Abstract
We describe the design, construction, and application of an instrument combining dual-trap, high-resolution optical tweezers and a confocal microscope. This hybrid instrument allows nanomechanical manipulation and measurement simultaneously with single-molecule fluorescence detection. We present the general design principles that overcome the challenges of maximizing optical trap resolution while maintaining single-molecule fluorescence sensitivity, and provide details on the construction and alignment of the instrument. This powerful new tool is just beginning to be applied to biological problems. We present step-by-step instructions on an application of this technique that highlights the instrument’s capabilities, detecting conformational dynamics in a nucleic acid-processing enzyme.
1. INTRODUCTION
Single-molecule techniques have become powerful tools to study fundamental biological processes. Two broadly defined categories are force-based manipulation and detection techniques (e.g., optical tweezers, magnetic tweezers, AFM, nanopores) and single-molecule fluorescence imaging and spectroscopy. In recent years, a new generation of tools combining both categories has emerged. For example, new hybrid instruments combining optical trapping with single-molecule fluorescence (Bianco et al., 2001; Heller et al., 2013; Hohng et al., 2007; Lang, Fordyce, Engh, Neuman, & Block, 2004; Lee, Balci, Jia, Lohman, & Ha, 2013; van Mameren et al., 2006) have allowed new avenues of investigation, making possible measurement of multiple biomolecular parameters simultaneously. In this chapter, we describe an instrument combining dual-trap optical tweezers with a confocal microscope (Figs. 1 and 2) (Comstock, Ha, & Chemla, 2011). This instrument has the ability to resolve mechanical signals at subnanometer spatial resolution (with the optical traps) and to detect simultaneously the emitted light from a single fluorophore (with the confocal microscope). Applications of this method have just begun to emerge (Comstock et al., 2015; Suksombat, Khafizov, Kozlov, Lohman, & Chemla, 2015), with new results on conformational dynamics of nucleoprotein complexes detected with optical traps and single-molecule Förster Resonance Energy Transfer (smFRET). Below, we provide a general overview of optical traps and single-molecule fluorescence, the challenges in combining them, the design principles of our instrument, and its alignment procedures. We end with protocols for replicating a recently reported experiment on the DNA helicase UvrD and the relationship between its conformational state and unwinding activity enabled by this instrument (Comstock et al., 2015).
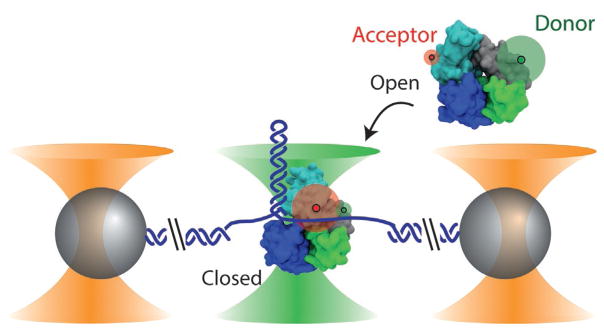
Fig. 1.
Combined high-resolution optical tweezers and confocal microscope. Dual optical traps (outer cones) hold polystyrene microspheres (spheres) tethered by a DNA construct (here a DNA hairpin), while a confocal microscope (middle cone) detects fluorescence from a single molecule. In this example, the conformational and unwinding dynamics of E. coli UvrD helicase are investigated. UvrD helicase exists in two conformational states—“open” (shown in the free protein) and “closed” (shown in the bound protein)—that are differentiated by smFRET between a donor–acceptor pair labeling the protein (green and red disks, respectively). The proteins in this figure were prepared with VMD (Humphrey, Dalke, & Schulten, 1996) from PDB entries 2IS2 and 3LFU. Figure reproduced from Comstock, M. J., Whitley, K. D., Jia, H., Sokoloski, J., Lohman, T. M., Ha, T., & Chemla, Y. R. (2015). Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science, 348(6232), 352–354 with permission from AAAS.
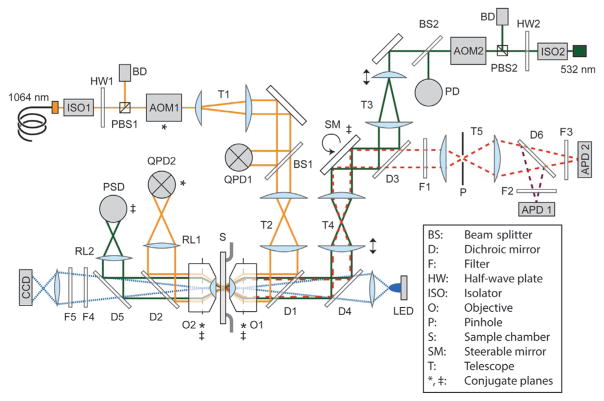
Fig. 2.
Detailed layout of the instrument (not to scale). The instrument consists of three modules: Optical trap (yellow solid lines), confocal excitation (green solid lines) and emission (red dashed lines), and bright-field imaging (blue dotted lines). The asterisk (*) denotes planes conjugate to AOM1, the double cross (‡) those conjugate to the steerable mirror (SM). Arrows indicate adjustable translational or rotational stages. Dotted lines indicate the back-focal planes of the objectives. Refer to text for details.
2. OPTICAL TRAPPING AND SINGLE-MOLECULE FLUORESCENCE
2.1 Principles of Optical Trapping
Optical tweezers utilize the momentum carried by light to exert forces on microscopic objects. An infrared (IR) laser tightly focused to a diffraction-limited spot by a high-numerical aperture (NA) microscope objective generates optical forces that can trap a dielectric object—such as a μm-sized polystyrene or glass bead—stably in three dimensions (Ashkin, 1986). Near the focus of light, the optical trap behaves as a linear spring, exerting a force on the trapped object proportional to its displacement. This displacement is typically detected by back-focal-plane interferometry (Gittes & Schmidt, 1998), in which the interference pattern between the incident light and that forward-scattered by the trapped object is imaged onto a position-sensitive photodetector. With proper calibration of the instrument, this signal can be converted into a displacement in nanometers and a force in piconewtons.
The sensitivity of optical tweezers has made them a powerful tool to investigate biomolecules at the single-molecule level. By tethering molecules to beads held in traps and applying force, optical tweezers have provided new insights on mechanical, structural, and dynamic properties of biomolecules (Bustamante, Bryant, & Smith, 2003; Heller, Hoekstra, King, Peterman, & Wuite, 2014; Ritchie & Woodside, 2015). They have also been well suited to studying the mechanisms of molecular motors involved in a range of functions—cytoskeletal transport, the central dogma, and beyond (reviewed in Bustamante, Cheng, & Mejia, 2011; Heller et al., 2014; Veigel & Schmidt, 2011). Nucleic acid-processing motors in particular are studied by monitoring the extension of the DNA or RNA molecules tethered by the trapped beads (for example, Fig. 1). These molecular tethers often serve an additional role to position the systems of interest away from the high light intensity of the optical traps.
Advances in instrument design over the last dozen years have increased optical tweezers sensitivity remarkably. Instruments with active stage stabilization (Carter et al., 2007) and others incorporating dual traps formed from the same laser (Abbondanzieri, Greenleaf, Shaevitz, Landick, & Block, 2005; Moffitt, Chemla, Izhaky, & Bustamante, 2006) have improved instrument stability to such an extent that it is now possible to detect subnanometer signals. These so-called high-resolution optical tweezers have enabled investigations of nucleic acid-processing molecular motors in extreme detail (Abbondanzieri et al., 2005; Cheng, Arunajadai, Moffitt, Tinoco, & Bustamante, 2011; Moffitt et al., 2009; Qi, Pugh, Spies, & Chemla, 2013).
2.2 Single-Molecule Fluorescence Detection
Another powerful technique to study biological processes relies on the detection of light emitted by individual fluorophores (Moerner, 2007). Labeling proteins, nucleic acids, or small molecules with a fluorophore—commonly, organic dyes such as Cy3, Cy5, and TMR—allows measuring their spatiotemporal dynamics at the single-molecule level. Single-molecule fluorescence detection can be incorporated into the standard types of optical microscopy—bright field, confocal, or total internal reflection (TIR)—requiring only an excitation light source at a wavelength tuned to the absorption spectrum of the fluorophore and a sensitive charged-coupled device (CCD) camera or avalanche photodiode (APD) to detect the emitted light efficiently.
Fluorescence provides several measurable quantities that can provide information on biomolecular conformational states and their dynamics: intensity, lifetime, position, and orientation. These can be used for tracking diffusion or directed motion of a labeled molecule, to detect the binding of labeled molecules to a system of interest, or to determine the stoichiometry or composition of multicomponent complexes (Joo, Balci, Ishitsuka, Buranachai, & Ha, 2008). A powerful tool for probing conformational dynamics of biomolecules is smFRET. Here, a “donor” molecule excited to a high-energy state transfers its energy to a neighboring “acceptor” molecule via induced-dipole interactions, which then emits light of a longer wavelength. The efficiency of energy transfer (or FRET efficiency), E, is strongly dependent on the distance R between the donor and acceptor, varying as E = 1/[1 + (R/R0)6], where the Förster radius R0 is the distance at which 50% of the energy is transferred (R0 = 60 Å for the Cy3-Cy5 pair). Thus, smFRET is a spectroscopic technique that measures the distance between a single donor–acceptor fluorescent dye pair (Ha, 2001; Ha et al., 1996) and is sensitive to conformational changes typically in the 30–80 Å distance range (Forster, 1965; Stryer & Haugland, 1967). This powerful technique has been used to study a wide variety of biological systems (Kim & Ha, 2013).
2.3 Combined Optical Tweezers/Single-Molecule Fluorescence Microscope
In recent years, a number of hybrid instruments combining optical tweezers with fluorescence detection have been developed. This work has been motivated by the desire to detect biomolecular dynamics with multiple orthogonal probes simultaneously. Optical tweezers have been combined with many standard forms of fluorescence microscopy—bright-field (Bianco et al., 2001; van Mameren et al., 2006), TIRF (Lang et al., 2004; Lee et al., 2013), confocal (Hohng et al., 2007), and even STED microscopy (Heller et al., 2013). Example applications of these new techniques include the tracking of singly labeled biomolecules on DNA stretched with optical traps, imaging of nucleoprotein complexes on extended DNA, and measurement of biomolecular conformational dynamics by smFRET as a function of force (reviewed in Chemla, 2016; Heller et al., 2014). The ability to measure and/or control multiple variables simultaneously has provided many new insights not available when using each method independently.
An important technical challenge to combining optical traps with single-molecule fluorescence detection is that many fluorophores photobleach very rapidly (~1–2 s) when located in the light field of the optical trap (van Dijk, Kapitein, van Mameren, Schmidt, & Peterman, 2004). This phenomenon results from a two-photon process in which a fluorophore absorbs a fluorescence excitation photon to its excited state followed by an IR optical trap photon to a higher energy level. Rather than decaying to the ground state and emitting light, the fluorophore instead decays to an ionized dark state. In the applications mentioned above, this problem was circumvented by imaging fluorescence far from the optical trap light field. This approach requires using long, compliant molecules to separate the trap(s) from the fluorescence imaging region spatially. Unfortunately, this reduces instrument resolution, which is proportional to the stiffness of the molecular tether (Moffitt et al., 2006). An alternative is to strobe the two light sources out of phase, separating them temporally (Brau, Tarsa, Ferrer, Lee, & Lang, 2006). As shown by Brau et al., interlacing trapping and fluorescence excitation laser significantly increases the length of time before fluorophores photobleach, allowing spatially overlapping light sources.
Recently, we integrated these advances to develop an instrument combining high-resolution optical traps with a single-molecule fluorescence microscope (Comstock et al., 2011). The sensitivity of this instrument allows detection of fluorescence signals with single-molecule sensitivity simultaneously with mechanical displacements at sub-nm resolution. This approach is starting to be adopted by other groups (Duesterberg, Fischer-Hwang, Perez, Hogan, & Block, 2015; Sirinakis, Ren, Gao, Xi, & Zhang, 2012) and has begun to provide new insights into biomolecular systems (Comstock et al., 2015; Suksombat et al., 2015). In the following sections, we describe the design principles and construction of this hybrid instrument. We also provide detailed protocols for its application, recapitulating measurements of conformational dynamics in a nucleic acid-processing enzyme (Fig. 1) (Comstock et al., 2015).
3. INSTRUMENT DESIGN
3.1 Overview
In this section we provide a general overview of the instrument design. Due to their exceptional stability, we utilize dual-trap optical tweezers in which both traps are formed from the same IR laser (Bustamante, Chemla, & Moffitt, 2008; Moffitt et al., 2006). In the dual-trap design, the traps are formed microns from the sample chamber surface. As a result, TIR microscopy, in which fluorescence excitation occurs only in the exponentially decaying evanescent field ~100 nm off the surface, is not well suited for this configuration. In our design, we use confocal microscopy instead, where we position the confocal excitation spot in the same plane as the two trapped beads (Fig. 1).
High-resolution measurements have so far been made using short (~1 μm), stiff tethers (e.g., Abbondanzieri et al., 2005; Cheng et al., 2011; Moffitt et al., 2009; Qi et al., 2013). In such a configuration, the optical traps and confocal excitation spot occupy the same volume and the light sources overlap significantly. To avoid fast photobleaching of fluorophores by absorption of IR and visible photons, we interlace the two light sources. Thus, the instrument incorporates and adapts design elements of previous setups: (1) dual-trap high-resolution optical tweezers (Moffitt et al., 2006), (2) combined optical trap/confocal microscope (Hohng et al., 2007), and (3) interlaced trap and fluorescence excitation light sources (Brau et al., 2006).
3.2 Interlacing and Time-Sharing
3.2.1 Interlacing With Acousto-Optic Modulators
A critical component of the instrument design is the interlacing of the trapping and fluorescence excitation light sources. As shown in previous work(Brau et al., 2006), the interlacing rate is an important consideration. If the rate is too low, the trap stiffness will be lower than that expected from the average trapping laser power (i.e., peak power × interlacing duty ratio) because the trapped bead moves beyond the trapping region when the beam is turned OFF for long durations. (A free 1-μm-diameter bead experiencing a 1 pN force will move by 100 nm in 1 ms.) Thus, it is critical to modulate the trapping beam at a sufficiently high frequency fm. A good practice is to operate above the characteristic frequency of the bead in the trap, fc =κ/2πγ, where κ is the stiffness and γ =6πηr is the Stokes drag of the bead (η is the viscosity and r is the bead radius). Typically fm needs to be >10 kHz.
These interlacing frequencies are feasible with acousto-optic devices. In an acousto-optic modulator (AOM), a radio frequency (RF) sound wave generated by a piezoelement propagates inside a crystal. An incident beam passing through this crystal is diffracted by the periodic modulation of the index of refraction generated by the sound wave. An AOM behaves essentially as a tunable diffraction grating, where the sound wave amplitude determines the intensity of the diffracted laser beam and its frequency the diffraction angle. The maximum frequency at which the AOM can modulate a laser beam is approximately given by the speed of sound in the crystal divided by the beam diameter. In our instrument, a 1.5-mm beam waist corresponds to ~2 MHz, and we interlace at a frequency of fm = 66 kHz, wellabove fc. Two separate AOMs modulate the trapping (Fig. 2, AOM1; IntraAction, ATM-803DA6B) and fluorescence excitation beams (AOM2; IntraAction, AOM-802AF1) out of phase. This requires synchronizing the interlacing cycles of both AOMs with μs-level precision, achieved through a data acquisition and instrument control architecture that we discuss below (see Section 3.4).
The RF source that drives the AOM sound wave is an important component of the setup, as precise and stable RF generation is critical for stable trap positioning. We chose to build a custom RF synthesizer rather than use an integrated commercial source, as the former exhibits significantly lower noise (Comstock et al., 2011). This RF source consists of a temperature-compensated crystal oscillator (TCXO; Conner-Winfield, HTFL5FG5-049.152M) mounted on a direct digital synthesis RF synthesizer board (Analog Devices, AD9852/PCBZ or AD9854/PCBZ). The TCXO provides a 49.152 MHz fixed output reference frequency with 1 ppm stability. The stability of this clock frequency has a direct effect on the stability of the RF output frequency and the stability of the trap positions. We configure the RF synthesizer board to multiply the clock frequency 6 × to a final value of ~300 MHz. Finally, a low-noise, fixed-gain RF power amplifier (Mini-Circuits, ZHL-5 W-1) amplifies the synthesizer output signal to drive the AOM. A commercial RF source (IntraAction, ME-801.5–6) is used to drive the AOM modulating the fluorescence excitation, since an ultra-stable RF frequency is not as critical in this case.
3.2.2 Formation of Dual Traps by Time-Sharing
In many dual-trap instruments, the two traps are generated by separating the trapping laser into orthogonally polarized beams. One of the two beams is then independently controlled by an actuated tip–tilt mirror to displace one trap relative to the other in the specimen plane (Bustamante et al., 2008; Moffitt et al., 2006). In our instrument, we use the interlacing AOM both to generate and steer the traps by controlling the beam diffraction angle with the RF drive frequency. We exploit the high speed of acousto-optic devices to generate the dual traps by time-sharing, deflecting the trapping beam between two angles (corresponding to two positions in the specimen plane) at fast rates (Visscher, Brakenhoff, & Krol, 1993; Visscher, Gross, & Block, 1996). Although acousto-optic deflectors (AODs) are optimized to provide a larger deflection range and are more commonly used for steering than AOMs, we found that they affect beam shape and quality. In addition, we have observed that beams deflected by AODs exhibit larger fluctuations in power over small angles than those deflected by AOMs. We thus use an AOM instead of an AOD despite the smaller deflection range.
Time-sharing the two traps has advantages over more common polarization-based designs. When splitting the trapping light by polarization, the orthogonally polarized beams travel along separate paths and must pass through different optical components in order to be independently controllable. Prior studies (Bustamante et al., 2008) demonstrated that the longer this differential optical path is, the more susceptible the instrument can be to environmental noise, because each beam is subject to different local environments. In contrast, our approach keeps the differential path between traps to an absolute minimum, since the time-shared beams share identical optical components and a virtually identical beam path. Another issue affecting polarization-based designs is that microscope objectives mix polarization, which leads to cross-talk and interference between the two traps that is separation dependent (Bustamante et al., 2008). These artifacts are eliminated in the time-sharing approach.
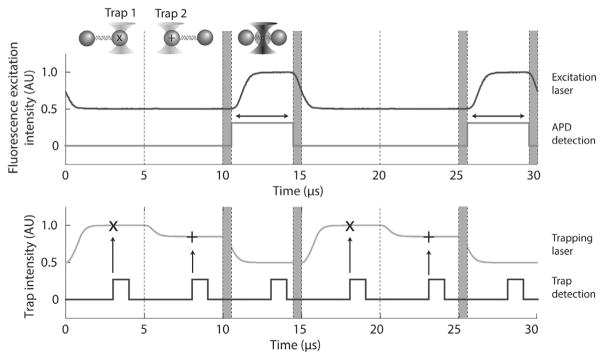
In summary, we use AOM1 and AOM2 to interlace the trapping and fluorescence excitation lasers, respectively, where AOM1 is also responsible for both time-sharing and steering the traps. During one 15-μs cycle (1/66 kHz), each trap is turned ON for 1/3 of the cycle, and then both remain OFF for the final 1/3 cycle when the fluorescence excitation is ON (Fig. 3).
Fig. 3.
Interlacing and time-sharing timing. Trap 1 and trap 2 are ON each for 1/3 of the cycle and fluorescence excitation is ON during the final 1/3. Plotted are the laser intensities measured by feedback photodetectors QPD1 (traps 1 and 2; gray, bottom panel) and PD (fluorescence excitation; black, top panel). Traps 1 and 2 are set to different intensities for clarity. 625-ns delays (shaded regions) between turning OFF (ON) the optical traps and turning ON (OFF) the fluorescence excitation ensure no overlaps between light sources. Trap data acquisition occurs at the time points “x” and “+” for trap 1 and 2, respectively. A digital pulse (black, bottom panel) synchronous with trap data acquisition is output from the DAQ card to make temporal adjustments during alignment. The rising edge of a digital pulse is synchronous with the trap data acquisition (vertical arrows). A digital pulse (dark gray, top panel) synchronous with the APD fluorescence data acquisition is also output from the DAQ card. Emission photons are counted only during this time interval (horizontal arrow). Figure adapted from Comstock, M. J., Ha, T., & Chemla, Y. R. (2011). Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nature Methods, 8(4), 335–340 with permission from Nature Publishing Group.
3.3 Optical Layout
In the following section we describe the overall optical layout. The instrument is organized around three “modules” described below: (1) dual-trap optical tweezers (Fig. 2; yellow lines), (2) a fluorescence confocal microscope (green lines for excitation, red and pink dashed lines for emission), and (3) a bright-field imaging system (blue lines). Each module has its own illumination source and detectors, and is partitioned with blackout enclosures to minimize stray light. Please note that some details on the assembly of this instrument are omitted, as optical trap designs and construction protocols have been discussed extensively in the literature (Block, 1998; Bustamante et al., 2008; van Mameren, Wuite, & Heller, 2011).
3.3.1 Trapping Module
A single-mode, polarization-maintaining fiber laser (IPG Photonics, YLR-5-1064-LP; λ =1064 nm, 5 W) generates the trapping beam. This type of laser provides an ideal Gaussian mode and excellent pointing stability. After an optical isolator (ISO1), we make coarse adjustments to the trapping beam power by passing the laser through a rotary half-wave plate (HW1) and a polarizing beam splitter (PBS1) cube, diverting a fraction of the power into a high-power beam dump (BD).
The trapping beam next passes through AOM1. The beam entering the AOM has a diameter smaller than the 3-mm “active area” of the sound field in this device (~1.5 mm) to maximize the modulation rate of the AOM. The first-order diffraction beam out of the AOM is next collimated and expanded in two stages by telescope T1 (expanding the beam by 3 ×) and T2 (by 2 ×) to a final beam diameter of ~9 mm, overfilling the back aperture of the front objective (8 mm) (Neuman & Block, 2004). The telescopes also serve as imaging systems that make the pivot plane for the trapping beam inside the AOM conjugate to the back-focal plane of the objective (denoted by * in Fig. 2). Thus, deflecting the trapping beam with the AOM displaces the traps in the sample plane without clipping the beam on the objective aperture. In practice, there is no single pivot plane inside the AOM about which the trapping beam rotates, so this can only be achieved approximately.
Between the two telescopes, we deflect a small percentage (1–2%) of the trapping beam with a high-quality wedged beam sampler (BS1; Newport, 10Q20NC.3) to a power feedback stage. In all acousto-optic devices, the intensity of the diffracted beam varies as a function of deflection angle, resulting in a trap stiffness that changes as the trap is steered in the sample plane. The power feedback stage eliminates these unwanted changes in the trapping beam intensity. Measuring the intensity of the sampled beam with a photodetector (QPD1; First Sensor, QP154-Q-HVSD), the instrument control system (see Section 3.4) modulates the RF amplitude driving AOM1 to ensure a constant trapping beam intensity. This power feedback stage reduces intensity noise in the trapping beam by up to 6 orders of magnitude across a broad frequency band (Comstock et al., 2011).
We next reflect the trapping beam into the front objective (O1) using a short-pass dichroic mirror (D1; reflected wavelength = 1064 nm, transmitted wavelength = 415–700 nm). Similar to a prior high-resolution optical tweezers design (Bustamante et al., 2008; Moffitt et al., 2006), we use water-immersion objectives (Nikon 1.2 NA, CFI Plan APO VC 60XWI). Although they generate slightly weaker traps and have a lower fluorescence collection efficiency than oil-immersion objectives due to their smaller NA, water-immersion objectives have the advantage of focusing independently of sample depth. Thus, the optical traps are better decoupled from surface drift, improving stability (Bustamante et al., 2008). O1 forms the optical traps inside the sample chamber by focusing the trapping beam, and an identical back objective (O2) collects the trapping light. The sample chamber is mounted on a three-axis motorized translation stage (Newport, Ultralign 562-XYZ) controlled by motorized actuators (Newport, ESP301-3 N and TRA12CC), so the traps can be moved to different areas of the sample (see Section 5.5).
A second dichroic (D2) reflects the trap light collected by O2 into the detection stage, where the trapped bead positions are monitored by a quadrant photodetector (QPD2; First Sensor, QP154-Q-HVSD). Both QPD1 and QPD2 are IR-enhanced photodetectors and do not suffer from the parasitic, low-pass (~10 kHz) filtering exhibited at 1064 nm by more conventional silicon-based position-sensitive detectors (PSDs) (Huisstede, van Rooijen, van der Werf, Bennink, & Subramaniam, 2006). This choice of photodetector is particularly important because of the fast interlacing and time-sharing frequency, 66 kHz. Note that a single detector, QPD2, monitors the positions of both trapped beads. Since the traps are time-shared, a separate signal from each bead is obtained during different phases of the cycle (Fig. 3). Thus, data acquisition must be tightly synchronized with the timesharing/interlacing cycle so that the appropriate signal is recorded at the correct time (see Section 4.1).
3.3.2 Fluorescence Confocal Module
We next describe the layout for the fluorescence microscope module. A 532-nm excitation laser beam (50 mW; Spectra-Physics Excelsior, 532-50-CDRH) passes through an optical isolator, a power stage, an AOM, a power stabilization stage, and two beam expansion stages (Fig. 2). We remind the reader that in contrast to the trap module, the AOM used here (AOM2; IntraAction, AOM-802AF1) only controls the intensity of the fluorescence excitation, not its deflection angle. We instead use a tip–tilt steerable mirror (SM; Mad City Labs, Nano-MTA2 Invar) to move this beam laterally (x–y) in the sample plane. The fluorescence excitation beam is collimated and expanded in two stages by telescope T3 (expanding the beam by 4 ×) and T4 (by 1.3 ×) to a final beam diameter of ~6 mm. We underfill the back aperture of the front objective (O1) slightly as this facilitates coalignment with the trapping beam. A short-pass, 532-nm dichroic mirror (CVI, SWP-43-RU532-TUVIS-PW-1025-C or Semrock, FF875-Di01-25) reflects the excitation beam into O1, which focuses it into the sample plane.
During measurements, the position of the confocal excitation spot is adjusted with SM to match those of the trapped beads in the sample plane. Telescope T4 images the pivot plane of SM at the back-focal plane of O1 (denoted by ‡ in Fig. 2), such that lateral adjustments can be made without clipping the beam. We adjust the axial (z) position of the confocal excitation spot by two translation stages on the first and second lenses of T3 and T4, providing coarse and fine adjustments, respectively. Each stage moves its lens along the optical axis, changing the beam collimation. It is important to note that chromatic aberrations in O1 mean that collimated trapping and fluorescence beams will not be focused in the same plane. Therefore, axial adjustment of the fluorescence excitation is essential for proper alignment (see Section 4.2).
The fluorescence emitted at the confocal excitation spot is collected by O1 and travels back through T4. A long-pass dichroic mirror (D3; Semrock, LPD02-532RU-25) between T3 and T4 diverts the emission light into a confocal pinhole stage where out-of-focus light is rejected. All the lenses that comprise the fluorescence emission pathway are achromatic doublet lenses (430–700 nm antireflection coated) to minimize chromatic aberrations across the range of fluorescence excitation and emission wavelengths. The first lens of telescope T5 focuses the emitted light into the pinhole (20-μm diameter) and the second lens collects the transmitted light and focuses it onto two APDs (Excelitas, formerly PerkinElmer, SPCM-AQRH-14). A 590-nm edge dichroic mirror (D6; Chroma, 590dcxr) splits the emission into a donor channel (APD1) and an acceptor channel (APD2) for smFRET. We use a notch filter (F1; Chroma, HQ545lp) to cut out 532-nm excitation light from the confocal pinhole stage, and emission filters (F2 and F3; Chroma, HQ580/60m and HQ680/60m) for the donor and acceptor channels, respectively. For alignment purposes, we also collect the fluorescence excitation beam passing through the sample chamber with O2 and divert this light onto a standard silicon-based PSD (First Sensor, DL100-7-PCBA3).
3.3.3 Bright-Field Imaging
The last module consists of a video imaging system to visualize the sample chamber during measurements and to image the optical traps and the fluorescence excitation during alignment. The sample plane is illuminated by a blue LED using Köhler illumination and imaged onto an IR-enhanced CCD camera (Watec, WAT-902-B). Band-pass filters on motorized flip mounts remove the 1064-nm trapping light (F4, 1000-nm short-pass filter, Thorlabs, FES1000; Fig. 2) and 532-nm fluorescence excitation light (F5, 532-nm single-notch filter, Semrock NF01-532U-25) from the video image during typical measurements. The video signal is recorded and displayed on the instrument computer using a PCI-express video frame-grabbing card (Matrix Vision, mvDELTAe-BNC).
3.4 Data Acquisition and Instrument Control
The interlacing and time-sharing scheme of the instrument requires coordination between components that control the trapping and fluorescence excitation beams—AOM1 and AOM2—and detectors that collect data—feedback detectors QPD1 and PD, bead position detectors QPD2 and PSD, and single-photon-counting detectors APD1 and APD2. We use a multifunction Field Programmable Gate Array (FPGA) PC data acquisition (DAQ) card (National Instruments, NI PCIe-7852R) to control these components with 25 ns-level synchrony. FPGA-based DAQ cards have an advantage over traditional cards because all key timing operations are programmed directly onto a chip on the card itself. This DAQ architecture avoids any operating system interference from the PC and runs consistently with 40 MHz (25 ns) timing resolution.
With the exception of the bright-field imaging system camera signal, all signals are acquired through the FPGA DAQ card (Fig. 4). Eight analog signals are sampled by the card: three inputs from the trap QPD2 monitoring the bead positions (x, y, and sum voltage), three inputs from the fluorescence PSD monitoring the fluorescence excitation beam (x, y, and sum voltage), and one input each for the trap and fluorescence feedback detectors QPD1 and PD monitoring beam intensity (sum voltage). Data from these inputs are acquired in sequence and transferred from the FPGA to the host PC memory using Direct Memory Access (DMA) every 5 μs (1/3 cycle). The DAQ card also has two digital inputs that collect single-photon digital pulse trains generated by APD1 and APD2. These signals are counted, integrated, and transferred via DMA (typically every 1 ms).
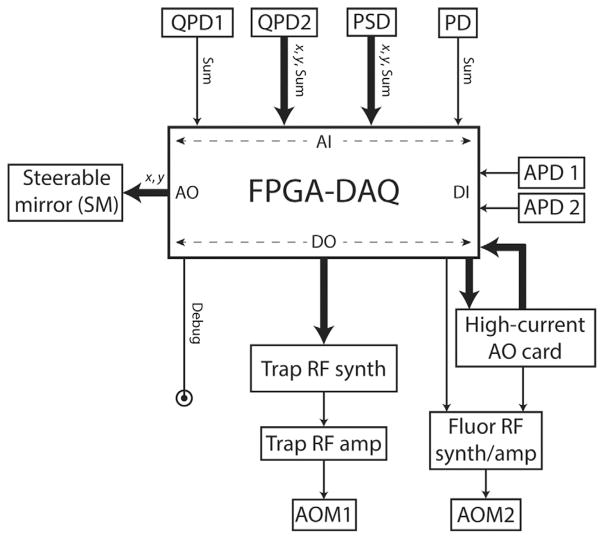
Fig. 4.
Input/output architecture of the FPGA-based DAQ card. The DAQ card receives analog inputs (AI) from four photodetectors (QPD1, QPD2, PSD, PD; see Fig. 2) and digital inputs (DI) from two avalanche photodiodes (APD1, APD2). It communicates via digital output (DO) lines to the RF synthesizer board, the output of which is sent to an amplifier and then to the trap AOM (AOM1). “Debugging” DO lines are used for synchronizing detection input timing with the interlacing cycle (see Fig. 3). The DAQ card also uses analog output (AO) lines to control the steerable mirror (SM) for the fluorescence excitation. To control the commercial RF synthesizer that drives the fluorescence AOM (AOM2), a DO line from the DAQ card is used along with an AO line from an external high-current analog output card.
The FPGA outputs are predominantly dedicated to communication with AOM1 and AOM2. (The exceptions are the analog outputs that drive the piezoactuated stage (SM) that steers the confocal excitation in the sample plane.) We use FPGA digital output channels to communicate with the custom-built RF synthesizer driving AOM1 that modulates the optical trap. Every cycle, the FPGA writes RF frequency and amplitude output values to the RF synthesizer chip buffered memory through these digital lines. There are many instrument-specific subtleties in communicating with the RF synthesizer, and detailed protocols for doing so are now in the literature (Whitley, Comstock, & Chemla, 2017). Control of AOM2 that modulates fluorescence excitation is simpler due to the commercial driver for this AOM: an analog output channel controls the beam intensity, and a digital output gates the RF. We note that this commercial driver requires an external high-current analog output card (Fig. 4; National Instruments, NI 9265), as the DAQ card cannot provide sufficient current to drive it. This input/output architecture allows feedback loops to be run directly on the FPGA at 66 kHz: trap and fluorescence laser power stabilization and force clamping during measurements.
4. INSTRUMENT ALIGNMENT
4.1 Temporal Alignment
Since switching between the two traps and fluorescence excitation occurs at high rates (66 kHz), several components must be carefully synchronized for optimal functioning. Timing considerations generally fall into two categories: synchronization between light sources and synchronization between data acquisition and light source. The former category includes (1) modulating the traps and fluorescence excitation out of phase with each other, and (2) synchronizing changes in trapping beam position and intensity during time-sharing. In the latter category, (1) trapped beam signals acquired by the QPDs must coincide with the appropriate trap ON interval; (2) fluorescence excitation measurements by the feedback PD must occur during the fluorescence ON interval; and (3) fluorescence emission photons must be collected by the APDs in the appropriate ON interval.
In this section we describe these timing adjustments for optimal performance. The adjustments performed here are robust, and only need to be made when the instrument is first set up. We make adjustments by viewing signals from various detectors directly on an oscilloscope with the appropriate bandwidth (e.g., a Tektronix MDO4000B, which is a combined DPO oscilloscope and RF spectrum analyzer). In addition, we configure the FPGA with several “debugging” DO lines that output digital pulses synchronous with the data acquisition timing and view these on the oscilloscope (Fig. 3). Detailed protocols are provided elsewhere (Whitley et al., 2017).
4.1.1 Interlacing the Trapping and Excitation Lasers
We align the trapping laser intervals with the excitation laser interval by viewing the total intensity signals (sum voltages) from the feedback QPD1 for the trapping beam and PD for the fluorescence beam. The phase of the fluorescence excitation AOM2 is then adjusted programmatically until the fluorescence ON interval is positioned between trap ON intervals (Fig. 3). We set additional 625-ns delays between turning OFF (ON) the optical traps and turning ON (OFF) the fluorescence excitation to avoid any overlap between light sources (Brau et al., 2006) (Fig. 3, gray-shaded regions).
4.1.2 Synchronizing Beam Position and Intensity Modulation During Time-Sharing
The position and intensity of the trapping laser during time-sharing are controlled by the frequency and amplitude of the RF signal sent to the trap AOM. We find there is a delay (typically 125–250 ns) between switches in beam position and intensity when the FPGA is programmed to switch both simultaneously. Such a delay results in an unwanted sudden change in one trap stiffness. In order to prevent this artifact, we configure the FPGA so that the timing for RF frequency and amplitude changes are controlled independently, and adjust the delay between trigger signals to make the switches synchronous by viewing the RF signal directly on an oscilloscope (Whitley et al., 2017).
4.1.3 Synchronizing Data Acquisition and Interlacing: Trapping Laser
Measurements of trapping beam intensity and bead position must be synchronized with the corresponding trap ON interval. Moreover, it is important to realize that each trapped bead moves toward its trap center during the ON interval and away from it during the OFF interval due to the imbalances in force during interlacing. This bead motion can span several nanometers at a 66 kHz interlacing rate, depending on the force (Fig. 5). Since we care about measuring the average force on the bead, position data must be acquired precisely at the time point of each ON interval that corresponds to the average bead position. For interlacing rates fm much faster than the characteristic frequency fc of the bead, the bead displacement is close to linear in time and the average bead position corresponds to the center of the ON interval. To synchronize the trap intervals with the input timing of the DAQ card, we output from the DAQ card a digital pulse synchronous with the analog input timing (Fig. 3, black trace, bottom panel) and view this pulse alongside the trapping laser intensity measured by the feedback QPD1. We adjust the phase of the input timing programmatically until it is centered on each trapping laser ON interval.
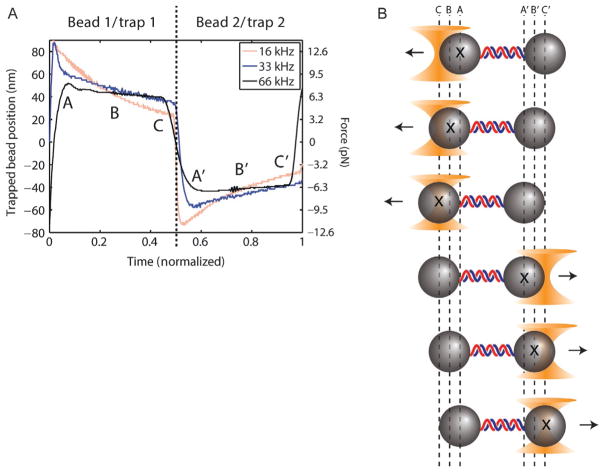
Fig. 5.
Bead motion during time-sharing period. Measurements of trapped beads’ positions during one time-sharing–interlacing cycle for three different time-sharing–interlacing rates (16, 33, 66 kHz). The time axis is normalized to 2/3 of the interlacing cycle duration, i.e., the time-sharing period (see Fig. 3) where trap 1 is ON for half and trap 2 is ON for the second half. Bead oscillations due to imbalanced forces (schematic) are minimized by faster time-sharing–interlacing rates. Data acquired in the middle of each trap ON period (points B and B′) give the correct average displacement and force, compared to data before (A and A′) or after (C and C′). Figure reproduced from Comstock, M. J., Ha, T., & Chemla, Y. R. (2011). Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nature Methods, 8(4), 335–340 with permission from Nature Publishing Group.
4.1.4 Synchronizing Data Acquisition and Interlacing: Fluorescence Excitation
As with the trapping laser, detection of the excitation laser by its feedback PD must be synchronized with its ON interval. Due to time delays in the detector analog input, the DAQ card may not sample the excitation laser intensity at the appropriate time even though the two lasers are synchronized. The procedure for synchronizing the excitation laser to its detectors is identical to that for the trapping laser, where a digital pulse synchronous with the analog input timing of the excitation laser detector is viewed alongside the excitation laser intensity signal from PD, and then adjusted until the digital pulse is centered on the laser’s ON interval.
4.1.5 Synchronizing Data Acquisition and Interlacing: Fluorescence Emission
A final temporal alignment is synchronizing the excitation laser ON interval with fluorescence emission detection by the APDs. The best way we have found to do this is to leave the APDs ON continuously and program the FPGA to record photon counts only during the excitation laser ON interval. As above, synchronizing the FPGA recording interval with the excitation laser ON interval is eased by creating a digital pulse synchronous with the APD measurement ON time and viewing it (Fig. 3, dark gray trace, top panel) alongside the excitation laser ON interval.
We typically make this timing adjustment by trapping a fluorescent bead (e.g., 1-μm, 575-nm emission bead; Thermo-Fisher, F-8819) and centering the confocal excitation spot on it (see Section 4.2). The APD measurement ON time is then adjusted until the observed fluorescence emission signal just begins to increase. From this time point, we shift the phase of the APD recording interval forward by 25% of the interlacing cycle. We use the increase of the fluorescence signal for this alignment rather than its decay, because the beads exhibit phosphorescence, leading to a fluorescence signal that does not decay instantly when the excitation laser is turned OFF.
4.2 Co-Alignment of Trapping and Excitation Lasers
Procedures for aligning optical tweezers and confocal microscopes have been described in detail elsewhere, and will not be duplicated here. Instead we describe coaligning the optical traps and confocal excitation spot both laterally and axially to the same depth as the trapped beads. As mentioned above, the instrument objectives are not perfectly achromatic, focusing a collimated IR trapping beam at a different depth than a collimated fluorescence excitation beam.
The coalignment of the two lasers is done in several stages. A first, coarse alignment is done by imaging the beam spots onto the CCD camera of the bright-field imaging system (Fig. 2). With both beams well-collimated and centered on the back aperture of O1, we adjust the front objective stage until the trapping laser and excitation laser beam spots are both visible on the CCD camera. It is necessary to adjust the intensities of the lasers to ease visualization. The traps should appear as two focused spots on the CCD image. We then align the fluorescence excitation beam spot’s lateral (x–y) and axial (z) position to match those of the traps by manually adjusting the rotary stage beneath the SM and the translational stage on the first lens of telescope T3, respectively.
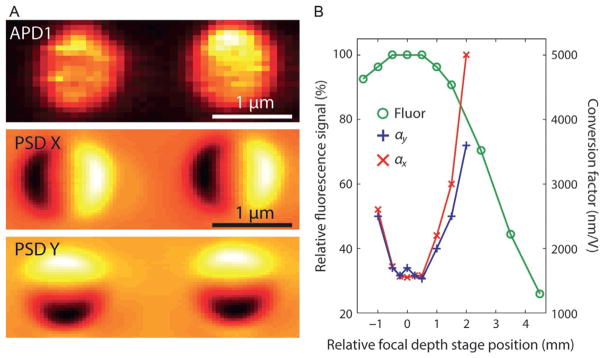
Fine adjustments to the x–y and z positions of the fluorescence excitation are made using the piezomotors of SM and the second, movable lens in telescope T4, respectively. Although the axial alignment only needs to be done once, we find that the lateral alignment must be repeated at the start of each experiment. We have devised two procedures for fine lateral and axial alignment: (1) taking an image of a trapped fluorescent bead by confocal raster scan, or (2) using the fluorescence excitation laser as a “detection beam” (Neuman & Block, 2004) for a trapped nonfluorescent bead. In the first method, a fluorescent bead is trapped and its fluorescence signal from APD1 is collected. By scanning the confocal spot with SM over the trapped bead in the x and y directions (Fig. 6A), an image of the bead is taken, from which an optimal lateral alignment can be obtained. For axial alignment, the confocal spot lateral position is set to the center of the bead, and the fine adjustment telescope lens stage position is adjusted incrementally while recording the fluorescence intensity at each position (Fig. 6B). The optimal axial position is the one that maximizes the fluorescence signal.
Fig. 6.
Spatial alignment of confocal excitation spot and optical traps. Two methods of alignment are shown. (A) Top panel: Lateral alignment by scanning the confocal spot (using SM) over two trapped beads. Signals resulting from bead fluorescence are recorded by APD1 (yellow: higher fluorescence intensity). Middle and bottom panels: Lateral alignment by scanning confocal spot laser over two trapped beads. Here, the PSD measures the deflection in x (middle) and y (bottom) of the laser as it is scanned [black (yellow) denotes negative (positive) deflections]. (B) Axial alignment by two methods. Circles: Axial alignment by scanning the focal depth of the confocal spot (using the translational stage in T4) over a trapped fluorescent bead. The fluorescence intensity measured by APD1 is maximized when the spot is aligned in the plane of the trapped bead. Crosses and exes: Axial alignment by scanning the confocal spot depth over a trapped bead. Here, the PSD voltage-to-bead position conversion factors, α, derived from trap calibration are minimized when the spot is aligned in the plane of the trapped bead.
In the second method, we use the fluorescence excitation laser as a detection beam, measuring the position of a trapped nonfluorescent bead (e.g., 810-nm streptavidin bead; Spherotech, SVP-08-10) by back-focal plane interferometry with the PSD. Here it is convenient to use a high beam intensity (~3 mW) for a larger signal, although the APDs must be turned off to prevent damage. Lateral alignment is achieved by measuring the beam deflection as the confocal spot is scanned through the trapped bead in x and y using SM (Fig. 6A). For axial alignment, the confocal spot lateral position is set to the center of the bead and the Brownian motion of the bead is measured, similar to when the trap is calibrated (Berg-Sørensen & Flyvbjerg, 2004; Neuman & Block, 2004). By fitting the power spectral density of the bead position, the conversion factors αx and αy between bead position (in nm) and PSD output (in V) are obtained. After plotting αx and αy vs stage position (Fig. 6B), the optimal position is found from the minimum in αx and αy. This position corresponds to the highest detection sensitivity, where the confocal spot is aligned in the same plane as the trapped bead.
5. COMBINED OPTICAL TRAP/smFRET ASSAY
In the following section, we describe an application of this instrument combining optical trapping and smFRET. The protocols below recapitulate an experiment carried out by Comstock et al. investigating the structure–function relationship of the E. coli DNA helicase UvrD (Comstock et al., 2015). UvrD is involved primarily in DNA repair and utilizes the energy of ATP hydrolysis to translocate on single-stranded DNA in a 3′–5′ direction and to separate the strands of the duplex. It is a prototype for the structural superfamily 1 (SF1) of helicases. Crystal structures of UvrD and other SF1 helicases reveal two distinct conformational states, termed “closed” and “open” (Fig. 1), where one of its four domains is rotated ~160 degrees with respect to the other three (Jia et al., 2011; Lee & Yang, 2006). These two states were long proposed to regulate protein activity, but their exact roles remained highly debated. To understand the relationship between these two structures and UvrD function, Comstock et al. used the hybrid instrument described here to correlate the open/closed conformation of fluorescently labeled protein (measured by smFRET) with its helicase activity (measured by optical tweezers). Below we describe the steps to making these measurements, the analysis of the data, and interpretation of the results.
5.1 Protein Expression and Fluorescent Labeling
We prepare UvrD labeled with a FRET donor–acceptor pair at positions on the protein shown to exhibit a large change in FRET efficiency upon transition from “closed” to “open” state (Fig. 1) (Jia et al., 2011; Lee & Yang, 2006). We use standard maleimide chemistry for fluorescence labeling (Joo & Ha, 2008), where the cysteines (Cys) in the protein react specifically to the maleimide form of the dyes AlexaFluor555 (donor) and AlexaFluor647 (acceptor) (Molecular Probes, Eugene, OR). A UvrDΔCys(A100C, A473C) mutant was constructed with all six native cysteine residues replaced with serine and with two separate mutations at position 100 and 473 substituting alanine to cysteine. The protein was labeled stochastically with a 1:1 ratio of donor and acceptor dyes. Details on protein expression and purification are reported elsewhere (Jia et al., 2011).
5.2 DNA Hairpin Construct
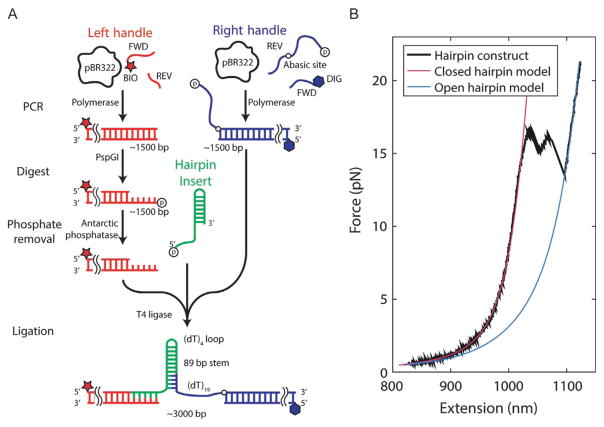
We utilize a hairpin assay (Dumont et al., 2006) to monitor helicase unwinding at high resolution. As shown in Fig. 1, a molecule containing a DNA hairpin is tethered between two optically trapped beads. To monitor helicase-catalyzed unwinding, the assay is performed at a constant force below that required to unfold the hairpin mechanically and the extension of the tethered molecule is monitored with the instrument in force clamp. For every 1 bp unwound by the helicase, 2 nt are released and extend the tethered molecule, mechanically amplifying the unwinding signal (Dumont et al., 2006). For example, at a force of 10 pN, each bp unwound generates a 0.8-nm extension change signal.
The DNA hairpin construct consists of three DNA fragments that are ligated together (Fig. 7): a “hairpin” (HP) flanked by a “right handle” (RH) and a “left handle” (LH) that serve as functionalized linkers that attach to the trapped beads. RH is made from a 1.5-kb PCR-amplified section of the pBR322 plasmid (New England Biolabs) using a 5′-digoxigenin-modified forward primer and a reverse primer containing one abasic site and a long 5′ overhang (Table 1). The digoxigenin moiety is used to link this end of the construct to an antidigoxigenin-coated bead. The overhang consists of a 19-nt poly-dT loading site for helicase binding immediately adjacent to the abasic site, followed by 29 nt that anneal to a complementary sequence in HP. LH is synthesized from a different PCR-amplified section of pBR322 using a 5′-biotin-modified primer. The biotin moiety allows this end of the construct to bind to a streptavidin-coated bead. HP is a single long oligonucleotide containing the complementary sequence to the LH overhang on its 5′ end, followed by the complementary sequence to the RH overhang and a 153-nt self-complementary sequence (Table 1). When self-annealed and ligated to LH and RH, HP makes an 89-bp hairpin stem capped by a (dT)4 tetraloop.
Fig. 7.
Construction of DNA hairpin substrate. (A) Schematic depicting the major steps involved in preparing the DNA construct. (B) A representative force–extension curve of the final construct (black), showing models of the folded (magenta) and unfolded (blue) states.
Table 1.
Oligonucleotides for Constructing Hairpin
| Oligonucleotide | Sequence (IDT Format, 5′ to 3′) |
|---|---|
| LH forward primer | /5Biosg/TGA AGT GGT GGC CTA ACT ACG |
| LH reverse primer | CAA GCC TAT GCC TAC AGC AT |
| RH forward primer | /5Phos/TTG AAA TAC CGA CCG CTC AGC TAT CAG CCT TTT TTT TTT TTT TTT TTT/idSp/CTC TGA CAC ATG CAG CTC CC |
| RH reverse primer | /5DigN/CAA CAA CGT TGC GCA AAC T |
| Hairpin insert | /5Phos/CCT GGG GCT GAT AGC TGA GCG GTC GGT ATT TCA AAA GTC AAC GTA CTG ATC ACG CTG GAT CCT AGA GTC AAC GTA CTG ATC ACG CTG GAT CCT ATT TTT AGG ATC CAG CGT GAT CAG TAC GTT GAC TCT AGG ATC CAG CGT GAT CAG TAC GTT GAC TT |
5.2.1 Synthesis and Purification of LH and RH
For PCR synthesis of LH, mix 35 μL of nuclease-free water, 5 μL of forward primer (10 μM concentration), 5 μL of reverse primer (10 μM), 2 μL of pBR322 template DNA (10 ng/μL) (NEB), 3 μL DMSO, and 50 μL 2 × Phusion HF Master Mix (NEB) for a final volume of 100 μL.
For PCR synthesis of RH, mix 35 μL of nuclease-free water, 5 μL of forward primer (10 μM concentration), 5 μL of reverse primer (10 μM), 2 μL of pBR322 template DNA (10 ng/μL), 3 μL DMSO, and 50 μL 2 × Phusion HF Master Mix for a final volume of 100 μL.
Run PCR on both reaction mixes. We use the following program: (1) 98°C for 30 s, (2) 98°C for 10 s, (3) 59°C for 10 s, (4) 72°C for 33 s, (5) repeat steps 2–4 30 ×, (6) 72°C for 5 min, and (7) 4°C forever.
Purify PCR products following the QIAquick PCR purification kit “spin protocol” (Qiagen). Add 30 μL of elution buffer instead of 50 μL for a more concentrated solution. These PCR products can be verified by gel electrophoresis.
5.2.2 Digestion of LH and Removal of the 5′-Phosphate
Add the following to 30 μL of LH: 2 μL of PspGI restriction enzyme (20 units total; NEB) and 3.5 μL of CutSmart 10 × buffer (NEB).
Incubate the LH reaction mix at 75°C for 1 h.
Add the following to the ~35 μL of digested LH: 4 μL 10 × Antarctic phosphatase buffer (NEB), 1 μL Antarctic phosphatase (5 units; NEB).
Incubate the new LH reaction mix at 37°C for 30 min, then 80°C for 2 min to inactivate the phosphatase.
Purify the digested LH using the QIAquick PCR purification kit as before.
5.2.3 Ligation of LH and RH to HP
Measure the concentrations of LH and RH using a Nanodrop UV–vis spectrophotometer (Thermo Scientific), which requires only 1 μL of each solution.
Mix the three DNA components in an equimolar ratio (1:1:1) to a final volume of 32 μL.
Add 4 μL of 10 × T4 DNA ligase buffer (NEB) and 4 μL of T4 DNA ligase (1600 units; NEB) for a final volume of 40 μL.
Ligate at RT (~22°C) for 1 h and then heat to 65°C for 15 min to inactivate T4 ligase.
Run the final product on a 1% agarose gel with no ethidium bromide for ~70 min.
Incubate the gel in a solution of 50 mL 0.5 × TBE and 15 μL 10,000 × GelGreen (Biotium) for ~20 min.
Image the gel using Dark Reader Transilluminator (Clare Chemical Research) and cut out the appropriate band (~3.4 kb) using a clean razor blade. Place this gel slice in a previously weighed 1.5-mL tube.
Purify the final ligated construct from the agarose gel slices using QIAEX II gel extraction kit (Qiagen). Add 30 μL of elution buffer instead of 50 μL for a more concentrated solution.
5.3 Sample Flow Chamber
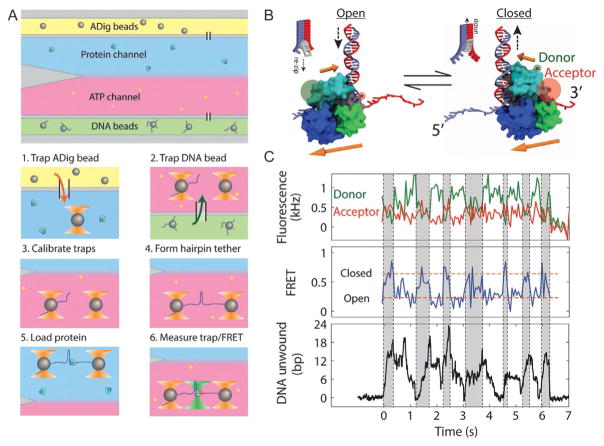
During measurements, we use custom laminar flow sample chambers to assemble protein–DNA complexes in situ. Chambers consist of a central channel with two adjacent laminar fluid streams that do not mix (Qi et al., 2013) (Fig. 8). In the experiments described below (Fig. 9), the lower stream contains ATP (10 μM) but no protein, while the upper stream contains UvrD (10 nM) but no ATP. By moving the sample chamber stage, we can move the optical traps from one stream to the other in a few seconds (typically ~2 s). This allows us to assemble the protein–DNA complex first (in the upper stream), then initiate unwinding (in the lower stream) in a controlled fashion.
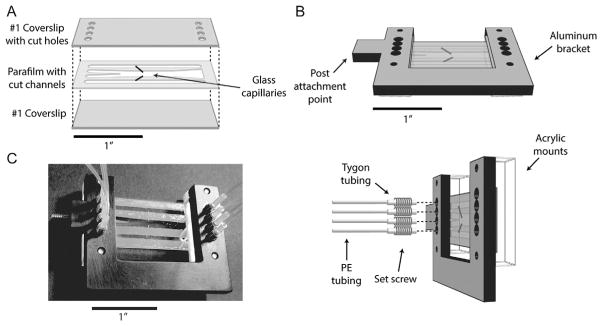
Fig. 8.
Laminar flow cell layout. (A) Chambers are assembled from a piece of parafilm melted between two #1 coverslips. The top coverslip has inlet holes cut into it. The parafilm has patterned into it two outer channels for flowing in beads, and a central channel consisting of two laminar flow streams for assembling complexes in situ. Two glass capillaries connect the top and bottom channels to the central channel and are used to dispense beads. (B) An assembled flow chamber is mounted on a custom aluminum bracket into which tubing mates with the inlet holes. The tubing assembly consists of PE tubing inserted into short lengths of Tygon tubing threaded through set screws screwed into the bracket. (C) Photograph of a flow chamber.
Fig. 9.
Simultaneous measurement of UvrD conformation and activity. (A) Steps for in situ nucleoprotein complex assembly. The flow chamber consists of three channels. The top (yellow) and bottom (green) channels contain antidigoxigenin (ADig) and DNA-coated streptavidin (DNA) beads, respectively, and the central measurement channel consists of two parallel laminar flow streams containing 10 nM protein (blue) and 10 μM ATP (red). During an experiment, we carry out the following steps in sequence: (1) trap an ADig bead dispensed out of the top capillary, (2) move to the bottom capillary and trap a DNA bead, (3) calibrate the traps, (4) form a tether by bringing the beads into contact and pulling apart (and optionally taking a F–x curve), (5) move to the protein stream and load UvrD by incubating for 15 s, and (6) move to the ATP stream, turning on the fluorescence excitation. (B) Model of UvrD conformational switching, in which the closed (open) state corresponds to translocation into (away from) the DNA fork, on opposite strands of the hairpin. (C) Simultaneous measurement of donor (green, top panel) and acceptor (red) fluorescence intensity, FRET efficiency (middle panel), and hairpin base pairs unwound (bottom panel) during UvrD unwinding. A correlation is seen between the “closed,” high FRET state and hairpin unwinding (gray-shaded time intervals) and “open,” low FRET state and hairpin rezipping (unshaded time intervals). Figure adapted from Comstock, M. J., Whitley, K. D., Jia, H., Sokoloski, J., Lohman, T. M., Ha, T., & Chemla, Y. R. (2015). Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science, 348(6232), 352–354 with permission from AAAS.
The laminar flow chambers are made by melting a piece of parafilm between two #1 microscope cover glasses (Fig. 8; Fisher Scientific) as described previously (Qi et al., 2013). Briefly, we use a CO2 laser engraver (VSL2.25; Universal Laser Systems) to cut four inlet and outlet holes into one cover glass. We use the same laser engraver to cut three channels into the parafilm. The top and bottom channels are used to flow in anti-digoxigenin- and streptavidin-coated beads, respectively. A small flow of beads enter into the central channel via glass capillaries (ID = 0.0250 mm, OD = 0.10 mm; King Precision Glass) that are embedded in the melted parafilm and shunt the top and bottom channels to the central channel. The central channel consists of two separate streams from two central inlets, which merge smoothly and maintain a sharp interface due to the laminar, nonturbulent flow.
In order to prevent proteins from adsorbing to the cover glasses, we passivate the glass surfaces with polyethylene glycol (PEG) in situ using the following protocol modified from Ha et al. (2002):
Clean and sonicate cover glasses in 3 M KOH for 20 min prior to assembling the flow cell.
Assemble the flow cell.
Flow a solution of 1% (v/v) N-(2-aminoethyl)-3-aminopropyl-trimethoxysilane (United Chemical Technologies, Bristol, PA) and 5% (v/v) acetic acid in methanol through all channels and let incubate for 15 min.
Rinsethe channels with methanol andwater, flow asolution of 25%(w/v) methoxy-PEG-succinimidyl valerate (MW 5000; Laysan Bio, Arab, AL) in 0.1 M sodium bicarbonate, and let incubate for 4 h.
Rinse the flow cell with copious amounts of Type 1 deionized water and dry with nitrogen gas. Flow chambers are stored dry in the dark at 4°C.
The sample chamber is mounted on a custom bracket with screw-in adapters (Fig. 8) for flowing in buffers and samples via polyethylene (PE) tubing (427406, BD Intramedic). During measurements, we use motorized syringe pumps (PHD Ultra Nanomite 703601 and PHD Ultra Remote Infuse/Withdraw Programmable 703107, Harvard Apparatus) to inject buffers and samples loaded in glass syringes (PTFE Luer Lock, 81320, Hamilton) into the chamber. Each inlet is controlled by a separate pump. We use a programmable syringe pump for the central channel for better control over the laminar flow. Typical volumetric flow rates are 100 μL/h, corresponding to a linear flow rate of ~140 μm/s.
5.4 Sample and Buffer Preparation for Optical Trap–smFRETAssay
Here we describe the sample preparation steps for our example experiment. First, we incubate the DNA hairpin construct with streptavidin-coated polystyrene microspheres (810-nm diameter, SVP-08-10, Spherotech) in a 1:1 DNA:bead ratio for ~1 h to allow biotin–streptavidin linkages to form. Second, we prepare antidigoxigenin antibody-coated polystyrene beads as follows:
Add 40 μL of 1% w/v Protein G microspheres (880-nm diameter, PGP-08-5, Spherotech) into 160 μL 1 × PBS buffer+0.01% Tween 20. Tween helps prevent bead aggregation.
Wash the beads twice by centrifuging at 6000 rcf for 1 min, remove the supernatant, and resuspend with PBS+Tween solution.
Add 10 μL of 1 mg/mL antidigoxigenin antibody (11333089001, Roche) into the bead solution after the last resuspension.
Rotate or shake the solution for 30–60 min.
Wash the beads 2 × as in step 2 to remove excess antibody. Store in final wash (PBS+Tween).
Measurements are conducted in 35 mM Tris (pH 8.0), 20 mM NaCl, 5 mM MgCl2, and 2% glycerol. We use an oxygen scavenging system to increase the lifetimes of the fluorophores and the tethers (Ha, 2001; Landry, McCall, Qi, & Chemla, 2008)—1.2% glucose, 1 mg/mL glucose oxidase (Sigma-Aldrich, St. Louis, MO), and 0.13 mg/mL catalase (EMD Millipore, Billerica, MA)—and a triplet-state quencher to prevent fluorophore blinking (Rasnik, McKinney, & Ha, 2006)—1 mg/mL Trolox (Sigma-Aldrich, St. Louis, MO).
5.5 Optical Trap–smFRET Measurement
The first step for this experiment is to tether the DNA hairpin between two trapped beads. We first trap an antidigoxigenin bead flowing out of the top capillary (Fig. 9A), then translate the sample stage such that the traps are near the bottom capillary and capture a streptavidin–DNA bead in the second trap. The traps are then calibrated by the standard method (Berg-Sørensen & Flyvbjerg, 2004; Neuman & Block, 2004), measuring the beads’ Brownian motion to extract the conversion factors αx,y between bead position and QPD output (in nm/V) and the trap stiffnesses κx,y (in pN/nm). The QPD outputs a zero-force offset voltage that we find depends on trap position; we map this offset voltage, moving the trapped beads together after calibration. We then form a tether by bringing the beads into contact, waiting a short time, and moving them apart. The detection of a force (>4 pN) indicates a tether has been formed. We often take a force–extension curve to ensure that a single molecule is tethering the beads and to examine the quality of the DNA construct (Fig. 7B). The tethered DNA molecule is held at a constant force (5–13 pN) as it is moved into the channel containing UvrD to load protein onto the DNA construct. After incubating in the UvrD channel for ~15 s, the tether is then moved back into the channel containing ATP, and the excitation laser is turned ON. Trap and fluorescence data are acquired.
Fig. 9C shows a time trace obtained from such an experiment. The raw QPD bead position data are collected at the interlacing rate (66 kHz) and boxcar averaged to 133 Hz. As the helicase unwinds the DNA hairpin, the released ssDNA extends the tethered molecule. Under a constant force, this “slack” is taken up by moving the traps further apart. To determine the number of base pairs that are unwound, we first convert the change in the AOM1 RF drive frequency (in MHz) into a trap displacement (in nm). This conversion factor is obtained a priori by taking images of trapped beads at different RF frequencies with the visible system CCD camera, and determining the positions of the bead centers. The conversion factor for our setup is 123 nm/MHz, but will be different depending on the details of the setup. Next, we convert the trap displacement (in nm) to base pairs (bp) unwound by dividing by the extension of the 2 released nucleotides per bp unwound at the force of the measurement. We use the worm-like chain model, with interphosphate distance 0.59 nm/nt, persistence length 1 nm, and stretch modulus 1000 pN, which fits well to our force–extension curve of the mechanically unzipped DNA hairpin (Fig. 7B).
The simultaneous fluorescence signal (Fig. 9C) is analyzed as follows. We integrate the photon counts from the APDs over a set time window (typically 10 ms) and determine photon emission rates (in kHz; top panel) by dividing the integrated counts by the time window. We determine the FRET efficiency E (middle panel) from the standard formula: E = IA/(IA + ID), where IA and ID are the fluorescence intensities (photonrates) of the acceptor and donor dyes, respectively (Ha, 2001). We subtract the background fluorescence signals obtained after both dyes have photo-bleached to obtain IA and ID.
Fig. 9C illustrates the power of using multiple simultaneous methods to measure biomolecular dynamics. The data trace of FRET efficiency reveals that UvrD does not remain in one conformational state during its activity but rather switches between open and closed states (compare shaded and unshaded regions). Meanwhile, the trap data (bottom panel) show not only periods of unwinding of the hairpin (shaded regions), but also periods during which the duplex rezips (unshaded regions). The simultaneous measurement of conformational state and UvrD activity demonstrates a clear correlation between the two types of dynamics. In the closed (high FRET) state the helicase is on average unzipping DNA, while in the open (low FRET) state the DNA is rezipped.
This correlation provides new insights into the functional roles of the open and closed states of UvrD. Based on prior structural and single-molecule work, we believe the two conformational states correspond to UvrD translocating on opposing strands of the hairpin (Comstock et al., 2015). In the closed state, the helicase translocates 3′–5′ into the DNA fork, unwinding it, while in the open state it moves 3′–5′ on the opposing strand, away from the fork, allowing the duplex to rezip in UvrD’s wake (Fig. 9B).
Acknowledgments
We thank members of the Chemla and Comstock laboratories for scientific discussion. Funding was provided by NSF grants MCB-0952442 (CAREER to Y.R.C.), PHY-1430124 (Center for the Physics of Living Cells to Y.R.C.), RC-105094 (to M.J.C.), and NIH grant R21 RR025341 and R01 GM120353 (to Y.R.C.).
References
- Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438(7067):460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkin A. Observation of a single-beam gradient force optical trap for dielectric particles. Optics Letters. 1986;11(5):288–290. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- Berg-Sørensen K, Flyvbjerg H. Power spectrum analysis for optical tweezers. The Review of Scientific Instruments. 2004;75(3):594–612. [Google Scholar]
- Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409(6818):374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- Block SM. Constructing optical tweezers. In: Spector D, Goldman R, Leinward L, editors. Cells: A laboratory manual. New York: Cold Spring Harbor Press; 1998. [Google Scholar]
- Brau RR, Tarsa PB, Ferrer JM, Lee P, Lang MJ. Interlaced optical force-fluorescence measurements for single molecule biophysics. Biophysical Journal. 2006;91(3):1069–1077. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421(6921):423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Chemla YR, Moffitt JR. High resolution dual trap optical tweezers with differential detection. In: Selvin PR, Ha J, editors. Single-molecule techniques: A laboratory manual. Woodbury, New York: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Bustamante C, Cheng W, Mejia YX. Revisiting the central dogma one molecule at a time. Cell. 2011;144(4):480–497. doi: 10.1016/j.cell.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AR, King GM, Ulrich TA, Halsey W, Alchenberger D, Perkins TT. Stabilization of an optical microscope to 0.1 nm in three dimensions. Applied Optics. 2007;46(3):421–427. doi: 10.1364/ao.46.000421. [DOI] [PubMed] [Google Scholar]
- Chemla YR. High-resolution, hybrid optical trapping methods and their application to nucleic acid processing proteins. Biopolymers. 2016;105:704–714. doi: 10.1002/bip.22880. [DOI] [PubMed] [Google Scholar]
- Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333(6050):1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock MJ, Ha T, Chemla YR. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nature Methods. 2011;8(4):335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock MJ, Whitley KD, Jia H, Sokoloski J, Lohman TM, Ha T, Chemla YR. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015;348(6232):352–354. doi: 10.1126/science.aaa0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesterberg VK, Fischer-Hwang IT, Perez CF, Hogan DW, Block SM. Observation of long-range tertiary interactions during ligand binding by the TPP riboswitch aptamer. eLife. 2015;4:e12362. doi: 10.7554/eLife.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439(7072):105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster T. Modern quantum chemistry. New York: Academic; 1965. [Google Scholar]
- Gittes F, Schmidt CF. Interference model for back-focal-plane displacement detection in optical tweezers. Optics Letters. 1998;23(1):7–9. doi: 10.1364/ol.23.000007. [DOI] [PubMed] [Google Scholar]
- Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25(1):78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(13):6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419(6907):638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- Heller I, Hoekstra TP, King GA, Peterman EJG, Wuite GJL. Optical tweezers analysis of DNA-protein complexes. Chemical Reviews. 2014;1(14):3087–3119. doi: 10.1021/cr4003006. [DOI] [PubMed] [Google Scholar]
- Heller I, Sitters G, Broekmans OD, Farge G, Menges C, Wende W, … Wuite GJL. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nature Methods. 2013;10(9):910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- Hohng S, Zhou R, Nahas MK, Yu J, Schulten K, Lilley DM, Ha T. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318(5848):279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisstede JH, van Rooijen BD, van der Werf KO, Bennink ML, Subramaniam V. Dependence of silicon position-detector bandwidth on wavelength, power, and bias. Optics Letters. 2006;31(5):610–612. doi: 10.1364/ol.31.000610. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. Journal of Molecular Graphics. 1996;14(1):33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jia H, Korolev S, Niedziela-Majka A, Maluf N, Gauss G, Myong S, … Lohman TM. Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. Journal of Molecular Biology. 2011;411(3):633–648. doi: 10.1016/j.jmb.2011.06.019. http://dx.doi.org/10.1016/j.jmb.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annual Review of Biochemistry. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- Joo C, Ha T. Single-molecule FRET with total internal reflection microscopy. In: Selvin PR, Ha T, editors. Single-molecule techniques: A laboratory manual. Woodbury, New York: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Kim H, Ha T. Single-molecule nanometry for biological physics. Reports on Progress in Physics. 2013;79(1):016601. doi: 10.1088/0034-4885/76/1/016601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry MP, McCall PM, Qi Z, Chemla YR. Characterization of photoactivated singlet oxygen damage in single-molecule optical trap experiments. Biophysical Journal. 2008;97(8):2128–2136. doi: 10.1016/j.bpj.2009.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nature Methods. 2004;1(2):1–7. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127(7):1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Balci H, Jia H, Lohman TM, Ha T. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nature Communications. 2013;4(1878):1–9. doi: 10.1038/ncomms2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE. New directions in single-molecule imaging and analysis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(31):12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, Anderson DL, Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457(7228):446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(24):9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman KC, Block SM. Optical trapping. The Review of Scientific Instruments. 2004;75(9):2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Pugh RA, Spies M, Chemla YR. Sequence-dependent base pair stepping dynamics in XPD helicase unwinding. eLife. 2013;2:1–23. doi: 10.7554/eLife.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nature Methods. 2006;3(11):891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Ritchie DB, Woodside MT. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Current Opinion in Structural Biology. 2015;34:43–51. doi: 10.1016/j.sbi.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirinakis G, Ren Y, Gao Y, Xi Z, Zhang Y. Combined versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Review of Scientific Instruments. 2012;83(9):093708. doi: 10.1063/1.4752190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L, Haugland RP. Energy transfer: A spectroscopic ruler. Proceedings of the National Academy of Sciences of the United States of America. 1967;58(2):719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suksombat S, Khafizov R, Kozlov AG, Lohman TM, Chemla YR. Structural dynamics of E. coli single-stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. eLife. 2015;4:1–23. doi: 10.7554/eLife.08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk MA, Kapitein LC, van Mameren J, Schmidt CF, Peterman EJG. Combining optical trapping and single-molecule fluorescence spectroscopy: Enhanced photobleaching of fluorophores. Journal of Physical Chemistry B. 2004;108:6479–6484. doi: 10.1021/jp049805+. [DOI] [PubMed] [Google Scholar]
- van Mameren J, Modesti M, Kanaar R, Wyman C, Wuite GJL, Peterman EJG. Dissecting elastic heterogeneity along DNA molecules coated partly with Rad51 using concurrent fluorescence microscopy and optical tweezers. Biophysical Journal. 2006;91(8):L78–L80. doi: 10.1529/biophysj.106.089466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mameren J, Wuite GJL, Heller I. Introduction to optical tweezers: Background, system designs, and commercial solutions. Methods in Molecular Biology. 2011;783:1–20. doi: 10.1007/978-1-61779-282-3_1. [DOI] [PubMed] [Google Scholar]
- Veigel C, Schmidt CF. Moving into the cell: Single-molecule studies of molecular motors in complex environments. Nature Reviews Molecular Cell Biology. 2011;12(3):163–176. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- Visscher K, Brakenhoff GJ, Krol JJ. Micromanipulation by multiple optical traps created by a single fast scanning trap integrated with the bilateral confocal scanning laser microscope. Cytometry. 1993;14:105–114. doi: 10.1002/cyto.990140202. [DOI] [PubMed] [Google Scholar]
- Visscher K, Gross SP, Block SM. Construction of multiple-beam optical traps with nanometer-resolution position sensing. Journal of Selected Topics in Quantum Electronics. 1996;2(4):1066–1076. [Google Scholar]
- Whitley KD, Comstock MJ, Chemla YR. High-resolution “fleezers”: Dual-trap optical tweezers combined with single-molecule fluorescence detection. Methods in Molecular Biology. 2017;1486:183–256. doi: 10.1007/978-1-4939-6421-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]