Abstract
The genome is organised via CTCF/Cohesin binding sites, which partition chromosomes into 1-5Mb topologically associated domains (TADs), and further into smaller sub-domains (sub-TADs). Here we examined in vivo an ~80kb sub-TAD, containing the mouse α-globin gene cluster, lying within a ~1Mb TAD. We find that the sub-TAD is flanked by predominantly convergent CTCF/cohesin sites which are ubiquitously bound by CTCF but only interact during erythropoiesis, defining a self-interacting erythroid compartment. Whereas the α-globin regulatory elements normally act solely on promoters downstream of the enhancers, removal of a conserved upstream CTCF/cohesin boundary extends the sub-TAD to adjacent upstream CTCF/cohesin binding sites. The α-globin enhancers now interact with the flanking chromatin, upregulating expression of genes within this extended sub-TAD. Rather than acting solely as a barrier to chromatin modification, CTCF/cohesin boundaries in this sub-TAD delimit the region of chromatin to which enhancers have access and within which they interact with receptive promoters.
Introduction
Whereas previous work has intensively studied the role of enhancers and promoters in regulating gene expression, it is becoming increasingly clear that their dynamic interactions in 3-dimensions within the nucleus provide a fundamentally important third component for switching genes on and off. We now know that this chromosomal topology is determined by a third class of regulatory elements defined by their binding of CCCTC-binding factor (CTCF) and components of the structural maintenance of chromosome (SMC) Cohesin complex1,2. These elements appear to organize chromosomes into a series of increasingly complex topological structures (chromosome loops, sub-TAD, TAD, etc.)3. However, not all CTCF/Cohesin sites appear equivalent, and a variety of different functions have been attributed to such elements in different contexts, including; acting as boundaries to chromatin modifications4–6, facilitating interactions between regulatory elements7,8, and insulating genes from tissue-specific enhancers9–11. However, at present, how they interact with each other and with other regulatory elements in their natural chromosomal context is poorly understood. To address this, we have examined the interactions between CTCF/Cohesin sites, enhancers and promoters, and determined their functional role(s) using chromosome engineering of an ~80kb sub-TAD containing the well characterised mouse α-globin cluster, in its natural chromosomal environment, in vivo. We find that CTCF/Cohesin sites in this sub-TAD play a key role in regulating gene expression by delimiting the region of chromatin within which active enhancers can interact with receptive promoters.
Changes in chromatin and gene expression across the 1Mb TAD containing the α-globin locus in erythroid cells
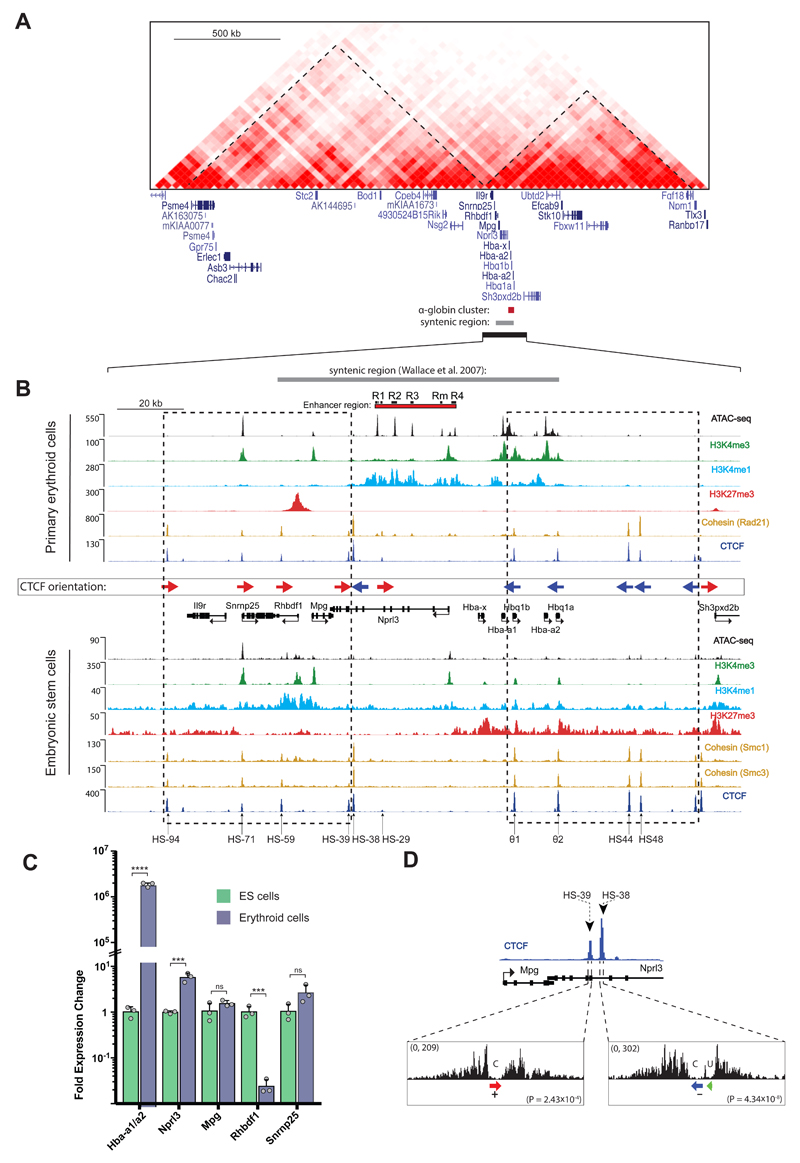
The mouse α-globin cluster is located in close proximity to a cluster of 10 widely expressed genes near the boundary of a ~1Mb TAD as defined by Dixon et al (Fig. 1A)12. We have previously characterised the chromatin in and around the α-globin cluster and noted that activation of α-globin expression in erythroid cells is associated with the appearance of a broad domain of histone acetylation and modification by H3K4me1 spanning ~80kb including the α-globin genes and their regulatory elements (Fig. 1B)13,14. Using ATAC-seq, DNase-seq and ChIP-seq, we have identified all promoters and enhancers in this region via their characteristic chromatin signatures (Fig. 1B). We have shown that the α-globin genes are regulated by four conserved erythroid-specific enhancers (R1-R4) and a mouse specific element (Rm) located 14-38 kb upstream of the promoters15–17. Four of these enhancers (R1, R2, R3 and Rm) lie within the introns of an adjacent widely expressed gene (Nprl3)15. We show here that the remaining regions of open chromatin within and around the gene cluster, identified in all cell types analysed, correspond to binding sites for CTCF/Cohesin (Fig. 1B).
Figure 1. Regulation of the α-globin cluster in mouse ES and primary erythroid cells.
A. Heat map of Hi-C chromatin interactions surrounding the α-globin gene cluster in mouse ES cells. TADs are annotated by dashed lines as defined by Dixon et al. 201212. Gene annotation is Refseq. B. The α-globin locus with enhancer elements (R1, R2, R3, Rm, and R4), gene promoters and CTCF binding sites marked by peaks of open chromatin as indicated by ATAC-seq tracks (RPKM) in mouse ES and primary erythroid cells. All normalised ChIP-seq read-densities (RPKM) represent an average of 2 independent experiments, in which 2 animals (erythroid cells) or 2 biologically independent samples (ES cells) were analysed in total, except for ES and erythroid H3K4me1 which represent single experiments. CTCF binding orientation is annotated with forward (red arrows) and reverse (blue arrows). Dashed boxes indicate clusters of convergent CTCF binding sites. Gene annotation is Refseq. The following datasets were previously published: ES SMC1 and SMC331; erythroid H3K4me314; ES CTCF, H3K27me3, H3K4me138; ES H3K4me339; ES ATAC-seq40. C. Relative gene expression of mouse primary erythroid versus ES cells measured by real-time qPCR and representing n=3 independent experiment in which animals (erythroid cells) or n=3 biologically independent samples (ES cells) were analysed in total [AU: OK?]. Bars represent the mean and the error bars represent the standard deviation (S.D.). Grey dots represent individual data points. P-values are obtained via an unpaired, two-tailed student t-test. ns P>0.05, *** P<0.001, **** P<0.0001. D. DNaseI footprints of HS-38 and HS-39 CTCF binding sites. CTCF motifs are indicated by arrows; red arrow: forward core motif, blue arrow: reverse core motif, and green arrow: upstream motif. P-values indicate the significance of the match of the HS-38 and HS-39 sequence to the core consensus motif (derived with the FIMO tool as described41). The DNaseI data was previously published in Hosseini et al (2013)42.
The α-globin genes and the closely linked, widely expressed gene (Nprl3), which lie together within the H3K4me1-marked domain are expressed at high levels in erythroid cells (Fig 1C). Surrounding genes within the larger (1Mb) TAD are unaffected by activation of the strong erythroid-specific enhancers and we show here that one of these genes (Rhbdf1) is marked by high levels of the Polycomb-mediated repressive mark (H3K27me3) and completely silenced in erythroid cells (Figs 1B and C). In this way we have accounted for all regions of open chromatin, the corresponding regulatory elements, and the pattern of gene expression within and surrounding the α-globin locus.
An α-globin sub-TAD is surrounded by convergent CTCF/Cohesin binding sites which interact with each other specifically in erythroid cells
While the role of CTCF/cohesin binding sites at boundaries between TADs are starting to be established12, less is known about the CTCF/Cohesin sites that are dispersed within TADs. These CTCF/Cohesin sites are thought to contribute to the formation of smaller self-interacting domains that have been termed sub-TADs18, contact domains19, and insulated neighborhoods11,20. In contrast to TADs, sub-TADs (40kb to 3Mb, median size of 185kb19) often appear in a cell specific manner and, as in the case of the 80kb sub-TAD corresponding to the H3K4me1 and histone acetylation-marked α-globin domain, may be identified via an increased density of chromatin interactions (as seen by Capture–C, Fig. 2A) and specific histone modifications in a specific cell type. Of interest, recent studies have shown a strong preference for interactions to occur between CTCF sites lying in a convergent orientation with respect to each other21–24. We therefore established the orientation of CTCF binding sites surrounding and within the α-globin sub-TAD (Fig. 1B). Motif orientations were predicted by inspecting each CTCF consensus core and flanking motifs and subsequently validated by analyzing the directionality of associated DNaseI footprints in vivo (Fig. 1D and Supplementary Fig. 1). This analysis revealed a striking pattern of CTCF orientations in which the regions flanking the α-globin genes and their enhancers were shown to contain clusters of largely convergently orientated CTCF binding sites (Fig. 1B).
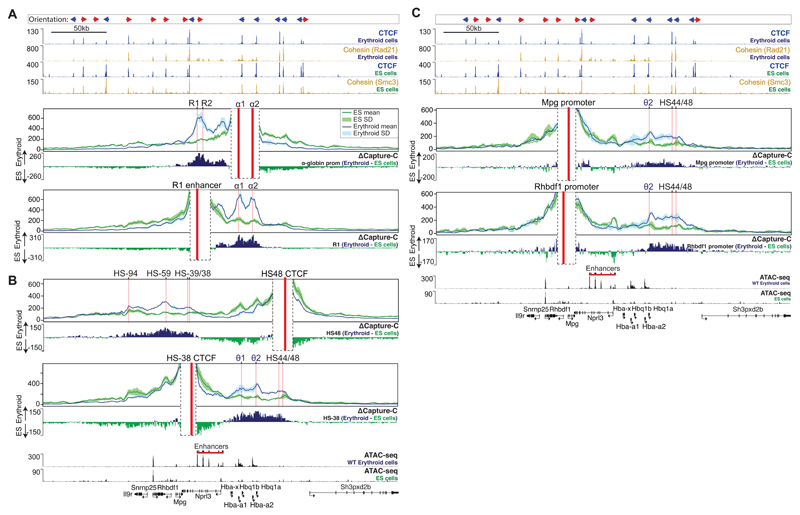
Figure 2. Differential interactions of α-globin regulatory elements between mouse ES and primary erythroid cells.
A. Panels show overlaid, normalised Capture-C data for the α-globin promoter (α1, α2) and the R1 enhancer viewpoints in mouse ES and primary erythroid cells merged from 3 independent experiments, in which 3 animals (erythroid cells) or 3 biologically independent samples (ES cells) were analysed in total. Each of the α-globin promoters interacts with the enhancers independently, resulting in the expression of both genes (α1, α2)17,43. The mean, plus and minus one standard deviation (S.D.), of sliding 5kb windows are visualised. Differential tracks (ΔCapture-C) show a subtraction (erythroid - ES) of the mean number of meaningful interactions per restriction fragment. Red vertical bars indicate the position of the viewpoint. Also shown are normalised CTCF and Cohesin (Rad21 or Smc3) ChIP-seq (RPKM) and ATAC-seq (RPKM) tracks for both ES and primary erythroid cells, all merged from 2 independent experiments, in which 2 animals were analysed in total. Gene annotation is Refseq. B. Data presented as described in A for HS48 and HS-38 CTCF site viewpoints. C. As described in A, data for Mpg and Rhbdf1 promoter viewpoints.
To investigate the mechanisms by which CTCF/Cohesin-mediated domains may form, interact and influence the activity of strong tissue-specific enhancers and promoters, we performed next-generation Capture-C in mouse erythroid and non-erythroid, embryonic stem (ES) cells17,25. In ES cells, α-globin is not expressed whereas transcription of flanking genes (Snrnp25, Mpg, Rhbdf1 and Nprl3) occurs in the absence of the erythroid-specific enhancer activity (Figs 1B and C). Using viewpoints from the enhancer region (R1) and the α-globin promoters (α1 and α2), in ES cells we observed broad, diffuse interactions extending across the entire gene cluster (Fig 2A). By contrast, in erythroid cells, we observed much stronger interactions throughout the sub-TAD but especially between the enhancers and promoters (Fig 2A).
Interaction profiles from nearby viewpoints located at the CTCF/Cohesin binding sites directly flanking the α-globin cluster were very different. Despite their proximity to the α-globin enhancer elements, these sites clearly do not interact with the enhancers within the sub-TAD: rather, they interact with the domains of chromatin containing convergent CTCF/cohesin sites extending from the θ-globin promoters to the 3’ flanks of the α-globin sub-TAD (Fig. 2B). Of particular interest, despite the near-identical CTCF/Cohesin binding landscape across the α-globin sub-TAD in erythroid and non-erythroid cells (Fig. 1B), we observed significantly increased interactions between flanking CTCF/Cohesin clusters in erythroid cells, suggesting the development of a hairpin-like structure of the sub-TAD in erythroid cells. While this proposed structure would exclude interactions between flanking sequences and α-globin enhancers, it would not prevent interactions between the two CTCF sites directly adjacent to the enhancers (HS-38 and HS-39) and the α-globin promoters. Such interactions may occur with the CTCF binding sites at the θ-globin promoters. Profiles from viewpoints located at the promoters of flanking genes (Mpg and Rhbdf1) are consistent with this topological model (Fig. 2C), and suggest that CTCF-mediated chromatin interactions between domains flanking the α-globin cluster may insulate promoters contained within these regions from the activity of the α-globin enhancers by constraining their interactions with these strong enhancers. Consistent with this model, the Nprl3 gene, whose promoter is located within the α-globin sub-TAD, shows a 6-fold increase in expression in erythroid cells compared to non-erythroid cells, whereas the expression of Mpg and Rhbdf1 lying outside the sub-TAD is unchanged and repressed in erythroid cells respectively (Fig. 1C).
Deletion of CTCF/Cohesin sites alters multiple chromatin interactions within the α-globin sub-TAD
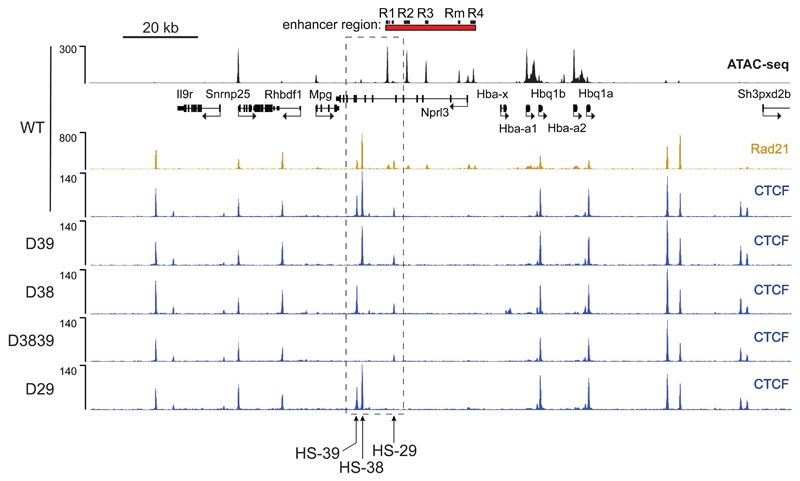
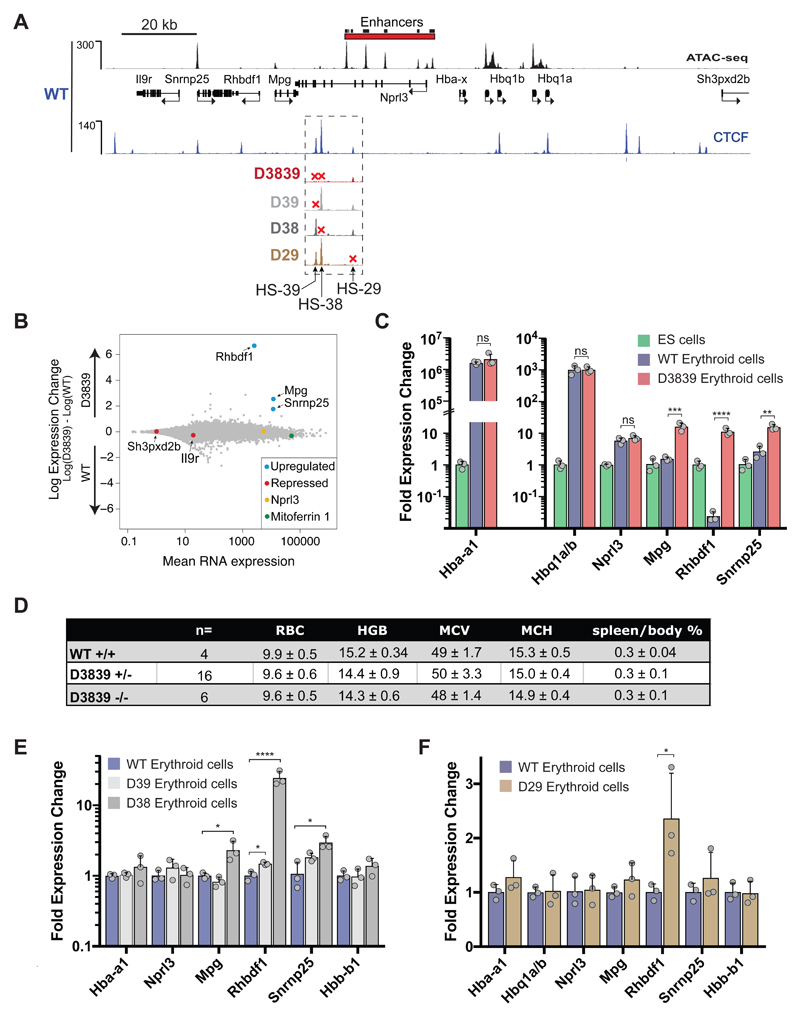
Close inspection of the chromatin profile identified two prominent CTCF/cohesin binding sites (HS-38 and HS-39) and a less prominent site (HS-29) lying close to and directly upstream of the α-globin enhancers at the boundary of the erythroid-specific sub-TAD defined by histone acetylation and H3K4me1 enrichment. These CTCF/cohesin sites are positioned in between the erythroid enhancers and the widely expressed upstream genes (Mpg, Rhbdf1 and Snrnp25) suggesting that they may act individually or together as an insulator, shielding these upstream genes from enhancer activity. To test this hypothesis in vivo, we used TALEN26 and CRISPR-mediated mutagenesis27 to generate mice with small deletions in the binding sequences of these three CTCF/cohesin sites, singly and in combination (Fig. 3 and Supplementary Fig. 2).
Figure 3. Disruption of CTCF binding motifs results in loss of CTCF binding at the α-globin locus in primary erythroid cells derived from mutant mice.
ATAC-Seq (RPKM) in wild-type (WT) primary erythroid cells shows chromatin accessibility across the α-globin locus. Local genes (Refseq) and the α-globin enhancers are annotated. Normalised CTCF ChIP-seq reads (RPKM, 2 independent experiments in which 2 animals were analysed in total) across the α-globin locus are shown for WT and each of the generated CTCF binding site mutants; D39: HS-39 mutant, D38: HS-38 mutant, D3839: combined HS-38 and HS-39 mutant, D29: HS-29 mutant. Also shown is normalised Rad21 ChIP-seq (RPKM, 2 independent experiments in which 2 animals were analysed in total) for WT primary erythroid cells. The dashed box indicates the genomic region within which the generated CTCF binding mutations are located.
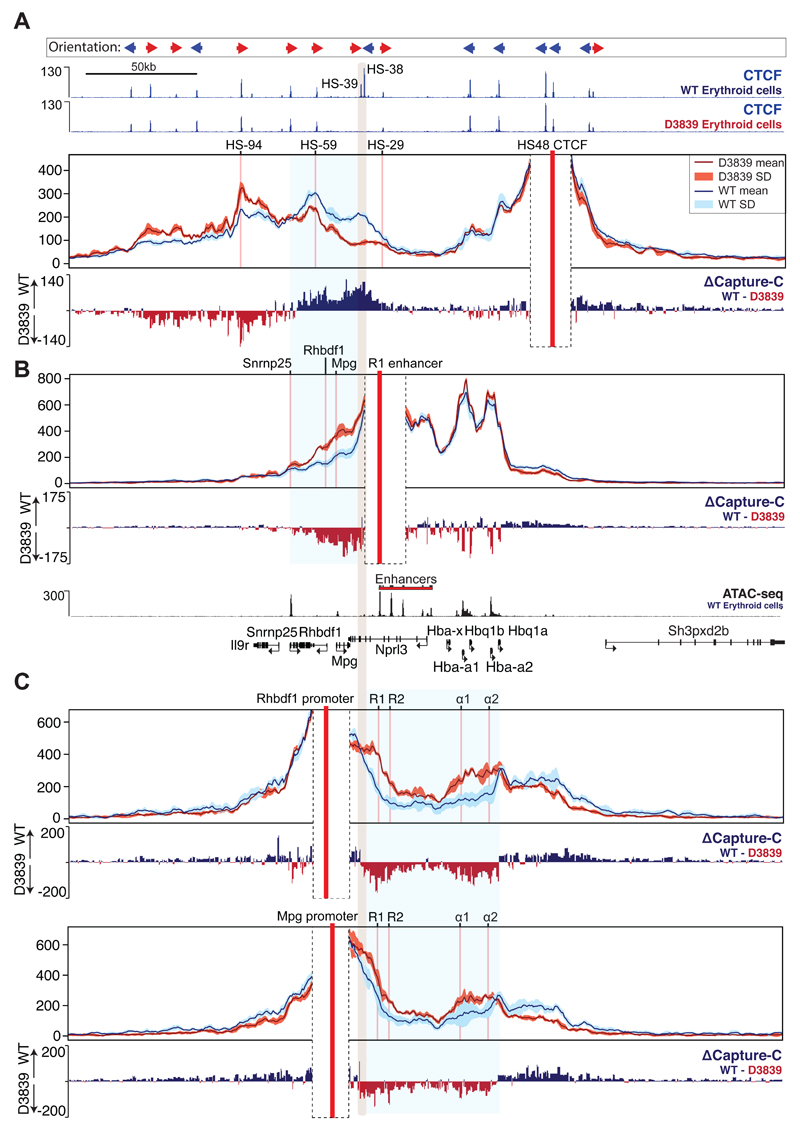
We first analysed erythroid cells of a mouse lacking both HS-38 and HS-39 (D3839). Mutation of the two CTCF core sequences resulted in a complete loss of CTCF binding at these sites (Figs 3, 4A, and 5A), but in contrast to previous reports28,29 did not affect binding of CTCF to other, nearby sites. To investigate whether mutation of both HS-38 and HS-39 altered interactions between the regions of chromatin flanking the sub-TAD, we used the downstream CTCF binding site (HS48) as a Capture-C viewpoint. Although interactions between flanking domains remain intact in the D3839 mutant, the upstream boundary of the domain in erythroid cells shifts from the deleted HS-38/-39 sites to the next adjacent upstream site (HS-59) site within the Rhbdf1 gene (Fig. 4A). Capture-C using the R1 enhancer as a viewpoint shows that, while interactions between R1 and the α-globin promoters appear to be unchanged, ablation of CTCF binding in the D3839 mutant results in increased interactions between R1 and a region of chromatin, directly upstream, containing the Mpg, Rhbdf1, and Snrnp25 genes (Fig. 4B). This is further confirmed by the interaction profiles obtained from the Rhbdf1 and Mpg promoters which show a strong increase of interactions with the R1 and R2 enhancers and the α-globin genes, while losing interactions with the downstream genomic region flanking the cluster (Fig. 4C). Thus, the elimination of CTCF binding in the D3839 mutant is associated with an entirely new set of contacts between the α-globin enhancers and the Rhbdf1 and Mpg genes. Importantly, these interactions occur with non-erythroid promoters and involve interactions with promoters located upstream, in the opposite direction to those normally seen from the α-globin enhancers. In the proposed hairpin analogy of the sub-TAD, contacts within the CTCF/Cohesin stem of the hairpin have shifted to increase the region of chromatin within its loop which now includes the Mpg, Rhbdf1 and Snrnp25 genes.
Figure 4. Differential interactions of α-globin regulatory regions and flanking genes between wild-type and D3839 primary erythroid cells.
Capture-C data for the indicated viewpoints (red vertical bars) in wild-type (WT) and CTCF mutant D3839 primary erythroid cells are shown as described in Fig 2A. Data represent 3 independent experiments in which 3 animals were used in total. Differential tracks (ΔCapture-C) show a subtraction (WT - D3839) of the mean number of normalised meaningful interactions per restriction fragment. Mutated CTCF sites are indicated with a shaded grey vertical bar. Also shown are normalised CTCF ChIP-seq (RPKM) for both WT and D3839 primary erythroid cells and ATAC-seq (RPKM) for WT erythroid cells, all merged from 2 independent experiments in which 2 animals were used in total. Gene annotation is Refseq. A. HS48 CTCF viewpoint. The shaded blue box indicates a chromatin region with altered CTCF-CTCF interactions B. R1 enhancer viewpoint. The shaded blue box indicates altered R1 enhancer interactions. C. Rhbdf1 and Mpg promoter viewpoints. The shaded blue box highlights the chromatin region with altered interactions.
Figure 5. Effects of individual and combined CTCF binding site deletions near the α-globin enhancers on local gene expression in primary erythroid cells.
A. The α-globin locus annotated with enhancer elements and genes (Refseq). Shown are normalised ATAC-seq (RPKM) data for WT and CTCF ChIP-seq (RPKM) for indicated WT and mutant mouse erythroid cells merged from 2 independent experiments in which 2 animals were analysed in total. B. MA plot of RNA-seq data derived from WT and D3839 primary erythroid cells. Data represent n=3 independent experiments in which 3 animals were analysed in total. Mean RNA abundance is plotted on the x-axis and enrichment is plotted on the y-axis. Significant upregulation of local genes in the D3839 mutant is highlighted in blue and was calculated using a Wald test (DEseq2): Snrnp25: P=1.46e-46, Rhbdf1: P<9.99e-99, Mpg: P=6.71e-64. Controls are indicated in different colours: Mitoferrin-1 (Slc25a37, green), a highly expressed erythroid gene; Sh3pxd2b and Il9r (red), repressed in erythroid cells; Nprl3 (yellow), a housekeeping gene within the α-globin locus, unaffected by deletions. C. Relative gene expression in WT and D3839 erythroid cells versus ES cells was measured by real-time qPCR. Data represent n=3 independent experiments in which 3 animals (erythroid cells) or 3 biologically independent samples (ES cells) were analysed in total. Bars represent the mean and the error bars represent the standard deviation (S.D.). Grey dots represent individual data points. P-values are obtained with an unpaired, two-tailed student t-test. ns P>0.05, * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. For significance of gene expression changes between ES and WT erythroid cells, see Fig 1C. D. Table summarising the haematological parameters of erythroid cells in WT and D3839 mutant mice. RBC = red blood cell count, HGB = haemoglobin count, MCV = mean corpuscular volume (fL), MCH = mean corpuscular haemoglobin (g/dl), Spleen/body% = spleen weight as a percentage of body weight, WT+/+ = wild-type, D3839+/- = heterozygous for D3839, D3839-/- = homozygous for D3839. E. Relative gene expression in D38 and D39 versus WT erythroid cells (as in Fig. 5C, n=3 independent experiments in which 3 animals were analysed in total). F. Relative gene expression in WT vs D29 erythroid cells (as in Fig. 5C, n=3 independent experiments in which 3 animals were analysed in total). No substantial difference in expression of local genes is detected (Rhbdf1: P=0.03, unpaired, two-tailed student t-test).
Mutation of CTCF/Cohesin sites alter gene expression in the α-globin sub-TAD
To examine whether the changes in local topology caused by the deletion of CTCF binding sites influence transcription in erythroid cells, we performed RNA sequencing (RNA-seq) on D3839 and wild-type primary erythroid cells. We found that expression of the three genes whose promoters are located in the genomic region that shows increased interactions with the R1 enhancer (Fig 4B), Mpg, Rhbdf1 and Snrnp25, is strongly up-regulated in D3839 mutant mice (Figs 5A, B, and C). Housekeeping genes Mpg and Snrnp25 are normally expressed in wild-type erythroid cells but in the absence of HS-38 and HS-39 their expression increases by 12- and 6-fold, respectively. Interestingly, the Rhbdf1 gene, which is normally silenced by Polycomb group complexes in wild-type erythroid cells, increases its expression ~600-fold in the D3839 mutant. In ES cells, Rhbdf1 is transcribed at relatively high levels. Even when compared to this active gene regulatory state, expression was significantly increased under the influence of the α-globin enhancers in the absence of CTCF insulation in D3839 erythroid cells (Fig. 5C). By contrast, the Il9r gene, whose promoter is located within the chromatin region in which interactions between flanking CTCF/cohesin domains are retained in the D3839 mutant (Fig. 4A), remained inactive (Fig. 5B) and insulated from the influence of the α-globin enhancers. We also detected no significant changes in expression of the α- or θ-globin genes (Fig. 5C), consistent with the identical interaction profile of the R1 enhancer with the α-like globin promoters in D3839 mutant mice (Fig. 4) and the lack of any detectable change in the haematological phenotype (Fig. 5D).
Not all CTCF sites in the sub-TAD are equivalent
Clearly, mutation of both HS-38 and HS-39 sites causes a change in the functional interactions between the α-globin enhancers and the promoters of the surrounding genes in the TAD. It has been suggested that effective chromatin boundaries are formed by two directly adjacent divergent CTCF binding sites30. Therefore, we next made and analysed mice with single deletions of CTCF binding at either the HS-38 (D38) or HS-39 (D39) elements (Supplementary Fig. 2). Loss of HS-38 alone led to an up-regulation of the upstream Mpg, Rhbdf1, and Snrnp25 genes, although to a lesser extent than that observed in the D3839 mice (Fig. 5E). Rhbdf1 was ten-fold less upregulated in the D38 mutant than in the double D3839 mutant, suggesting that the presence of HS-39 in the D38 mutant prevents a complete loss of enhancer insulation. However, deletion of HS-39 alone did not have a strong effect on local gene expression (Fig. 5E). The observation that loss of HS-39 does not result in gene expression changes is consistent with the fact that only HS-38 is conserved across mammals including human and bound by higher levels of CTCF/cohesin, suggesting this site is sufficient for adequate enhancer insulation. These data do not exclude the opposite orientation of HS-38 and HS-39 or the difference in the composition of their CTCF binding motif as possible causes for this difference in insulator activity (Supplementary Fig. 1). Finally, we generated a mouse lacking the HS-29 CTCF binding site (D29), located between the R1 and R2 enhancers. While the loss of CTCF binding at HS-29 resulted in increased interactions between the enhancers and the α-globin promoters (Supplementary Fig. 3), these changes did not result in any detectable changes in local gene expression or histone modifications (Fig. 5F and Supplementary Fig. 4). However, minor changes in α-globin gene expression may not have been detected by qPCR, which cannot detect fractional changes in expression. Notably, the HS-29 CTCF site binds lower levels of CTCF and Cohesin than HS-38, possibly providing an explanation for its lack of boundary activity.
The CTCF/Cohesin boundary constrains enhancer interactions rather than encroachment of chromatin modifications
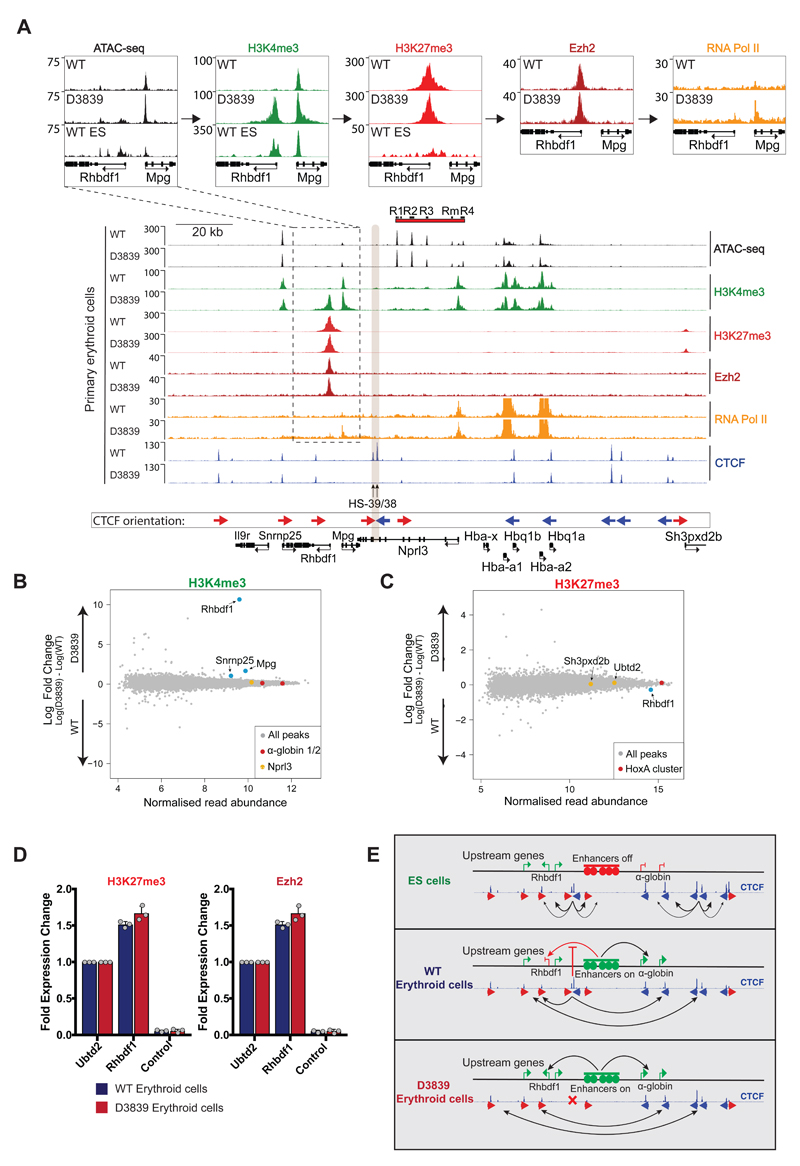
Our results demonstrate that the removal of a bona fide insulator lying within a TAD does not simply cause encroachment of one chromatin state into another, but rather extends the range of interactions from a strong enhancer to flanking promoters. This suggests that enhancer/promoter interactions may be promiscuous rather than specific and that such interactions are normally constrained, in some way, by CTCF/cohesin binding sites. In this respect, it was of interest that the normally silent Rhbdf1 promoter acquired high levels of H3K4me3 in D3839 erythroid cells, consistent with its ectopic expression in these mutant cells (Figs 6A and B). Similar increases in H3K4me3 were also noted at the Mpg promoter, and, to a lesser extent, at the Snrnp25 promoter, consistent with their transcriptional up-regulation in D3839 erythroid cells. Following the smaller effects on gene transcription, the single disruption of HS-38 CTCF binding resulted in the recruitment of lower levels of H3K4me3 to the Rhbdf1 promoter, whereas loss of HS-39 had no detectable effect on local deposition of H3K4me3 (Supplementary Fig. 5). Changes in H3K4me3 in the D3839 mutant are accompanied by higher levels of RNA Polymerase II (PolII) recruitment and increased chromatin accessibility (ATAC-seq) at the gene promoters. Surprisingly, we could not detect a decrease in the levels of H3K27me3 at the Rhbdf1 gene promoter despite its strong transcriptional activation (Figs 6A, C, and D). To exclude the possibility that H3K27me3 was retained despite the loss of PRC2 recruitment upon α-globin enhancer activation, we verified that binding of the PRC2 complex component Ezh2 is retained in the D3839 mutant (Figs 6A and D). Thus, it appears that insulation of the Rhbdf1 promoter from the α-globin enhancers by CTCF/Cohesin is required for effective Polycomb-mediated transcriptional repression. This is not consistent with a model of simple chromatin encroachment.
Figure 6. Effects of combined deletion of HS-38 and HS-39 on the local chromatin state in primary erythroid cells.
A. Normalised ATAC-seq and ChIP-seq read-densities (RPKM; 2 independent experiments in which 2 animals were analysed in total) for H3K4me3, H3K27me3, Ezh2, RNA Pol II, and CTCF at the α-globin locus, both in WT and D3839 primary erythroid cells. Shaded grey bar indicates the position of HS-38 and HS-39. The dashed box highlights the region over the Rhbdf1 and Mpg genes, magnified in top panels for ease of data visualisation. B. MA plot of H3K4me3 ChIP-seq data derived from WT and D3839 erythroid cells (2 independent experiments in which 2 animals were analysed in total). Mean read abundance is plotted on the x-axis and enrichment on the y-axis. Changes in H3K4me3 detected as indicated on the plot by the genes highlighted in blue: Snrnp25: FDR<0.1, Rhbdf1: FDR<0.05, Mpg: FDR<0.05. Controls are shown in red (α-globin promoters) and yellow (Nprl3, unaffected by the combined disruption of HS-38 and HS-39). The FDR was calculated with the Diffbind package using DEseq2. C. MA plot of H3K27me3 ChIP-seq data (2 independent experiments in which 2 animals were analysed in total) derived from WT and D3839 erythroid cells. Mean read abundance is plotted on the x-axis and enrichment on the y-axis. Highlighted on the plot are Ubtd2 and Sh3pxd2b (yellow), Polycomb-repressed genes directly downstream of the α-globin cluster, and the HoxA cluster (red) as a control. Rhbdf1 (blue) is unchanged (FDR=0.19). The FDR was calculated with the Diffbind package using DEseq2. D. ChIP-qPCR for H3K27me3 and Ezh2 in WT and D3839 primary erythroid cells (n=3 independent experiments in which 3 animals were analysed in total). Grey dots represent individual data points. Data displayed as fold change relative to Ubtd2, a Polycomb-repressed gene within 200kb downstream of the α-globin cluster. An amplicon within the Nprl3 gene is used as an α-globin locus control. No significant changes are detected by two-tailed student t-test. E. Model for α-globin cluster gene regulation. Interactions between clusters of flanking CTCF sites prevent contacts between the α-globin enhancers and upstream genes from forming. Upon deletion of HS-38 and HS-39 CTCF binding sites (D3839), CTCF interactions shift towards more distally located upstream sites, allowing bidirectional α-globin enhancer interactions and upregulation of upstream genes.
Discussion
The above results indicate that upon activation of the α-globin enhancers, selective convergent CTCF/Cohesin binding sites act as boundary elements and create an erythroid-specific chromatin structure, delimiting enhancer interactions and consequently ensuring an erythroid-specific transcriptional program (Fig. 6E). The ability of active enhancers to interact with an unexpectedly wide range of receptive promoters was revealed when critical CTCF/Cohesin elements were removed. More widespread and bidirectional enhancer interactions appeared and were associated with the up-regulation of three genes; in one case (Rhbdf1) overcoming Polycomb-mediated repression. While genetic perturbation of CTCF binding has been shown to result in misregulation of gene expression in various cell lines and cancer9–11,28, our more detailed investigation involving precise CTCF-site disruptions, and high-resolution chromatin conformation analysis (Capture-C) clearly link gene activation and acquisition of H3K4me3 to the establishment of aberrant enhancer contacts within a perturbed, tissue-specific sub-TAD. Importantly, we have shown that not all CTCF/Cohesin sites subserve the same functions in the sub-TAD: HS-38 acts as a strong boundary element, HS-39 as a weaker element and HS-29 has no apparent insulator activity. The molecular basis of this is currently unknown.
In addition, we show that interactions between the two flanking clusters of CTCF sites are weaker or absent in ES cells despite the presence of CTCF and Cohesin binding (Fig. 6E). This raises the question of what regulatory mechanisms drive the tissue-specificity of these CTCF interactions. As cohesin is loaded at active enhancer-promoter junctions31, one intriguing possibility is that additional cohesin recruitment in erythroid cells results in stabilisation of flanking CTCF-CTCF interactions via a recently described loop extrusion mechanism22,32,33. In addition, the enhancer-promoter interactions which occur in erythroid cells may further stabilize the interactions between flanking CTCF binding sites.
In conclusion, our findings suggest that rather than enhancers having inherent specificity for their cognate promoters34, this communication is at least in part driven by the CTCF-mediated chromatin architecture which normally shields genes flanking a sub-TAD from the influence of enhancers in a tissue-specific manner35,36. However, of interest, we have previously shown that the human α-globin enhancers may influence the expression of a gene (NME4) lying 400kb away and outside of the orthologous region described here37. Therefore, insulation may not be absolute. Nevertheless, given the dynamic genome partitioning through development and differentiation described here, it seems likely that in addition to variants in enhancers and promoters, intergenic variants within critical CTCF/Cohesin binding sites will underlie changes in gene expression associated with a wide variety of complex traits and diseases.
Method
Animal procedures
C57BL/6J mice were sourced from MRC Harwell/Charles River Laboratories. The mutant mouse strains reported in this study were maintained on a C57BL/6J background and were generated and phenotyped in accordance with Animal [Scientific Procedures] Act 1986, with procedures reviewed by the clinical medicine Animal Welfare and Ethical Review Body (AWERB), and conducted under project licences PPL 30/2966 and PPL 30/3339. Animals were housed in specific pathogen free conditions, with the only reported positives on health screening over the entire time course of these studies being Entamoeba spp. All animals were singly-housed, provided with food and water ad-libitum and maintained on a 12h light:12h dark cycle (150-200 lux cool white LED light, measured at the cage floor. No statistical method was used to predetermine sample size. Experiments to determine haematological parameters were blinded. Mice were given neutral identifiers and analysed by research technicians unaware of mouse genotype during outcome assessment. Experiments for Capture-C, gene expression and ChIP-seq analysis were not randomised and the investigators were not blinded to allocation during these experiments and outcome assessments. No statistical method was used to predetermine sample size. No samples or animals were excluded from the analysis.
Isolation and selection of ter119+ cells from mice
Mature primary erythroid cells were obtained from young adult mice of both genders between 2 and 6 months of age that were pre-treated with acetylphenylhydrazine (APH) as described45. Spleens of APH-treated mice were mechanically disrupted to single cell suspension in RPMI media (Thermo Fischer Scientific) supplemented with 10% fetal bovine serum (FBS, Gibco). To isolate late-stage erythroid cells, cells from a single spleen were resuspended in 5 mL of cold PBS/2% BSA and stained with 120 μL PE anti-ter119 antibody (Ly-76, BD Biosciences) at 4°C for 15 minutes45. After washing stained cells in PBS/0.5% BSA, cells were resuspended in 1.6 mL of PBS/0.5% BSA and 400 μL of anti-PE magnetic beads (Miltenyi Biotec) and incubated for 20 minutes at 4°C. Ter119 positive cells were isolated via auto-magnetic-activated cell sorting (autoMACS, Miltenyi Biotec) and processed for downstream applications. Purity of the isolated erythroid cells was routinely verified by FACS.
Cell lines
The embryonic stem cell line ES-E14TG2a was used for gene expression and Capture-C analysis and cultured according to standard conditions (ATCC CRL-1821). The E14TG2a line was a kind gift from Andrew Smith and was tested negative for mycoplasma and has been extensively authenticated by blastocyst injection. This cell line is not found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample.
Preparation of TALEN expression constructs
For TALEN construction, a 500bp sequence centred around the HS-38 CTCF consensus sequence was submitted to the TALE-NT Targeter using NN for G recognition (Golden Gate TALEN and TAL Effector Kit 1.0, Addgene)26. Two TALEN pairs with a differential spacer region that targeted the HS-38 CTCF binding sequence were selected and constructed via the Golden Gate assembly method26. TALEN-AF targeted the sequence 5’-TCCTGGGTAGGCCTCT-3’ with the RVD array HD-HD-NG-NN-NN-NN-NG-NI-NN-NN-HD-HD-NG-HD-NG and TALEN-AR targeted the sequence 5’-GAGTCCCACGTATCGT-3’ on the reverse strand with the RVD array NN-HD-NG-NI-NG-NN-HD-NI-HD-HD-HD-NG-NN-NI-NN. The vector RCIscript-Goldytalen (38142, Addgene) were used as the scaffold vector in the final step of the Golden Gate cloning protocol.
Preparation of CRISPR-Cas9 expression constructs
To generate the single guide-RNAs (sgRNAs) used to target CTCF binding sequences, oligonucleotides corresponding to the target protospacers were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid #42230, pX330) or pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10) (Addgene plasmid #42335, pX335) vectors as described previously46. pX330 and pX335 were modified to contain a puromycin and neomycin selection cassette respectively. DNA oligos containing the 20nt protospacer sequences are shown in Supplementary Table 1.
Preparation and injection of TALEN mRNA and CRISPR sgRNA
TALEN micro-injections were performed as previously described47. DNA templates for use in in vitro transcription reactions were generated from CRISPR-Cas9 expression constructs by PCR. The forward, sgRNA-specific primer was modified with a 5’ extension that contained a T7 polymerase binding site, and used to amplify the gRNA with a reverse primer binding downstream of the mature gRNA sequence (gRNA-R) (see Supplementary Table 1). The MEGAshortscript™ T7 Transcription Kit (Thermo Scientific) was used for in vitro transcription of the gRNAs. In vitro transcribed RNAs were purified with the MEGAclear Kit (Thermo Scientific) and diluted in 10 mM Tris-HCl pH 7.5, 0.1 mM EDTA pH 8.0 before microinjection. Manipulations using wild-type Cas9 were performed using a Cas9 expressing mouse line to provide maternal supply of Cas9 to zygotes as previous described48. Briefly, female mice homozygous or heterozygous for the CAG-Cas9 transgene insertion were superovulated and mated with C57BL/6 or, for the production of double (D3839) mutants, with DEL38 studs. Oocytes were prepared for microinjection from plugged females and 20 ng/µl of gRNA was injected into the pronucleus. Depending on the experiment, ssODN templates for HDR (Eurogentec) were added at a final concentration of 20 ng/µl (see Supplementary Table 1). For the single mutation of HS-39, D10A Cas9 protein (PNA Bio) was injected with two sgRNAs at a concentration of 40 ng/µl into C57BL/6 oocytes. The microinjected zygotes were immediately transferred to pseudopregnant CD1 foster mothers.
Next-Generation Capture-C
Next-Generation Capture-C was performed as previously described17. 2 × 107 mouse ES cells or isolated ter119+ mouse spleen cells were used per biological replicate experiment and processed in parallel. To visualize differences in Capture-C profiles, normalised interactions in ES cells or erythroid cells of CTCF binding site mutants were subtracted from wild-type erythroid interactions to generate a differential Capture-C track. For plotting of multiple interaction profiles simultaneously, Capture-C interactions were binned in 250bp bins and a sliding 5 kb window was used. The mean of three biological replicates and standard deviation were plotted in R.
ATAC-sequencing
ATAC-seq was performed on 65,000 ter119+ cells isolated from APH-treated mouse spleens and mouse ES cells as previously described49.
RNA expression analysis
Isolation of total RNA was performed by lysing 1 × 107 mouse ES or purified ter119+ cells in TRI reagent (Sigma) according to the manufacturer’s instructions. To remove genomic DNA from RNA samples, samples were treated with TURBO™ DNase with the DNA-free™ DNA removal kit (Ambion). DNase-treated RNA samples were stored at -80°C. To assess relative changes in gene expression by qPCR, 1 μg of total RNA was used for cDNA synthesis using the Superscript III first-strand synthesis SuperMix (Invitrogen). The ΔΔCt method was used for relative quantitation of RNA abundance using the primers in Supplementary Table 1. For RNA-seq libraries, rRNA and globin mRNA species were removed using the Globin-Zero Gold kit (Illumina) with 5 μg of total RNA according to the manufacturer’s instructions. To further enrich for mRNA, poly(A)+ were isolated using the NEBNext Poly(A) mRNA magnetic isolation module (New England Biolabs) followed by a NEBNext Ultra™ directional RNA library preparation (New England Biolabs) according to the manufacturer’s instructions. Fragmentation of mRNA was achieved by incubating samples at 95°C for 12 min. To achieve strand specificity, Actinomycin D was added (5 μL of 0.1 μg/μL) to the first strand cDNA synthesis reaction. Poly(A)+ libraries (4 nM) were sequenced on the Illumina NextSeq platform. All RNA-seq datasets were aligned to the mm9 mouse genome build using STAR50. Deeptools bamCoverage was used to calculate normalised (RPKM) and strand-specific read coverage which was visualised in the UCSC genome browser. Mapped RNA-seq reads were assigned to genes using Subread featureCounts using RefSeq gene annotation. Normalised differential gene expression, between biological triplicate data from litter-mate wild-type and CTCF binding site mutant mice extracted in parallel, was calculated with the DESeq2 R package.
De novo CTCF motif analysis in Ter119+ cells
Motif analysis was performed as previously described51. Briefly, 2000 CTCF peak sequences from ter119+ cells were retrieved and used for de novo motif discovery using the MEME suite. The motif with the highest score matched the previously published consensus CTCF core binding motif. Significant matches (p < 10-3) for the CTCF core motif within all CTCF peak regions were identified using fimo. When multiple core motifs were detected within the same peak region, only the best match was retained. Motifs up- and downstream of the core motif were identified from 6000 randomly selected 20 bp sequences of up- and downstream flanks and were similar to those previously identified51. Analysis of spacing between the core and flanking motifs revealed a preferential spacing for both up- and downstream motifs. Significant upstream or downstream motif matches (P-value threshold of 10-2) were added to CTCF peak annotation only if the motifs were present at the preferred spacing (5-6 bp for upstream motif, 4-6 for downstream motif).
DNaseI footprint analysis
DNaseI footprints and meta-plots at CTCF binding sites were generated using a custom perl script based on Samtools using previously published C57BL/6 DNaseI-seq data41. DNaseI-seq cuts were counted as the 5’ end of the first read and the 3’ end of the second read of DNA fragments. For meta-plots of CTCF peaks with different combinations of core and auxiliary motifs, average cut-counts relative to the start of the CTCF core sequence were calculated for each category.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on purified ter119-positive primary erythroid cells (1 × 107 cells/ChIP) using the ChIP Assay Kit (Cat. No. 17-295, Millipore). For ChIP of Cohesin component Rad21, cells were subjected to dual cross-linking with 2 mM disuccinimidyl glutarate (DSG, Thermo Fischer Scientific) for 50 min and 1% (v/v) formaldehyde for 10 min, whereas a single 10 min 1% formaldehyde fixation was used for all other antibodies (see details in the Reporting Summary). Chromatin fragmentation was performed with the Bioruptor sonicator (Diagenode) for 15 min at 4°C to obtain an average fragment size between 200 and 500bp. Immunoprecipitated DNA fragments analysed by qPCR or sequencing. Primers used for qPCR are listed in Supplementary Table 1. DNA libraries for sequencing were prepared with the NEBNext Ultra™ II DNA library prep kit (New England Biolabs) and sequenced on the Illumina platform.
Analysis of ChIP-seq data
The MACS (Model based analysis of ChIP-seq) peak finding algorithm was used to identify regions of ChIP-seq enrichment over background in an unbiased manner. The MACS2 callpeak function was used on biological duplicate ChIP-seq data of CTCF (-q 10-5) and H3K4me3 (-q 10-3). For H3K27me3, the MACS2 callpeak function was used with the --broad-cutoff option (--broad-cutoff 0.05) on biological duplicate ChIP-seq data. To identify regions that were differentially enriched between wild-type and CTCF binding site mutant mice, the R package DiffBind was used. Two biological duplicate datasets and independent peak calls of CTCF, H3K4me3 and H3K27me3 were used to identify differentially enriched regions with a false discovery rate (FDR) of 0.05. Differential analysis within the DiffBind package was performed with DESeq2.
Statistics and reproducibility
Statistical analysis was carried out with Graphpad Prism (version 7.0c) unless otherwise indicated. All gene expression experiments (both RT-qPCR and RNA-seq) were performed on three biological replicates with similar results (standard deviation (SD) is shown for all measurements). Statistical analysis was performed using multiple unpaired, two-tailed t-tests and corrected for multiple comparisons using the Holm-Sidak method where appropriate. The statistical analysis of RNA-seq data was performed in R using the Bioconductor DEseq2 package. All Capture-C experiments were performed on three biological replicates with similar results. The standard deviation of 250bp bins was calculated in R and visualised to illustrate the reproducibility of this chromatin interaction analysis. All ChIP experiments newly generated for this study were performed at least in biological duplicate with similar results. The analysis of ChIP-seq data was performed with Bioconductor package DiffBind, using DEseq2 to determine false discovery rate (FDR). P-values are represented as *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Supplementary Material
Acknowledgements
The authors would like to thank Jelena Telenius for her help in submitting genome-wide datasets to the GEO database, Deborah Hay, Richard Gibbons, and Andrew King for critically reading the manuscript. L.L.P.H. and A.M.O. would like to thank the Wellcome Trust for funding (Chromosome and Developmental Biology PhD Programme, grant code 099684/Z/12/Z; and Wellcome Trust Genomic Medicine and Statistics PhD Programme, grant code 083323/Z/07/Z respectively). The work was further supported by the Wellcome Trust core award 203141/Z/16/Z and the Medical Research Council (MRC Core Funding and Centenary Award reference 4050189188).
Footnotes
Author contributions
L.L.P.H. planned and carried out experiments, analysed the data, carried out bioinformatic analysis, and wrote the manuscript. M.T.K. coordinated and advised the project and revised the manuscript. A.M.O. carried out Capture-C experiments and revised the manuscript. M.T.K. and A.M.O. contributed equally to this work. D.B. carried out cell culture and mouse experiments. C.P. carried out mouse micro-injection experiments. D.J.D. and M.G. carried out ATAC-seq experiments. J.A.S and J.A.S-S carried out mouse maintenance and haematology experiments. J.R.H. coordinated the project and gave technical and bioinformatics advice throughout the project. B.D. planned and coordinated transgenic mouse experiments and wrote the manuscript. D.R.H conceived, planned and coordinated the project and wrote the manuscript. B.D. and D.R.H. share senior authorship.
Competing financial interests
The authors have no competing financial interests
Data availability
RNA-sequencing, ChIP-sequencing, ATAC-sequencing, and Capture-C data generated for this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE97871. Previously published ChIP-seq data that were re-analysed here are available under the following accession codes: GSE27921, GSE31039, GSE30203. Previously published Capture-C data that were re-analysed here are available under the accession code GSE67959. Previously published DNaseI- and ATAC-seq data that were re-analysed here are available under the accession codes GSE49460 and GSE94249.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Ghirlando R, Felsenfeld G. CTCF: making the right connections. Genes Dev. 2016;30:881–891. doi: 10.1101/gad.277863.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nichols MH, Corces VG. A CTCF Code for 3D Genome Architecture. Cell. 2015;162:703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. doi: 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 4.Weth O, et al. CTCF induces histone variant incorporation, erases the H3K27me3 histone mark and opens chromatin. Nucleic Acids Research. 2014;42:11941–11951. doi: 10.1093/nar/gku937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuddapah S, et al. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Research. 2008;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handoko L, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of Embryonic Stem Cell Lineage Commitment by Core Promoter Factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirayama T, Tarusawa E, Yoshimura Y, Galjart N, Yagi T. CTCF is required for neural development and stochastic expression of clustered Pcdh genes in neurons. Cell Rep. 2012;2:345–357. doi: 10.1016/j.celrep.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowen JM, et al. Control of Cell Identity Genes Occurs in Insulated Neighborhoodsin Mammalian Chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anguita E, Johnson CA, Wood WG, Turner BM, Higgs DR. Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc Natl Acad Sci USA. 2001;98:12114–12119. doi: 10.1073/pnas.201413098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowalczyk MS, et al. Intragenic Enhancers Act as Alternative Promoters. Molecular Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Hay D, et al. Genetic dissection of the α-globin super-enhancer in vivo. Nat Genet. 2016;48:895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anguita E. Deletion of the mouse alpha-globin regulatory element (HS -26) has an unexpectedly mild phenotype. Blood. 2002;100:3450–3456. doi: 10.1182/blood-2002-05-1409. [DOI] [PubMed] [Google Scholar]
- 17.Davies JOJ, et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Meth. 2016;13:74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips-Cremins JE, et al. Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D, Day DS, Young RA. Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell. 2016;167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Wit E, et al. CTCF Binding Polarity Determines Chromatin Looping. Molecular Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 24.Rudan MV, et al. Comparative Hi-C Reveals that CTCF Underlies Evolution of Chromosomal Domain Architecture. CellReports. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes JR, et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nature Publishing Group. 2014;46:205–212. doi: 10.1038/ng.2871. [DOI] [PubMed] [Google Scholar]
- 26.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82–e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran FA, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narendra V, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang R, et al. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Research. 2016;44:3082–3094. doi: 10.1093/nar/gkv1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez-Marín C, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci USA. 2015;112:7542–7547. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. CellReports. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dekker J, Mirny L. The 3D Genome as Moderator of Chromosomal Communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zabidi MA, Stark A. Regulatory Enhancer–Core- Promoter Communication via Transcription Factors and Cofactors. Trends in Genetics. 2016;32:801–814. doi: 10.1016/j.tig.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oti M, Falck J, Huynen MA, Zhou H. CTCF-mediated chromatin loops enclose inducible gene regulatory domains. BMC Genomics. 2016;17:252. doi: 10.1186/s12864-016-2516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narendra V, Bulajić M, Dekker J, Mazzoni EO, Reinberg D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016;30:2657–2662. doi: 10.1101/gad.288324.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lower KM, et al. Adventitious changes in long-range gene expression caused by polymorphic structural variation and promoter competition. Proc Natl Acad Sci USA. 2009;106:21771–21776. doi: 10.1073/pnas.0909331106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 39.Consortium TEP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;488:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon CS, et al. Functional characterisation of cis-regulatory elements governing dynamic Eomesexpression in the early mouse embryo. Development. 2017;144:1249–1260. doi: 10.1242/dev.147322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosseini M, et al. Causes and Consequences of Chromatin Variation between Inbred Mice. PLoS Genetics. 2013;9:e1003570. doi: 10.1371/journal.pgen.1003570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leder A, Daugherty C, Whitney B, Leder P. Mouse zeta- and alpha-globin genes: embryonic survival, alpha-thalassemia, and genetic background effects. Blood. 1997;90:1275–1282. [PubMed] [Google Scholar]
- 44.Spivak JL, Toretti D, Dickerman HW. Effect of phenylhydrazine-induced hemolytic anemia on nuclear RNA polymerase activity of the mouse spleen. Blood. 1973;42:257–266. [PubMed] [Google Scholar]
- 45.Kina T, et al. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol. 2000;109:280–287. doi: 10.1046/j.1365-2141.2000.02037.x. [DOI] [PubMed] [Google Scholar]
- 46.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies B, et al. Site Specific Mutation of the Zic2 Locus by Microinjection of TALEN mRNA in Mouse CD1, C3H and C57BL/6J Oocytes. PLoS ONE. 2013;8:e60216–7. doi: 10.1371/journal.pone.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cebrian-Serrano A, et al. Maternal Supply of Cas9 to Zygotes Facilitates the Efficient Generation of Site-Specific Mutant Mouse Models. PLoS ONE. 2017;12:e0169887–20. doi: 10.1371/journal.pone.0169887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Meth. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakahashi H, et al. A Genome-wide Map of CTCF Multivalency Redefines the CTCF Code. Cell Reports. 2013;3:1678–1689. doi: 10.1016/j.celrep.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.