Humans serve as both host and reservoir for Mycobacterium tuberculosis (M. tuberculosis), making tuberculosis a theoretically eradicable disease. How M. tuberculosis alternates between host-imposed quiescence and sporadic bouts of replication to complete its life cycle, however, remains unknown. Here, we identify a metabolic adaptation that is triggered upon entry into hypoxia-induced quiescence but facilitates subsequent cell cycle re-entry. Catabolic remodeling of M. tuberculosis's cell surface trehalose mycolates specifically generates metabolic intermediates reserved for re-initiation of peptidoglycan biosynthesis. These adaptations reveal a metabolic network with the regulatory capacity to mount an anticipatory response.
Mycobacterium tuberculosis (M. tuberculosis), the causative agent of tuberculosis (TB), kills more humans than any other bacterium. Yet, humans remain its only major natural reservoir. Replication of the bacillus is slowed or arrested in most hosts, but resumes in some only decades later with a host response that is frequently fatal yet linked to the generation of infectious aerosols that enable completion of its life cycle1. Such delayed replication has allowed infected hosts to have offspring before transmitting the infection2. M. tuberculosis has thus come to depend on regulation of cell cycle exit and re-entry as a critical mediator of pathogenicity and survival as a species.
Hypoxia is a physiologic feature common to many of M. tuberculosis's intra- and extracellular niches in humans and experimentally infected animals3. In vitro, hypoxia arrests M. tuberculosis's replication and renders phenotypic tolerance to nearly all clinically used TB drugs3-5. However, M. tuberculosis also re-encounters atmospheric oxygen when inflammatory host tissue damage has become extensive enough to erode into the airways and facilitate re-entry into cell cycle1. Hypoxia and atmospheric oxygen have thus co-evolved as highly correlated but temporally sequential features of M. tuberculosis's physiology.
Studies of M. tuberculosis's response to hypoxia have revealed a broad and diverse range of responses, most of which have been considered in the physiologic context of situational adaptation6-10. Here, we report a functionally distinct set of metabolic adaptations that are triggered by, but facilitate survival through and recovery from, hypoxia. These adaptations thus reveal a predictive regulatory capability of M. tuberculosis's metabolic network in which the temporal relationship between hypoxia and re-aeration has been encoded into an anticipatory response that links entry into and exit from dormancy.
Using our previously described in vitro system7, we noted that exposure to hypoxia (1% O2, 5% CO2) triggered discrete accumulations of intermediates in the early portion of glycolysis, the pentose phosphate pathway, and aminosugar biosynthesis. These accumulations occurred with glucose or acetate as a carbon source and reversed with re-aeration (Fig. 1; Supplementary Fig. 1A, B). These changes were further linked to reciprocal, hypoxia-dependent decreases in levels of the downstream glycolytic intermediate, phosphoenolpyruvate (PEP), and upstream disaccharide, trehalose, but not further upstream glycogen polysaccharide stores (Supplementary Fig. 2A, B). These changes, albeit to varying degrees, were also durable to experimental limit of this system (4d) without a measurable loss of viability, and dissociated from TCA cycle activity, as tested by inclusion of nitrate, a physiologic alternate terminal electron acceptor capable of supporting near aerobic levels of TCA cycle activity in hypoxic environments (Supplementary Fig. 2C, D)7.
Fig 1. Hypoxia induces remodeling of upper glycolysis, amino sugar and nucleotide sugar biosynthesis and pentose phosphate pathways in M. tuberculosis.
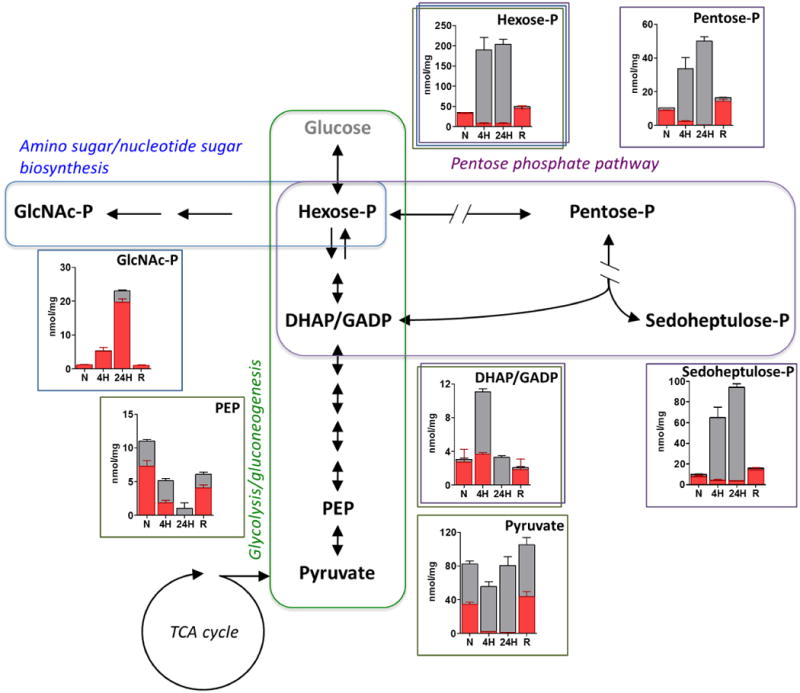
Intrabacterial pool sizes (nmol/mg protein) and isotopic labeling of M. tuberculosis intermediates in: upper glycolysis (green box); amino sugar biosynthesis (blue box); and pentose phosphate pathways (purple box), incubated in [U-13C] acetate-containing media for 24 h at 20% O2 (normoxia; N); 4 h (4H) or 24 h (24H) at 1% O2 (hypoxia; H); and 24 h at 20% O2 following pre-incubation at 1% O2/5% CO2 for 24 h (re-aerated; R). Total bar heights indicate relative intrabacterial concentrations (nmol/mg protein), whereas the red area of each bar denotes the enrichment of 13C labeling achieved following transfer to [U-13C] acetate-containing m7H10 media under the condition indicated. All values are the average of three biological replicates (n=3) ±SEM and representative of 2 independent experiments. DHAP, dihydroxyacetone phosphate; GADP, glyceraldehyde phosphate; GlcNAc-P, N-acetyl glucosamine phosphate; hexose-P, glucose-6-phosphate and its isomers; pentose-P, ribose-5-phosphate and its isomers; PEP, phosphoenolpyruvate; sedoheptulose-P, sedoheptulose-7-phosphate and its isomers.
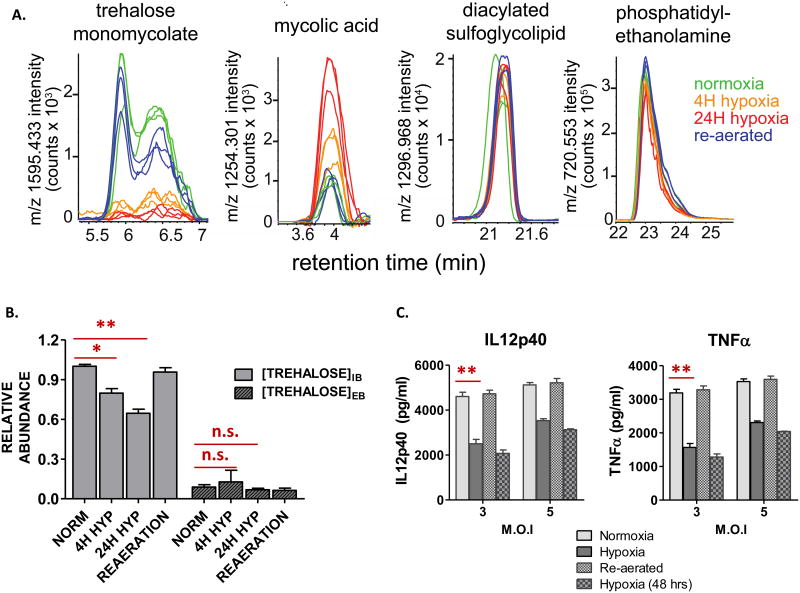
Upon transferring M. tuberculosis from unlabeled to uniformly 13C labeled ([U-13C]) glucose or acetate at the time of exposure to hypoxia, we noted that these metabolite accumulations predominantly and unexpectedly occurred within the unlabeled fraction of each pool (Fig. 1; Supplementary Fig. 1A, B), suggesting catabolism of a pre-existing metabolic store. Trehalose is a highly abundant, non-reducing disaccharide of glucose that can serve as both carbohydrate store and bioprotectant11. In M. tuberculosis, trehalose is also a core component of mycolyl glycolipids. Trehalose mono- (TMM) and di-mycolate (TDM), widely known as cord factor, form the outer lipid barrier of M. tuberculosis's cell envelope and serve as potent triggers of host inflammation12. We therefore hypothesized that the foregoing metabolic accumulations arose from hypoxia-induced catabolism of M. tuberculosis's cell surface trehalose mycolates. Consistent with a recent lipidomic study13, we observed a rapid and extreme depletion of M. tuberculosis's TMMs and TDMs during hypoxia, while levels of sulfolipids and the plasma membrane phospholipid, phosphatidylethanolamine, were unchanged (Fig. 2A; Supplementary Fig. 3). Moreover, we observed linked changes in intra-, but not extra-, cellular levels of unlabeled trehalose (which decreased) and free mycolates (which increased), consistent with the release, re-uptake, and catabolism of mycolyl glycolipid-derived trehalose (Fig. 2A, B).
Fig 2. Hypoxia induces selective alterations in M. tuberculosis cell envelope lipid levels and immunoreactivity.
Time and O2-specific levels of: (A) α-trehalose monomycolate (TMM); free mycolic acid; diacylated sulfolipids; plasma membrane phosphatidylethanolamine (depicted as extracted ion chromatograms of a single, abundant molecular species representative of each lipid class); and (B) intrabacterial (IB) and extracellular (EB) trehalose (expressed as units of relative abundance compared to those of normoxia). (C) Secreted levels of IL-12p40 and TNF following stimulation of mouse bone marrow derived macrophages (BMM) with formaldehyde-fixed M. tuberculosis recovered from aerobic cultures, 24 h and 48 h incubation at 1% O2/5% CO2, or 24 h re-aeration following 24 or 48 h of hypoxic incubation. MOI: multiplicity of infection. All values are the average of three biological replicates (n=3) ± SEM and representative of 2 independent experiments. *p≤0.01; **p≤0.001; n.s. not significant; by Student's unpaired t-test.
TDM is a ligand of the Mincle receptor of macrophages and potent inducer of the pro-inflammatory cytokines IL-12 and TNF12. We hypothesized that the hypoxia-induced decreases in TDM and TMM levels might be accompanied by changes in levels of M. tuberculosis-elicited IL-12p40 and TNF secretion. Incubation of mouse bone marrow derived macrophages with equivalent numbers of paraformaldehyde-fixed M. tuberculosis cells revealed that hypoxia reversibly halved M. tuberculosis's ability to induce both cytokines (Fig. 2C), suggesting a potential functional link between M. tuberculosis's metabolism and immunoreactivity during exit from and re-entry into cell cycle.
To gain further insight into the dynamics of this cell envelope remodeling, we incubated M. tuberculosis in [U-13C] glucose for 2 days to enable labeling of glycolipids, and then transferred to unlabeled glucose for 18h to ‘wash out’ 13C label from glycolytic and pentose phosphate pathway intermediates but maintain 13C enriched pools of labeled trehalose (Supplementary Fig. 4A). We then subjected M. tuberculosis to hypoxia in the presence of unlabeled acetate as the carbon source. As shown in Fig. S4B and C, we observed the same hypoxia-induced changes in trehalose and hexose phosphate pools but now in the 13C labeled fraction of each pool, indicating their origin in pre-existing depots of trehalose mycolates and free trehalose, respectively.
M. tuberculosis encodes 3 annotated enzymatic pathways capable of metabolizing trehalose: OtsA-OtsB, TreS and Rv2402 (Supplementary Fig. 5A)11. Biochemical and genetic studies indicate that the OtsA-OtsB pathway functions primarily in the biosynthetic direction14, while an Rv2402 deletion mutant exhibited no defect in trehalose utilization (Supplementary Fig. 5B). In contrast, both biochemical and genetic studies indicate that TreS is a catalytically competent trehalose synthase capable of reversibly interconverting maltose and trehalose, enabling conversion of trehalose to glucose in vivo14,15. We therefore generated a treS-deletion strain of M. tuberculosis (ΔtreS) and genetically complemented counterpart to test the role and essentiality of treS in the foregoing response (Supplementary Fig. 5C). As expected, ΔtreS M. tuberculosis exhibited a selective growth defect in media containing trehalose, but not glucose, as the sole exogenous carbon source (Supplementary Fig. 5D).
Metabolic profiling studies showed that ΔtreS M. tuberculosis failed to exhibit the same metabolite accumulations observed in wild type M. tuberculosis and treS complemented strain at early time points after exposure to hypoxia (Fig. 3). ΔtreS M. tuberculosis was also unable to adapt to, or recover from, incubation at 1% O2 (Supplementary Fig. 5D). We could similarly reproduce these effects using a validated chemical inhibitor of TreS, validamycin A (Supplementary Fig. 6A-C)14. Thus, while wild type M. tuberculosis exhibited a modest, hypoxia-induced repression of treS transcription in response to hypoxia (Supplementary Fig. 6D), these data identify a functionally essential, though not exclusive, role for TreS.
Fig. 3. Biochemical essentiality of TreS in hypoxia-triggered anticipatory metabolic remodeling of M. tuberculosis.
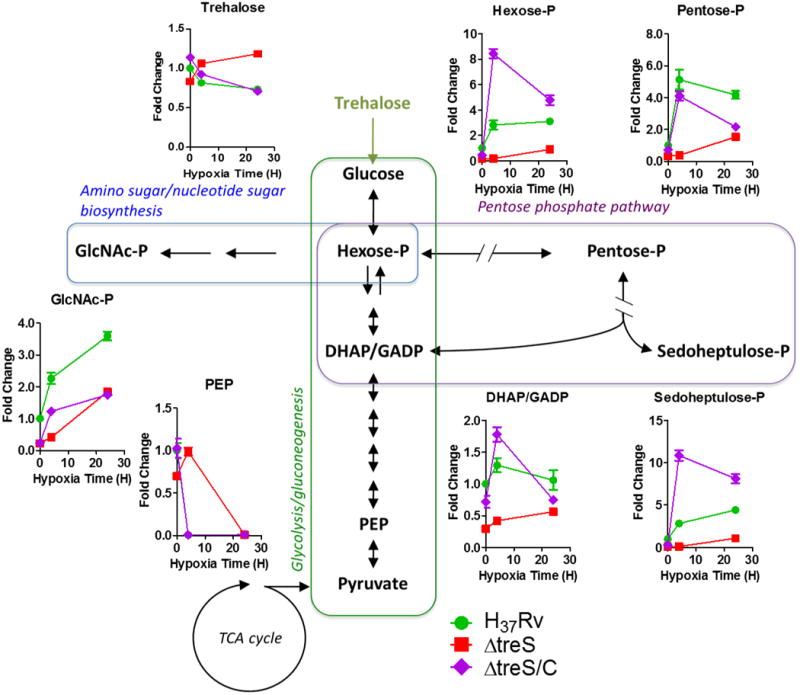
Metabolic time-course responses of WT, ΔtreS or ΔtreS/C M. tuberculosis following either 4 or 24 h of incubation under hypoxia. Changes are expressed relative to the pool sizes of WT under 20% O2. All values are the average of three biological replicates (n=3) ± SEM and representative of 2 independent experiments. DHAP, dihydroxyacetone phosphate; GADP, glyceraldehyde phosphate; GlcNAc-P, N-acetyl glucosamine phosphate; hexose-P, glucose-6-phosphate and its isomers; pentose-P, ribose-5-phosphate and its isomers; PEP, phosphoenolpyruvate; sedoheptulose-P, sedoheptulose-7-phosphate and its isomers.
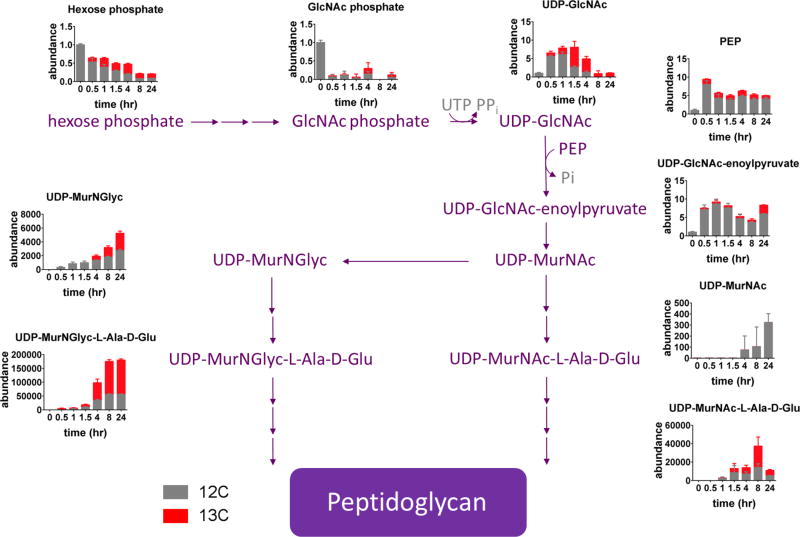
The foregoing results indicate that hypoxia triggers a catabolic remodeling of M. tuberculosis's immunostimulatory cell envelope, part of which is shunted into a stable accumulation of pentose phosphate pathway intermediates. The pentose phosphate pathway furnishes reducing equivalents for reductive biosynthesis and anti-oxidant defense and precursors for nucleotide and aromatic amino acid synthesis16. Metabolomic profiling, however, revealed marked and specific accumulations of pentose phosphates and sedoheptulose phosphates in the absence of significant (>2 fold) changes in NADP(H) levels or ratios, or aromatic amino acids (Supplementary Fig. 7); and a metabolically unrelated accumulation of N-acetylglucosamine phosphate (Fig. 1; Supplementary Fig. 1A, B). These changes suggested a potential role for pentose phosphate pathway intermediates in de novo peptidoglycan biosynthesis. To investigate this possibility, we tracked the fate of these accumulated unlabeled intermediates during re-entry into cell cycle by transferring M. tuberculosis into 13C labeled media upon re-aeration. As expected, the majority of monitored metabolites exhibited time-dependent increases in 13C labeling7. However, there were specific increases in total levels of PEP, UDP-N-acetylglucosamine and UDP-N-acetylglucosamine enoylpyruvate 30 minutes after re-aeration that were primarily mediated by increases in their unlabeled pools, that derive, in part from the previously accumulated pool of N-acetylglucosamine phosphate (Fig. 4). PEP is a thermodynamically trapped enol that furnishes the carboxyvinyl moiety bridging the glycan and peptide portions of mature peptidoglycan. UDP-N-acetylglucosamine enoylpyruvate is the first committed intermediate in peptidoglycan biosynthesis and arises from the enol ether transferase, MurA17,18. These results thus link the hypoxia-induced catabolism of M. tuberculosis's trehalose mycolates to a metabolic program that poises M. tuberculosis to re-initiate peptidoglycan biosynthesis upon re-aeration.
Fig. 4. Biochemical shunting of hypoxia-induced metabolic stores into de novo peptidoglycan biosynthesis during re-aeration.
Metabolic tracing of hypoxia-induced 12C metabolic stores of glycolytic, pentose phosphate pathway and amino sugar biosynthesis into de novo peptidoglycan biosynthesis following re-aeration at 20% O2 for 0h, 0.5h, 1h, 1.5h, 4h, 8h, and 24h. Changes are shown in stacked bar graph format relative to accumulated levels observed following incubation at 1% O2 for 48h (designated 0h) for either 12C (unlabeled species) or total (12C + all 13C species) of indicated metabolite. Gray font indicates undetected intermediates. D-Glu, D-glutamate; GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; L-Ala, L-alanine; MurNGlyc, glycolyl N-acetylmuramic acid; PEP, phosphoenolpyruvate; Pi, inorganic phosphate; UDP, uridine diphosphate. All values are the average ± SEM of three experimental replicates (n=3) and representative of 2 independent experiments.
Based on these findings, we tested the functional role of this response during recovery from hypoxia. To do so, we compared the susceptibilities of aerobic, hypoxic and re-aerated M. tuberculosis to a discrete (24h) exposure of ampicillin, a covalently reactive inhibitor and functional probe of peptidoglycan biosynthesis. As expected, hypoxic M. tuberculosis exhibited a phenotypic tolerance to ampicillin and streptomycin, compared to aerobic counterparts (as reported by time to outgrowth following treatment and subculture into fresh antibiotic-free media). Re-aerated M. tuberculosis, in contrast, exhibited a return to near aerobic levels of susceptibility to ampicillin, but not streptomycin, during the first 24h of re-aeration (Supplementary Fig. 8A, B). These results thus link the observed biochemical response to an early, specific, and essential role for de novo peptidoglycan biosynthesis during recovery from hypoxia.
Growing evidence indicates that genetic regulatory systems have evolved the capacity to encode anticipatory responses to conserved spatio-temporal features of the environment that can prepare organisms for a given condition in advance of its arrival19-21. The capacity of non-genetic molecular networks to encode similar anticipatory responses has not been described. Our studies now expand this regulatory capability to include a metabolic network.
Metabolic enzymes catalyze biochemical reactions that typically occur on the order of seconds to minutes but serve physiologic activities that operate across a far broader range of time scales. Metabolic enzymes are a well-documented determinant of M. tuberculosis's pathogenicity22. However, the specific roles they play remain incompletely described. Ascribed roles have chiefly consisted in reactive situational responses. Yet, survival of M. tuberculosis ultimately depends on its ability to re-enter and complete cell cycle following prolonged, and often unpredictable, periods of host-induced quiescence, posing a distinct regulatory challenge.
Traditional stimulus-specific responses help meet the challenge of unpredictability, in part, through probabilistic, bet-hedging mechanisms19. In contrast, our studies identify a deterministically encoded mechanism in which the temporal relationship between hypoxia and environmental oxygen in M. tuberculosis's lifecycle has been encoded into an anticipatory metabolic response, wherein hypoxia serves as a cue for the subsequent arrival of oxygen.
Peptidoglycan is an essential component of the cell wall required for both structural integrity and growth of virtually all bacteria, including M. tuberculosis. De novo synthesis of peptidoglycan begins with the condensation of 2 thermodynamically activated substrates, PEP and UDP-N-acetylglucosamine, by MurA, the first committed enzyme of the pathway. However, hypoxic M. tuberculosis maintains ATP at levels approximately five times lower than those of replicating counterparts7,23. The hypoxia-induced shunting of catabolized trehalose mycolates into activated pentose phosphate intermediates may thus help M. tuberculosis overcome a key thermodynamic barrier to cell cycle re-entry and completion of its life cycle, consistent with recently emerging reports of additional O2-sensitive mechanisms of MurA regulation8,24.
The mechanisms used to accumulate, store and release trehalose mycolate-derived intermediates in M. tuberculosis remain to be determined. Existing evidence implicates the Antigen 85 complex, which generates trehalose as a byproduct of the biosynthesis of the mycolic acid-based outer cell envelope; the LpqY-SugA-SugB-SugC ABC transporter, which catalyzes the specific, retrograde transport of extracellular trehalose; the TreS-encoded trehalose synthase, described here; and glyceraldehyde-3-phosphate dehydrogenase, which was recently shown to regulate flux between upper and lower glycolysis via its universally conserved requirement for high Pi concentrations11,25,26.
Mechanism notwithstanding, growing evidence indicates that the lesions in which M. tuberculosis resides within the lung are both diverse and dynamic27. The evolution of anticipatory responses may thus help M. tuberculosis bridge the challenge of navigating an often decades-long life cycle on an intermediary basis.
We previously reported that, unlike other bacteria, replicating M. tuberculosis operates its intermediary metabolism in a modular and compartmentalized manner by co-catabolizing different carbon sources to distinct metabolic fates28. Our discovery of metabolism of cell surface-derived trehalose to functionally compartmentalized pentose phosphate intermediates now expands this view to another pathway and M. tuberculosis in a hypoxic, non-replicating state. This work more broadly demonstrates a metabolically-encoded regulatory capability that enables a critical transition point in a pathogen's life cycle.
Materials and Methods
M. tuberculosis filter culture and metabolite extraction
M. tuberculosis strains H37Rv, treS knock-out (ΔtreS KO) and the complemented strain (ΔtreS::ptreS) were cultured in a biosafety level 3 facility at 37 °C in Middlebrook 7H9 broth (m7H9) or on 7H10 agar (m7H10) (Difco) supplemented with 0.2% acetate (or 0.2% glucose in case of requirement), 0.04% Tyloxapol (broth only), 0.5 g/L BSA, and 0.085% NaCl. M. tuberculosis-laden filters used for metabolic profiling were generated as previously described 7 and incubated at 37 °C for 5 days to reach the mid-log phase of growth. M. tuberculosis-laden filters were then transferred onto chemically identical medium containing fresh 12C acetate (or glucose) or [U-13C] acetate (or glucose)-m7H10. M. tuberculosis-laden filters were metabolically quenched by plunging filters into a mixture of acetonitrile/methanol/H2O (40:40:20) precooled to −40 °C; metabolites were extracted by mechanical lysis with 0.1 mm Zirconia beads in a Precellys tissue homogenizer for 3 min (6,500 rpm) twice under continuous cooling at or below 2 °C. Lysates were clarified by centrifugation and then filtered across a 0.22 μm filter. Residual protein content of metabolite extracts (BCA Protein Assay kit; Thermo Scientific) was determined to normalize samples to cell biomass. Trehalase, amyloglucosidase, and trehalose synthase specific inhibitor (validamycin A) were purchased from Sigma-Aldrich. Validamycin A was used at either 5 or 10 mM concentration. Ampicillin and streptomycin were purchased from Sigma-Aldrich and used at 100 ug/ml (8X MIC) and 1.5 μg/ml (2X MIC), respectively. All data obtained by metabolomics were average of independent triplicates.
In vitro hypoxia model
In vitro hypoxia was achieved using the type A Bio-bag environmental chamber (Becton Dickinson) as specified by the manufacturer's instructions and validated by a resazurin indicator, which decolorized at oxygen concentrations of 1.0 % or less. M. tuberculosis-laden filters were transferred to fresh chemically equivalent m7H10 immediately before insertion or after recovery from sealed hypoxic chambers. M. tuberculosis-laden filters recovered from hypoxic chambers were immediately plunged into the precooled quenching solution as described above 7. This system depletes oxygen gradually and generates CO2 via a palladium catalyst, achieving a final atmosphere of ∼1% O2 (as reported by a resazurin-based indicator strip) and ∼5% CO2, levels similar to those encountered in the tuberculous lungs of infected animals, within 4 h. We previously showed that, in this system, M. tuberculosis neither underwent net replication nor death in response to incubation at 1% O2 and resumed growth upon reaeration. We further showed that this system was associated with both the reversible biphasic induction of dosR, a previously validated transcriptional marker of hypoxia, and accompanying reductions of levels of ATP and NAD.
LC-MS Metabolomic profiling
LC-MS based metabolomics was used as described 7. Extracted metabolites were separated on a Cogent Diamond Hydride type C column (gradient 3) and the mobile phase consisted of solvent A (ddH2O with 0.2% formic acid) and solvent B (acetonitrile with 0.2 % formic acid). The mass spectrometer used was an Agilent Accurate Mass 6220 time of flight (TOF) coupled to an Agilent 1200 liquid chromatography (LC) system. Detected ions were deemed metabolites on the basis of unique accurate mass-retention time identifiers for masses exhibiting the expected distribution of accompanying isotopologues. Metabolite identities were searched for using a mass tolerance of <0.005 Da. The relative concentration of metabolites was determined by using a calibration curve generated with varying concentrations of chemical standard spiked into homologous mycobacterial extract to correct for matrix-associated ion suppression effects. The abundance of extracted metabolite ion intensities was extracted using Profinder 6.0 and Qualitative Analysis 6.0 (Agilent Technologies). The clustered heat map and hierarchical clustering trees were generated using Cluster 3.0 and Java TreeView 1.0.
LC-MS lipidomic analysis
Total lipid extraction was performed with minor modification as described previously 29. Total lipids were extracted from the M. tuberculosis-laden filters that were used for metabolomics profiling. Three M. tuberculosis-laden filters were combined for an experimental condition that enables to acquire enough amount of total lipid that LC-MS further detects. After harvest, cells were washed twice in ice-cold PBS, transferred to a 50 ml amber glass bottle, and contacted with 15 ml CHCl3/CH3OH (2:1, v/v) overnight to sterilize bacteria and extract total lipids, simultaneously. After centrifugation, lipid extracts were decanted, and bacteria pellets were then subjected to two additional extractions using CHCl3:CH3OH (1:1, v/v) and CHCl3:CH3OH (1:2, v/v) with pooling of all extracts and evaporation with N-EVAP (Organomation Associates Inc.) using the low boiling point mixture setting. Dried lipids were resuspended in a minimum volume of CHCl3:CH3OH (1:1, v/v) and dried under nitrogen in preweighed vials, then reweighed in triplicate on microbalance (Mettler Toledo, XP205), and values were reported when fully dried as shown by replicate measurements showing less than 1% variance. Using 2 mg of lipid extract, replicate measures showed variance of 20 μg, providing mass errors below 1% for in vitro-derived samples. Extracts were then redissolved in CHCl3:CH3OH (1:1, v/v) at 1 mg/mL. LC-MS grade solvents (Fisher) and clean borosilicate glassware (Fisher), amber vials (Supelco), and Teflon-lined caps (Fisher) were used.
Intrabacterial trehalose and glycogen assays
Intrabacterial trehalose and glycogen concentrations under varying metabolic states (Replicating, Non-replicating and Reaeration) were measured by using Glucose (GO) Assay Kit (Sigma-Aldrich). The GO assay kit allows quantitative determination of glucose content present in bacterial extracted materials. Extracted glucose is oxidized to gluconic acid and hydrogen peroxide by glucose oxidase. Hydrogen peroxide then reacts with o-dianisidine in the presence of peroxidase to form a colored product, reaction with sulfuric acid with which forms a more stable colored product. The intensity of the colored final product can be measureable at 540 nm which is proportional to the glucose amount extracted. Extractions of intrabacterial trehalose and glycogen were similarly performed as does for the extraction of M. tuberculosis metabolites. M. tuberculosis-laden filters were incubated for 5 days on m7H10, transferred to fresh chemically identical m7H10 under either ambient, hypoxic or reaerated after incubation under 24 hr hypoxia, and harvested the cytosolic materials by mechanical lysis with 0.1 mm Zirconia beads in a Precellys tissue homogenizer for 3 min (6,500 rpm) twice under continuous cooling at or below 2 °C. PBS was used for both washing twice and extracting cytosolic fraction. Extracted cytosolic fraction containing both trehalose and glycogen was used as a substrate for intermediate enzyme mixture containing either trehalase or amyloglucosidase to produce glucose, proportional to the amount of either trehalose or glycogen, respectively. Intrabacterial glucose produced was quantitated by using Glucose (GO) Assay kit. Known concentration of authentic trehalose or glycogen was used for generation of standard curves and a positive control.
13C pulse chase labeling of cell envelope TMM
To selectively 13C label M. tuberculosis trehalose mycolates, we applied the following experimental approach (Fig. S4A). To fully 13C label all intermediates in cells, we first incubated M. tuberculosis-laden filters on m7H10 containing [U-13C] glucose for 2 days (all metabolites detected in our setting were greater than 99% 13C labeled). We then transferred the M. tuberculosis-laden filters to fresh m7H10 containing unlabeled glucose for 18 hrs to replace 13C labeled metabolites in the central carbon metabolism with unlabeled intermediates (around 67% trehalose and 9% hexose phosphate were remained as labeled with 13C; reverse-washing step) (Fig. S4A lower panel). Under these condition, we transferred M. tuberculosis-laden filters to fresh m7H10 containing either unlabeled or [U-13C] acetate for up to 12 hrs of hypoxia and metabolites were sampled in a time dependent manner as described in previous section. The quantity and 13C labeling pattern were analyzed by LC-MS as previously described 7.
Mutant strain generation
The rv0126 deficient knock-out mutant (ΔtreS) was generated in the M. tuberculosis H37Rv background as previously used. ΔtreS was constructed via a suicide plasmid approach (Fig. S5). Approximately 500 bp fragments corresponding to regions upstream (primer set; treS #1 and treS #2) and downstream (primer set; treS #3 and treS #4) of treS gene were amplified by PCR and cloned into a temperature-sensitive vector pDE43-XSTS to flank the hygromycin resistance cassette and generate pKO-XSTS- M. tuberculosis treS. H37Rv was first transformed with pKO-XSTS- M. tuberculosis treS and plated on m7H10 with hygromycin at the permissive temperature of 30 °C. Transformants were then grown at 30 °C to OD580=∼1.0 and plated on m7H10 with hygromycin, 10% sucrose, 0.2% glycerol and 0.2% glucose at the restrictive temperature of 40 °C. For complementation of ΔtreS, we generated pGMEK-phsp60- M. tuberculosis treS (primer set; Compl #1 and Compl #2), a vector that expresses treS under the control of the hsp60 promoter and wild type TreS are constitutively expressed episomally. The rv2402 deficient knock-out mutant (Δrv2402) was generated similarly (Fig. S5B). The treS or rv2402 deletion was confirmed by PCR amplification (primer set of pcr#1 and pcr #2) comparing with wild type H37Rv counterpart.
Primer sequences corresponding to Supplementary Fig. 5B and 5C are as follows: SFig. S5B: rv2402 #1: GGGGACAACTTTGTATAGAAAAGTTGACCAGGGTGATGCTCGTATGC; rv2402 #2: GGGGACTGCTTTTTTGTACAAACTTG GGCGAACTGGACGATAGAGC; rv2402 #3: GGGGACAGCTTTCTTGTACAAAGTGG CACCTGGCACTGATCAAC; rv2402 #4: GGGGACAACTTTGTATAATAAAGTTG CTGTTTGCCGTGTTCCTC; pcr #1: CAGCTCAATGGGTGAGAAG; pcr #2: TCCTCGGTGAAGGTGGATG; SFig. S5C: treS #1: GGGGACAACTTTGTATAGAAAAGTTGGGCCACAGGTGGTCA ACATC; treS #2: GGGGACTGCTTTTTTGTACAAACTTGCTGCCTCGTTCATTGCTTCC; treS #3: GGGGACAGCTTTCTTGTACAA AGTGGACACGGGTTCTACTGGTTCC; treS #4: GGGGACAACTTTGTATAATAAAGTTGTCGATCGTTGGCGCGTA CTC; pcr #1: GACAGGTGGTCGGTATGAAC; pcr #2: CTGTTCGGCGTCACATACCC; Compl #1: GGGGACAA GTTTGTACAAAAAAGCAGGCTGATTCCTGGCGGCAAGTCTC; Compl #2: GGGGACCACTTTGTACAAG AAAGCTGGGTTCGTTGGCGGACGACATTG.
Antibiotic susceptibility of replicating and non-replicating M. tuberculosis
The relative sensitivity of aerobic, hypoxic and re-aerated M. tuberculosis to ampicillin (covalent inhibitor of peptidoglycan biosynthesis) or streptomycin was determined by measuring the time to outgrowth in 96 well plates following a 24 h challenge with ampicillin at 8X MIC (100 ug/ml) or streptomycin at 2X MIC (1.5 ug/ml) after which bacteria were diluted 1:100 dilution and monitored for growth by optical density. Experiments consisted in biological triplicates and were repeated in at least 2 independent experiments.
Isotopologue analysis using isotope-labeled carbon sources
The extent of isotopic labeling for metabolites was determined by dividing the summed peak height ion intensities of all labeled isotopologue species by the ion intensity of both labeled and unlabeled species, expressed in percentage. Label-specific ion counts were corrected for naturally occurring 13C species (i.e., [M+1] and [M+2]). The relative abundance of each isotopically labeled species was determined by dividing the peak height ion intensity of each isotopic form (corrected for naturally occurring 13C species as above) by the summed peak height ion intensity of all labeled species. Ion intensities were converted into molar abundances using standard curves generated by addition of chemical standards and serial dilution of samples to establish the co-linearity of ion intensity and molar abundance 29.
M. tuberculosis viability Assay
M. tuberculosis viability was determined using liquid cultures manipulated under experimentally identical conditions as filter-grown cultures used for metabolomics profiling, which we previously demonstrated to be microbiologically similar. CFUs were determined by plating on m7H10 with supplements; 0.2% glycerol, 0.2% glucose, 0.5 g/L BSA, and 0.085% NaCl. All cell viability analyses were performed in triplicate in 2 independent experiments.
Macrophage cytokine ELISAs
Mouse bone marrow-derived macrophages (BMM) were collected from 8-10 week-old C57BL/6 mice and differentiated in cell culture as described 30. Total 2 × 105 macrophages per well in 24 well plates were exposed to single cell suspensions of paraformaldehyde-fixed M. tuberculosis recovered from replicating, non-replicating (hypoxic) or re-aerated cultures taken from the early log phase of growth at multiplicity of infection (MOI) of 3 and 5. Four hours following exposure, cells were washed twice with PBS to remove extracellular bacteria and replaced with fresh media. Supernatants from M. tuberculosis-exposed macrophages were collected 24 hours later and cytokines IL-12 and TNF-α were measured by ELISA (R&D Systems, Minneapolis). Supernatant from mock treated macrophages was used as a control. Three independent experiments were performed for each infection condition. Bacterial input was determined by plating serial dilutions on m7H10 plates at 37 °C and enumerating CFUs after 2 week-incubation. We fixed M. tuberculosis under replicating, non-replicating, and re-reaerated state by treatment of 4% formaldehyde followed by washing with PBS twice before macrophage infection. Bacterial input was determined before fixing.
Measurement of trehalose secretion from M. tuberculosis culture
As previously described, the filter culture system used for metabolomics studies was modified by replacing the underlying m7H10 agar medium with a plastic inset containing chemically equivalent m7H9 liquid medium (without Tyloxapol) in direct contact with the underside of the M. tuberculosis-laden filter 7. Growth atop this new device enabled timed start-stop measurements of trehalose secretion by sampling the cell-free liquid medium. A time-dependent trehalose secretion was sampled in the medium following exposure to and incubation under hypoxia and measured by using LC-MS.
Measurement of intrabacterial NADPH/NADP Ratios
M. tuberculosis-laden filters as used for metabolomics profiling were separately generated to measure intrabacterial NADPH and NADP content and its ratio. NADPH and NADP concentrations were measured using a FluroNADP/NADPH detection kit (Cell Technology). Metabolism of M. tuberculosis was rapidly quenched by plunging bacilli in the first solvent in the kit cooled to <4°C.
Statistical Analysis
Analyses were performed using the ANOVA test. A p value of less than 0.05 was considered statistically significant.
Data availability
The data that support the findings of this study are either available within the paper and its supplementary information files or upon reasonable request to the corresponding author.
Supplementary Material
Acknowledgments
We thank Carl Nathan, Julien Vaubourgeix, Sabine Ehrt and Saeed Tavazoie for critical discussions and reading of the manuscript, Steven Fischer for expert mass spectrometric support, and the Bill and Melinda Gates Foundation Grand Challenges Exploration Program (OPP1068025) and NIH Tri-I TBRU (U19-AI11143) for support.
Footnotes
Author contributions: H.E. and Z.W. designed, conducted and analyzed metabolomic profiling studies. E. L. and D. B. M. conducted and analyzed lipidomic profiling studies. H. E. and P. R. conducted macrophage cytokine release assays. H. E., Z. W. and R. M. conducted antibiotic susceptibility assays. K. Y. R. initiated and directed this research.
References and Notes
- 1.Russell DG, Barry CE, 3rd, Flynn JL. Tuberculosis: what we don't know can, and does, hurt us. Science. 2010;328:852–856. doi: 10.1126/science.1184784. doi:328/5980/852 [pii] 10.1126/science.1184784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser MJ, Kirschner D. The equilibria that allow bacterial persistence in human hosts. Nature. 2007;449:843–849. doi: 10.1038/nature06198. [DOI] [PubMed] [Google Scholar]
- 3.Rustad TR, Sherrid AM, Minch KJ, Sherman DR. Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 2009;11:1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. doi:CMI1325 [pii] 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 4.Lakshminarayana SB, et al. Comprehensive physicochemical, pharmacokinetic and activity profiling of anti-TB agents. J Antimicrob Chemother. 2015;70:857–867. doi: 10.1093/jac/dku457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–570. doi: 10.1128/CMR.00060-05. doi:19/3/558 [pii]10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe S, et al. Fumarate Reductase Activity Maintains an Energized Membrane in Anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega C, et al. Mycobacterium tuberculosis Ser/Thr protein kinase B mediates an oxygen-dependent replication switch. PLoS Biol. 2014;12:e1001746. doi: 10.1371/journal.pbio.1001746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega C, et al. Systematic Survey of Serine Hydrolase Activity in Mycobacterium tuberculosis Defines Changes Associated with Persistence. Cell Chem Biol. 2016;23:290–298. doi: 10.1016/j.chembiol.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert OT, et al. Absolute Proteome Composition and Dynamics during Dormancy and Resuscitation of Mycobacterium tuberculosis. Cell Host Microbe. 2015;18:96–108. doi: 10.1016/j.chom.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 11.Kalscheuer R, Koliwer-Brandl H. Genetics of Mycobacterial Trehalose Metabolism. Microbiol Spectr. 2014;2 doi: 10.1128/microbiolspec.MGM2-0002-2013. [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galagan JE, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalscheuer R, et al. Self-poisoning of Mycobacterium tuberculosis by targeting GlgE in an alpha-glucan pathway. Nat Chem Biol. 2010;6:376–384. doi: 10.1038/nchembio.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miah F, et al. Flux through trehalose synthase flows from trehalose to the alpha anomer of maltose in mycobacteria. Chem Biol. 2013;20:487–493. doi: 10.1016/j.chembiol.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stincone A, et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc. 2014 doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh CT, Benson TE, Kim DH, Lees WJ. The versatility of phosphoenolpyruvate and its vinyl ether products in biosynthesis. Chem Biol. 1996;3:83–91. doi: 10.1016/s1074-5521(96)90282-3. [DOI] [PubMed] [Google Scholar]
- 18.De Smet KA, Kempsell KE, Gallagher A, Duncan K, Young DB. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology. 1999;145(Pt 11):3177–3184. doi: 10.1099/00221287-145-11-3177. [DOI] [PubMed] [Google Scholar]
- 19.Venturelli OS, Zuleta I, Murray RM, El-Samad H. Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol. 2015;13:e1002042. doi: 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell A, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 21.Tagkopoulos I, Liu YC, Tavazoie S. Predictive behavior within microbial genetic networks. Science. 2008;320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warner DF. Mycobacterium tuberculosis metabolism. Cold Spring Harb Perspect Med. 2015;5 doi: 10.1101/cshperspect.a021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. doi:0711697105 [pii] 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boutte CC, et al. A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. Elife. 2016;5 doi: 10.7554/eLife.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Backus KM, et al. The three Mycobacterium tuberculosis antigen 85 isoforms have unique substrates and activities determined by non-active site regions. J Biol Chem. 2014;289:25041–25053. doi: 10.1074/jbc.M114.581579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Heerden JH, et al. Lost in transition: start-up of glycolysis yields subpopulations of nongrowing cells. Science. 2014;343:1245114. doi: 10.1126/science.1245114. [DOI] [PubMed] [Google Scholar]
- 27.Lenaerts A, Barry CE, 3rd, Dartois V. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev. 2015;264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Carvalho LP, et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. doi:S1074-475 5521(10)00347-9 [pii] 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Layre E, et al. A Comparative Lipidomics Platform for Chemotaxonomic Analysis of Mycobacterium tuberculosis. Chem Biol. 2011;18:1537–1549. doi: 10.1016/j.chembiol.2011.10.013. doi:S1074-5521(11)00398-X [pii] 10.1016/j.chembiol.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrt S, et al. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J Exp Med. 2001;194:1123–1140. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are either available within the paper and its supplementary information files or upon reasonable request to the corresponding author.