbamate and ammonia, which is used by H. pylori to neutralize the cytoplasmic and periplasmic pH under acid shock conditions.7, 8
Urease is abundant in H. pylori under both acidic and neutral conditions, making up to 10% of the total protein synthesized.9 However, excessive urease activity under neutral conditions can lead to an increase in internal pH and subsequent cell death. Therefore, the activity of the enzyme is carefully regulated through multiple post-translational mechanisms.10–12 One such mechanism is through modulation of urease maturation via nickel insertion into apo-urease.12 In addition to the urease specific UreEFGH accessory proteins, which are required for the assembly of the di-nickel urease active site, activity is also regulated by the hydrogenase maturation proteins HypAB.13 The deletion of the hypA or hypB gene from H. pylori results in loss of urease activity. This can be partially restored through nickel supplementation in the growth media,14 suggesting that HypAB function as metallochaperones in the transport and delivery of nickel to the urease maturation pathway. Indeed, loss of nickel binding in the H2A-hypA variant of H. pylori results in a severely urease-deficient phenotype.15 Additionally, HypA has been shown to directly interact with HypB15 and UreE16 in vitro, establishing a potential pathway to deliver nickel to the urease maturation pathway through HypA-UreE protein-protein interactions.
The structure of the HypA Ni-site is shrouded in controversy. Multiple sequence alignments of HypA homologues reveal a rigidly conserved MHE-motif at the N-terminus (SI Fig. S1). N-terminal sequencing of purified WT-HypA protein showed that the motif is intact in the mature protein.15 Mutagenesis confirmed that His2 is a Ni-ligand and mutation to Ala (H2A) resulted in loss of Ni binding in the purified protein.15 However, it was unclear whether the purified H2A-HypA protein variant retained the N-terminal Met residue; this is due to the propensity for N-terminal processing of Met when followed by a small hydrophobic residue in the E. coli17 overexpression system. Nevertheless, Ni-binding clearly occurs at the N-terminal MHE-motif, involving His2 as a ligand. The structure of the WT-HypA Ni-binding site was initially characterized by X-ray absorption spectroscopy (XAS) and found to be consistent with a 6-coordinate site that was best modeled with exclusively N/O-donor ligands, of which one or two were modeled as imidazole ligands (presumed to include His2).18 The coordination geometry was corroborated by Evans magnetic susceptibility (NMR) that confirmed a high-spin paramagnetic S = 1 nickel center.18 In contrast, a diamagnetic Ni(II) center was found for an N-terminally modified HypA, which had extra Gly-Ser residues on the N-terminus that remained after proteolytic cleavage of an affinity tag. That study employed a Ni(II) titration that was monitored by 2D 1H-15N HSQC spectra as part of effort to solve the solution NMR structure (PDB ID: 2KDX).19 The lack of paramagnetic broadening in neighboring cross-peaks was consistent with a diamagnetic planar Ni-site, which was proposed to bind four protein ligands in a planar geometry including the side chain imidazole ring of His2 as well as the backbone amide N-donor atoms of His2, Glu3, and Asp40.19 Subsequently, the same technique was used to characterize Ni(II) titration into the unmodified WT-HypA protein, and found paramagnetic broadening of cross-peaks throughout the proposed Ni binding region.20 It is possible that the differences in the geometry and number of ligands in the Ni-site stems from the N-terminal modification of the HypA protein used in the NMR study, leading to our hypothesis that the N-terminal amine of HypA is involved in proper Ni ligation and therefore affects the function of the protein in vitro and in vivo.
Additional evidence of N-terminal Ni-ligation in HypA is found in the crystal structure of a thermostable form of HypA (from T. kodakarensis) in a complex with an ATPase analog of HypB with ADP- or ATP-bound (PDB ID: 5AUO and 5AUN).21 The Ni-sites of the HypA-HypBAT complex show four-coordinate sites with the ligation of the N-terminal amine from Met1, backbone amide and side chain imidazole of His2, and the imidazole side chain from His98. Although this conformation of the Ni-site encompasses both the N-terminal ligation and the four-coordinate planar site, it is unlikely to exist in H. pylori HypA. Firstly, this conformation of the Ni-site is formed only in complex with HypBAT. Additionally, it involves ligation of His98, which is found in an extended loop in the Zn-binding domain that is absent in the H. pylori version of HypA. However, the other three ligands derived from part of the conserved HypA Ni-binding motif (Met1 and His2) and are likely to be involved in formation of all conformations of the HypA Ni-site in support of the N-terminal Ni-ligation in unmodified HypA.
To test the role of the N-terminus in the structure and function of the Ni-site, an alternate N-terminal extension variant protein was created in which a Leu residue was inserted into the second position of the HypA protein sequence, resulting in the Met1_His2insLeu variant, hereafter known as the L2*-HypA variant (SI Fig. S1). Insertion of Leu into the conserved MHE-motif allowed for Ni-ligation at the His2 and Glu3 position, but extends the N-terminal amine so that it is too remote for binding to Ni(II). Direct comparison of the WT-HypA and the L2*-HypA variant in vitro and in vivo definitively identifies the N-terminal amine in HypA as an essential nickel ligand that is required for its function in urease maturation and the formation of the proper Ni-site.
EXPERIMENTAL PROCEDURE
Detailed descriptions of materials and methods can be found in supporting information.
RESULTS
The L2* mutation does not affect the protein secondary or quaternary structure of HypA
To ensure that the insertion of Leu into the N-terminal MHE-motif did not disrupt the overall fold of the purified HypA protein, the secondary and tertiary structures of purified WT- and L2*-HypA were investigated. Circular dichroism (refer to the supporting information Experimental section) of purified WT-HypA and L2*-HypA proteins show that the N-terminal mutation has not disrupted the mostly α-helical characteristics of the purified protein (SI Fig. S2A) either with or without Ni bound. These results show that the L2*-HypA variant has not altered its overall fold as compared to WT.
To further analyze the overall size and shape of the purified HypA, size exclusion chromatography in combination with multi-angle light scattering (SEC-MALS) was used to analyze both the molecular weight and the hydrodynamic radius of the purified HypA proteins. Previously, SEC was used to estimate the “size” of HypA in solution based on its elution volume in comparison to known molecular weight standards. However, elution volume from a SEC column is dependent on the hydrodynamic radius of the eluted molecule, and the estimated “molecular weight” calculated from standard curves is based on the assumption that the eluted molecule is globular shape. HypA has been reported to have an elongated shape with two distinct lobes in the solution NMR structure (PDB ID: 2KDX)19 and in the crystal structure of T. kodakarensis (PDB ID: 3A43).22 Therefore, elution volume alone is an unreliable method for estimating “size” of the HypA protein, and have led to the HypA protein being reported as a solution dimer in the past.15, 20 Using elution volume alone, HypA was estimated to be ~23 kDa for both WT and L2*-HypA, based on a calibration curve using known molecular weight standards, which is consistent with a solution dimer (with a calculated monomeric molecular weight of 13.2 kDa for WT-HypA). MALS calculates absolute molecular mass based on light scattering properties of the molecule, independent of the hydrodynamic radius or shape.23 Using MALS analysis, the absolute masses of WT- and L2*-HypA were calculated as 13.01 ± 0.007 and 13.37 ± 0.021 kDa, respectively (SI Fig. S2B), which is consistent with a monomeric solution structure for both WT- and L2*-HypA. Comparison of the relative intensity of the different angles of light scattering (from 18 different angles available on the instrument) shows that WT- and L2*-HypA scatter light similarly to one another (data not shown) and therefore likely have similar tertiary structures in solution.
L2* N-terminal extension of HypA results in coordination change at the Ni site
The controversy over the HypA Ni(II) binding site structure in the literature points to the unmodified WT-HypA Ni(II) site having a six-coordinate, and therefore paramagnetic site,18, 24 and the N-terminally modified variant (PDB: 2KDX) having a four-coordinate planar and diamagnetic site.19 Direct comparison of the structures of the Ni(II) sites in WT- and L2*-HypA by XAS was made to clarify the role of the N-terminal amine as a ligand in the Ni(II) site of WT-HypA.
Comparison of the Ni K-edge x-ray absorption near-edge structure (XANES) region of the XAS spectra obtained from WT-HypA and L2*-HypA Ni(II) complexes in NaBr buffer show a clear difference that corresponds to a change in the coordination number/geometry of the nickel site (Figure 1). Typically, two types of high-energy bound-transitions may be observed in the pre-edge XANES region of the XAS spectra of Ni(II) complexes.25 These features are associated with transitions involving promotion of a 1s electron into the 3d manifold (1s ➔ 3d) or into the 4pz orbital (1s ➔ 4pz) of the Ni(II) complex and are observed near 8331 and 8336 eV, respectively.25 In the case of Ni-WT-HypA, no discernable pre-edge features were observed. The small (unresolved) 1s ➔ 3d (plus shakedown) transition is consistent with a centrosymmetric Ni(II) site, and the absence of a peak associated with a 1s ➔ 4pz transition rules out a four-coordinate planar site.25 The XANES analysis is therefore consistent with a 6-coordinate nickel site in WT-HypA, in agreement with the paramagnetism associated with the site by Evans susceptibility measurements (vide infra). In contrast, a clear feature corresponding to a 1s ➔ 4pz transition (plus shakedown) is observed on the Ni-L2*-HypA K-edge that indicates the loss of at least one axial ligand from the Ni site compared to WT-HypA (Fig. 1, inset).25 The 1s ➔ 4pz feature in the Ni-L2*-HypA spectrum appears as an unresolved shoulder on the Ni K-edge, and is therefore more consistent with a five-coordinate pyramidal geometry than with a four-coordinate planar complex, where a resolved maximum is expected.25 A second set of Ni K-edge XAS data were collected on a Ni,Zn-L2*-HypA in NaCl buffer (20 mM HEPES, 200 mM NaCl, 1mM TCEP, pH 7.2) to rule out buffer effects on the geometry of the Ni-site. The XANES spectrum in NaCl buffer is very similar to the spectrum obtained in NaBr buffer (SI Fig. S3), and confirms the five-coordinate pyramidal Ni(II) structural assignment in L2*-HypA.
Figure 1.
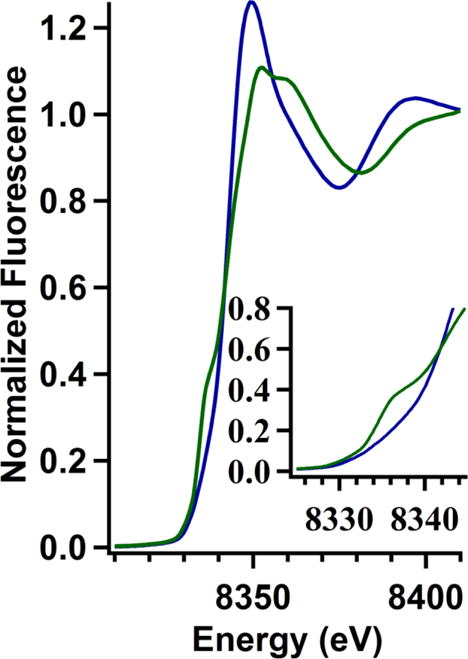
Overlay of the Ni K-edge XANES region of the XAS spectrum of Ni,Zn-WT-HypA (in blue) and Ni,Zn-L2*-HypA (in green) in 20 mM HEPES, 200 mM NaBr, 1mM TCEP, pH 7.2 with the 1s ➔ 4pz region enlarged in the inset.
Six-coordinate Ni(II) complexes with octahedral geometry are invariably high-spin with S = 2, while four-coordinate planar Ni(II) complexes are invariably low-spin and diamagnetic.26 Five-coordinate complexes may be either high- or low- spin depending on the relative strengths of the axial and in-plane ligand fields.25 Magnetic susceptibility measurements using Evans’ technique27, 28 show that the WT-HypA Ni complex is high-spin (μ = 3.52 BM, well within the range typically observed for Ni(II) complexes: μ = 2.8 – 4.0 BM26). In contrast, the Ni(II) complex of L2*-HypA protein is diamagnetic.
Loss of two Ni protein ligands occurs in L2*-HypA
To further characterize the changes in the Ni site structure in WT- and L2*-HypA proteins, the EXAFS regions of the XAS spectra were analyzed. One limitation of EXAFS analysis is that the models cannot distinguish scattering atoms with Z ± 1, which means the structural models cannot easily distinguish between a N- vs. O-donor ligand, or a S-donor vs. a Cl− ligand.29 Fits were generated using parameters for N-donors and are represented as N/O in Table 1 to emphasize this lack of distinction. To address the issue of S vs. Cl ligation, samples were prepared in a buffer containing NaBr, rather than NaCl. This practice increases the confidence of models containing S ligand donors, and also can identify solvent accessible metal sites in those cases where Br− becomes a ligand.
Table 1.
| XANES analysis | EXAFS analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HypA | K-edge Energy (eV) | 1s ➔3d peak area (x10−2 eV) | 1s ➔ 4pz observed | Coordination Number | Shell | r (Å) | σ2 (x10−3 Å2) | ΔEo (eV) | R factor (%) |
| WT | 8342.8 | < 0.05 | No | 6 | 1 N/O | 2.21 (5) | 4 (8) | 2 (1) | 3.53 |
| 3 N/O (1 Imid0°) | 2.08 (2) | 3 (2) | |||||||
| BBAm | 1.92 (3) | 6 (3) | |||||||
| 1.97 (3) | |||||||||
| L2* | 8342.5 | < 0.05 | Yes | 5 | 1 N/O | 2.04 (3) | 4 (4) | 0 (1) | 3.49 |
| 1 Br | 2.42 (2) | 11 (3) | |||||||
| 1 Imid7° | 1.94 (4) | 5 (5) | |||||||
| BBAm | 1.82 (1) | 1 (1) | |||||||
| 1.87 (1) | |||||||||
Uncertainties in the fitted parameters are shown in parentheses.
Models to interpret the EXAFS data were developed beginning with the coordination number determined by XANES and magnetic studies (vide supra). Models were developed by first using single-scattering analysis. Multiple-scattering scaffolds were added to the best single-scattering models to account for features in the FT-EXAFS spectra arising from the second and third coordination sphere scattering atoms resulting from the coordination of His imidazole ligands (see SI Experimental section, SI Fig. S4) as previously described.30, 31 Imidazoles were fit as rigid rings with a single adjustable distance. Integer numbers of rings were added to “count” the number of imidazole ligands. A separate model was developed that incorporated a rigid, bidentate 1,2-ethylenediammine-like unit (see SI Experimental section, SI Fig. S4) similar to the bidentate coordination of α-ketoglutarate Fe-binding in nonheme Fe(II) enzymes used in prior models.32, 33 This model simulates the formation of five-membered chelate rings that form when the binding of the N-terminal amine is accompanied by backbone amide binding, which lead to the ordering of second coordination sphere C atoms similar to prior models of backbone amidate ligation in the active site of NiSOD.34, 35
The best fit for Ni-WT-HypA EXAFS is a six-coordinate complex (Fig. 2A, Table 1, SI Table S1) with exclusively N/O-donor ligands at distances ranging from 1.92 – 2.21 Å, and includes multiple-scattering paths that describe an imidazole with 0° tilt (Imid0°) and binding of one bidentate backbone amide (BBAm) unit. The inclusion of the BBAm was required to generate a fit with a R-factor < 5 %. Although both imidazole ligands and five-membered chelate rings feature carbon atoms in the second coordination sphere at similar Ni-C distances, the two ligands are distinguished by scattering from atoms in the third coordination sphere (C and N) that contribute to scattering intensities between 3 – 4 Å in the phase uncorrected FT-EXAFS (SI Fig. S4), and exist only for imidazole ligation. The imidazole in the Ni-WT-HypA fit likely arises from Ni binding to the His2 sidechain, which has been shown to be critical to Ni-binding and function.15
Figure 2.
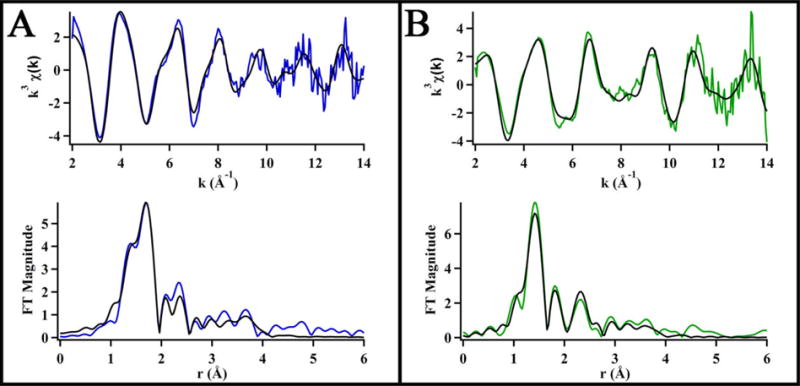
Data (in color) and best fit models (in black) of the Ni K-edge EXAFS represented in k3-weighted unfiltered data (top) and Fourier transformed (k = 2 – 14 Å−1) data uncorrected for phase shifts (bottom) for (A) Ni,Zn-WT-HypA and (B) Ni,Zn- L2*-HypA in 20 mM HEPES, 200 mM NaBr, 1mM TCEP, pH 7.2.
The L2*-HypA Ni-EXAFS obtained in buffer NaBr was modeled as a 5-coordinate Ni(II) site that featured one Br− ligand at a Ni-Br distance of 2.42 ± 0.2 Å and four N/O-donor ligands ranging from 1.82 – 2.04 Å, and included multiple-scattering paths that were consistent with one imidazole ligand with a 7°tilt (Imid7°) and one bidentate BBAm unit (Fig. 2B, Table 1, SI Table S2). The multiple-scattering paths used to fit the WT-HypA EXAFS are mostly preserved in the L2*-HypA Ni site (although at slightly different distances and angles), including the bidentate backbone N coordination and a His imidazole. However, the BBAm unit is not required in order to generate a fit to the L2*-RcnR data with an R-factor < 5 %, though the alternative fit requires the presence of two His imidazole ligands (see SI Table S2). It is not clear where the second His imidazole would come from, and the single His fit is consistent with the nickel occupying the same locus (the N-terminus) of the protein as in WT-HypA and preserving the coordination of the His2 sidechain. In contrast to the WT-HypA Ni site structure, for L2*-HypA one N/O-donor ligand was lost and another substituted by a Br− ligand originating from the buffer. The bromide scattering atom is required to generate a fit with an R-factor < 5 % when BBAm is used to model backbone N coordination. Additionally, a systematic shortening of the average distances of Ni-N/O ligation by 0.14 Å in the Ni-L2*-HypA complex is consistent with the observed spin-state change measured by Evans’ method (vide supra).
To confirm the 5-coordinate fitting model with exogenous Br− ligand, the EXAFS of a Ni-L2*-HypA sample in buffer with NaCl was analyzed. The best 5-coordinate model for this data set include the bidentate BBAm scaffold, as well as a S/Cl-donor at a reasonable distance of 2.25 Å, consistent with the XANES analysis and the best fits in NaBr buffer. A S/Cl donor is required to generate a fit with R-factor < 5 % (see SI Fig. S5, SI Table S3).
The Zn site in L2*-HypA is unchanged from WT-HypA
In addition to the Ni K-edge, the Zn K-edge XAS spectra were also obtained for Ni,Zn-WT- and Ni,Zn-L2*-HypA and confirm that Zn-site coordination involving two conserved CXXC motifs is unperturbed in L2*-HypA. The Zn K-edge XANES spectra of WT- and L2*-HypA overlay well, indicating no significant change in geometry at the Zn site has occurred (Fig. S6A). The zinc EXAFS spectra for both WT- and L2*-HypA were best fitted with a model featuring four S-donors with Zn-S distances of ~2.34 Å and an imidazole ligand with a 2° tilt (Imid2°) at a distance of 2.18 Å, although acceptable fits with exclusively Cys ligands are also consistent with the data (SI Figure S6BC, SI Table S4). Possible coordination of the flanking His residue demonstrates that the dynamic nature of the HypA Zn-site is preserved in WT- and L2*-HypA.20, 36
L2* hypA mutant strain of H. pylori exhibits acid sensitivity and urease deficiency
To investigate the in vivo effect of the N-terminal extension of the HypA protein in H. pylori, the chromosomal hypA gene was replaced with the coding sequence for L2*-HypA. This mutant strain (DSM1475) expresses the L2*-HypA protein under the control of the endogenous promoter (SI Table S5). As the hypA gene has been shown to be vital for urease activity in H. pylori,14 and therefore an integral part of the acid survival mechanism, the urea-dependent acid survival was assessed for this variant strain. H. pylori strains were challenged for one hour at mild (pH 6) or harsh (pH 2.3) acidic shock conditions either with or without urea supplement, and then plated to assess for surviving colony-forming units (CFU). In addition to the L2*-hypA strain, the parental WT strain, the ΔureB strain where a subunit of the urease enzyme has been deleted, the hypA∷kan-sacB strain where the hypA gene was interrupted, and the hypA-R strain where the hypA gene was restored after deletion were tested concurrently as controls (Fig. 3). Under mild acid shock conditions, all strains tested were able to survive with or without urea supplement (Fig. 3AB). Without urea supplement, harsh acid shock was lethal for all strains tested (Fig. 3C). At harsh acid shock conditions with urea supplement, distinct phenotypes for each strain were revealed with WT and hypA-R strains each exhibiting unimpaired acid survival, whereas the acid survival of L2*-hypA strain along with the hypA∷kan-sacB and ΔureB strains were significantly reduced compared to WT (p < 0.0001) (Fig. 3D). To further confirm the results obtained with the L2*-hypA mutant, the strain was independently remade and tested in a single replicate of the acid survival assay; this mutant strain displayed similar acid survival phenotype as the original L2*-hypA mutant strain (data not shown).
Figure 3.
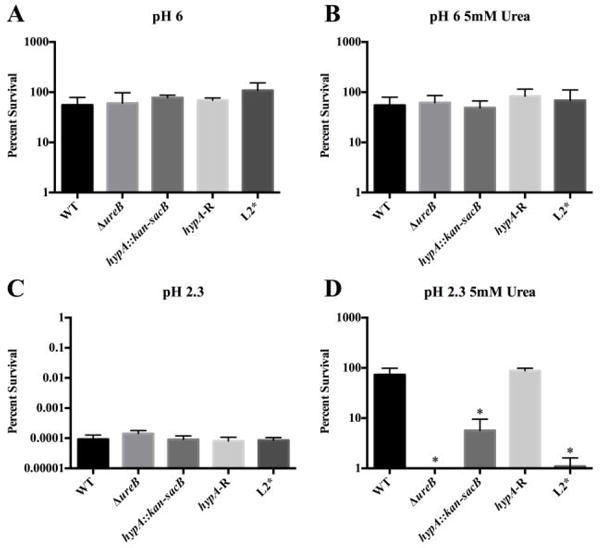
Acid viability results for the various HypA mutants in Helicobacter pylori. Wild type G27 (WT), a ureB knockout (ΔureB), the hypA interrupted mutant (hypA∷kan-sacB), the hypA restorant (hypA-R), and the L2* HypA mutant were exposed to PBS for one hour at pH 6 or pH 2.3 with or without 5 mM urea: pH 6 (A), pH 6 containing 5mM urea (B), pH 2.3 (C), and pH 2.3 containing 5 mM urea (D). Each bar represents mean percent survival ± standard deviation. * = acid viability was significantly reduced when compared to wild type (p < 0.0001, one-way ANOVA followed by Dunnett’s test for multiple comparisons).
To confirm that the acid sensitive phenotype in L2*-hypA strain was a result of urease deficiency, urease activity of H. pylori strains was measured using a modified phenol-hypochlorite assay for NH3 production. In 30 minutes, the clarified lysates of H. pylori strains produced 109, 0.47, 0.77, 86, and 0.33 nmol NH3 per μg of total protein for WT, ∆ureB, hypA∷kan-sacB, hypA-R, and L2*-hypA strains respectively. These results confirmed the trend observed in acid survival tests, with hypA-R strain having WT-like urease activity, whereas L2*-hypA strain along with negative control strains ΔureB and hypA∷kan-sacB had less than 1% of WT relative urease activity (Fig. 4). These results show that insertion of a single Leu residue into the N-terminal Ni-binding motif of HypA (L2*-hypA variant) is sufficient to nearly eliminate urease activity and to severely impair urea-dependent acid survival in vivo.
Figure 4.
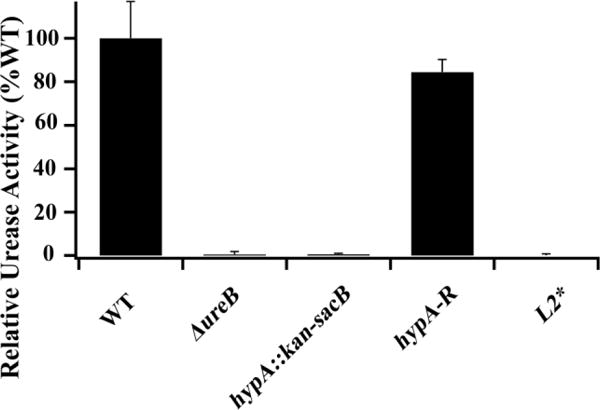
Relative urease activity of whole cell extract of Helicobacter pylori strains: Wild type G27 (WT), a ureB knockout (ΔureB), the hypA interrupted mutant (hypA∷kan-sacB), the hypA restorant mutant (hypA-R), and the L2* HypA mutant. Each bar represents the averaged relative activity measured in three technical replicates from two independent growths normalized against WT ± standard deviation.
L2*-HypA retains protein-protein interaction properties but loses Ni-binding affinity
The L2*-hypA strain could induce in vivo urease deficiency by two potential mechanisms. Firstly, the L2*-HypA mutation could disrupt protein-protein interactions between HypA and the known interaction partner, UreE, in the urease maturation pathway.16
Alternatively, insertion of a Leu at the N-terminal MHE-motif could disrupt the Ni-binding properties of HypA directly. These mechanisms are not mutually exclusive, and both would result in impaired Ni delivery and the loss of urease activity and acid viability phenotype that is observed for the L2* hypA strain.
To assess whether the L2* mutation of HypA affected protein-protein interaction between HypA and UreE, binding of purified HypA to UreE was assessed using ITC. WT- or L2*-HypA protein and WT-UreE protein were purified and buffer exchanged into the same Buffer GF (20 mM HEPES, 200 mM NaCl, 1 mM TCEP, pH 7.2). Concentrated WT- or L2*-HypA proteins were injected into WT-UreE dimers30, 37 and the change in heat from each injection was fitted to OneSite models using the MicroCal Analysis module in Origin7.0 (Fig. 5). The number of sites (N) in each case was fitted to be ~1.5 HypA per UreE dimer. The larger than expected value of N (HypA:UreE dimer >1) is due to the tendency of H. pylori UreE protein to degrade slowly over time at the C-terminal Ni-binding motif.30, 38 This degradation, which can be observed in SDS-PAGE as resulting in a lower molecular weight band (SI Fig. S7), makes the concentration of holo-UreE in a sample uncertain, thus complicating an accurate determination of N. The amount of degradation was carefully monitored using SDS-PAGE post-titration to ensure that holo-UreE is comparable in each titration (SI Fig. S7). The binding of monomeric WT-HypA or L2*-HypA to WT-UreE dimer had very similar affinities (with apparent Kd values of 0.96 ± 0.09 and 0.69 ± 0.11 μM respectively, Table 2). This finding suggests that the L2* variant did not disrupt the protein-protein interactions with the urease maturation pathway through UreE.
Figure 5.
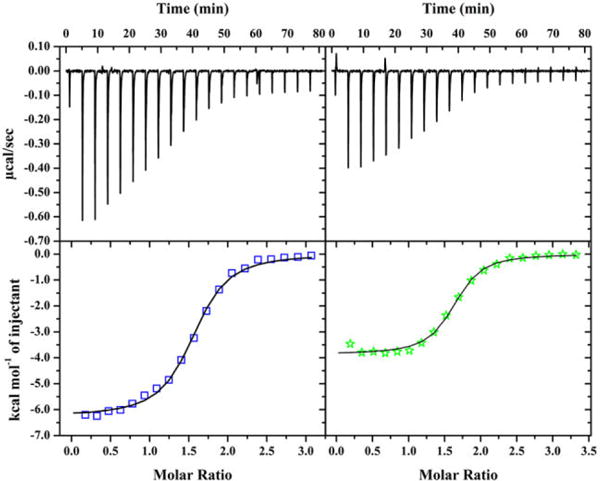
ITC binding curves of WT- (left) or L2*-HypA (right) into UreE WT dimer in buffer with 20mM HEPES, 200mM NaCl, 1mM TCEP, pH 7.2. The top panels show the heat response from each injection, which are integrated to generate the enthalpy of binding plotted on the bottom panels (WT = circles; L2* = stars). Binding of WT- and L2*-HypA to UreE WT dimer are each fitted to OneSite models as described in the text.
Table 2.
Apparent binding to HypA as measured by ITC
| Binding Partner | HypA | Kd (μM) | N | ΔH (kcal/mol) |
|---|---|---|---|---|
| UreE (dimer) | WT | 0.96 ± 0.09 | 1.5 ± 0.1 | −6.27 ± 0.06 |
| L2* | 0.69 ± 0.11 | 1.6 ± 0.2 | −3.86 ± 0.06 | |
|
| ||||
| Ni | WT | 1.0 ± 0.2 | 0.88 ± 0.01 | −4.99 ± 0.07 |
| L2* | 59 ± 12 | 0.39 ± 0.07 | −1.30 ± 0.29 | |
Another mechanism by which L2*-HypA could hinder urease maturation is through lowering the ability of HypA to bind Ni. This was also addressed using ITC to measure the difference in Ni binding affinity between WT- and L2*-HypA. ITC titrations involving addition of NiCl2 solutions (~ 1 – 2 mM, see SI experimental section) to the HypA protein solutions were performed with the both the titrant and the titrand in the same Buffer GF. The relatively concentrated Ni(II) solution was titrated into either WT- or L2*-HypA protein (~ 150 μM, where the Zn-sites were occupied and the Ni-sites were vacant, as confirmed by metal analysis, see SI Experimental section) and the heat of binding was monitored directly. WT-HypA bound one nickel per protein and had an apparent Kd = 1.0 ± 0.2 μM (Fig. 6 left, Table 2), which is consistent with prior studies.24 NiCl2 titration into L2*-HypA at the same concentrations resulted in curves that did not reach saturation after the addition of 2 equivalents of Ni (data not shown), indicating a much lower Ni affinity for L2*-HypA. A more concentrated NiCl2 solution was subsequently used for the L2*-HypA titration to push the titration to apparent saturation. The resulting heat of binding was best fitted with a OneSite model with an apparent Kd = 59 ± 12 μM and N ~ 0.5 (Fig. 6 right, Table 2).
Figure 6.
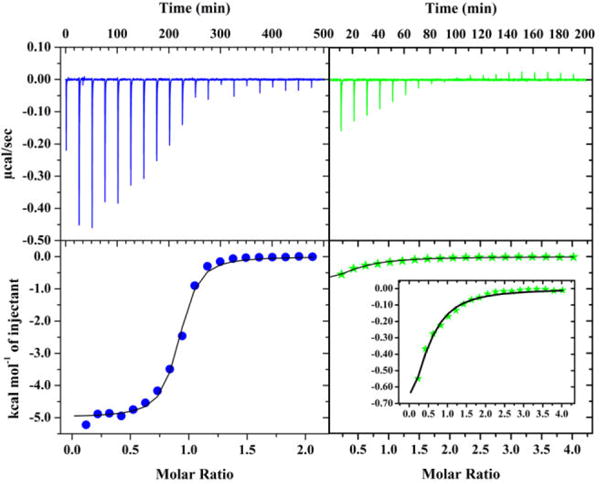
ITC binding curves of Ni titration into WT- (blue) and L2*-HypA (green) in buffer with 20mM HEPES, 200mM NaCl, 1mM TCEP, pH 7.2. The top panels show the heat response of each injection, which are integrated to generate the enthalpy of binding plotted on the bottom panels (WT = blue circles; L2* = green stars). Binding curves are graphed to the same scale on the y-axes for WT and L2* to emphasize the difference in enthalpy response with more appropriately scaled y-axis for Ni-L2* binding shown in the inset. Ni binding to HypA WT and L2* are each fitted to a OneSite model as described in the text.
However, it clear that the amount of heat generated by Ni binding to L2*-HypA is very small, making Kd and N difficult to measure. For this reason, the Ni-binding affinities of WT- and L2*-HypA, were also measured by Ni2+ titration into protein solutions monitored by changes in absorbance in the UV-Vis spectra. The absorption spectra of samples with a constant concentration of WT or L2*-HypA mixed with increasing concentrations of Ni2+ were measured, and the apparent Kd was calculated from the change in absorption maxima in response to [Ni] (see SI Experimental section, SI Fig. S8). The apparent Kd was calculated to be 1.1 ± 1.0 μM for Ni-binding to WT-HypA, corroborating the ITC data. The apparent Kd was calculated to be 27 ± 4 μM for Ni-binding to L2*-HypA, which is of the same order of magnitude as the value found using ITC, and ~ 20-fold greater than WT. Thus, inserting a Leu residue into the Ni-binding motif of HypA (L2*-HypA) significantly decreases its ability to bind Ni, rendering it unable to deliver this metal efficiently to the urease maturation pathway.
DISCUSSION
In vitro and in vivo studies of the N-terminal insertion variant, L2*-HypA, lead to the conclusion that proper Ni-binding by HypA is critical for its function as a Ni-chaperone in the urease maturation pathway. Proper Ni-binding requires the unmodified MHE-motif at the N-terminus of HypA (likely by coordinating the N-terminal amine). Deficiency in urease activity and acid viability of the L2*-hypA mutant strain of H. pylori does not appear to result from interruption of protein-protein interactions with the urease maturation pathway, as the apparent affinities of WT-HypA and L2*-HypA binding to WT-UreE dimer were found to be similar (vide supra). Instead, the apparent affinity of L2*-HypA for Ni(II) ions was found to be reduced by at least 20-fold compared to WT-HypA (vide supra). This “loss of Ni-binding affinity” mechanism to account for the observed loss of function in urease maturation is corroborated by the structural changes found for the Ni-binding site in L2*-HypA (vide infra).
The identification of the proper Ni-binding site in HypA has been controversial. From multiple sequence analysis to mutagenesis studies, it is clear that Ni binds to HypA at the N-terminal MHE-motif, involving His2 as one of the ligands.15, 16, 19 However, the geometry of the Ni-binding site and the identity of the other Ni ligands from this Ni metallochaperone were not clear. One report identified the unmodified WT-HypA as having a 6-coordinate all N/O site involving 1 – 2 imidazole ligands.18, 24 Another group, working with a N-terminal extended HypA but containing an intact MHE sequence (resulting from a 2-residue cleavage overhang of N-terminal affinity tag) proposed a 4-coordinate planar site involving the His2 side-chain and three additional backbone amides from His2, Glu3, and Asp40.19 In the studies presented here, the unmodified WT-HypA protein is consistent with the 6-coordinate site with one His imidazole and where the scattering attributed to a putative second histidine is now attributed to chelate ring formation involving backbone amide coordination. The N-terminal extension variant protein, L2*-HypA, bound Ni at the same locus as WT-HypA, but having lost two N/O-donor protein ligands it adopts a pyramidal five-coordinate geometry with the additional ligand being a Br− contributed from the buffer system. The direct comparison between the Ni sites in unmodified WT-HypA and the variant protein with an N-terminal extension (L2*-HypA) demonstrates that the N-terminal amine of HypA is clearly involved in Ni binding.
The L2*-HypA Ni site structure described here is also consistent with the data from the N-terminally modified HypA protein (PDB: 2KDX), which was proposed to have a planar four-coordinate Ni-site based on the fact that it is diamagnetic (lack of paramagnetic line-broadening) and on chemical shift perturbations observed by 1H15N-HSQC experiments upon Ni(II) titration into a sample that identified four ligands.19 In the case of the L2*-HypA Ni(II) complex, the same set of ligands that were proposed in the 2KDX structure could be used to describe the best-fit for the Ni-site with the addition of a Br− in the axial position from the buffer system. While the NMR experiment clearly establishes that the Ni(II) site is diamagnetic in the N-terminally modified protein, it does not establish a four-coordinate planar geometry because the technique is blind to non-protein ligands and, despite the 5-coordinate pyramidal geometry, Ni(II) binding to the L2*-HypA protein variant results in a diamagnetic low-spin site. In determining the ligands involved in Ni binding, Xia et al. would not have been able to determine the involvement of non-protein ligands in coordinating the Ni site using NMR and therefore concluded that the N-terminally modified HypA Ni site is planar in geometry.19 Xia et al. also concluded that all other conserved Glu, Asp, and His residues are quite far away from the N-terminus of the protein in the 2KDX structure and therefore are unlikely to be involved in formation of the 6-coordinate Ni site in WT-HypA.19 However, the proximity of Glu3 and Asp40 was established in their experiments through backbone amide binding in forming the planar Ni site.19 Having established the N-terminal amine as one of the ligands in WT-HypA, in addition to the His2 side chain and backbone amide, the missing three ligands are proposed to be the sidechains of Glu3 and Asp40 as well as an additional backbone amide residue (most likely from Glu3). The binding of Glu3 and Asp40 sidechains and neutral amides would result in a charge neutral complex. The use of the N-terminal amine, and both the sidechain and backbone amides of His2 and Glu3 would firmly establish the rigidly conserved MHE-motif for Ni binding in HypA.
One crystallographically characterized example of the HypA Ni-binding site is available from the T. kodakarensis KOD1 (PDB ID: 5AUN and 5AUO) in a complex with the ATP-binding form of HypB from the same organism in two distinct nucleotide-bound forms (ATP and ADP analogs).21 Each Ni-site features HypA in planar four-coordinate geometry with ligands comprised of the N-terminal amine and backbone amide of His2 and the His2 and His98 sidechains (His79 in H. pylori HypA), the latter of which is pushed into the Ni-binding site by HypB binding close to the HypA Zn-site. Since the H. pylori HypA Zn-binding region is much more compact compared to the T. kodakarensis site, it is unlikely that the His residues in the Zn-site would ever be able to coordinate the distant Ni site.
The planar, and therefore diamagnetic, T. kodakarensis HypA Ni-site resembles other classic N-terminal Ni-binding motifs. Use of the N-terminal amine as a metal ligand is unusual in metalloproteins, but more common in Ni and Cu proteins. Coordination of Ni(II) in the N-terminus of proteins and peptides has been most extensively studied in the classic amino terminal Cu(II) and Ni(II) (ATCUN)-motifs. The ATCUN-motif was first described in albumin, which is amongst the major transporters of Cu(II) and Ni(II) in serum.39 The motif has since been described in several other peptides such as human protamine 2, histatin 5, heuromedin C, and hepcidin, of which the physiological evidence of Cu(II) and Ni(II) binding has yet to be confirmed.39, 40 Additionally, small molecule mimics and protein/peptide engineering studies have shown that ATCUN-motifs are associated with DNA-cleavage activities.39, 41, 42 Despite the high stability and tight binding of the ATCUN metal site (Kd ~1 pM for Cu(II) and 150 nM for Ni(II)), the ATCUN metal site structure has yet to be crystallized in albumin and is presumed to contain multiple conformations even in crystals.40 Instead, most of the structural information on ATCUN motifs has been derived from small molecule and oligopeptide mimics,40–43 although a high-resolution crystal structure of the Ni-bound ATCUN motif at the N-terminus of a protein is available in SH3 mutant domain of c-SRC histidine kinase from G. gallus (PDB ID: 4OMO).44 The classic ATCUN-motif has a strictly conserved His-residue in the third position and binds metals using the N-terminal amine, backbone amide N-donors from the second and third amino acid, and the His-imidazole sidechain, forming two chelate rings, one 5-membered and one 6-membered, with the metal in a distorted four-coordinate planar geometry.39, 40, 42 This stable planar structure excludes sidechain coordination from the first two amino acids of the motif.40
Despite binding Ni(II) at the N-terminus, the WT-HypA Ni-site structure is distinct from the classic ATCUN-motif in several ways. Like the ATCUN-motif, Ni(II) binds to HypA with the N-terminal three-residue motif (MHE-) through the N-terminal amine and likely the backbone amides of His2 and Glu3 as well as the imidazole sidechain of His2. However, Ni-coordination to the MHE-motif forces one of these N/O-donor ligands into an axial position, instead of favoring the rigid planar structure found in the classic Ni-ATCUN site. In five-coordinate Ni(II) complexes, the relative ligand field of the ligands in the basal plane vs. the axial ligand determines the spin-state. The axial coordination of an N/O-donor, such as His imidazole, may favor a high-spin configuration, which also favors a 6-coordinate Ni(II) site, instead of the planar four-coordinate low-spin Ni-ATCUN structure. Like the ATCUN-motif, the HypA MHE-motif is associated with known Ni-transporter activity, although with a much more modest Kd (~ 1 μM, vide supra), which allows for more appropriate Ni metallochaperone activity inside the cell. With the L2*-HypA variant, the conserved His2 residue is shifted to the third position (MLHE-) to more closely resemble the classic ATCUN motif.
Despite having a more ATCUN-like metal binding motif, the L2*-HypA variant did not exhibit the tight Ni(II) binding that is associated with the ATCUN complexes like albumin,40 although albumin Ni-binding has not been measured under similar conditions. The reason for the decreased affinity is not clear; one possibility lies with the difference in protein structure. In HypA proteins, the N-terminus is associated with a helical structure that would be perturbed by Ni binding, effectively lowering the affinity relative to the unstructured N-termini in albumin structures.40 Another possibility is that the protonation state of the amide N-donors is important. If one or more amides is left protonated in the L2*HypA structure, this would decrease Ni affinity and enhance the binding of an additional anion, such as Br− or Cl−. In any case, the L2*-HypA Ni-site is low-spin, like the Ni-ATCUN site, indicating that it can adopt a similar planar coordination of the four protein ligands. Addition of a Br− ligand, a weak field ligand, leads to a pyramidal geometry, but retains the low-spin configuration.
In addition to the ATCUN-motif, there are several examples of proteins that bind Ni(II) at the N-terminus, specifically coordinating the N-terminal amine. One such example is the N-terminal high affinity site in E. coli HypB (absent in H. pylori HypB), which binds Ni(II) through a CXXCGC motif at the N-terminus, coordinating the N-terminal amine and the three S-Cys in a planar geometry.45 Another such example is S. coelicolor nickel superoxide dismutase (NiSOD), which binds Ni(II) through the N-terminal amine of His1, the backbone amide of Cys2, and the sidechains of Cys2 and Cys6, with the sidechain imidazole of His1 also bound in the Ni(III) state.46–48 Both the E. coli HypB N-terminal motif and NiSOD Ni binding sites have been referred to as “ATCUN-like” in literature due to the binding of the N-terminal amine and planar geometry.35, 45, 46 Another example of Ni-binding at the N-terminus can be found in E. coli RcnR, a Ni(II)-/Co(II)-responsive transcriptional regulator, which binds Ni(II) with the N-terminal amine of Ser2 (Met1 is cleaved in E. coli) in a 6-coordinate site, where there is no evidence of backbone amide binding thus far.49 Thus, Ni(II) binding at the N-terminal amine has been found in proteins with vastly different functions and metal coordination: metal transporters/chaperones (e.g., albumin (planar)40 and HypA (6-coordinate)), both redox-active and inactive enzymes (e.g., NiSOD (planar/pyramidal)46 and EcHypB N-terminal Ni-motif (planar)45), and a metal-responsive transcriptional regulator (RcnR (6-coordinate)49), demonstrating the N-terminal amine is a common and important ligand for Ni(II)-binding in biology overall.
One mechanism for controlling cellular urease activity in H. pylori is through modulation of urease maturation via nickel insertion into apo-urease.12, 50, 51 Apo-urease is produced in large quantities (10% of all synthesized protein9) but remains largely in the apo form.12, 14 Increases in enzyme activity are correlated with Ni available in the media.12, 50, 51 Beyond Ni availability, acid conditions also strongly enhance urease activity, but urease transcription is only marginally increased.52, 53 Thus, nickelation of the apo-protein is employed to boost urease activity under acidic conditions.12 With respect to the accessory proteins involved in Ni incorporation, the roles of the hydrogenase proteins HypAB and the urease protein UreE are quite distinct. Both the Ni-binding deficient hypA H2A mutant15, 16 and the nucleotide-binding deficient hypB K59A mutant54 were reported to lack urease activity, but this defect could be complemented by Ni supplementation in the media, consistent with their roles in Ni(II) transport and delivery, and implying that under high Ni concentration some Ni is available to UreE. In contrast, Ni supplementation could not rescue the urease-deficient phenotype in either the ΔureE mutant16 or the ureG K14A mutant (nucleotide-binding deficient UreG).54 Hence urease maturation specifically requires the urease cascade, but not HypAB when excess Ni is present.
With Ni-binding to WT-HypA having only a modest Ni-binding affinity at (Kd ~1 μM) (vide supra) and H. pylori having a full cascade of urease maturation accessory proteins UreEFGH,55 why would the full maturation of urease require the hydrogenase maturation accessory proteins HypA and HypB? Specifically, why is HypA Ni-binding necessary for Ni transport and delivery to the urease maturation pathway? One possibility is that HypAB provide an additional pathway for Ni incorporation that is triggered under acidic conditions that supplements a low basal level provided by H. pylori UreE alone. In fact, H. pylori appears to restrict Ni incorporation into urease by UreEFGH by lowering in the Ni-binding properties of UreE. The poor Ni-sequestration ability of H. pylori UreE compared to homologues (such as K. aerogenes UreE, which has a poly-His tail) has been studied.56 Incorporation of a poly-His tail into H. pylori UreE has been shown to increase the Ni-sequestration ability and the urease activity in the absence of HypA or HypB proteins.56 Under acidic conditions, HypAB could provide a higher level of Ni incorporation into UreE by 1) increasing the local concentration of Ni by formation of a complex with HypAB, or 2) by simply enhancing the affinity of UreE for Ni in a complex with HypAB. The former mechanism is suggested by the known HypA-UreE complex formation,16, 57 and by protein structural changes that accompany changes in the Zn site structure at pH 6.3.18, 24 The latter mechanism is supported by the increase in Ni affinity that occurs in the HypAB complex in T. kodakaraensis.21
These prior studies along with our own study point to the role of the hydrogenase maturation accessory proteins HypA and HypB in the acquisition and delivery of Ni to the urease maturation pathway under non-abundant Ni conditions (without Ni supplementation). Instead of enhancing the Ni-sequestration ability of the urease cascade, HypA and HypB are recruited from the hydrogenase maturation pathway to sequester Ni for the urease maturation pathway; in doing so they assert a higher level of control over Ni availability to the urease maturation pathway. Due to the high level of Apo-urease synthesized under both acidic and neutral conditions9, the higher level of Ni control could hypothetically be employed to finely tune the urease activity of H. pylori to prevent an alkaline internal pH or depletion of urea in the cytoplasm under neutral conditions. However, this fine-tuning mechanism would impose acid sensitivity to the strains where the ability of HypA to bind Ni is impaired as is the case in the H2A hypA strain15, 16 and the L2* hypA strain.
Supplementary Material
Tables S1 – S3: supporting fitting tables for Ni K-edge EXAFS
Table S4: Zn K-edge EXAFS
Table S5: a list of H. pylori strains, plasmids and primers referenced in this work
Figure S1: amino acid sequence alignments for HypA proteins
Figure S2: CD spectra and SEC-MALS results for WT- and L2*-HypA proteins
Figure S3: overlay of Ni K-edge XANES for L2*-HypA complexes in NaBr and NaCl buffer
Figure S4: a pictorial representation of multiple scattering scaffolds for used in EXAFS fits
Figure S5: data and best EXAFS fits for L2*-HypA samples in NaCl buffer
Figure S6: Zn K-edge EXAFS data best fits for WT- and L2*-HypA
Figure S7: an example of SDS-PAGE of proteins after ITC experiments involving UreE
Figure S8: changes in UV-Vis absorption and titration curve upon Ni-titration into HypA
Acknowledgments
XAS data collection at the National Synchrotron Light Source at Brookhaven National Laboratory was supported by the U.S. Department of Energy, Division of Materials Science and Division of Chemical Sciences. Beamline X3B at NSLS was supported by the NIH Grant P30-EB-009998 from the National Institute of Biomedical Imaging and Bioengineering. Portions of this research were conducted at the Stanford Synchrotron Radiation Light (SSRL) Source, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structure Molecular Biology Program is supported by the Department of Energy, Office of Biological Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program.
ITC experiments were performed at the Biophysical Characterization Core Facility, part of the Institute for Applied Life Sciences (IALS) at Univ. Massachusetts Amherst. The authors would like to thank Hsin-Ting Huang for performing mass spectrometry to confirm exact protein masses using the UMass Institute for Applied Life Sciences mass spectrometry facility.
Funding Sources:
This work was supported by the National Institute of Health Grants R01-GM69696 to M.J.M., and R56-AI065529 to D.S.M.
ABBREVIATIONS
- XAS
X-ray Absorption Spectroscopy
- XANES
X-ray absorption near-edge structure
- EXAFS
extended X-ray absorption fine structure
- ITC
isothermal titration calorimetry
- ICP-OES
inductively coupled Plasma Optical Emission Spectroscopy
Footnotes
Supporting Information (SI) is available free of charge on the ACS publication website DOI #: Experimental section includes detailed descriptions of all procedures
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Marshall BJ, Warren JR, Goodwin CS. Duodenal ulcer relapse after eradication of Campylobacter pylori. Lancet. 1989;1:836–837. doi: 10.1016/s0140-6736(89)92287-3. [DOI] [PubMed] [Google Scholar]
- 3.Eusebi LH, Zagari RM, Bazzoli F. Epidemiology of Helicobacter pylori infection. Helicobacter. 2014;19(Suppl 1):1–5. doi: 10.1111/hel.12165. [DOI] [PubMed] [Google Scholar]
- 4.Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. International journal of cancer. Journal international du cancer. 2015;136:487–490. doi: 10.1002/ijc.28999. [DOI] [PubMed] [Google Scholar]
- 5.Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential Role of Urease in Pathogenesis of Gastritis Induced by Helicobacter-Pylori in Gnotobiotic Piglets. Infection and Immunity. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton KA, Krakowka S. Effect of Gastric Ph on Urease-Dependent Colonization of Gnotobiotic Piglets by Helicobacter-Pylori. Infection and Immunity. 1994;62:3604–3607. doi: 10.1128/iai.62.9.3604-3607.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clinical microbiology reviews. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastro Hepat. 2010;7:629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauerfeind P, Garner R, Dunn BE, Mobley HLT. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: The link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 11.Sachs G, Weeks DL, Wen Y, Marcus EA, Scott DR, Melchers K. Acid acclimation by Helicobacter pylori. Physiology. 2005;20:429–438. doi: 10.1152/physiol.00032.2005. [DOI] [PubMed] [Google Scholar]
- 12.Stingl K, De Reuse H. Staying alive overdosed: how does Helicobacter pylori control urease activity? International journal of medical microbiology: IJMM. 2005;295:307–315. doi: 10.1016/j.ijmm.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Farrugia MA, Macomber L, Hausinger RP. Biosynthesis of the Urease Metallocenter. Journal of Biological Chemistry. 2013;288:13178–13185. doi: 10.1074/jbc.R112.446526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori (vol 39, pg 176, 2001) Molecular microbiology. 2001;40:270–270. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 15.Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori Nickel Metabolism Accessory Proteins Needed for Maturation of both Urease and Hydrogenase. Journal of bacteriology. 2003;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benoit SL, Mehta N, Weinberg MV, Maier C, Maier RJ. Interaction between the Helicobacter pylori accessory proteins HypA and UreE is needed for urease maturation. Microbiology. 2007;153:1474–1482. doi: 10.1099/mic.0.2006/003228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirel PH, Schmitter JM, Dessen P, Fayat G, Blanquet S. Extent of N-Terminal Methionine Excision from Escherichia-Coli Proteins Is Governed by the Side-Chain Length of the Penultimate Amino-Acid. P Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kennedy DC, Herbst RW, Iwig JS, Chivers PT, Maroney MJ. A dynamic Zn site in Helicobacter pylori HypA: A potential mechanism for metal-specific protein activity. J Am Chem Soc. 2007;129:16–17. doi: 10.1021/ja066958x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia W, Li HY, Sze KH, Sun HZ. Structure of a Nickel Chaperone, HypA, from Helicobacter pylori Reveals Two Distinct Metal Binding Sites. J Am Chem Soc. 2009;131:10031–10040. doi: 10.1021/ja900543y. [DOI] [PubMed] [Google Scholar]
- 20.Herbst RW. Department of Chemistry. University of Massachusetts Amherst; UMASS Amherst: 2010. Structure and Function in a Nickel Metallochaperone, HypA and Nickel Dependent Superoxide Dismutase; p. 184. [Google Scholar]
- 21.Watanabe S, Kawashima T, Nishitani Y, Kanai T, Wada T, Inaba K, Atomi H, Imanaka T, Miki K. Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc Natl Acad Sci U S A. 2015;112:7701–7706. doi: 10.1073/pnas.1503102112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe S, Arai T, Matsumi R, Atomi H, Imanaka T, Miki K. Crystal structure of HypA, a nickel-binding metallochaperone for [NiFe] hydrogenase maturation. Journal of molecular biology. 2009;394:448–459. doi: 10.1016/j.jmb.2009.09.030. [DOI] [PubMed] [Google Scholar]
- 23.Andersson M, Wittgren B, Wahlund KG. Accuracy in multiangle light scattering measurements for molar mass and radius estimations. Model calculations and experiments. Anal Chem. 2003;75:4279–4291. doi: 10.1021/ac030128+. [DOI] [PubMed] [Google Scholar]
- 24.Herbst RW, Perovic I, Martin-Diaconescu V, O’Brien K, Chivers PT, Pochapsky SS, Pochapsky TC, Maroney MJ. Communication between the Zinc and Nickel Sites in Dimeric HypA: Metal Recognition and pH Sensing. J Am Chem Soc. 2010;132:10338–10351. doi: 10.1021/ja1005724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colpas GJ, Maroney MJ, Bagyinka C, Kumar M, Willis WS, Suib SL, Baidya N, Mascharak PK. X-Ray Spectroscopic Studies of Nickel-Complexes, with Application to the Structure of Nickel Sites in Hydrogenases. Inorg Chem. 1991;30:920–928. [Google Scholar]
- 26.Lewis J, Wilkins RG. Modern coordination chemistry: principles and methods [Google Scholar]
- 27.Evans DF. The Determination of the Paramagnetic Susceptibility of Substances in Solution by Nuclear Magnetic Resonance. J Chem Soc. 1959:2003–2005. [Google Scholar]
- 28.Schubert EM. Utilizing the Evans Method with a Superconducting Nmr Spectrometer in the Undergraduate Laboratory. J Chem Educ. 1992;69:62–62. [Google Scholar]
- 29.Yano J, Yachandra VK. X-ray absorption spectroscopy. Photosynthesis research. 2009;102:241–254. doi: 10.1007/s11120-009-9473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banaszak K, Martin-Diaconescu V, Bellucci M, Zambelli B, Rypniewski W, Maroney MJ, Ciurli S. Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni(2)(+) and Zn(2)(+) reveals a role for the disordered C-terminal arm in metal trafficking. The Biochemical journal. 2012;441:1017–1026. doi: 10.1042/BJ20111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Diaconescu V, Bellucci M, Musiani F, Ciurli S, Maroney MJ. Unraveling the Helicobacter pylori UreG zinc binding site using X-ray absorption spectroscopy (XAS) and structural modeling. Journal of biological inorganic chemistry: JBIC: a publication of the Society of Biological Inorganic Chemistry. 2012;17:353–361. doi: 10.1007/s00775-011-0857-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giri NC, Sun H, Chen H, Costa M, Maroney MJ. X-ray absorption spectroscopy structural investigation of early intermediates in the mechanism of DNA repair by human ABH2. Biochemistry. 2011;50:5067–5076. doi: 10.1021/bi101668x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giri NC. Chemistry. University of Massachusetts Amherst; UMASS Amherst: 2013. Structural Investigations of Early Intermediates and Nickel Inhibition Complexes of Human DNA and Histone Demethylases; p. 343. [Google Scholar]
- 34.Ryan KC. Chemistry. University of Massachusetts Amherst; UMASS Amherst: 2013. Investigation of the Structure/Function Relationship in Nickel Containing Superoxide Dismutase; p. 259. [Google Scholar]
- 35.Campecino JO, Dudycz LW, Tumelty D, Berg V, Cabelli DE, Maroney MJ. A Semisynthetic Strategy Leads to Alteration of the Backbone Amidate Ligand in the NiSOD Active Site. J Am Chem Soc. 2015;137:9044–9052. doi: 10.1021/jacs.5b03629. [DOI] [PubMed] [Google Scholar]
- 36.Johnson RC, Hu HQ, Merrell DS, Maroney MJ. Dynamic HypA zinc site is essential for acid viability and proper urease maturation in Helicobacter pylori. Metallomics: integrated biometal science. 2015;7:674–682. doi: 10.1039/c4mt00306c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellucci M, Zambelli B, Musiani F, Turano P, Ciurli S. Helicobacter pylori UreE, a urease accessory protein: specific Ni(2+)- and Zn(2+)-binding properties and interaction with its cognate UreG. The Biochemical journal. 2009;422:91–100. doi: 10.1042/BJ20090434. [DOI] [PubMed] [Google Scholar]
- 38.Shi R, Munger C, Asinas A, Benoit SL, Miller E, Matte A, Maier RJ, Cygler M. Crystal structures of apo and metal-bound forms of the UreE protein from Helicobacter pylori: role of multiple metal binding sites. Biochemistry. 2010;49:7080–7088. doi: 10.1021/bi100372h. [DOI] [PubMed] [Google Scholar]
- 39.Harford C, Sarkar B. Amino terminal Cu(II)- and Ni(II)-binding (ATCUN) motif of proteins and peptides: Metal binding, DNA cleavage, and other properties. Accounts Chem Res. 1997;30:123–130. [Google Scholar]
- 40.Bal W, Sokolowska M, Kurowska E, Faller P. Binding of transition metal ions to albumin: Sites, affinities and rates. Bba-Gen Subjects. 2013;1830:5444–5455. doi: 10.1016/j.bbagen.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 41.Neupane KP, Aldous AR, Kritzer JA. Macrocyclization of the ATCUN motif controls metal binding and catalysis. Abstr Pap Am Chem S. 2014;247 doi: 10.1021/ic302820z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sankararamakrishnan R, Verma S, Kumar S. ATCUN-like metal-binding motifs in proteins: Identification and characterization by crystal structure and sequence analysis. Proteins. 2005;58:211–221. doi: 10.1002/prot.20265. [DOI] [PubMed] [Google Scholar]
- 43.Kritzer JA, Neupane KP, Aldous AR. Design of Macrocyclic ATCUN Peptides as Redox Catalysts. Biopolymers. 2013;100:281–281. [Google Scholar]
- 44.Bacarizo J, Martinez-Rodriguez S, Martin-Garcia JM, Andujar-Sanchez M, Ortiz-Salmeron E, Neira JL, Camara-Artigas A. Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation. PloS one. 2014;9 doi: 10.1371/journal.pone.0113224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung KCC, Cao L, Dias AV, Pickering IJ, George GN, Zamble DB. A High-Affinity Metal-Binding Peptide from Escherichia coli HypB. J Am Chem Soc. 2008;130:14056–+. doi: 10.1021/ja8055003. [DOI] [PubMed] [Google Scholar]
- 46.Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED. Nickel superoxide dismutase structure and mechanism. Biochemistry. 2004;43:8038–8047. doi: 10.1021/bi0496081. [DOI] [PubMed] [Google Scholar]
- 47.Choudhury SB, Lee JW, Davidson G, Yim YI, Bose K, Sharma ML, Kang SO, Cabelli DE, Maroney MJ. Examination of the nickel site structure and reaction mechanism in Streptomyces seoulensis superoxide dismutase. Biochemistry. 1999;38:3744–3752. doi: 10.1021/bi982537j. [DOI] [PubMed] [Google Scholar]
- 48.Wuerges J, Lee JW, Yim YI, Yim HS, Kang SO, Djinovic Carugo K. Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc Natl Acad Sci U S A. 2004;101:8569–8574. doi: 10.1073/pnas.0308514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higgins KA, Chivers PT, Maroney MJ. Role of the N-terminus in determining metal-specific responses in the E. coli Ni- and Co-responsive metalloregulator, RcnR. J Am Chem Soc. 2012;134:7081–7093. doi: 10.1021/ja300834b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Vliet AH, Kuipers EJ, Waidner B, Davies BJ, de Vries N, Penn CW, Vandenbroucke-Grauls CM, Kist M, Bereswill S, Kusters JG. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect Immun. 2001;69:4891–4897. doi: 10.1128/IAI.69.8.4891-4897.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Contreras M, Thiberge JM, Mandrand-Berthelot MA, Labigne A. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Molecular microbiology. 2003;49:947–963. doi: 10.1046/j.1365-2958.2003.03621.x. [DOI] [PubMed] [Google Scholar]
- 52.Bury-Mone S, Skouloubris S, Labigne A, De Reuse H. The Helicobacter pylori UreI protein: role in adaptation to acidity and identification of residues essential for its activity and for acid activation. Molecular microbiology. 2001;42:1021–1034. doi: 10.1046/j.1365-2958.2001.02689.x. [DOI] [PubMed] [Google Scholar]
- 53.Scott DR, Marcus EA, Weeks DL, Sachs G. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology. 2002;123:187–195. doi: 10.1053/gast.2002.34218. [DOI] [PubMed] [Google Scholar]
- 54.Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microbial Pathogenesis. 2003;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- 55.Carter EL, Flugga N, Boer JL, Mulrooney SB, Hausinger RP. Interplay of metal ions and urease. Metallomics: integrated biometal science. 2009;1:207–221. doi: 10.1039/b903311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Benoit S, Maier RJ. Dependence of Helicobacter pylori Urease Activity on the Nickel-Sequestering Ability of the UreE Accessory Protein. Journal of bacteriology. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benoit SL, McMurry JL, Hill SA, Maier RJ. Helicobacter pylori hydrogenase accessory protein HypA and urease accessory protein UreG compete with each other for UreE recognition. Biochimica et biophysica acta. 2012;1820:1519–1525. doi: 10.1016/j.bbagen.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 – S3: supporting fitting tables for Ni K-edge EXAFS
Table S4: Zn K-edge EXAFS
Table S5: a list of H. pylori strains, plasmids and primers referenced in this work
Figure S1: amino acid sequence alignments for HypA proteins
Figure S2: CD spectra and SEC-MALS results for WT- and L2*-HypA proteins
Figure S3: overlay of Ni K-edge XANES for L2*-HypA complexes in NaBr and NaCl buffer
Figure S4: a pictorial representation of multiple scattering scaffolds for used in EXAFS fits
Figure S5: data and best EXAFS fits for L2*-HypA samples in NaCl buffer
Figure S6: Zn K-edge EXAFS data best fits for WT- and L2*-HypA
Figure S7: an example of SDS-PAGE of proteins after ITC experiments involving UreE
Figure S8: changes in UV-Vis absorption and titration curve upon Ni-titration into HypA


