Abstract
Evidence suggests many neurological disorders emerge when normal neurodevelopmental trajectories are disrupted, i.e. when circuits or cells do not reach their fully mature state. Microglia play a critical role in normal neurodevelopment and are hypothesized to contribute to brain disease. We used whole transcriptome profiling with Next Generation sequencing of purified developing microglia to identify a microglial developmental gene expression program involving thousands of genes whose expression levels change monotonically (up or down) across development. Importantly, the gene expression program was delayed in males relative to females and exposure of adult male mice to LPS, a potent immune activator, accelerated microglial development in males. Next, a microglial developmental index (MDI) generated from gene expression patterns obtained from purified mouse microglia, was applied to human brain transcriptome datasets to test the hypothesis that variability in microglial development is associated with human diseases such as Alzheimer’s and autism where microglia have been suggested to play a role. MDI was significantly increased in both Alzheimer’s Disease and in autism, suggesting that accelerated microglial development may contribute to neuropathology. In conclusion, we identified a microglia-specific gene expression program in mice that was used to create a microglia developmental index, which was applied to human datasets containing heterogeneous cell types to reveal differences between healthy and diseased brain samples, and between males and females. This powerful tool has wide ranging applicability to examine microglial development within the context of disease and in response to other variables such as stress and pharmacological treatments.
Keywords: Microglia, development, sex differences, whole transcriptome analysis
Graphical Abstract
INTRODUCTION
Increasing evidence suggests that many neurological disorders emerge because of a disrupted normal developmental trajectory (Marin, 2016). For instance, studies have documented disruptions in the development or maturation of fast-spiking (FS) interneurons in the pathophysiology underlying disorders like schizophrenia (Lewis, Hashimoto, & Volk, 2005; Morishita, Kundakovic, Bicks, Mitchell, & Akbarian, 2015). Similarly, one of the primary pathologies in attention-deficit disorder is a delay in normal white matter development. Notably, it is only in the assessment of multiple individuals over multiple time points that typical developmental maturation – and thus abnormal development – can be determined (Cao et al., 2015). At a molecular level, whole transcriptome analysis (e.g. Next Generation RNA Sequencing) is a powerful technique that reveals insight into the global/regional, maturational, and/or time-dependent changes in gene expression of a given tissue, and thus can provide similar insight into normal or altered development. A few studies have looked at developmental gene expression patterns in a cell-type specific manner, including fast-spiking GABAergic interneurons, neural crest stem cells, Schwann cells, astrocytes, and oligodendrocytes (Buchstaller et al., 2004; Cahoy et al., 2008; Gandal, Nesbitt, McCurdy, & Alter, 2012; Okaty, Miller, Sugino, Hempel, & Nelson, 2009), all within animal models that are easily amenable to cell sorting strategies. Such an approach is often very difficult for human tissue, and is impossible for the large number of existing databases generated from human postmortem brain tissues.
To overcome this barrier, we used a novel data simplification strategy to analyze whole transcriptome profiling of purified developing male and female mouse microglia. Next, we applied this same approach to heterogenous human tissue datasets to provide insight into cell specific gene program changes. We show, using a wide range of validations, that when the same developmentally regulated genes identified in mice are measured in human datasets from heterogeneous brain tissue, the average of thousands of developmentally co-regulated genes provide a robust measure of cell-type specific developmental maturity. This approach has demonstrated previously that fast-spiking interneurons (FS interneurons) are developmentally immature in multiple neuropsychiatric diseases including schizophrenia, autism, and bipolar disorder (Gandal et al., 2012), a finding that has been supported by several studies (Canetta et al., 2016; Hamburg et al., 2016; Oskvig, Elkahloun, Johnson, Phillips, & Herkenham, 2012; Schmalbach et al., 2015; Wang et al., 2011) and thus points to a potential common mechanism underlying disease pathogenesis.
The goal of this study was to analyze microglial development over time in purified male and female mice; to assess potential sex differences in the response to immune challenge; and to adopt and validate via multiple measures the same strategy for analyzing microglia in heterogeneous human brain tissue. Microglia are increasingly implicated for their roles in normal neurodevelopment (e.g. synaptic pruning, (Schafer et al., 2012; Schafer, Lehrman, & Stevens, 2013), as well as their contributions to neural dysfunction during development and the risk of neuropsychiatric disorders (Blank & Prinz, 2013; Garden & Campbell, 2016). Thus, we now appreciate that it is because immune molecules and microglia are important for normal development, that they are also implicated in abnormal brain development (e.g. in response to inflammatory signals) and the increased susceptibility to neurodevelopmental and neurological disorders. This recognition presents an urgent need to better understand the mechanisms underlying microglial development in health and disease.
We isolated microglia from male and female hippocampi of mice from different developmental ages ranging from embryonic day 18 (E18) to postnatal day 60 (P60). Hippocampus was selected based on its importance to mental health, its marked vulnerability relative to other brain regions in response to diverse threats to homeostasis (epilepsy, stroke, and cardiac arrest (Fujioka et al., 2000; Petito, Feldmann, Pulsinelli, & Plum, 1987; Salmenpera, Kalviainen, Partanen, & Pitkanen, 1998; Sapolsky, Uno, Rebert, & Finch, 1990)), and due to evidence of morphological sex differences in microglial development in this region (Schwarz, Sholar, & Bilbo, 2012). Following whole transcriptome analysis in purified mouse microglia, we developed a microglia-specific developmental index (MDI) based on global gene expression patterns. MDI tracked with chronological age as expected, and strikingly predicted the well characterized change in structural morphology that occurs in microglia over development. Interestingly, as microglia developed, the expression of immune-related moleclules increased. Thus, we demonstrate that microglial maturation is closely related to its immune reactive state. Moreover, we discovered that the percentage of genes that increase over normal development (a proxy for maturation) is delayed in males relative to females, and that acute immune stimulation accelerated this normal microglial maturation only in males. Next, we mined the same developmentally-regulated genes identified in mouse microglia from several human brain transcriptome datasets to test the hypothesis that variability in microglial maturity is associated with human disease. Microglial maturity was significantly increased in both Alzheimer’s disease and in autism compared to age-matched healthy controls, suggesting that accelerated or altered microglial development, and not simply “inflammation”, critically contributes to neuropathology. Finally, the same developmentally regulated genes enriched in both autism and in human microglia show accelerated maturity in males compared to females, which is striking given the 4:1 male bias in autism. Thus, we identified a microglia-specific gene expression program in mice that was used to assess microglial developmental trajectory in heterogeneous human datasets to reveal differences between healthy and diseased brain samples, and between males and females.
MATERIALS AND METHODS
Animals
In-house bred adult male and female C57BL/6 mice (Charles River Laboratories Raleigh, NC, USA; http://www.criver.com/products-services/basic-research/find-a-model/c57bl-6n-mouse) were used as breeders and housed in pathogen-free vivarium conditions with ad libitum access to food and filtered water on regular light cycle (12h ON/12h OFF). For the embryonic day 18 (E18) time point, time-mated females were sacrificed at 18 days post vaginal plug discovery. For all other time points, pregnant females were allowed to give birth and pups were sacrificed at appropriate time points. For the adult time point, pups were weaned at postnatal day 24 (P24), separated based on sex with a minimum of 3 animals per cage (maximum of 5), and sacrificed at P60. All experiments were conducted with protocols approved by Duke University Animal Care and Use Committees.
LPS treatments and tissue collection
For all time-points, littermates were equally distributed across sexes and treatment groups. 4–6 litters represented each time point to control for litter variability. For E18 and P4, offspring were sacrificed by rapid decapitation. Hippocampus (HP) was isolated and processed for CD11b isolations (see below). For E18 and P4, HP samples were collected from four litters, and tail clips were processed for sex determination using SRY genotyping. Due to the smaller number of cells obtained from each HP of E18 and P4 animals, samples were pooled based on sex prior to RNA extraction. Hence a total of 4 male and 4 female samples were present for E18 and P4 time points. For P14 and P60 (n = 10 females and 10 males per time point), animals were euthanized using a ketamine-xylazine (1:1) mixture and perfused intracardially using cold 0.9% saline. Hippocampus was rapidly isolated and processed for CD11b isolations. For P60 animals, prior to sacrifice, males and females were injected intraperitoneally with 330 μg/kg Lipopolysaccharide (LPS, from Escherichia coli serotype 0111:B4, Sigma-Aldrich, St. Louis, MO, USA) dissolved in 0.9% sterile saline, or with 0.9% saline control (n = 8 saline-treated females, 8 saline-treated males, 10 LPS-treated females and 9 LPS-treated males). Animals were euthanized 2 hours after injections and tissue was isolated as described above. For P60, one brain hemisphere was post-fixed in 4% paraformaldehyde (PFA, Sigma-Aldrich, St. Louis, MO) for immunohistochemical analysis while hippocampus from the other hemisphere was processed for CD11b isolations. For P60 females, estrous smears were performed at the time of sacrifice. Estrous cycle was determined by imaging slides containing estrous smears and matching vaginal cell morphology visually to established standards for rodents from published literature (Goldman, Murr, & Cooper, 2007).
Microglia isolations
Hippocampus isolated from animals was homogenized in enzyme digestion mix containing collagenase A (1.5 mg/ml, Roche, Indianapolis, IN, USA) and DNase I (0.4 mg/ml, Roche, Indianapolis, IN, USA) in Hank’s Buffered Salt Solution (HBSS, Thermo Fisher Scientific, NY, USA) for 45 min in water bath. Every 15 min during the incubation, samples were removed from the water bath and passed through glass Pasteur pipettes multiple times to ensure complete dissociation. Samples were then filtered through nylon filter and centrifuged at 1200 rpm for 10 min at 4°C. Cell pellets were resuspended in 30% Percoll (GE Healthcare, Uppsala, Sweden) in 1X sterile phosphate buffered saline prepared from isotonic percoll (ITP, containing 90% Percoll and 10% 10X PBS, Thermo Fisher Scientific, NY, USA) and underlayed carefully with 70% Percoll in 1X PBS prepared from ITP. Samples were centrifuged at 1400 rpm for 15 min at 23°C with no brake. The interphase containing mostly mononuclear phagocytes was isolated and incubated with CD11b antibody conjugated magnetic beads (MACS Miltenyi Biotec, San Diego CA) for 15 min at 4°C. After washing, cells were passed through a magnetic bead column (MACS Miltenyi Biotec, San Diego CA) and CD11b positive (CD11b+) and negative (CD11b−) populations were separated. Samples were centrifuged, washed with 1X PBS and stored at −80° until RNA extraction.
RNA extraction, quantification and purity determination
Frozen samples were homogenized in 800 μl TRIzol R (Thermo Fisher Scientific, NY) followed by vortexing at 2000 rpm for 10 min. 160 μl of chloroform (Thermo Fisher Scientific, NY) was added to each tube and vortexed for additional 2 min. Samples were then centrifuged at 11,900 rpm for 15 min at 4°C, after which the top clear aqueous phase was separated into a fresh tube. The pink later (Trizol-chloroform mix) was saved at −80 for protein extraction (see below). 2 μl of Glycogen (Thermo Fisher Scientific, NY) was added followed by 360 μl of Isopropanol (Thermo Fisher Scientific, NY). Samples were vortexed for 1 min and centrifuged. Supernatant was discarded carefully without disturbing RNA pellet. Pellets were washed two times with 1 ml of ice cold 75% ethanol, air dried and resuspended in 6–8 μl of RNase-free water.
Preliminary RNA quantification and purity determination was done using NanoDrop Spectrophotometer (Thermo Scientific, Wilmington DE, USA). 1 μl of RNA sample was loaded on the spectrophotometer for measurement. RNA amount was recorded at 260 nm wavelengths and RNA purity was determined by the 260/230 and 260/280 ratios. RNA was considered pure if 260/280 (RNA:protein contamination) ratio was in the range of 1.8–2.0, and 260/230 (RNA:Ethanol Contamination) was between 2.0–2.2.
NextGen RNA sequencing and whole transcriptome analysis
All RNA samples were coded numerically prior to shipping to University of Pennsylvania and sequencing was performed blind to sample identity. Illumina TruSeq dual-indexed libraries were prepared from total RNA using the Illumina mRNA High-throughput RNA-Seq kit (Illumina Inc., San Diego, CA). Libraries were pooled and sequenced on a HiSeq 2500 (Illumina Inc., San Diego, CA) in 100SR rapid mode. The mean number of reads per sample after ribosomal and repeat removal was 2,510,105. Reads were aligned to the genome and gene expression values tallied using RUM (Grant et al., 2011). The mean unique alignment rate was 86.5% or 2,159,926 reads per sample. All RNA sequencing data in the current mouse microglia study is deposited and available at Gene expression Omnibus (GEO) using accession number GSE99622.
Microglial developmental indices
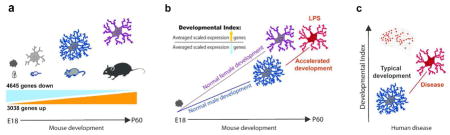
Cell-type specific developmental indices represent a data simplification strategy, which takes known information about cell-type specific developmental gene expression patterns and uses that information to measure the maturity of specific cell types in heterogeneous tissue using gene expression profiling (Gandal et al., 2012). Time course developmental gene expression profiling data from microglia (current study) were used to generate a microglial developmental index (MDI). Specifically, genes were divided into groups based on whether they were significantly up-regulated or down-regulated during development, that is from E18 to P60. Genes that were up-regulated at P60 compared to E18 were considered as “up-regulated over development” whereas those that were down-regulated at P60 vs. E18 were considered as “down-regulated over development”. These genes were mined from the RNAseq dataset for each sample, and the expression value for each gene was scaled between 0 and 1 in order to equally weigh all genes. For any given gene the scaled expression value was equal to the (expression value of the gene in a sample – minimum expression for the gene across all samples)/(maximum expression for the gene across all samples – minimum expression for the gene across all samples), which means that for every gene the minimum expression value will equal zero and the maximum will equal one. This assured that the index would vary only if a sufficiently large number of genes changed their expression levels in a non-random manner with respect to microglial development. An index was calculated for a sample by taking the ratio of the average of the scaled expression of all up-regulated genes and the average of the scaled expression of all the down-regulated genes. As most genes changed monotonically (up or down) across development, the ratio of the average of developmentally up- or down-regulated genes also changed monotonically with time (Figure 1).
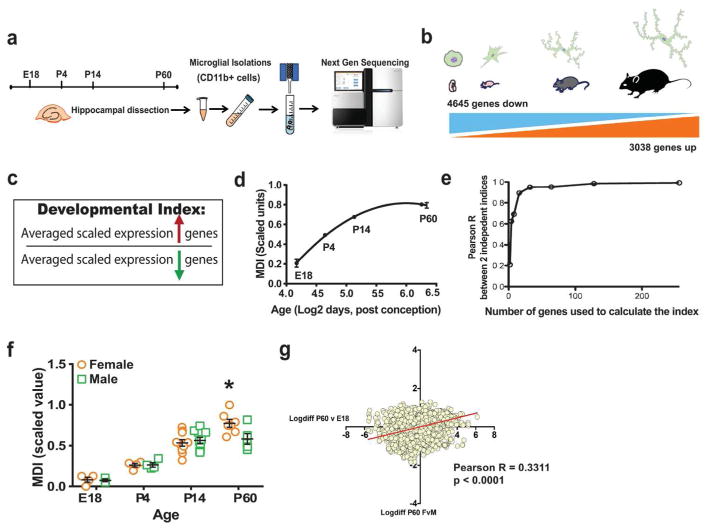
Figure 1. Sex differences in immune modulation of microglial developmental programs.
(a) Hippocampal microglia isolated from male and female mice at different developmental time points were subjected to RNA extraction and Next Generation sequencing. (b) A total of 4645 genes were found to be down-regulated over development whereas 3038 genes were up-regulated from E18 to P60 in mouse microglia. (c) Developmental indices were created by taking the ratio of the average scaled expression levels of genes that were up-regulated during development in microglia, divided by the average expression levels of all down-regulated genes. (d) Line graph plots microglia index (MDI) across development against log2 age in weeks post conception (Non-linear fit, MDI R2 = 0.8525). (e) In order to validate the robustness of the index, gene group size was progressively increased 2-fold from 2 to 256 up- and down-regulated genes. (f) MDI was calculated from transcriptome data of mouse male and female microglia obtained from different developmental time points (E18, P4, P4 and P60, n = 4–10 per group, two-way ANOVA, post-hoc * p < 0.05). (g) Log fold-changes in gene expression between males and females at P60 were compared to those of P60vsE18 (developmental gene expression changes) to obtain positive correlation (Linear regression, Pearson’s r = 0.3311, *** p < 0.0001).
Gene functional annotation clustering analysis
The top 1000 genes that were significantly down- or up-regulated between P60 and E18; P60 and P4; P14 and E18; and P60 females, males, LPS, and SAL were uploaded into DAVID functional gene ontology software. All genes that were expressed in the RNAseq data for all groups served as “background”. Functional clustering was run and the top 7 enriched annotation groups were chosen to represent the following group comparisons: P60 > E18, P60 >P 4, P14 > P14, P60 F > P60 M, P60 F LPS > F SAL and P60 M LPS > M SAL.
Immunohistochemical analyses
Following 0.9% saline perfusion, P60 half-brains were post-fixed in 4% PFA overnight and cryoprotected in 30% sucrose for at least 3 days. Brains were then frozen in bubbling isopentane and embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, USA). Frozen blocks were sliced at 40 μm thickness and floating sections stained using a standard immunohistochemical protocol. Briefly, slices were washed three times in 1X PBS, incubated with 0.3% hydrogen peroxide for 30 min, followed by incubation in 0.5% sodium borohydride for 30 min to reduce background staining. Slices were incubated in blocking solution containing 5% normal goat serum (NGS), 0.3% Triton X in 1X PBS for 1 hour.
For Iba1 microglial staining, slices were incubated in Rabbit anti-Iba1 primary antibody (Wako Chemicals, Richmond, VA, USA, 1:20,000, 2 overnights at 4°C with shaking) following which they were washed and incubated with Biotinylated Goat Anti-Rabbit IgG Secondary Antibody (1:200, Vector Laboratories, Burlingame, CA, USA) for 2 hours at 4°C with shaking. Signal amplification was achieved using the standard Vectastain ABC kit (Vector Laboratories, Burlingame, CA, USA) and after which slices were incubated in in-house prepared 3′-3′-diaminobenzidine tetrahydrochloride (10 mg tablet, Sigma Aldrich, St. Louis, MO, USA) solution for 20–30 mins. Slices were washed and mounted on slides, following which alcohol dehydration was performed. Slides were coverslipped and imaged at 10X magnification on a Zeiss laser microdissection and capture microscope. Images were then processed in ImageJ software and integrated density of the signal was measured over different subregions of the hippocampus and normalized to area.
For Imaris 3D reconstruction of microglia, sections were first incubated on a shaker plate overnight at 4 °C in a 1:2000 dilution of rabbit anti-P2Y12 polyclonal antibody (AS-55043A, AnaSpec, Fremond, CA). Sections were washed 3 times in 1X PBS and incubated for approximately 18 hours in a 1:500 dilution of secondary antibody, donkey anti-rabbit IgG (Alexa FluorR 568 nm, ThermoFisher, Waltham, MA) in blocking buffer at 4 °C on a shaker plate, in darkness, and wrapped in foil. After the 18 hour incubation, slices were again washed. Sections were mounted onto gelatin-coated slides, allowed to dry and then coverslipped with VectaShield Antifade Mounting Medium containing DAPI (Vector Laboratories, Burlingame, CA). Slides were stored at 4°C until ready for confocal imaging.
Confocal Microscopy, Image Deconvolution and Imaris 3D Reconstruction of P2Y12 stained microglia
All confocal imaging, image deconvolution and Imaris 3D reconstructions were performed by separate researchers that were blind to the identities of the samples. Three independent coronal brain sections per mouse, containing area CA3 (approximately between bregma −1.555 mm and −2.155 mm) were imaged on a Leica SP8 confocal laser-scanning microscope. The 16-μm-thick confocal scans (optical section depth 0.5 μm, 32 sections/scan) of the CA3 stratum oriens in the hippocampus were performed at 100× magnification. CA3 subregion was chosen for analysis because preliminary data suggested significant sex difference microglial density in this region (Supp. Fig. 4).
Confocal Z-stacks were deconvolved with Huygens Essential (Scientific Volume Imaging, The Netherlands) software. The deconvolution wizard tool was used to deconvolve the DAPI and P2Y12 channel. Images were cropped to include only the microglia of interest. Although settings for the deconvolution process varied as dictated by the parameters of the Z-stack, parameters close to the following were used for the DAPI channel: a signal to noise ratio of approximately 5, 40 iterations, and a quality threshold of 0.05. For the P2Y12 channel, a signal to noise ratio of approximately 12, 40 iterations, and a quality threshold of 0.05 was utilized. After deconvolution, images were loaded onto the Imaris software VERSION (Bitplane, Zurich, Switzerland) to create 3D filament traces of every cell. The FilamentTracer module was used for this purpose, and a region of interest was selected to include all possible branches of the microglia. The AutoPath algorithm (with no loops) was used, wherein a starting point marker was placed at the cell body of the microglia. The seed point threshold was adjusted to fill the tips of the smaller processes of the microglia. Spines were not detected. After the FilamentTracer created a structure of the branching of the microglia, mistakes in the structure were corrected manually. Duplicate branches were removed, connections that did not follow processes were deleted, and if two microglia were present in the same Z-stack, an attempt was made to determine where the branching processes of one cell began and the processes of the other cell ended. Small processes that the algorithm missed were added in by creating a new starting point, and following the algorithm generated pathways to add in small processes or extend processes. In manually adding and removing processes, the image was consulted to ensure that processes that were separated by a large enough gap were not connected. Because the P2Y12 staining is diffuse in the cell body, compared to the processes, larger gaps were connected if they were close to the cell body. Smaller gaps at the end of processes were not connected. The Sholl analysis with 1 μm concentric spheres and total filament length statistical parameters were exported from Imaris into Microsoft Excel.
Application of MDI to human brain heterogeneous cell population
In order to generate an index to measure microglial development in human brain tissue samples, genes common between our purified microglial transcriptome and the human brain transcriptome datasets were identified. A sub-index was created from the common genes by identifying the genes that were developmentally regulated in purified microglial transcriptome. For a given sample, MDI was equal to the average of developmentally up-regulated genes divided by the average of down-regulated genes. Because brain tissue is heterogeneous each measurement of a microglial development gene contains information about the developmental state of microglia in the brain tissue sample in addition to noise and expression variability for that gene in cells other than microglia. By averaging measurements from thousands of different genes, information about microglia development is enriched to provide an estimate of the developmental maturity of microglia in a given tissue sample.
Statistical analyses
RNAseq and morphology data were graphed and analyzed using GraphPad Prism software licensed to Duke University, Durham, NC (GraphPad Software, La Jolla, CA, USA). Each value is expressed as the mean ± standard error of the mean (SEM). For morphological analyses, data were averaged from 3 microglia per slice and 3 slices per mouse brain. Secondary analysis was performed on human transcriptome datasets, hence the authors had no control over the “n” values for these data. The data were tested using either an unpaired t-test or two-way ANOVA, followed by Bonferroni’s’s multiple comparison tests wherever necessary. Multiple linear regression was used wherever necessary and specified in the results and/or figure legend. Filament length data were analyzed using a two-way ANOVA in JMP Pro (version 12.0, SAS, Cary, NC), comparing sex and treatment (independent variables) and length (μm, depedent variable) of all filaments in the reconstruction. Sholl intersections and number of branches were averaged for every 10μm, such that intervals averaged from 0μm to 10μm, 10μm to 20μm, 20μm to 30μm, 30μm to 40μm, and 40μm to 50μm were obtained. Two-way ANOVA was performed followed by Bonferroni’s post-hoc test for multiple comparisons. p <0.05 were considered significant.
RESULTS
Generation of a developmental index using gene expression programs in isolated mouse microglia over time
The development of specific cell types involves stereotypical changes in thousands of genes, called gene expression programs. For any given cell, the developmental state of its transcriptome will be the product of its specific cell type and the position of the cell along its cell type-specific developmental gene expression program, which represents the transition of a cell type to its fully functional and mature state (Alter, 2013; Gandal et al., 2012). Because most genes change monotonically up or down across development the state of a developmental program can be measured by taking the average of developmentally up-regulated genes divided by down-regulated genes. The approach of using thousands of genes is much more robust than picking one or a few representative developmental genes because the measurement of any individual gene is subject to multiple sources of noise including biological, stochastic, and technical. Whereas, when the measurements of thousands of co-varying genes are combined these sources of noise tend to cancel out. Applying this general principal to microglia gene expression data we developed a “microglial developmental index” or MDI to quantify the relative position of transcriptomes along the microglial developmental program. Such an approach provided a robust quantitative measure of microglial maturity allowing us to ask interesting questions about how the maturity of microglia varied in different contexts such as between sexes, in association with disease, and in response to treatment with LPS.
Briefly, hippocampi were isolated from male and female mice at E18, P4, P14, or P60 (Fig. 1a). Microglia were isolated and the extracted RNA was subject to whole transcriptome profiling using Next Generation Sequencing. Only genes detected in all samples (12,452) were used to assess developmental gene expression changes in microglia. A majority of genes (61.7%) were significantly developmentally regulated (7,683 genes) with 37.3% down-regulated (4,645 genes) and 24.4% up-regulated (3,038 genes) across development, i.e in P60 vs. E18 (Fig. 1b). Prior to the calculation of the microglia specific developmental index, gene expression levels were scaled across all samples such that for all genes the maximum expression level of each gene was 1 and the minimum was 0, and thus an equal weight in the calculation of index measures was given to each gene. This assured that the behavior of highly expressed genes could not dominate the behavior of the index and thus that the index would vary only if a sufficiently large number of genes changed their expression levels in a non-random manner with respect to microglial development. Next, to calculate the index, scaled expression levels of genes up-regulated from E18 to P60 were averaged and were divided by the average of scaled expression levels of down-regulated genes (Fig. 1c). As most genes changed monotonically (up or down) across development, the ratio of the average of developmentally up- or down-regulated genes also changed monotonically with time (Fig. 1d). Interestingly, a relatively small number of genes could be used to quantify the aggregate behavior of the entire group of developmentally-regulated genes. To demonstrate this, gene group size was progressively increased 2-fold from 2 to 256 up- and down-regulated genes. MDI was calculated separately on two independent sets of genes and results were correlated. A steep increase in concordance was seen with a 0.9 Pearson R correlation at 32 (16 up and 16 down) genes and > 0.98 Pearson R by 128 genes, which is less than 2% of the total number of developmentally regulated genes and indicates a high degree of redundancy in information about the microglial developmental gene expression program (Fig. 1e). Thus, in our enriched set of microglia we can use a relatively small number of genes to measure broad transcriptional changes over development, an important validation of this index that is critical for the application to heterogeneous human tissue, which we describe in detail later (Fig. 6).
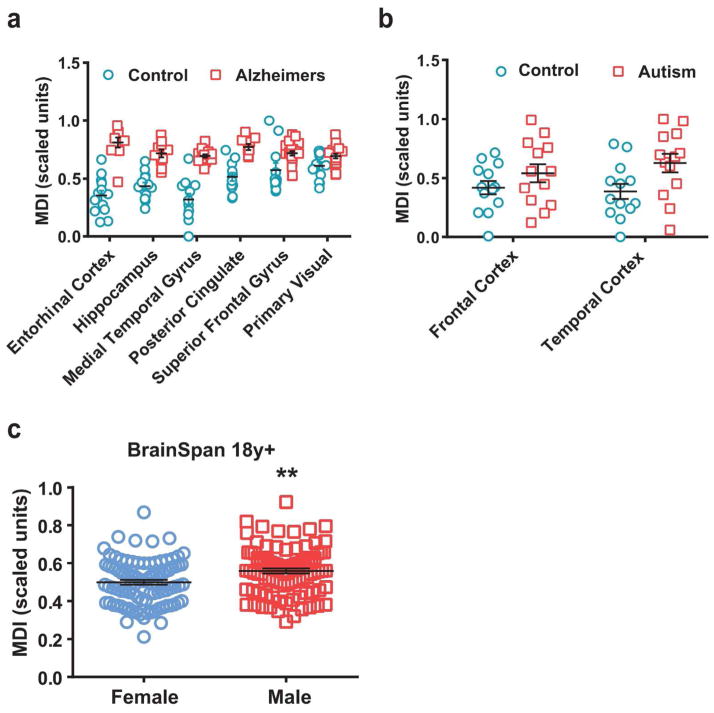
Figure 6. MDI is accelerated in brains of Alzheimer’s disease and autism patients.
MDI was quantified from gene expression data of patients with (a) Alzheimer’s disease (GSE5281, two-way ANOVA, Region p< 0.0001; Disease p < 0.0001) and (b) autism (GSE28521, two-way ANOVA, MDI * p < 0.05) and compared to controls as discussed above. For both, results remained significant after controlling for age-related and regional variability (p<0.05). (c) MDI was calculated using data from adult males and females (18y+, BrainSpan developmental dataset).
Sex differences in the microglial developmental index
Next, we looked at sex differences in microglial development. We found that male and female microglial development is similar during early developmental time points, but by early adulthood, P60, male microglial development plateaus, whereas in females it does not, resulting in a significant sex difference in MDI at this time point (Fig. 1f). When the log difference in expression levels of genes that were changed over development (from E18 to P60) were plotted against the log difference in expression levels of the same genes that were changed between females and males at P60, there was a strongly significant positive correlation, indicating that the difference in global gene expression pattern in females and males was similar to the difference in global gene expression over development (Fig. 1g, Pearson r = 0.3311, p < 0.0001).
Sex difference in microglial developmental index response to LPS challenge
There are known sex differences in several neuropsychiatric disorders which we and others hypothesize are linked to microglial function, at baseline and/or in response to the immune challenge inherent in disease (Hanamsagar & Bilbo, 2015). Lipopolysaccharide (LPS) is a potent immune modulator which we to determine the effect of systemic immune challenge on male and female microglia. P60 male and female mice were injected with a moderate dose of LPS (330 μg/kg) and sacrificed 2 hours later (Fig. 2a). RNA sequencing was performed on the microglia collected from one hippocampal hemisphere, and the opposite hemisphere was used to perform immunohistochemical analysis of microglia in the same mice. Both LPS-treated males and females had extensive transcriptional responses with 21% of genes (10% up and 11% down) significantly changing in males, and 18% (6% up and 12% down) in females relative to sex-matched saline controls (Fig. 2b). The magnitude of transcriptional responses to LPS was similar between males and females and responses were highly correlated (Table 1). Male transcriptional changes to LPS explained 61.5% of the variance in significant gene expression changes in females (Pearson R2 = 0.615 – Table 1b – LPS - Model 1). Similarly, female transcriptional changes to LPS explained 51.4% of the variance in the male LPS response (Pearson R2 = 0.514 – Table 1a – LPS - Model 1). However, despite strong similarities between males and females in the magnitude of the gene expression response to LPS, there was a marked sex difference at the level of microglial development, indicating that while there was a subset of genes regulated by LPS irrespective of sex, males and females also had sex-specific responses. Specifically, male microglia significantly increased their MDI in response to LPS, whereas females did not, such that LPS-stimulated male microglia now had the same MDI as female microglia (Fig. 2c), thereby “maturing” to the level of females in response to challenge.
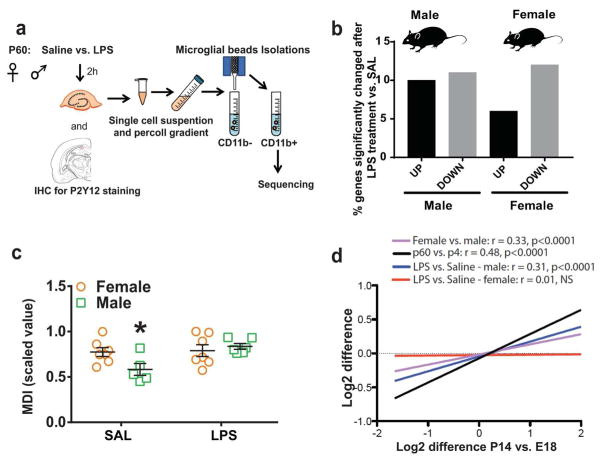
Figure 2. Sex differences in microglial response to LPS.
(a) C57BL/6 male and female P60 mice were sacrificed 2 hours after saline or LPS (330 ug/kg, i.p.) injections following which microglia were isolated from the hippocampus and subject to RNA sequencing. (b) Out of the total number of genes changed in males and females following LPS treatment, about 21% (10% up and 11% down) were changed significantly in males (vs. male SAL), and 16% (5% up and 11% down) were changed significantly in females (vs. female SAL). (c) Microglial developmental index was calculated for P60 samples as described above (two-way ANOVA, post-hoc * p < 0.05, n = 6–8 per group). (d) Gene expression changes between P14 and E18 were compared with those in P60 vs. P4 (black line, linear regression, Pearson’s r = 0.48, *** p < 0.0001), P60 females vs. males (purple line, linear regression, Pearson’s r = 0.33, *** p < 0.0001), P60 male LPS vs. SAL (blue line, linear regression, Pearson’s r = 0.31, *** p < 0.0001) and P60 female LPS vs. SAL (red line, linear regression, Pearson’s r = 0.0.1, not significant).
Table 1.
Sex difference in effect of LPS on microglia development
| a. Dependent variable: LPS male - significant log2 expression changes | |||
|---|---|---|---|
| Model 1 (R2=0.514***) | Model 2 (R2=0.12***) | Model 3 (R2=0.602***) | |
| Variable | Std. Coeff. | Std. Coeff. | Std. Coeff. |
| LPS female | 0.717*** | 0.696*** | |
| Development | 0.347*** | 0.298*** | |
| b. Dependent variable: LPS female - significant log2 expression changes | |||
|---|---|---|---|
| Model 1 (R2=0.615***) | Model 2 (R2=0.00) | Model 3 (R2=0.621***) | |
| Variable | Std. Coeff. | Std. Coeff. | Std. Coeff. |
| LPS male | 0.784*** | 0.791*** | |
| Development | −0.014 | −0.078 | |
Because LPS-induced changes in gene expression were measured against the same samples that were used to measure developmental changes, P60 Saline, this represented a potential confound in comparing LPS and developmental changes in gene expression. For this reason, we used developmental gene expression changes between P14 and E18 in analyses where developmental changes were related to LPS-induced changes. As expected, gene expression changes between E18 and P14 were highly correlated with changes between P4 and P60 even though differences were calculated using independent samples, thus supporting the use of P14 vs. E18 as a surrogate measure for P60 vs. E18 (Fig. 2d, black line, r = 0.48, p < 0.001). Multiple linear regression was used to examine the sex differential response more closely (Table 1a, b, bottom row). Using multiple linear regression, developmental changes in gene expression levels explained 12% of the variance in the male response to LPS (Pearson’s R2=0.12***, Table 1a, Development, Model 2) and 0% of the female response (Pearson’s R2=0.00, Table 1b, Development, Model 2). Thus, in addition to the transcriptional response shared with female microglia, male but not female microglia have a transcriptional response that moves along the developmental program in response to LPS, suggesting the possibility that movement along the developmental gene expression program may be necessary for an inflammatory male response to LPS. This was further confirmed by strong positive correlation between gene expression log differences in the male LPS response and microglial development (Fig. 2d, blue line, r = 0.31, p < 0.001). In contrast, no correlation was found between the female LPS response and development (Fig. 2d, red line, Pearson r = 0.01, NS).
Gene expression changes in microglia following an immune challenge are related to development
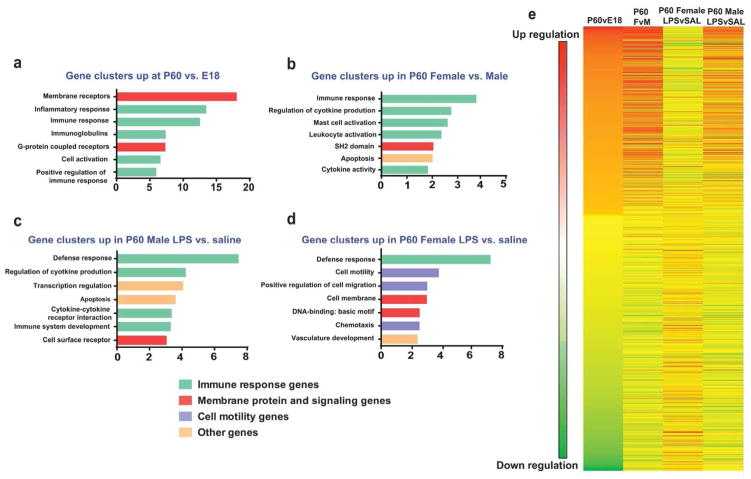
To determine the functional gene changes that occur over development and following LPS challenge in male and female microglia, we performed gene functional clustering analysis of RNA Sequencing data using DAVID software. The top 1000 genes that were up- or down-regulated over development, between males and females, and after LPS treatment were categorized based on common or shared functional groups. Many immune-related gene clusters (green bars) were up-regulated over development (Fig. 3a), and in P60 females vs. males at baseline (Fig. 3b). Functional clusters that change over development were also up-regulated in males following LPS treatment (Fig. 3c), whereas a very different group of gene clusters was up-regulated in LPS-treated females (Fig. 3d), supporting the analyses described in Figure 2. Heat map analysis illustrates that genes that are significantly up-regulated at P60 compared to E18 are very similar to genes that are significantly up-regulated at baseline in females compared to males at P60, and in males following LPS treatment; and vice versa for down-regulated genes (Fig. 3e), further supporting the hypothesis that the sex difference in microglial gene expression is a consequence of differential developmental maturity, and that this is closely related to immune or defense-related gene expression. Further, the changes in gene expression taking place between P60vE18, P14vE18 and P60vP4 are largely the same, further confirming that gene expression changes taking place over development are monotonic.
Figure 3. Gene expression changes in microglia following an immune challenge are related to development.
Top 1000 genes were selected between different group comparisons to input into DAVID gene functional annotation software (https://david.ncifcrf.gov/tools.jsp). Top 7 highly enriched gene functional groups were chosen for representation of group differences: (a) P60 vs. E18, (b) P60 females vs. males, (c) P60 male LPS vs. SAL, (d) P60 female LPS vs. SAL. Immune response genes are represented as green bars, membrane protein and signaling molecules as red bars, cell motility genes as purple bars and miscellaneous genes as orange bars. (e) Heat map of gene expression changes depicts up- or down-regulation of individual genes in different group comparisons. Red = up-regulation, blue = down-regulation.
Several of these immune function-related genes were validated using qPCR, demonstrating that indeed, immune gene expression over development was related to the sex difference at P60, as well as the sex-specific response to peripheral LPS challenge (Supp. Fig. 1). Similarly, developmental gene expression changes for select genes (up and down regulated gene fold changes) from the RNAseq dataset were plotted (Supp. Fig. 2) to illustrate that gene expression patterns of individual genes over development were similar to sex differences at baseline.
Microglial gene expression in females remains consistent across the estrous cycle
Next, we tested whether microglial gene expression changes varied over the estrous cycle of females. Estrous phase of cycling females was determined at P60 by performing vaginal swabs and visualizing the cells under a bright field microscope. Stage of estrous cycle was determined by identifying stereotypical morphology of cells obtained from the vaginal swab as described previously (Goldman et al., 2007). Three females belonging to each phase of the estrous cycle: proestrous, estrous and diestrous, were identified for sacrifice, hippocampal dissection and CD11b isolations. Microglia were also isolated from age-matched males for comparison. There was no significant effect of estrous phase on any gene examined in females (Supp. fig. 3). However, we did continue to observe the same significant sex (male vs. female) difference across all genes, assessed using ANOVA (Supp. table 1) and multiple linear regression (Supp. table 2), thus supporting our previous RNA-seq data and indicating that the sex differences we observe are not simply a consequence of changes in circulating sex steroid concentrations in young adult females.
Sex differences in MDI strongly correlate with morphology
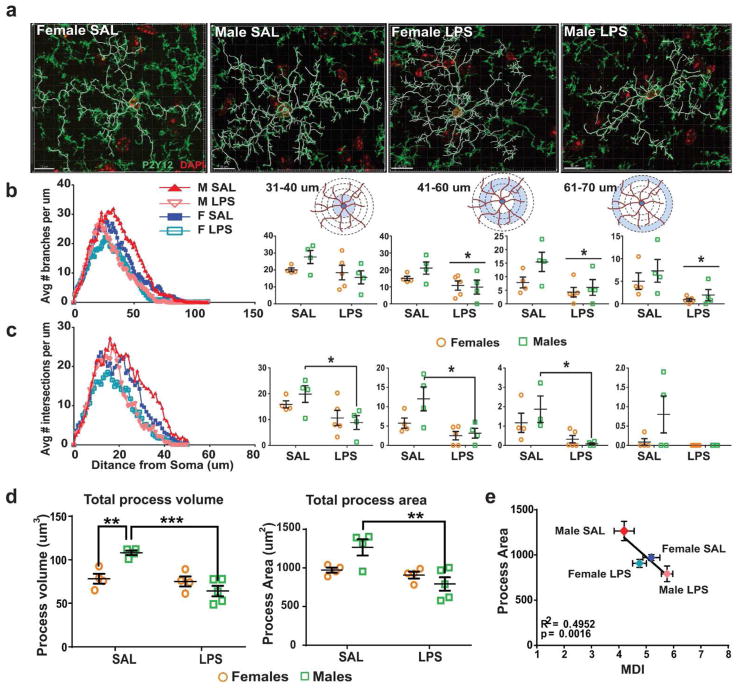
Thus far we have demonstrated in mice that (i) Microglial developmental index tracks with the chronological age of mice, (ii) At P60, female MDI is higher than male MDI, and (iii) Only male microglia are developmentally reactive to an immune challenge assessed at P60. In order to validate the gene expression data using an independent measure correlated with microglial function, we analyzed microglial morphology in the brains of the same mice used for transcriptome analysis at P60. We have previously reported sex differences in microglial cell number and morphology at distinct developmental time points in rats (Schwarz et al., 2012). However, microglial morphology has never to our knowledge been directly correlated to gene expression within the same brains, which is critical to understand function. We performed 3D morphometric reconstructions of individual immunolabeled microglia within the hippocampus of P60 mice. To select a sub-region of hippocampus to target for reconstructions, we first assessed global changes in microglial surface area using an integrated density/thresholding technique of the ionized calcium-binding adaptor molecule (Iba)-1 protein (Supp. fig. 4a). There was a significant sex difference in the CA3 sub-region with females having lower values than males (p = 0.0328) (Supp. fig. 4b); thus we focused on this area for 3D morphometric reconstructions. Alternate sections of the same brains were stained for the microglial-specific marker P2Y12, which is superior to Iba1 for visualizing processes (Fig. 4a) The average number of branches per μm and the average number of process intersections with 1 μm concentric spheres (Sholl analysis) were determined (Fig. 4b, c). Similar to the sex differences observed in MDI analysis at P60, we found significant differences in microglial morphology between males and females at baseline. Specifically, microglial process volume, process area, number of branches and number of intersections were higher in males compared to females at baseline. LPS significantly decreased branching and number of intersections at intermediate and distal lengths from the cell body (41–70 μm) in males. Similarly, LPS led to a reduction in total process volume and process area in males alone (Two-way ANOVA; interaction ** p < 0.01; Tukey’s multiple comparison, SAL males vs. LPS males *** p < 0.001; SAL female vs. SAL male (area) **p < 0.01; Fig. 4d). To determine whether microglial gene expression and morphology are related, we plotted the MDI of each sample with the averaged process area of all microglia analyzed for the same sample. There was a highly significant inverse correlation between the MDI, and total process area, which was segregated by group; i.e. the lower the MDI, the greater the process area, and LPS had a much larger impact on both measures in males than in females, by accelerating microglial development and reducing process area in males (Linear regression; R2 = 0.4952, p = 0.0016; Fig. 4e), but not in females. The same was true for other measures of morphology such as total process volume; that is, total process volume correlated inversely to MDI (Supp. fig. 5).
Figure 4. Sex differences in MDI strongly correlate with morphology.
(a) C57BL/6 male and female P60 mice were sacrificed 2 hours after saline or LPS (330 ug/kg, i.p.) injections following which brains were post-fixed in 4% PFA and processed for immunostaining. Brain slices (40 μm) were stained with the P2Y12 marker for microglia, and imaged on a confocal microscope at 100X magnification. 3D reconstruction was performed and filament tracings were carried out using Imaris software. 2–3 microglia per slice, 2–3 slices per brain and 3–4 brains per group were analyzed. (b, c) Using Sholl analysis, number of branches and intersections were measured at increasing distances from the soma. Values for every animal were averaged (two-way ANOVA, post hoc p < 0.05). (d) Total process volume and total process area were calculated using Imaris software for each microglia, and averaged across cells and slices for every sample (two-way ANOVA, post hoc p < 0.05). (e) Averaged microglial developmental index for a group was correlated to the averaged microglial process area for each sample (Correlation, Pearson’s R = 0.4952, ** p < 0.0016).
Microglial developmental index applied to transcriptomes from heterogeneous brain tissue
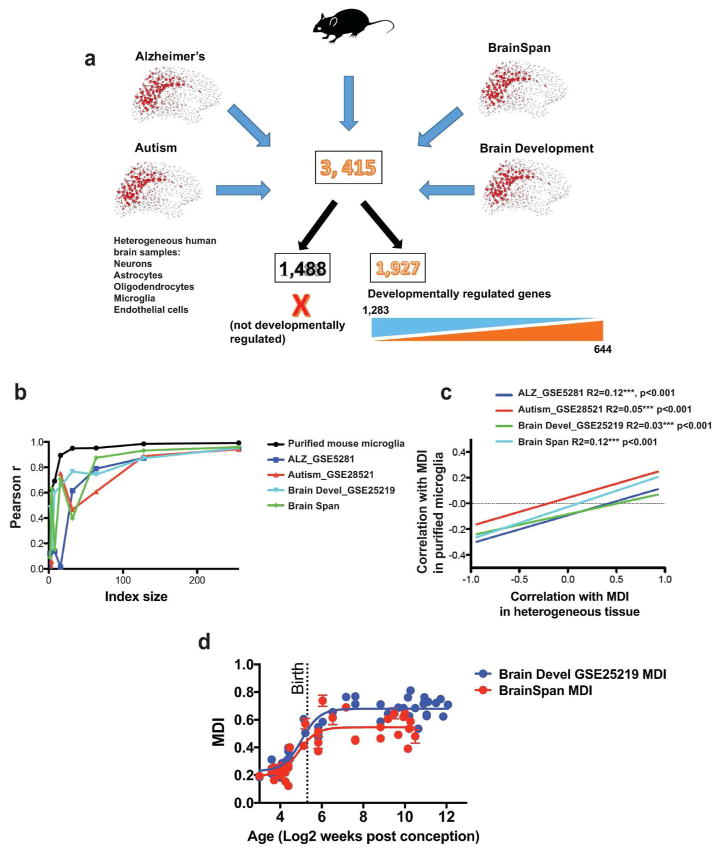
To apply our MDI to brain data gathered across multiple datasets, genes were limited to only those that were detected across all human datasets and in the current mouse study (Fig. 5a). Expression levels were averaged in cases where more than one measure was performed on a given gene as is frequently the case with microarray technology. Four different human brain datasets were selected for validation purposes: An Alzheimer’s disease dataset (Geo Accession Database: GSE5281), an autism dataset (Geo Accession Database: GSE28521), and 2 brain development datasets (GSE25219 - Spatio-temporal transcriptome of the human brain - https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse25219), and the BRAINSPAN dataset (http://www.brainspan.org/static/download.html). There were 3,415 genes detected across all platforms of which 644 were up-regulated in our full MDI, 1,283 were down-regulated, and 1,488 were not used to calculate MDI because they were not significantly developmentally regulated in purified microglia (Fig. 5a). To evaluate the reliability of the new MDI to detect a consistent measure of microglial development within each dataset from heterogeneous human tissue, the effect of MDI size on the correlation between independent indices was measured as described above in pure mouse microglia. Random non-overlapping sets of microglial developmental genes were used to calculate MDI for multiple datasets (Supp. Fig. 6). For each dataset 10 iterations were performed where index size increased by a factor of 2 from 2 to 256 developmentally up and down-regulated genes. As in the full index gene expression levels were scaled and averaged such that MDI equaled the average of developmentally up-regulated genes divided by the average of down-regulated genes. In all cases the correlation between MDI calculated with non-overlapping sets of genes increased as the size of the sets of genes increased with a Pearson r > 0.82 across all iterations and datasets when the number of genes reached 256 (Supp. Fig. 6). A representative iteration from each dataset was compared to the effect of index size in pure tissue (Fig. 5b – blue, red, cyan, green). Not surprisingly, concordance was slightly lower in heterogenous tissue than in purified cells (Fig. 5b, black line), but nonetheless concordance demonstrated a similar but slightly lower plateau between a Pearson r of 0.85 to 0.95 using independent sets of 256 up and down-regulated genes to calculate MDI. This strong concordance indicates that developmentally regulated microglial genes in the heterogeneous tissues are co-regulated in the same manner (i.e., with the same magnitude and in the same direction) as in the purified mouse microglia.
Figure 5. Microglial developmental index applied to transcriptomes from heterogeneous brain tissue.
(a) Four different human brain dataset platforms were selected for validation purposes: Brain Span dataset (http://www.brainspan.org/static/download.html), human brain developmental datasest (BRAIN DEVL_GSE25219), Alzheimer’s dataset (ALZ_GSE5281) and Autism dataset (AUTISM GSE28521). There were 3,415 genes detected across all platforms of which 644 were up-regulated in our full MDI, 1,283 were down-regulated, and 1,488 were not used to calculate MDI because they were not significantly developmentally regulated in purified microglia. (b) The effect of index size on the correlation between independent indices was measured. Gene group size was progressively increased by 2-fold from 2 to 256 up- and down-regulated genes. MDI was calculated on these independent sets of genes and results were correlated. (c) Common genes across 4 different platforms (and used in purified microglia index) were identified to generate the new indices for heterogeneous tissue. The gene expression was then correlated to its respective MDI for every sample in both pure microglia and heterogeneous human sample. The two correlates (gene expression with heterogeneous and pure microglia MDI) were further compared to obtain a highly significant positive correlation between pure and heterogeneous tissue (Pearson’s correlation; Alzheimer’s disease GSE5281 R2 = 0.12 ***, p < 0.001; Autism GSE28521 R2 = 0.05 ***, p < 0.001; Brain developmental dataset GSE2519 R2 = 0.03 ***, p <0.001; BrainSpan R2 = 0.12 ***, p < 0.001). (d) The subindex calculated above was used to track microglial development in two human brain developmental data sets. Data were fit using least squares fitting to a log(agonist) vs. response curve (Nonlinear fit, Brain Development GSE25219: MGLA IDX – R2 = 0.8057; BrainSpan: MGLA IDX – R2 = 0.5837).
A second way to demonstrate that the calculated MDI for human datasets indeed is a specific measure of microglial developmental gene expression patterns within a heterogeneous sample, is to examine how gene expression levels are related to MDI in heterogeneous and pure samples. If MDI calculated in heterogeneous human samples truly measures the development of microglia within the heterogeneous tissue, then gene expression levels should be related to MDI in heterogeneous tissue in a similar manner to that of purified microglia samples. That is, if the expression level of “gene X” is positively correlated with MDI in purified microglia then it should also be positively correlated with MDI in heterogeneous tissue. To perform this analysis, the expression levels of the identified common genes were correlated to MDI in each study, e.g. the expression level of gene X across all samples in a study was correlated with MDI across all samples in the same study, thus all genes have a Pearson r value for each study indicating the relationship of that gene to MDI in that study, which quantifies the direction and magnitude of the relationship. Pearson r values of genes in heterogeneous tissue were then related to the Pearson r values of the same genes in pure microglia. This directly tested the hypothesis that developmentally regulated microglial genes would be similarly related to MDI in pure and heterogeneous tissue and provide support that MDI is measuring microglial development in heterogeneous tissue. Indeed a highly significant positive correlation was found indicating that specific genes are related to MDI in the same way in pure and heterogeneous tissue (Fig. 5c).
Another interpretation however, could be that similar expression changes in other cell types in the heterogeneous sample were responsible for changes in index measurements for the entire sample. In other words, the gene expression patterns that we are detecting in the heterogenous human samples were not actually from microglia, because non-microglial cells might also have similar developmental gene expression changes. It would not be unexpected for the development of other cell types to covary with microglia development either through direct causality, e.g. microglial maturity influences the maturity of cell type X, or indirectly with another factor influencing both cell types such as in normal aging. However, it is important to know that changes in MDI do relate to microglial development and are not solely related to another cell type. To confirm that MDI was indeed estimating the maturity of microglia in heterogeneous samples we utilized the publicly available Brain RNA-seq database (http://web.stanford.edu/group/barres_lab/brain_rnaseq.html) to identify 20 highly enriched genes in microglia (in comparison with other brain cell types; 1st and 2nd columns in Table 2) that were also detected across all 4 human datasets, and were highly correlated with MDI in pure microglia. According to the Brain RNA-seq database, these genes are enriched in microglia anywhere from 136 fold (csf1r) to 23 fold (zfp36), compared to all other cell types. These microglial enriched genes were also developmentally regulated as indicated by a highly positive correlation with MDI in pure microglia (Table 2, Pearson r > 0.84) and developmental fold-changes > 1.2. Individual gene expression levels of these 20 genes were correlated with MDI in the 2 human brain development studies and 2 studies of human brain in the context of disease, autism and Alzheimer’s disease. Considering only these 20 genes in the 4 studies above, we found that, in 58 out of the 80 instances (20 genes × 4 studies) (72%), microglial enriched genes were positively correlated with MDI in contrast to 5,254/12,500 (42%) of the remaining genes, i.e. genes not used to generate the full MDI Chi-square = 29.01, p<0.0001). Thus, microglial-enriched developmentally regulated genes were highly correlated with MDI in all gene expression studies used in our analyses indicating that MDI predicted developmental variability in microglia.
Table 2.
Microglial enriched genes that are positively correlated with MDI in pure developing microglia are also positively correlated with MDI in heterogeneous tissue from multiple gene expression studies where microglia contribute only a fraction of total mRNA.
| Barres Brain RNA-seq | Fold-change P60/E18 (microglia RNAseq) | Correlation: (Pearson R) with MDI | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Enrichment in Microglia vs. other CNS cells (Barres Brain RNAseq database) | In pure microglia | BRAIN DEVELOP GSE25219 n=1340 | BRAIN SPAN n=524 | ALZHEIMER S GSE5281 n=161 | AUTISM GSE28521 n=51 | Avg. R across datasets | |
| csf1r | 136.58 | 1.26 | 0.89*** | 0.41*** | 0.26*** | 0.32*** | 0.29* | 0.36 |
| slc2a5 | 135.14 | 1.97 | 0.90*** | 0.58*** | 0.38*** | 0.13 | 0.43** | 0.32 |
| itgb2 | 114.17 | 1.4 | 0.91*** | 0.42*** | 0.17*** | −0.03 | 0.21 | 0.22 |
| cx3cr1 | 74.98 | 1.47 | 0.88*** | 0.26*** | −0.01 | −0.1 | −0.16 | −0.02 |
| inpp5d | 72.97 | 1.47 | 0.90*** | 0.67*** | 0.52*** | 0.47*** | 0.50*** | 0.52 |
| capn3 | 69.46 | 2.09 | 0.92*** | 0.78*** | 0.60*** | 0.17* | 0.56*** | 0.46 |
| hexb | 67.74 | 1.27 | 0.93*** | 0.63*** | −0.15*** | −0.32*** | −0.51*** | −0.32 |
| mlxipl | 51.74 | 2.17 | 0.85*** | 0.70*** | 0.41*** | −0.24** | 0.40** | 0.08 |
| ly86 | 49.28 | 1.22 | 0.89*** | 0.30*** | 0.22*** | −0.02 | 0.07 | 0.21 |
| itgb5 | 47.48 | 1.3 | 0.88*** | 0.60*** | 0.52*** | 0.60*** | 0.67*** | 0.57 |
| rgs10 | 45.87 | 1.4 | 0.84*** | 0.02 | 0.27*** | 0.11 | −0.07 | 0.22 |
| rab3il1 | 44.83 | 1.48 | 0.89*** | −0.10*** | 0.04 | 0.24** | 0.73*** | 0.3 |
| cd84 | 36.76 | 1.38 | 0.90*** | 0.53*** | 0.25*** | 0.31*** | 0.31* | 0.36 |
| sla | 35.76 | 1.59 | 0.94*** | −0.65*** | −0.59*** | 0.31*** | 0.40** | 0.13 |
| lair1 | 33.22 | 1.57 | 0.93*** | 0.49*** | 0.45*** | 0.06 | 0.37* | 0.34 |
| mafb | 31.36 | 1.77 | 0.92*** | −0.34*** | −0.32*** | 0.26** | −0.58*** | −0.18 |
| ier5 | 30.18 | 1.38 | 0.93*** | −0.38*** | −0.45*** | 0.03 | −0.57*** | −0.29 |
| hk2 | 29.44 | 1.85 | 0.94*** | −0.36*** | −0.30*** | 0.12 | 0.63*** | 0.25 |
| ptk2b | 24.97 | 1.6 | 0.85*** | 0.63*** | 0.31*** | −0.63*** | 0.11 | −0.17 |
| zfp36 | 23.13 | 1.42 | 0.93*** | 0.42*** | 0.22*** | 0.43*** | 0.50*** | 0.4 |
Table shows the Pearson R values for the correlation with MDI of the top 20 microglial enriched genes that were most correlated with MDI in pure microglia. Looking at 4 datasets from heterogeneous tissue 58 out of 80 (4 datasets × 20 genes) gene measurements from heterogeneous tissue were positively correlated (red) with MDI (72.5%) compared to 5,254/12,500 (42%) for the remaining genes (Chi-square = 29.01, p<0.0001). Significant correlations are indicated with asterisks
p<0.05,
p<0.01,
p<0.005.
Thus, these data support the utility of developmental indices for measuring the same biological construct–microglial development-in both pure and heterogeneous cell populations. Hence we measured microglial development across time in the two human developmental datasets using the subindex above. Data were fit using least squares fitting to a log(agonist) vs. response curve (Fig. 5d, Nonlinear fit, Brain Development GSE25219: MGLA IDX – R2 = 0.8057; BrainSpan: MGLA IDX – R2 = 0.5837). Plots demonstrate a logarithmic increase in the mid-to-late prenatal period plateauing toward the end of the first postnatal year in humans. All correlations remained highly significant (p < 0.001) in multiple linear regression models after controlling for brain region variability (data not shown). Thus, as expected MDI increased across development in heterogeneous human brain samples.
Microglial development is accelerated in brains of Alzheimer’s disease and autism patients
Previous work utilizing the developmental index approach has demonstrated that fast-spiking interneuron maturation is disrupted (delayed) in neuropsychiatric disorders (Gandal et al., 2012). To test whether microglial development is altered in disease, MDI was quantified using gene expression data of patients with Alzheimer’s disease (AD) and autism, and compared to controls as discussed above. MDI was significantly increased in AD samples compared to control samples in almost all brain regions tested (Fig. 6a, two-way ANOVA, Region p< 0.0001; Disease p < 0.0001). That is, the genes that were up-regulated over normal development in microglia showed enhanced up-regulation in AD samples (and vice versa for down-regulated genes) compared to controls. Results remained highly significant (p<0.0001) after controlling for age-related and regional variability. These data suggest that microglial development is accelerated in AD tissue compared to controls. Interestingly, the greatest disease-related effects on MDI were seen in the entorhinal cortex where pathologic changes in AD are first noted (Du et al., 2001; Khan et al., 2014; Velayudhan et al., 2013), suggesting that microglial development might be highly accelerated in brain regions of exacerbated pathology. MDI was also significantly increased in autism samples compared to controls (Fig. 6b, two-way ANOVA, MDI * p < 0.05). Results remained significant (p<0.05) after controlling for age-related and regional variability. These data collectively suggest that, similar to age effects in normal development, disease state is related to microglial development. In other words, in two samples of matched ages, diseased state is associated with increased microglial maturity – at least in the case of AD and autism. Due to the limited number of human samples, sex-dependent analyses were not possible in the AD and autism datasets, which is of significant interest given that each of these disorders is associated with a sexual dimorphism in severity or incidence.
However, a recent study reporting sexually dimorphic pathways in autism (Werling, Parikshak, & Geschwind, 2016), found that the genes expressed at higher levels in normal adult males (18y+, BrainSpan developmental dataset) compared to females, were also significantly enriched in post-mortem autistic brains; and these genes are primarily associated with astrocyte and microglia markers (Werling et al., 2016). We hypothesized that what these authors were detecting might be differences in microglial maturity, i.e. adult males in the BrainSpan study and autistic individuals both have more mature microglia than females and controls, respectively. To test this hypothesis we calculated MDI in the same samples (age: 18y+ of the human BrainSpan dataset) assessed by the Werling group, and looked at males and females separately. Strikingly, we found that MDI was significantly increased in male brains of 18+ years of age compared to females (Fig. 6c), suggesting the male microglia are more developmentally mature than female microglia in adulthood. Results remained highly significant (MDI: p<0.01) after controlling for age-related, regional, and individual variability.
DISCUSSION
An interesting recent study investigating transcriptional and epi-genomic regulation of microglia throughout brain development showed that microglial development proceeds through three distinct temporal stages: early microglia (until E14), pre-microglia (E14 to early postnatal weeks), and adult microglia (Matcovitch-Natan et al., 2016). At each stage of development, microglia evolve distinct pathways for processing relevant signals from the environment and to maintain homeostasis within the brain. This was supported by a recent study demonstrating that function of α5β1 integrin in microglia varies based on age during early colonization of the developing mouse cortex, based on microglial migration levels (Smolders et al., 2017). This is relevant because neurological disorders are increasingly viewed as originating from disrupted normal developmental trajectory or maturation of glial cells as well as neurons (Marin, 2016).
Recently, it was shown that maternal immune activation with the viral mimic poly I:C can disrupt the pre-microglial developmental stage in offspring and push the microglial transcription state to a more advanced developmental stage (Matcovitch-Natan et al., 2016). Similarly, we have demonstrated that LPS challenge in late adolescent mice accelerates microglial transcriptional maturation, but only in males. The developmental index described here was based on an approach previously published where a cell-type specific maturation index was generated from microarray datasets of fast-spiking interneurons, which was then applied to human transcriptome datasets to reveal reduced transcriptome maturation of FS interneurons in diseases such as bipolar disorder and schizophrenia (Gandal et al., 2012). In the current study, we generated a microglia-specific developmental index (MDI) using transcriptome profiling of purified mouse microglia across development and applied it to developmental and diseased human datasets. Overall, 7,683 genes were differentially expressed in mice across development between E18 and P60. Of the developmentally regulated genes, 4,645 genes were down-regulated and 3,038 genes were up-regulated over development. As the majority of genes changed monotonically up or down we found the average of developmentally up-regulated divided by down-regulated genes was an accurate estimate of microglial maturity in pure (mouse) and heterogeneous (human) tissue.
Using MDI we demonstrate that female microglia transcriptomes were developmentally advanced compared to male transcriptomes at P60 and that male but not female microglia transcriptomes had a developmental response to LPS. These gene expression changes were strongly corroborated by the sex difference in microglial morphology at baseline, as well as the change in microglial morphology specifically in males following LPS treatment, indicating that transcriptome maturity relates to morphological differences. Specifically, we saw that LPS treatment pushed male microglia to a more mature transcriptome state and at the same time, reduced their process complexity suggesting that morphological changes observed by others in activated microglia are likely associated with an underlying change in transcriptome maturity (and not necessarily “reactivity”). Because RNAseq and microglial morphology were performed on the same mice we were able to correlate these measures providing a strong level of support for the transcriptome-morphology relationship that would not be possible if measurements were done in different cohorts of mice. The linking of MDI to morphology gives a striking example of the power of this technique to inform observations with an understanding of gene expression system behavior: in this case, morphology linked to the development of the microglial transcriptome and, in humans, of microglial development linked to disease. In contrast to approaches that focus on changes in specific genes, MDI measurements provide information about the behavior of large groups of genes covarying as part of a developmental gene expression program. Thus, MDI and other global approaches to gene expression analysis aim to describe the state of the transcriptome and the cells in which transcriptomes operate. For instance, when we measured MDI in pure microglia and human developmental datasets, we found that MDI increased over time indicating that MDI increased with development as expected.
Another important observation in the current study was that, at least in the case of LPS treatment, there was a distinction between an LPS response that was shared between males and females and a LPS-induced transcriptome developmental response that occurred only in males. Importantly, morphological changes in response to LPS were also only seen in males suggesting that what has previously been reported as microglial activation in association with multiple conditions including disease state, may in fact represent variability in microglia maturity. Because microglia are immune cells, development will involve up-regulation of immune genes as microglia take on their mature cell-type specific immune characteristics. Thus researchers “looking under the lamp post” might assume microglia activation is occurring when they measure immune genes because before the current study microglia developmental genes have not been described and immune genes have been the focus of previous gene expression studies in microglia. Supporting our alternative interpretation of previous studies we demonstrate that all microglial developmental genes, not simply immune genes, are up-regulated in the context of autism and Alzheimer’s disease, which we show is because microglia are more transcriptionally mature in the brains of these individuals.
A steadily increasing publicly available set of gene expression studies of brain provides a tremendous resource for researchers asking important questions not addressed in the original studies during which data were collected. However, a serious limitation of these datasets is that transcriptional analyses are generally performed on heterogeneous tissue meaning that the transcriptomes of multiple cell types are mixed together. Approaches such as weighted gene co-regulatory network analysis (WGCNA) have demonstrated that even within these heterogeneous data it is nonetheless possible to identify patterns of gene expression co-regulation that appear in at least some cases to be associated with functional variability of specific cell types (Langfelder & Horvath, 2008, 2012). In our study, MDI tracked with development in heterogeneous tissue supporting an ability of MDI to estimate microglial development even when microglia are a small percentage of the total number of cells. Further validation was provided by using microglial-enriched genes obtained from the Barres Brain RNA-seq database to demonstrate that MDI tracked with developmentally regulated microglia-enriched genes. Next, having established that MDI was a robust measurement of microglial development in heterogeneous tissue, we used MDI calculations to test the hypothesis that reports of microglial involvement in brain diseases might be related to changes in microglial transcriptome during development. In support of this hypothesis, microglial development was exaggerated in brains of Alzheimer’s disease and autistic patients compared to the controls.
Indeed, the cell-type specific developmental index approach described for this study has been utilized successfully in the past to quantify developmental maturity of fast-spiking interneurons in human brain, and demonstrated disease-associated differences in the maturity of these important cells in autism and schizophrenia (Gandal et al., 2012). This technique was validated in microglia for the current study in multiple ways by demonstrating that (1) MDI increased with age in 2 large studies of human brain development, (2) microglial developmental genes used for the index covaried with each other in brain tissue, (3) MDI measurements in brain were correlated with the expression levels of multiple microglial enriched genes, and (4) microglial developmental genes covaried with MDI in the same direction and relative magnitude in pure microglia and heterogeneous brain tissue.
One of the striking findings in this study is that microglial transcriptional development is accelerated in a sex-dependent manner in adult mice and humans. When we applied our microglia-specific index to adult humans from the BrainSpan dataset, we saw that male microglial transcriptome was more developmentally mature than female microglia. In this same dataset, Werling et al showed that autism-susceptibility genes (which are mostly astrocyte and microglia specific) are also increased specifically in adult males (Werling et al., 2016), which might explain in part the male-biased autism incidence. The human data showing MDI in adult males vs. adult females is seemingly in contradiction to our mouse data, where during normal development, and in early adulthood, MDI in general is higher in females than in males. This could indicate a dissociation between microglial genes in general, and those that are associated with autism specifically. However, we also show in the current study that male microglial transcriptome profile is more developmentally sensitive to LPS stimulation. Thus one possibility is that because humans do not develop in artificially sterile conditions (in contrast to mice), that normal exposure to infectious agents may have a greater effect on males than females and thus account for the increased maturity of male microglia in the Werling study. This possibility is supported by growing evidence that inflammatory events during development (e.g., infections, toxin exposure) can alter central and/or peripheral immune function and therefore the risk of disorders such as autism (Goines & Ashwood, 2013; Schmidt, Lyall, & Hertz-Picciotto, 2014). In contrast, females have increased risk for disorders like AD that emerge later in life. Thus, future studies are needed to determine if sex differences in microglia sensitivity vary with age, and these studies are currently ongoing.
In conclusion, the current study utilizes a novel approach to assess microglial development in human development and disease, using gene expression data from purified and mixed cell types. These data point to the idea that the mechanisms driving development and immune reactivity in microglia are overlapping but potentially dissociable in males vs. females, and can be used to provide novel insight into healthy versus diseased human brain.
Supplementary Material
Main points.
Microglial development is related to its immune reactivity.
Male microglia become more developmentally mature in response to immune challenge.
Microglial developmental index can be used to examine microglial development in heterogeneous brain tissue samples across different platforms.
Acknowledgments
We would like to thank the Next Generation Sequencing Core at University of Pennsylvania for performing RNAseq and whole transcriptome profiling. Supported by R01 MH101183 to SDB.
Footnotes
The authors declare no competing financial interests.
References
- Alter MD. Studying gene expression system regulation at the program level. PLoS One. 2013;8(4):e61324. doi: 10.1371/journal.pone.0061324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank T, Prinz M. Microglia as modulators of cognition and neuropsychiatric disorders. Glia. 2013;61(1):62–70. doi: 10.1002/glia.22372. [DOI] [PubMed] [Google Scholar]
- Buchstaller J, Sommer L, Bodmer M, Hoffmann R, Suter U, Mantei N. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci. 2004;24(10):2357–2365. doi: 10.1523/JNEUROSCI.4083-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, … Barres BA. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canetta S, Bolkan S, Padilla-Coreano N, Song LJ, Sahn R, Harrison NL, … Kellendonk C. Maternal immune activation leads to selective functional deficits in offspring parvalbumin interneurons. Mol Psychiatry. 2016;21(7):956–968. doi: 10.1038/mp.2015.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B, Mwangi B, Hasan KM, Selvaraj S, Zeni CP, Zunta-Soares GB, Soares JC. Development and validation of a brain maturation index using longitudinal neuroanatomical scans. Neuroimage. 2015;117:311–318. doi: 10.1016/j.neuroimage.2015.05.071. [DOI] [PubMed] [Google Scholar]
- Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, … Weiner MW. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 2001;71(4):441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Nishio K, Miyamoto S, Hiramatsu KI, Sakaki T, Okuchi K, … Fujioka S. Hippocampal damage in the human brain after cardiac arrest. Cerebrovasc Dis. 2000;10(1):2–7. doi: 10.1159/000016018. [DOI] [PubMed] [Google Scholar]
- Gandal MJ, Nesbitt AM, McCurdy RM, Alter MD. Measuring the maturity of the fast-spiking interneuron transcriptional program in autism, schizophrenia, and bipolar disorder. PLoS One. 2012;7(8):e41215. doi: 10.1371/journal.pone.0041215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA, Campbell BM. Glial biomarkers in human central nervous system disease. Glia. 2016;64(10):1755–1771. doi: 10.1002/glia.22998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goines PE, Ashwood P. Cytokine dysregulation in autism spectrum disorders (ASD): possible role of the environment. Neurotoxicol Teratol. 2013;36:67–81. doi: 10.1016/j.ntt.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman JM, Murr AS, Cooper RL. The rodent estrous cycle: characterization of vaginal cytology and its utility in toxicological studies. Birth Defects Res B Dev Reprod Toxicol. 2007;80(2):84–97. doi: 10.1002/bdrb.20106. [DOI] [PubMed] [Google Scholar]
- Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, … Pierce EA. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27(18):2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburg H, Trossbach SV, Bader V, Chwiesko C, Kipar A, Sauvage M, … Korth C. Simultaneous effects on parvalbumin-positive interneuron and dopaminergic system development in a transgenic rat model for sporadic schizophrenia. Sci Rep. 2016;6:34946. doi: 10.1038/srep34946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamsagar R, Bilbo SD. Sex differences in neurodevelopmental and neurodegenerative disorders: Focus on microglial function and neuroinflammation during development. J Steroid Biochem Mol Biol. 2015 doi: 10.1016/j.jsbmb.2015.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan UA, Liu L, Provenzano FA, Berman DE, Profaci CP, Sloan R, … Small SA. Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer’s disease. Nat Neurosci. 2014;17(2):304–311. doi: 10.1038/nn.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46(11) [PMC free article] [PubMed] [Google Scholar]
- Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6(4):312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- Marin O. Developmental timing and critical windows for the treatment of psychiatric disorders. Nat Med. 2016;22(11):1229–1238. doi: 10.1038/nm.4225. [DOI] [PubMed] [Google Scholar]
- Matcovitch-Natan O, Winter DR, Giladi A, Vargas Aguilar S, Spinrad A, Sarrazin S, … Amit I. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353(6301):aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- Morishita H, Kundakovic M, Bicks L, Mitchell A, Akbarian S. Interneuron epigenomes during the critical period of cortical plasticity: Implications for schizophrenia. Neurobiol Learn Mem. 2015;124:104–110. doi: 10.1016/j.nlm.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty BW, Miller MN, Sugino K, Hempel CM, Nelson SB. Transcriptional and electrophysiological maturation of neocortical fast-spiking GABAergic interneurons. J Neurosci. 2009;29(21):7040–7052. doi: 10.1523/JNEUROSCI.0105-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskvig DB, Elkahloun AG, Johnson KR, Phillips TM, Herkenham M. Maternal immune activation by LPS selectively alters specific gene expression profiles of interneuron migration and oxidative stress in the fetus without triggering a fetal immune response. Brain Behav Immun. 2012;26(4):623–634. doi: 10.1016/j.bbi.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petito CK, Feldmann E, Pulsinelli WA, Plum F. Delayed hippocampal damage in humans following cardiorespiratory arrest. Neurology. 1987;37(8):1281–1286. doi: 10.1212/wnl.37.8.1281. [DOI] [PubMed] [Google Scholar]
- Salmenpera T, Kalviainen R, Partanen K, Pitkanen A. Hippocampal damage caused by seizures in temporal lobe epilepsy. Lancet. 1998;351(9095):35. doi: 10.1016/S0140-6736(05)78092-2. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM, Uno H, Rebert CS, Finch CE. Hippocampal damage associated with prolonged glucocorticoid exposure in primates. J Neurosci. 1990;10(9):2897–2902. doi: 10.1523/JNEUROSCI.10-09-02897.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, … Stevens B. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron. 2012;74(4):691–705. doi: 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Stevens B. The “quad-partite” synapse: microglia-synapse interactions in the developing and mature CNS. Glia. 2013;61(1):24–36. doi: 10.1002/glia.22389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmalbach B, Lepsveridze E, Djogo N, Papashvili G, Kuang F, Leshchyns’ka I, … Schachner M. Age-dependent loss of parvalbumin-expressing hippocampal interneurons in mice deficient in CHL1, a mental retardation and schizophrenia susceptibility gene. J Neurochem. 2015;135(4):830–844. doi: 10.1111/jnc.13284. [DOI] [PubMed] [Google Scholar]
- Schmidt RJ, Lyall K, Hertz-Picciotto I. Environment and Autism: Current State of the Science. Cut Edge Psychiatry Pract. 2014;1(4):21–38. [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Sholar PW, Bilbo SD. Sex differences in microglial colonization of the developing rat brain. J Neurochem. 2012;120(6):948–963. doi: 10.1111/j.1471-4159.2011.07630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolders SM, Swinnen N, Kessels S, Arnauts K, Smolders S, Le Bras B, … Brone B. Age-specific function of alpha5beta1 integrin in microglial migration during early colonization of the developing mouse cortex. Glia. 2017 doi: 10.1002/glia.23145. [DOI] [PubMed] [Google Scholar]
- Velayudhan L, Proitsi P, Westman E, Muehlboeck JS, Mecocci P, Vellas B, … dNeuroMed C. Entorhinal cortex thickness predicts cognitive decline in Alzheimer’s disease. J Alzheimers Dis. 2013;33(3):755–766. doi: 10.3233/JAD-2012-121408. [DOI] [PubMed] [Google Scholar]
- Wang AY, Lohmann KM, Yang CK, Zimmerman EI, Pantazopoulos H, Herring N, … Konradi C. Bipolar disorder type 1 and schizophrenia are accompanied by decreased density of parvalbumin- and somatostatin-positive interneurons in the parahippocampal region. Acta Neuropathol. 2011;122(5):615–626. doi: 10.1007/s00401-011-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling DM, Parikshak NN, Geschwind DH. Gene expression in human brain implicates sexually dimorphic pathways in autism spectrum disorders. Nat Commun. 2016;7:10717. doi: 10.1038/ncomms10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.