Abstract
Human brain development involves complex interactions between different areas, including long distance neuronal migration or formation of major axonal tracts. 3D cerebral organoids allow the growth of diverse brain regions in vitro, but the random arrangement of regional identities limits the reliable analysis of complex phenotypes. Here, we describe a co-culture method combining various brain regions of choice within one organoid tissue. By fusing organoids specified toward dorsal and ventral forebrain, we generate a dorsal-ventral axis. Using fluorescent reporters, we demonstrate robust directional GABAergic interneuron migration from ventral into dorsal forebrain. We describe methodology for time-lapse imaging of human interneuron migration that is inhibited by the CXCR4 antagonist AMD3100. Our results demonstrate that cerebral organoid fusion cultures can model complex interactions between different brain regions. Combined with reprogramming technology, fusions offer the possibility to analyze complex neurodevelopmental defects using cells from neurological disease patients, and to test potential therapeutic compounds.
Introduction
The cerebral cortex contains two main populations of neurons, excitatory glutamatergic pyramidal neurons, and inhibitory γ-Aminobutyric acid (GABA) producing interneurons1. Excitatory cortical neurons are predominantly generated by dorsal forebrain progenitors, while inhibitory GABAergic cortical interneurons are generated by ventral forebrain progenitors2. To integrate into cortical circuits, interneurons perform a long-distance migration from their ventral origin into their target dorsal cortical regions3. This long-range tangential migration is controlled by many signaling pathways4,5, and studies using animal models indicate that mutations in some neurological disease-associated genes can disrupt interneuron migration6,7. However, the relationship between patient-specific mutations and human brain development remains enigmatic without suitable experimental human model systems.
Three-dimensional (3D) organoid culture technology allows the development of complex, organ-like tissues reminiscent of in vivo development8. Importantly, cerebral organoids recapitulate many aspects of embryonic cortical development including the generation of diverse cell types corresponding to different brain regional identities9. For instance, cerebral organoids can produce dorsal and ventral forebrain progenitors that generate excitatory neurons and inhibitory interneurons, respectively9. Moreover, cerebral organoids can be generated from human patient-derived induced pluripotent stem cells (hiPSCs), and used for functional genomic studies of neurological disorders such as microcephaly9 and Autism10. Therefore, cerebral organoids represent an exemplary experimental system to study the role of neurological disease-associated genes in brain development.
In the current study, we focused on expanding the cerebral organoid model system by enhancing the phenotypic analyses to include neuronal migration. We developed an organoid co-culture “fusion” paradigm to successfully combine independently patterned organoids into a single tissue. Using this approach, we recreated the dorsal-ventral forebrain axis, and by labeling one organoid with a fluorescent reporter, we observed robust migration of cells between fused organoids. The molecular taxonomy and migratory dynamics of these cells resembles that of cortical interneurons. Therefore, we developed an organoid fusion assay that allows analysis of human cortical interneuron migration. This technology enhances the repertoire of phenotypic assays available for cerebral organoids, and in turn the complexity of phenotypes that can be used to study the developmental cell biology of human neurological diseases.
Results
Drug-patterning of cerebral organoids enhances the production of ventral forebrain identity
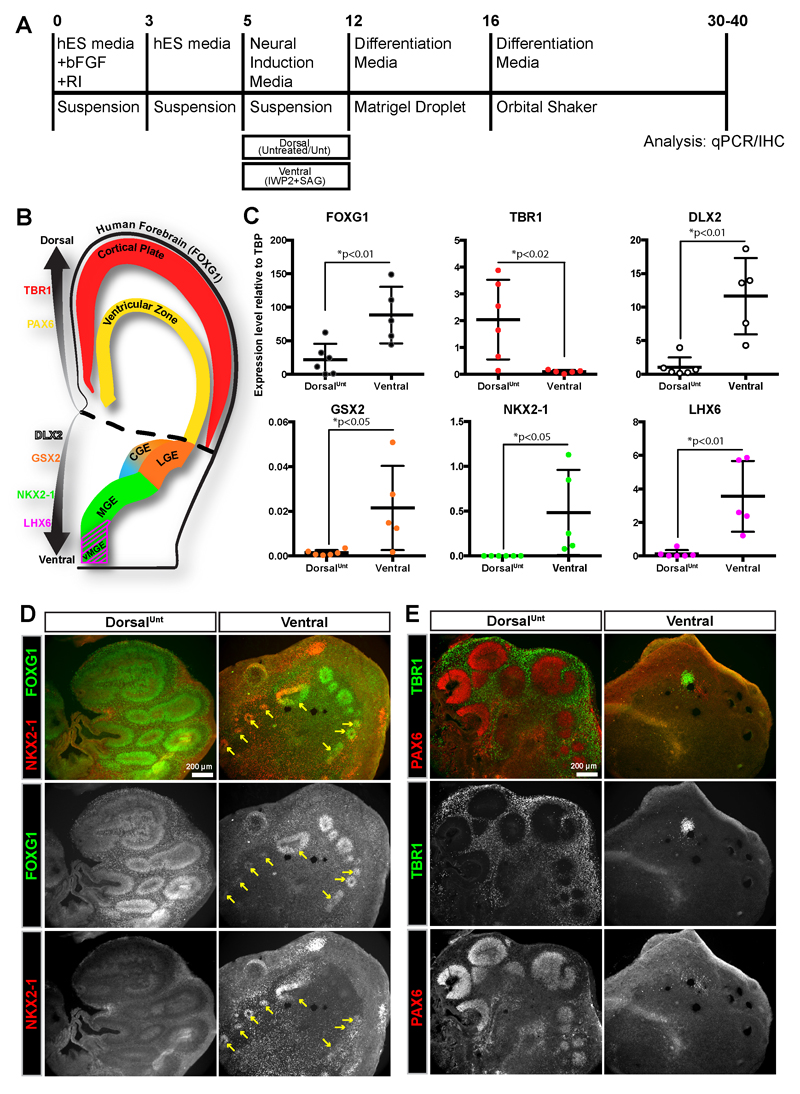
The cerebral organoid method9 is capable of producing many different brain regions, including dorsal and ventral forebrain. As it relies on intrinsic patterning, however, the production of some regions is variable and infrequent, especially more ventral (NKX2-1+) interneuron progenitor regions11. To increase the consistency and yield of ventral-forebrain interneuron progenitor regions, we modified the organoid protocol9,12 to include a ventral drug-treatment (Figure 1A). Based on previous 2D-neuronal differentiation13–15 protocols and recent 3D protocols 16,17, we utilized a combination of WNT-inhibition and enhanced SHH signaling to promote a rostro-ventral forebrain identity. qPCR analysis of the ventral organoids revealed a significant increase in expression of the forebrain marker FOXG118,19 (Figure 1B,C). The dorsal forebrain marker TBR120 became undetectable, while the ventral forebrain marker DLX221–23 was dramatically increased in ventral organoids compared to control organoids (Figure 1B,C). To further confirm the successful ventralization of cerebral organoid tissue, we examined the expression of specific markers of ventral forebrain ganglionic eminence (GE) subregions that produce interneurons of different subtypes3,24–26. GSX2 is expressed in the dorsal-lateral GE (LGE) and caudal GE (CGE), NKX2-1 is expressed in the ventral-medial GE (MGE), while LHX6 is expressed in a subregion of the MGE producing more ventral-derived MGE (vMGE) interneurons 3,23,24,27,28 (Figure 1B). GSX2 was expressed in control organoids, but further increased in ventral organoids (Figure 1C). Expression of NKX2-1 and LHX6 was undetectable in control organoids, but largely increased in ventral organoids (Figure 1C). Finally, immunostaining confirmed the qPCR results indicating widespread expression of FOXG1 in control and ventral organoids, but only ventral organoids expressed the ventral forebrain marker NKX2-1 (Figure 1D). Ventral organoids also highly expressed the ventral markers DLX2 and GSX2 (Supplementary Figure 1E-F). Intriguingly, control organoids widely expressed dorsal forebrain markers for both progenitors (PAX6)29 and early born neurons (TBR1)20 (Figure 1E). Therefore rather than referring to these organoids as “control,” we refer to them as “dorsalUnt” to accurately depict their dorsal patterning while also conveying the reliance on an intrinsic (untreated) patterning protocol. In contrast, ventral organoids contained only small regions of PAX6+ or TBR1+ tissue (Figure 1E). Additionally, we quantified the percentage of ventricular-zone (VZ) like progenitor regions expressing various dorsal and ventral markers in dorsalUnt and ventral organoids (Supplementary Figure 1). In both dorsalUnt and ventral organoids the number of FOXG1+ VZ-like regions were similar. The dorsalUnt organoids were nearly all dorsal tissue (96% TBR1+, and 76% PAX6+) with only small amounts of ventral tissue (0% NKX2-1+, 5% DLX2+, and 6% GSX2+). In contrast, the ventral organoid progenitor regions were highly positive for ventral markers (73% NKX2-1+, 97% DLX2+, and 100% GSX2+). Therefore, these results confirm the successful production of ventral cerebral organoids at the expense of dorsal tissue upon ventral drug-treatment.
Figure 1. Ventral drug-treatment produces ventral-forebrain containing cerebral organoids.
(A) Schematic of cerebral organoid protocol with ventral drug-patterning application (2.5µM IWP2 and 100nM SAG) during the neural induction step. (B) Schematic of a human coronal brain slice indicating the regional expression of the patterning markers used for qPCR/IHC analysis. (C) qPCR analysis of the expression of different brain regional markers showing an increase in ventral and decrease in dorsal forebrain identity in ventral cerebral organoids. Values are plotted as relative expression level (2-ΔCt) to the reference gene TBP. Each data point corresponds to a pooled batch of 8-10 organoids. Data is represented as mean±SD, and statistical significance was tested using the student’s t-test (df=9) for dorsalUnt (n=6 batches) versus ventral-(IWP2+SAG) (n=5 batches) treatment. (D-E) Widefield images of immunostaining analysis of dorsalUnt and ventral cerebral organoids indicating the ventral-forebrain identity of ventral organoids (D), and the dorsal-forebrain identity of dorsalUnt organoids (E). Scale bars are 200µm.
Fused cerebral organoids recapitulate a dorsal-ventral forebrain axis and long-distance cell migration
To recreate a dorsal-ventral identity axis in a single tissue, we developed an organoid co-culture method, which we termed organoid “fusion”. DorsalUnt organoids produced mostly dorsal forebrain tissue (Figure 1C,E, and Supplementary Figure 1). However, inhibition of SHH activity with the smoothened receptor inhibitor cyclopamine A (CycA) can enhance dorsal forebrain identity in 2D neuronal differentiation from hPSCs30. Therefore, to support dorsal identity, organoids were treated with CycA during the neural induction step of the cerebral organoid protocol (Figure 2A), and we refer to these organoids as “dorsalCycA.” In this approach, embryoid bodies (EBs) are individually patterned into either dorsal-(CycA) or ventral (IWP2+SAG) forebrain organoids (Figure 2A,B). After patterning treatments, a ventral and a dorsalCycA EB are embedded together within a single Matrigel droplet (Figure 2A), and over time the organoids grow together and become fused (Figure 2B). The successful fusion of organoids occurred in more than 90% of attempts, therefore the fusion process is highly efficient. The structural organization of organoid fusion tissue was not affected by the fusion procedure with ventricular zone (VZ) like regions present in fusions around 45 and 60 days old. Fewer VZ-like regions were observed in 80 day old organoid fusions (Supplemental Figure 2), as occurs in the normal organoid protocol 9,12. Immunostaining of fused ventral:: dorsalCycA organoids revealed the production of a continuous tissue where one side was highly positive for the ventral marker NKX2-1, while the opposite side was positive for the dorsal marker TBR1 (Figure 2C). Therefore, the organoid fusion method allows dorsal and ventral forebrain regions to be juxtaposed in an arrangement similar to that occurring during brain development.
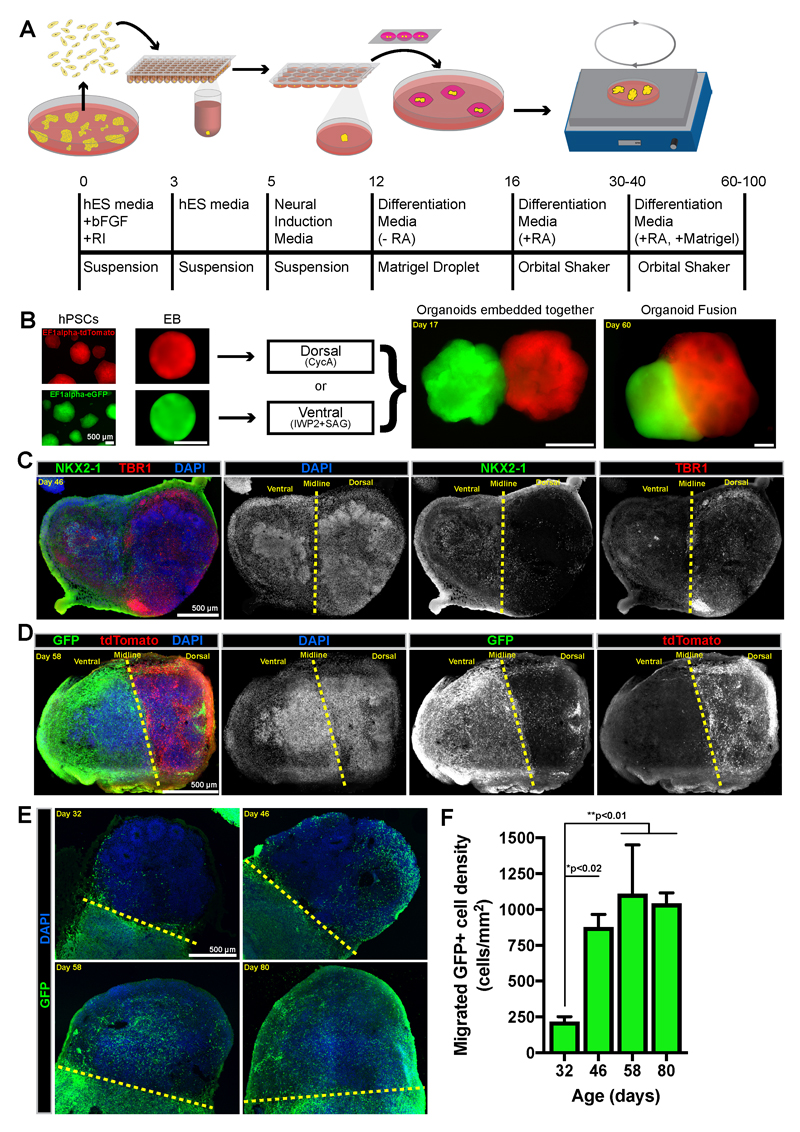
Figure 2. Fusion of cerebral organoids allows cell migration between ventral and dorsal forebrain tissue.
(A) The experimental outline of the cerebral organoid fusion co-culture method. (B) Representative widefield images at different stages throughout the organoid fusion procedure. (C) Tile-scan image of an immunostained cryosection of a ventral::dorsalCycA organoid fusion indicates the combination of ventral (NKX2-1+) and dorsal (TBR1+) regions. (D) Tile-scan image of an immunostained cryosection of a ventral/GFP+::dorsalCycA/tdTomato+ organoid fusion indicates robust migration from ventral (GFP+) to dorsal tissue, but little migration from dorsal (tdTomato+) into ventral tissue. (E) Immunostained ventral::dorsalCycA organoid fusion cryosections from organoids of different ages shows the time course of GFP+ cells migrating from ventral into dorsal tissue. (E) The GFP+ cell density in dorsal tissue was quantified in cryosections from 32 (n=3 organoids) 46 (n=3), 58 (n=4), and 80 (n=4) day-old organoids. The data is presented as mean±SD and statistical significance tested using the one-way ANOVA [F(3,10)=12.59, p=0.0010] with posthoc Tukey’s test for between group comparisons. Scale bars are 500µm.
To test whether cells could migrate between the fused organoids, we used cell lines containing either a ubiquitous GFP or tdTomato reporter to create ventral/GFP+::dorsalCycA/tdTomato+ organoid fusions. Many GFP+ cells from the ventral organoid were observed within the tdTomato+/GFP- dorsalCycA organoid (Figure 2D). In contrast, very few tdTomato+ cells were observed migrating from the dorsal to ventral organoid tissue (Figure 2D). Therefore, this data indicates a robust population of migrating cells that migrate in a ventral to dorsal direction. We next analyzed the time course of the ventral to dorsal migration using organoid fusions of different ages. Migrating cells were observed in small numbers around day 30 (Figure 2E,F). Their density drastically increased from day 30 to 46, but we did not observe a significant increase from day 46 to 80 (Figure 2F). Since organoids increase in size with age, the absolute numbers of migrated cells must increase over time to maintain a similar density. Alternatively, increased proliferation of the migrating cells could explain their increased cell density over time. To test this possibility, we examined the expression of a proliferating cell marker (Ki67) by the migrating GFP+ cells (Supplementary Figure 3), and we observed that few (≤1%) of the GFP+ cells expressed Ki67 at either day 46 or 80 (Supplementary Figure 3B). Therefore, the migrating GFP+ cells are largely postmitotic. Moreover, the cells appeared more dispersed throughout the dorsal regions in 80 day old organoids (Figure 2E). Therefore, based on these results, we focused our future analysis on organoids older than 60 days.
Mixing the tissue components of cerebral organoid fusions indicates directed ventral-to-dorsal cortical cell migration
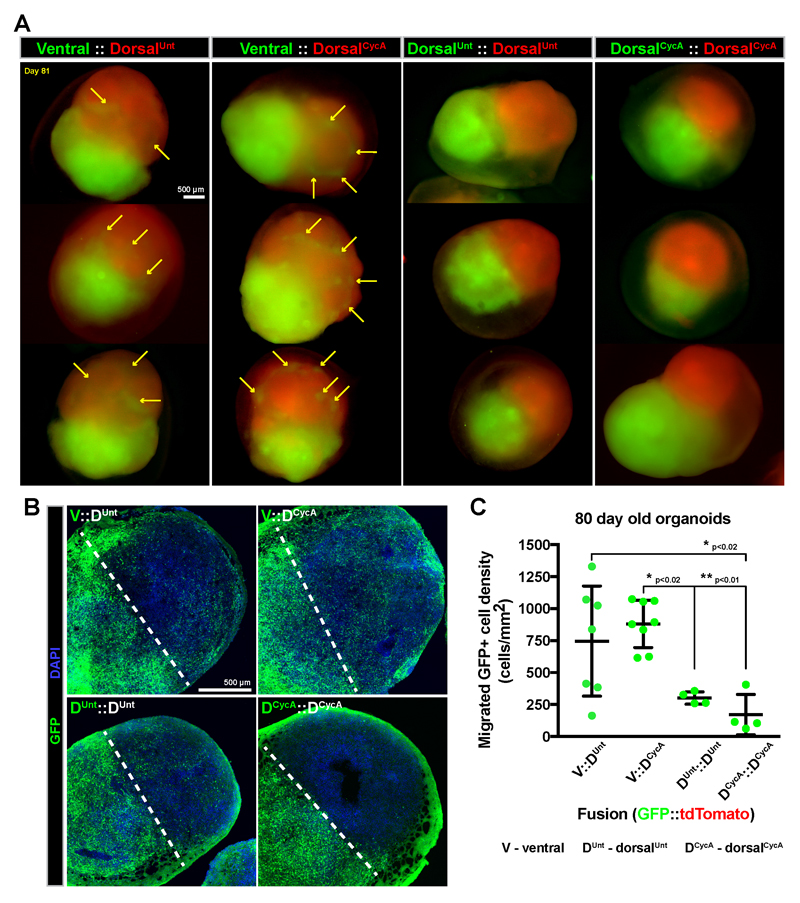
During in vivo brain development, GABAergic interneurons originate in ventral forebrain progenitor regions before migrating to their dorsal forebrain target4,5. To test whether this directionality is recapitulated in fused organoids, we varied the identities of their individual tissue components (Figure 3). We consistently labeled one organoid with GFP and the other with tdTomato, and delineated the fusions in an origin::target (GFP::tdTomato) arrangement. Using this paradigm, we analyzed the number of migrating cells when differentially controlling the dorsal/ventral identity of the origin and target regions of cerebral organoid fusions. Whole-mount imaging of ventral::dorsalUnt and ventral::dorsalCycA fusions indicated the occurrence of GFP+ spots within the tdTomato+ tissue (Figure 3A). Conversely, the spots were rarely observed in dorsalUnt::dorsalUnt or dorsalCycA::dorsalCycA fusions (Figure 3A). This difference was even more striking when analyzing immunostained cryosections of organoid fusion tissue. The highest amount of migrating cells occurred in ventral::dorsalUnt and ventral::dorsalCycA fusions, while the least migration occurred in dorsalCycA::dorsalCycA fusions (Figure 3B,C). Although the average density of migrating cells in ventral::dorsalUnt fusions was not significantly different than ventral::dorsalCycA fusions, the ventral::dorsalCycA fusions were less variable in the amount of migrating cells (Figure 3C). These data confirm our initial observation of directionally biased migration from ventral into dorsal organoid tissue, and strongly suggests that the migration between fused organoids resembles interneuron migration. Finally, this experiment shows that ventral::dorsalCycA fused organoids produce the most robust migration between organoids.
Figure 3. Mixing the tissue components of cerebral organoid fusions indicates the most robust migration from ventral into dorsal regions.
(A) Cerebral organoid fusions were created containing different combinations of ventral (V), dorsalUnt (Duntr) or dorsalCycA (DCycA) treated tissue. The components were labeled with either GFP (green) or tdTomato (red). (A) Whole mount images of ~80 day old organoid fusions show the emergence of GFP+ spots (arrows) in tdTomato+ tissue in ventral::dorsalUnt and ventral::dorsalCycA organoid fusions. (B) Tile-scan confocal images of immunostained mixed organoid fusion cryosections shows migration of GFP+ cells across the midline (dashed line) into the GFP- organoid. (C) Quantification of GFP+ cell density in GFP- tissue from tissue sections. Each data point corresponds to an individual organoid, and the data is represented as mean±SD with statistical significance tested using one-way ANOVA [F(3,19)=8.214, p=0.0010] with posthoc Tukey’s test for between group comparisons. The ventral::dorsalUnt (n=7 organoids) and ventral::dorsalCycA (n=8) fusions show the most migration of GFP+ cells compared to dorsalUnt::dorsalUnt (n=4) and dorsalCycA::dorsalCycA (n=4) fusions. Scale bars are 500µm.
GABAergic interneurons migrate between fused cerebral organoids
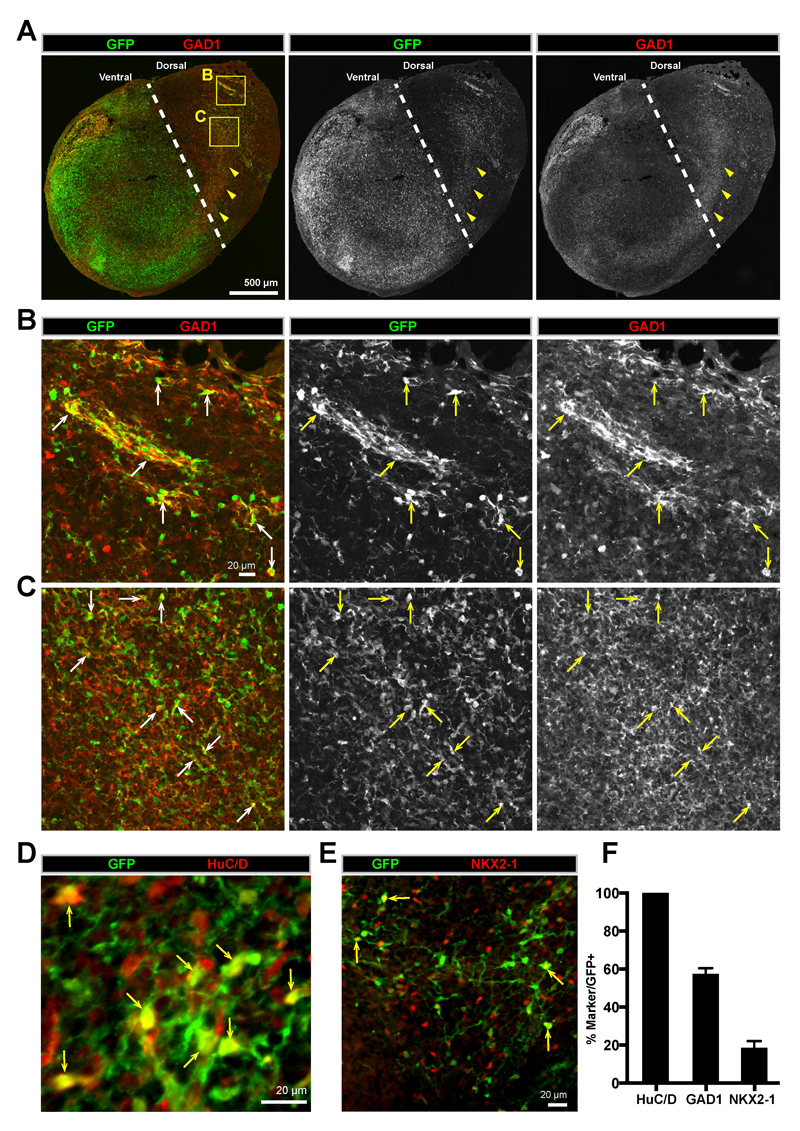
Since our previous experiments suggested that the migration in ventral::dorsalCycA organoid fusions resembles interneuron migration, we first tested whether the migrating cells were GABAergic by examining whether the migrating GFP+ cells expressed GAD1, one of the key enzymes for the synthesis of GABA31. Immunostaining revealed that the GFP+ cells that had migrated into the target organoid broadly expressed GAD1 (Figure 4A-C), and cell counting indicated that ~60% of the GFP+ cells also expressed GAD1 (Figure 4F). Strikingly, when visualizing the entire organoid, GAD1 was expressed in a similar pattern as the GFP+ migrating cells (Figure 4A). Additionally, the expression of GAD1 appeared stronger in regions near the edge of the organoid (Figure 4B), and farther away from the origin of the migrating cells. Therefore, interneurons can migrate from ventral into dorsal regions within organoid fusions.
Figure 4. GABAergic interneurons migrate between fused dorsal-ventral cerebral organoids.
(A) A whole organoid confocal tile-scan image of an immunostained 80-day old ventral::dorsalCycA organoid fusion cryosection. GFP+ cells can be observed migrating across the fusion midline (dashed line) from GFP+ ventral into GFP- dorsal tissue. The GABAergic marker GAD1 can be observed in a similar pattern as GFP (arrowheads). (B-C) A magnified view of peripheral (B) and internal regions (C) of the organoid fusion in (A). GFP+ cells expressing GAD1 can be observed in both regions (arrows). (D) A confocal image of GFP and HuC/D immunostaining in the dorsal region of an 80 day old ventral::dorsalCycA organoid fusion cryosection showing that migrating GFP+ cells express the neuronal marker HuC/D. Arrows point to cell bodies in focus in the single plane z-section. (E) A confocal image of GFP and NKX2-1 immunostaining in the dorsal region of an 80 day old ventral::dorsalCycA organoid fusion cryosection showing that some migrated GFP+ cells maintain NKX2-1 expression (arrows). (F) Quantification of the percentage (mean±SEM) of GFP+ migrated cells in dorsal organoid fusion tissue expressing HuC/D (100±0%, 1427 cells counted from n=4 organoids), GAD1 (57.5±3.0%, 1879 cells counted from n=4 organoids), or NKX2-1 (18.6±3.6%, 3067 cells counted from n=4 organoids). Scale bars are (A) 500µm, (B-E) 20µm.
Another major population of tangentially migrating cells in the developing human brain are the non-GABAergic Cajal-Retzius cells5. These cells are identified by the expression of Reelin (RELN)32,33. The migratory GFP+ cells in organoid fusions did not express RELN (Supplementary Figure 4), despite the substantial presence of GFP-, RELN+ cells in the dorsal target region of organoid fusions. Thus, our data indicate that the migratory GFP+ cells in organoid fusions are not Cajal-Retzius cells, and in combination with our previous results strengthens their identity as GABAergic interneurons.
Finally, we analyzed the expression of various neuronal markers by the migrating GFP+ cells in organoid fusions, including the pan-neuronal marker HuC/D34, the immature migrating neuron marker DCX35, the mature neuronal markers NeuN35, or MAP236. We observed that all the migrated GFP+ cells expressed HuC/D, indicating their neuronal identity (Figure 4D,F). In addition, many GFP+ cells expressed DCX (Supplementary Figure 5A), and a subset also expressed NeuN (Supplementary Figure 5A) or MAP2 (Supplementary Figure 5B). This finding indicates that the migrating GFP+ cells are neurons (HuC/D+ or DCX+), but also that they can become mature neurons (DCX+/NeuN+, or MAP2+). In addition, we visualized the morphology of GFP+ migrating interneurons. We observed cells with many branched processes and a circular cell body, which represents a more mature cell morphology (Supplementary Figure 6A). Other cells exhibited a migratory morphology with an elongated cell body as well as a branched leading process and trailing process (Supplementary Figure 6B-C). Therefore, the migrating GFP+ cells can express immature and mature neuronal markers, and exhibit both an immature, migratory morphology, or also an elaborate, mature morphology.
Since we observed that the migrating GFP+ cells had a range of maturity, and that not all of the GFP+ cells expressed GAD1, we hypothesized that some of the GFP+ cells may be immature, and therefore express markers of immature migrating interneurons. During normal mouse brain development, cortical interneurons downregulate the expression of NKX2-1 as they migrate toward dorsal cortical regions 37. In migrating GFP+ cells within organoid fusions we observed that ~20% of the GFP+ migrating cells in dorsalCycA organoid regions expressed NKX2-1 (Figure 4E-F). The maintained expression of NKX2-1 could indicate an immature status, and also a difference between the development of mouse and human interneurons. Interestingly, many NKX2-1+ or GAD1+ cells that were GFP- were present within the dorsalCycA region of organoid fusions (Figure 4E), indicating the production of ventral-derived interneurons within the dorsal organoid. DorsalUnt organoids were nearly entirely of dorsal forebrain identity when not fused (Figure 1C,E, and Supplemental Figure 1). Therefore, the tissue components of organoid fusions must be able to signal to each other. In support of this hypothesis, we observed NKX2-1+ cells within the dorsalCycA organoid near the fusion midline (Figure 2C), indicating that the ventral organoid can influence the patterning of the nearby dorsal tissue toward ventral forebrain tissue. Importantly, this does not affect the analysis of ventral to dorsal migrating cells, because these cells are uniquely labeled with GFP. Taken together, our results show that interneurons can migrate between fused ventral::dorsalCycA organoids, that many interneurons maintain expression of NKX2-1 after migrating into dorsal regions, and can become mature neurons.
Migrating interneurons produce various LGE/CGE and MGE-derived cortical interneuron subtypes in fused cerebral organoids
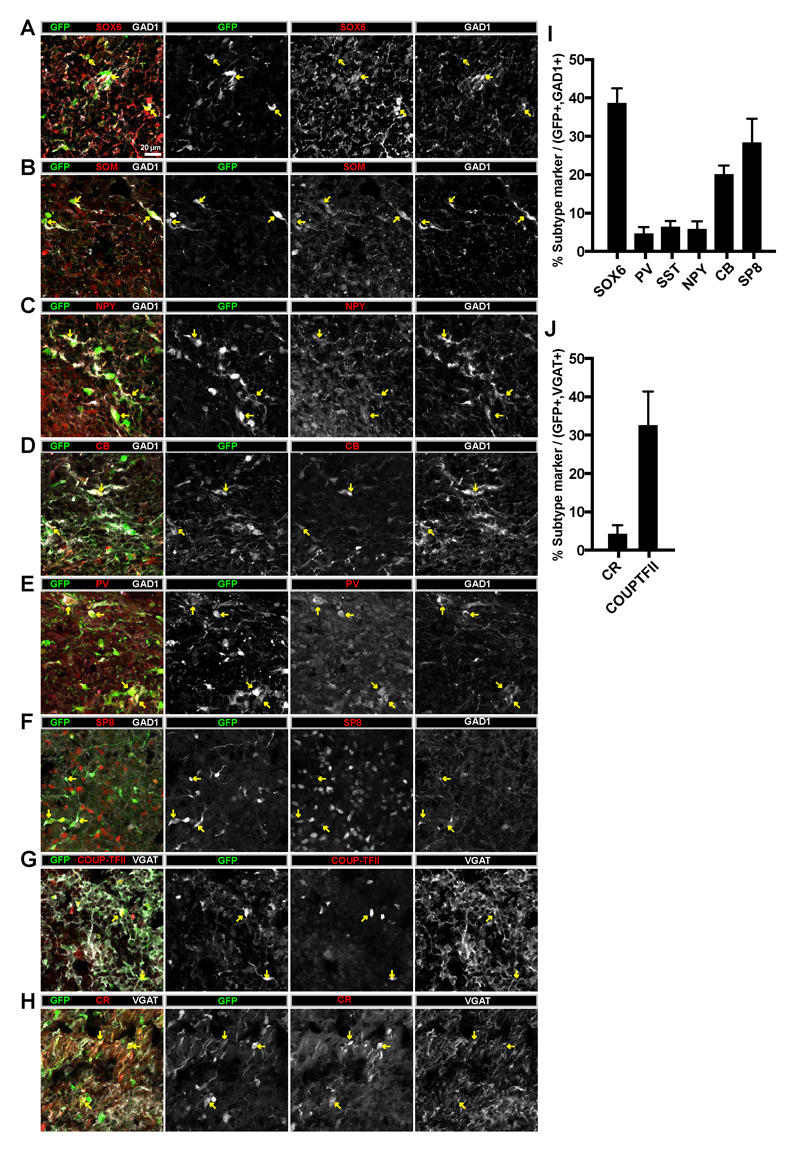
Since the migrating GFP+ cells can become mature neurons (Supplemental Figure 5), and appear to be predominantly GABAergic interneurons (Figure 4A,B,F), we next tested which interneuron subtypes are produced. Interneurons are particularly heterogeneous and multiple molecular markers can be used to identify various subtypes38,39 that are generated by distinct progenitor subpopulations within the ventral forebrain3,23–26,40. In humans, the majority of interneurons are generated from NKX2-1+ regions of the MGE41,42. Therefore, we tested whether the migrating GFP+ cells produce MGE-derived interneuron subtypes. In humans, SOX6 is expressed in the MGE and in immature and mature interneurons emerging from this region41. In organoid fusions, we observed that ~40% of the migrating GFP+/GAD1+ cells are SOX6+ MGE interneurons (Figure 5A,I). Therefore, MGE-derived interneurons can be generated within cerebral organoid fusions. To confirm this finding, we also examined the expression of markers for MGE-derived interneurons3,39,40. We observed expression of somatostatin (SOM) (Figure 5B,I) in 6%, neuropeptide Y (NPY) (Figure 5C,I) in 6%, calbindin D-28k (CB) (Figure 5D,I) in 20%, and parvalbumin (PV) (Figure 5E,I) in 5% of the migrated GFP+/GAD1+ interneurons. Therefore, in organoid fusions, multiple MGE-derived interneuron subtypes can be generated.
Figure 5. Migrating interneurons in ventral::dorsal cerebral organoid fusions express various interneuron subtype markers.
(A-H) Confocal images of immunostaining in dorsal regions of 80-day old ventral::dorsalCycA organoid fusion cryosections. Expression of the GABAergic markers GAD1 or VGAT were used to identify interneurons. Examples of various migrated GFP+ interneurons expressing either GAD1 or VGAT were observed expressing the MGE-derived interneuron marker SOX6 (A) or subtype markers SOM (B), NPY (C), CB (D), and PV (E). Migrated GFP+ interneurons also expressed the LGE/CGE-derived interneuron markers SP8 (F), COUP-TFII (G), or the subtype marker CR (H). (I) Quantification of the percentage of GFP+/GAD1+ migrating interneurons in the dorsal region of organoid fusions expressing various subtype markers. SOX6 (38.7±3.9%, 1002 cells counted from n=4 organoids), PV (4.7±1.7%, 1879 cells counted from n=4 organoids), SST (6.4±1.5%, 818 cells counted from n=3 organoids), NPY (5.9±2.0%, 748 cells counted from n=5 organoids), CB (20.1±2.3%, 1114 cells counted from n=4 organoids), SP8 (33.0±5.2%, 620 cells counted from n=4 organoids). (J) Quantification of the percentage of GFP+/VGAT+ migrating interneurons in the dorsal region of organoid fusions expressing various subtype markers. CR (4.3±2.2%, 898 cells counted from n=4 organoids), COUPTFII (38.7±7.8%, 781 cells counted from n=4 organoids. Scale bars are 20µm. Abbreviations: SOM=somatostatin, NPY=neuropeptide Y, CB=calbindin, PV=parvalbumin, CR=calretinin, VGAT=vesicular GABA transporter, GAD1=glutamate decarboxylase 1.
The remaining interneurons in the human brain arise from the LGE/CGE3,39,40. Similar to SOX6 for MGE, the transcription factors COUP-TFII/NR2F2 and SP8 can be used as LGE/CGE fate-mapping markers for cortical interneurons41. Both SP8 (Figure 5F) and COUP-TFII (Figure 5G) were expressed by ~30-40% of the GFP+ migrating interneurons (GAD1+ or VGAT+) in organoid fusions (Figure 5I-J). As confirmation of this result, we also analyzed the expression of markers for LGE/CGE-derived subtypes. Our previous data already indicated a lack of RELN expression by GFP+ migrating cells (Supplementary Figure 4), which in addition to non-interneuron Cajal-Retzius cells is also expressed by subpopulations of GABAergic MGE and CGE-derived interneurons3,40. We also did not observe any vasoactive intestinal peptide (VIP) expressing GFP+ interneurons (data not shown). However, we did observe calretinin (CR) expression in 4% of the GFP+ interneurons (VGAT+) in organoid fusions (Figure 5H,J). Collectively, these results indicate that organoid fusions contain many diverse interneuron subtypes originating from the major ventral forebrain subregions (MGE and LGE/CGE).
Neuronal migration in fused cerebral organoids resembles the features of migrating cortical interneurons
The migratory dynamics of migrating cortical interneurons has been extensively documented using time-lapse imaging experiments43–48. Interneurons utilize different modes of neuronal migration throughout development. For instance, radial migration refers to cells migrating along radial glia fibers. In contrast, tangential migration occurs independent of a radial glia scaffold, and in the case of interneurons, largely perpendicular to the radial glia fibers. Radially migrating cells exhibit a bipolar morphology with a single leading process, whereas tangentially migrating cortical interneurons contain branched leading processes which sample the extracellular environment for chemotaxis cues that regulate migration dynamics and direction. Cortical interneurons undergo tangential migration from ventral into dorsal telencephalic regions, but at some point upon arriving in dorsal regions, the cells decide to move radially into their desired cortical layer, followed by dispersion throughout that layer. Therefore, migrating interneurons undergo many different modes of migration throughout their development5,49.
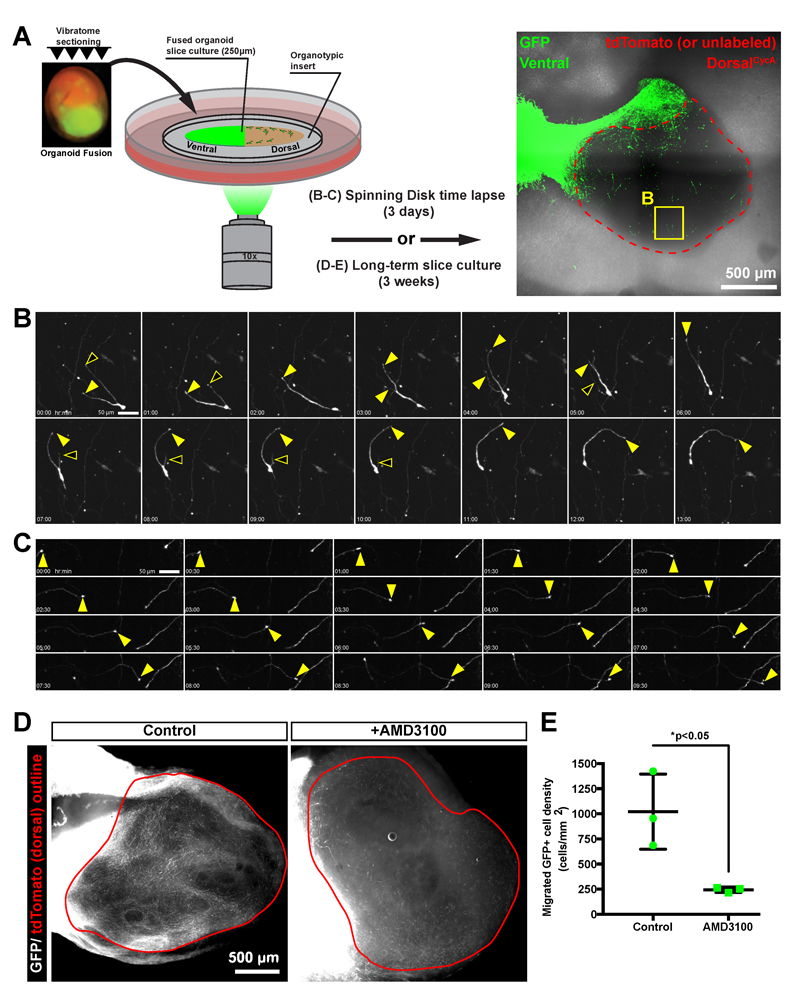
To determine whether the behavior of migrating GFP+ cells in organoid fusions resembles that of cortical interneurons, we performed time-lapse recordings of migrating GFP+ cells in ventral::dorsalCycA organoid fusion slice-cultures (Figure 6A). The GFP+ ventral origin region of the organoid fusion could easily be distinguished from the GFP- dorsal target region (Figure 6A). Thus, we could easily visualize the morphology of the sparsely labeled GFP+ cells that had migrated into the dorsal organoid fusion region. Over 3 days, we observed both stationary and motile cells (Figure 6B, Supplemental movies 1-5), and surprisingly we also observed dynamic neuronal processes resembling axons that extended long distances (Figure 6C, Supplemental movie 6). We clearly observed cells with a branched leading process that underwent extension and retraction followed by the cell body moving toward the extending branch (Supplemental Movie 1). Some cells with this behavior exhibited abrupt changes in direction (Supplemental Movie 2). This behavior appears very similar to that described for tangentially migrating interneurons48. A different behavior was observed in other cells containing a long leading process that extends in a single direction (Supplemental Movie 4), which resembles radial migration. Intriguingly, we also observed cells that exhibited multimodal behavior with branched leading processes, but then switched to a single leading process and unidirectional migration (Supplemental Movie 5). In summary, the GFP+ migrating cells within organoid fusions exhibit characteristics of different modes of migration resembling that of migrating cortical interneurons.
Figure 6. Migrating cells in cerebral organoid fusions exhibit the migratory dynamics of tangentially migrating interneurons and are sensitive to CXCR4 activity.
(A) A schematic representation of the organoid fusion slice culture assay for either short-term time-lapse imaging of migratory dynamics, or long-term drug-treatments. A representative tile-scan image of an entire ventral::dorsalCycA organoid fusion slice is shown with the ventral GFP+ regions labeled in green and the unlabeled or tdTomato dorsal regions outlined in red. A region containing the migrating cell shown in (B) is noted by a yellow box. (B) Still images from a 3-day time-lapse experiment showing a migrating GFP+ cell. The branched leading process exhibits both extending (closed arrowheads) and retracting (open arrowheads) branches as the cell body follows one of the leading processes. (C) An extending neurite (closed arrowhead) with a tuft that appears to be an axon growth cone travels in one direction across the field of view. (D) A widefield image of GFP+ cells that migrated into the tdTomato+ dorsal region (red outline) from long-term organoid fusion slice cultures that were either untreated (control) or treated with a the CXCR4 inhibitor (AMD3100). (E) Quantification of the migrated GFP+ cell density with data represented as mean±SD with statistical significance tested using the student’s t-test (df=4) comparing control (n=3 organoids) to AMD3100 (n=3) treatment. Slices from the same organoid were split between control and AMD3100-treatments, and 3 different organoids were used from 2 different independent differentiations. Each data point represents cell density counts from one slice. Fewer migrating cells are observed in AMD3100-treated slice cultures. Scale bars are (A) 500µm, (B-C) 50µm, and (D) 500µm. The images in (A-C) are from an organoid fusion created by fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid. The images and data in (D-E) are from iPSC-derived organoid fusions.
3D-culture systems can be effective experimental platforms for drug-screening50,51. Therefore, we developed a long-term slice culture assay (Figure 6A) to highlight the amenability of our organoid fusion system for small-molecule application. Since we observed a large increase in migration from day 30-50 in ventral::dorsalCycA organoid fusions (Figure2F), we applied a drug during this period. The chemoattractant SDF-1 (CXCL12) and its receptor CXCR4 regulate the migration of interneurons52. To test if the migration we observed in cerebral organoid fusions is CXCR4-dependent, we created long-term slice cultures of organoid fusions that were treated with the CXCR4 antagonist AMD3100. Compared to untreated control slices prepared from the same organoid fusion, the density of migrating GFP+ cells into GFP- dorsal regions was significantly reduced upon AMD3100 treatment (Figure 6D-E). Therefore, the cell migration in cerebral organoid fusions depends on CXCR4 activity. This experiment highlights the usefulness of the organoid fusion method as an initial drug-screening platform, and opens the possibility for larger scale screening in the future.
Discussion
In this study, we developed a cell migration assay using fused cerebral organoids. The concept of fusing organoids is based on classical co-culture experiments. However, our method represents an adaptation and extension of this technique to study human neuronal cell migration with the experimental control allowed by in vitro 3D cell culture. Ventral::dorsal organoid fusions exhibit robust long-distance cell migration resembling that of ventral forebrain-derived cortical inhibitory interneurons. This is based on multiple pieces of data. First, we observed substantially more migration from ventral into dorsal forebrain regions in cerebral organoid fusions. Second, the migrating cells within organoid fusions express GABAergic markers (GAD1/VGAT). Third, the migrated GFP+ cells can produce a variety of interneuron subtypes. In addition, the lack of RELN expression by migrating GFP+ cells supports a ventral forebrain-derived interneuron identity. Fourth, the migration dynamics of the GFP+ cells in fused organoids resembles the characteristic migratory behavior exhibited by cortical interneurons. Finally, the cell migration in organoid fusions was inhibited by CXCR4 inhibition, a known regulator of interneuron migration. Therefore, we have recapitulated the migration of human cortical interneurons from ventral into dorsal forebrain regions. Moreover, the observation that these cells exhibit both immature and mature neuronal markers indicates their ability to undergo maturation once arriving in their dorsal cortical forebrain target regions. This represents an exciting opportunity to recreate a developing human cortical circuit containing an enhanced repertoire of cellular diversity than was possible to achieve using the previous singular organoid method.
Cerebral organoid methodology exhibits variability, both between batches of differentiations and between different human pluripotent stem cell (hPSC) lines11,53. In the current study, we used an iPSC line that was previously used extensively for growing forebrain organoids9,12. As the organoid fusion method relies on proper patterning into dorsal and ventral brain regions for robust ventral to dorsal migration to occur, any future hPSC cell lines used should be tested for the ability to grow dorsal and ventral brain regions. The qPCR analysis we present can be used to address this caveat. Moreover, while we developed the organoid fusion method using a specific previously published cerebral organoid protocol9,12, the concept of organoid fusion should be compatible with any protocol which can grow 3D dorsal and ventral forebrain regions.
An interesting observation was the maintenance of NKX2-1 expression by GFP+ cells migrating from ventral into dorsal organoid fusion tissue. This differs from mouse development where NKX2-1 is downregulated by cortical interneurons as they leave the ventral ganglionic eminence regions and proceed to their dorsal forebrain target regions 37. However, NKX2-1+ cells have been observed within the dorsal forebrain of embryonic human brain tissue 54–56. This data has been interpreted as evidence for the generation of interneurons within dorsal forebrain progenitor regions. An alternative possibility is the prolonged expression of NKX2-1 by migrating cortical interneurons. Our data from human organoid fusions supports the latter possibility. In addition, the strong effect of CXCR4 inhibition on the migration of cells within organoid fusions is also intriguing. In CXCR4 knockout and AMD3100 treated mice, interneurons retain the ability to migrate from ventral into dorsal forebrain regions, but the invasion of interneurons into the cortical layers is altered 52,57. Whether the CXCR4 effect in human cerebral organoid fusions is another difference between mouse and human interneurons is an interesting avenue of future study. Nonetheless, these two findings highlight the potential usefulness of using the organoid fusion method to understand the various aspects of human interneuron development.
An exciting future application of this technique will be to study human developmental biology and its relationship to neurological diseases. With this in mind while characterizing this assay, we present different experimental paradigms, each of which can be used to investigate various aspects and scientific questions related to neuronal cell migration. For instance, the labeling of the cells within one of the fused organoids with a fluorescent reporter allows the visualization of long-distance neuronal migration. This can even be continuously monitored over time through whole-mount imaging of intact organoid fusions. In addition, the identity of the cells can be easily examined using immunostaining for identification of neuronal subtypes. This is useful because many psychiatric diseases, such as Schizophrenia, are thought to involve selective deficits in specific interneuron subpopulations58.
A second type of analysis is dissecting the role of specific molecules on neuronal migration to determine if they are acting in a cell-autonomous or cell non-autonomous manner. For instance, using our organoid mixing paradigm, organoids from either the origin of migration or the target can be genetically manipulated independently by deriving the origin and target organoids from distinct cell lines containing either mutant or wild-type alleles of a gene of interest. This is a powerful genetic tool routinely used in genetic model organisms, but can now also be utilized in 3D cultures of developing human brain tissue.
We also present an organoid fusion slice culture paradigm in which confocal time-lapse imaging can be used to analyze the short-term dynamics of neuronal cell migration. This is an important tool because some molecules such as GABA are known to affect the motility of migrating interneurons4, which may produce a subtle phenotype in a long-term migration assay, and therefore instead require a higher time-resolution analysis of the dynamics of migrating cells. In addition, long-term slice cultures can be used to test how different drug-treatments can affect cell migration. This presents the opportunity to perform drug-screens to determine the effect of many different molecules on interneuron migration.
Finally, the organoid fusion method also presents unique applications beyond cell migration, which can be developed further in the future. As one example, we observed substantial neurite dynamics with the appearance of axon projections. In addition, although we specifically focused on ventral::dorsal forebrain fusions, the organoid fusion paradigm allows flexibility in the brain regional identities that can be grown together. Combined with the additional brain-region specific organoid protocols59–62, the possible brain circuits that could be modeled using organoid fusions is vast. Therefore, organoid fusion technology greatly enhances the phenotypic analyses possible in cerebral organoids, and the flexibility of the method immensely expands the future development of in vitro models of human neurological diseases.
Methods
Cell culture
Feeder-dependent human induced pluripotent stem cells (hiPSCs) (Systems Biosciences, cat. no. SC101A-1) were obtained from System Biosciences with pluripotent verification and contamination-free. Feeder-dependent hiPSCs were cultured with irradiated mouse embryonic fibroblast (MEF) feeder cells (MTI-GlobalStem, cat. no. 6001G) on gelatin coated (0.1% gelatin in PBS) 6-well culture plates using human embryonic stem cell (hESC) medium: DMEM/F12 (Invitrogen) containing 20 ng/mL bFGF (produced by IMBA institute Molecular Biology Service core facility), 3.5µL/500mL media of 2-mercaptoethanol, 20% KnockOut Serum (Invitrogen), 1% GlutaMAX (Invitrogen), 1% MEM-NEAA (Sigma), and 3% FBS). Feeder free H9 human embryonic stem cells (hESCs) were obtained from WiCell with verified normal karyotype and contamination-free. Feeder-free hESCs were cultured on hESC-qualified Matrigel (Corning cat. no. 354277) coated 6-well plates using mTeSR1 (Stemcell Technologies). All stem cells were maintained in a 5% CO2 incubator at 37°C. Standard procedures were used for culturing and splitting hPSCs as explained previously12. All cell lines were routinely tested for contamination and confirmed mycoplasma-negative.
Cloning/Molecular biology/Generating hPSC lines
For ubiquitous fluorescent labeling of cells, a reporter construct was inserted into the safe-harbor AAVS1 locus in hPSCs as done previously with TALEN technology 63 using the AAVS1 SA-2A-Puro donor vector (gift from Dr. Rudolf Jaenisch) as a template. A modified backbone was created containing flanking tandem repeats of the core chicken HS4 insulator (2xCHS4). Fluorescent reporter expression cassettes were inserted between the flanking insulator sequences. The following expression constructs were inserted into iPSCs: 1) 2xCHS4-EF1α-eGFP-SV40-2xCHS4, 2) 2xCHS4-EF1α-tdTomato-SV40-2xCHS4. In feeder-free H9 hESCs, a 2xCHS4-CAG-eGFP-WPRE-SV40-2xCHS4 construct was inserted to enhance the GFP expression for time-lapse imaging experiments.
hPSCs were prepared for nucleofection as a single-cell suspension using the same cell dissociation procedures as for making EBs12. The Amaxa nucleofector (Lonza) was used with Stem Cell Kit 1 using 800,000 cells per nucleofection containing 1µg each of the TALEN guides, and 3µg of the donor plasmid DNA following manufacturer guidelines. After nucleofection, 200µL of the total 600 µL nucleofection solution was plated onto a 10 cm cell culture dish. Colonies from single cells were grown for 4 days, and then treated with puromycin (Puro) (Jena Bioscience, cat. no. NU-931-5). For feeder-dependent cells, 1.1 µg/mL puro was applied, and for feeder-free cells 0.7 µg/mL was applied. The puro treatment continued for 5-7 days until the surviving colonies were large enough to be picked manually and transferred into a 24-well plate. When splitting the colonies from a 24-well plate, half of the cells were used for genotyping, while the other half was expanded into 12 and then 6-well formats, and then used for further experiments. For genotyping, DNA was extracted using the QuickExtract solution (EpiCentre), and PCR was performed using the following primers to identify correctly targeted AAVS1 insertions: 1) Puro (AAVS1_Puro-fwd: tcccctcttccgatgttgag and AAVS1_Puro-rev: gttcttgcagctcggtgac), 2) eGFP (AAVS1_eGFP-fwd: GAACGGCATCAAGGTGAACT and AAVS1_eGFP-rev: cttcttggccacgtaacctg), and 3) tdTomato (AAVS1_tdTomato-fwd: ctacaaggtgaagatgcgcg) and (AAVS1_tdTomato-rev: tccagcccctcctactctag). To determine whether the insertion was heterozygous or homozygous, the presence of a WT allele was tested using additional PCR primers: 4) WT (AAVS1_WT-fwd: tcccctcttccgatgttgag, and AAVS1_WT-rev: tccagcccctcctactctag). Cell clones with correctly targeted heteroyzygous or homozygous insertions were archived by freezing with Cell Banker 2 solution (Amsbio, cat. no. 11891) and/or cultured for further experiments.
Cerebral organoid generation and fusion
Cerebral organoids were generated following the previous protocol developed in our lab 9,12 with slight modifications. The protocol for generating ventral organoids was tested using a feeder-dependent iPSC line (Systems Biosciences, cat. no. SC101A-1). A drug-patterning treatment was applied during the neural induction step of the protocol (~day 5-11) using one of the following treatments: 1) dorsalUnt, no drugs, 2) Ventral, 2.5µM IWP2 (Sigma, cat. no. I0536) and 100nM SAG (Millipore, cat. no. 566660), 3) dorsalCycA, 5µM CycA (Calbiochem, cat. no. 239803). Stock drug solutions were created as follows: IWP2 (5mM in DMSO), SAG (1 mM in H2O), and CycA (5 mM in DMSO). Following embedding in Matrigel (Corning, cat. no. 356235), organoids were grown in 10 cm cell culture dishes containing 25 mL of differentiation medium, and after the first media exchange maintained on an orbital shaker with medium exchange every 5-7 days. After day 40 of the protocol, when organoids begin to grow out of the Matrigel droplet, the differentiation medium was supplemented with 1% Matrigel.
To create organoid fusions, EBs were grown separately and individually patterned using either dorsalUnt, dorsalCycA, or ventral protocols as described above. During Matrigel embedding, two EBs were transferred into the same parafilm well and embedded in a single droplet (~30 µL) of Matrigel. The EBs were gently positioned as close together as possible using a 20 µL pipet tip to ensure the EBs would remain in close proximity within the middle of the solidified Matrigel droplet. The fusion process is very efficient, and occurs within a week when EBs are positioned as close as possible. If EBs were not positioned close enough, then visible space remained between organoids 1 week after embedding. These “failed” fusions were removed by aspiration. In addition, if the Matrigel droplet disassembles during the embedding process or during additional feedings before fusion has been completed, these organoids were also removed by aspiration.
RNA extraction/qPCR
For each drug-patterning treatment group, 8-12 organoids were collected at day 30-40 into 2 mL RNAse-free tubes and chilled on ice throughout the procedure. The organoids were washed 3x in cold PBS, then the Matrigel was dissolved by incubating the organoids in chilled Cell Recovery Solution (Corning, cat. no. 354253) for 1 hr at 4°C. The dissolved Matrigel was removed by rinsing 3x in cold PBS. RNA was extracted using the RNeasy mini kit (Qiagen). cDNA synthesis was performed using 2 µg of total RNA and the Superscript II (Invitrogen) enzyme according to the manufacturer protocols. qPCR reactions were performed using Sybr Green master mix (Promega) on a BioRad 384-well machine (CXF384) using the following reaction protocol: 1) 95°C for 3min, 2) 95 °C for 10s, 3) 62 °C for 10s, 4) 72 °C for 40s, 5) go to 2, 40 cycles, 6) 95°C for 1min, 7) 50 °C for 10s. Quantification was performed in excel by calculating the ΔCt value using TBP as a reference gene. Data is presented as expression level (2-ΔCt) relative to TBP.
Cerebral Organoid Slice Culture and Drug-treatment
Slice cultures were generated by vibratome sectioning of organoid fusions. The organoid fusion tissue was embedded in 4% low melt agarose (Biozym, cat. no. 850080), and sectioned in ice-cold PBS (without Ca2+/Mg2+) to create 200-250 µm sections. The sections were transferred onto Millicell organotypic inserts (Millipore, cat. no. PICM01250) in 6-well cell culture plates. For time-lapse imaging, sections were cultured for 1-2 days before mounting for imaging using a spinning disk confocal microscope (VisiScope). Sections were cut away from the culture insert membrane and inverted onto a glass-bottom dish. The section was immobilized by placing a cell culture insert on top of the section, and attaching it to the dish using vacuum grease. For drug-treatment, long-term slice cultures were initially cultured overnight in differentiation media (+5% FBS). Then the media was exchanged for fresh media (control) or media containing the CXCR4 inhibitor AMD3100 (Sigma, #A5602) and cultured for 3 additional weeks before tissue fixation and further immunofluorescent processing.
Histology/Cryosectioning/Immunofluorescence
Organoid tissue was collected at the desired age, rinsed 3x in PBS, and fixed in 4% PFA overnight at 4°C. The next day the tissue was rinsed 3x in PBS, and cryoprotected in 30% sucrose in PBS overnight at 4°C. Then the tissue was incubated 2-4 hours in a 1:1 mixture of 30% sucrose/PBS and O.C.T. cryoembedding medium (Sakura, cat. no. 4583). Next, groups of 2-4 organoids were transferred from the sucrose/OCT mixture into a cryomold and filled with O.C.T. The embedded tissue was frozen on dry ice, then stored at -80°C until cryostat sectioning. Frozen organoid tissue was sliced into 20 µm sections using a cryostat (Leica), and collected on superfrost Ultra Plus slides. Tissue sections were arranged such that every 10th slice was collected sequentially until each slide contained 8-10 sections per block of tissue. The sections were dried overnight, and then used for immunofluorescent labeling, or stored at -20°C until ready to stain.
Immunofluorescence was performed on tissue sections directly on slides. The O.C.T. was removed by washing 10 minutes in PBS. The sections were post-fixed directly on the slides using 4% PFA for 10 minutes at RT followed by washing 3x10 minutes in PBS. The tissue area was outlined using a hydrophobic PAP pen. Permeabilization/blocking was performed using 5% BSA/0.3% TX100 in PBS with 0.05% sodium azide and incubated 30 minutes at room temperature (RT) within a dark humidified slide box. Primary antibodies (a table of used primary and secondary antibodies can be found at the end of the method section) were added at the desired dilutions in antibody solution (5% BSA, 0.1% TX100 in PBS with 0.05% sodium azide) and incubated overnight at 4°C. Slides were rinsed 3x in PBS and then washed 3x10 minutes in PBST (PBS +0.1% Tween 20) at RT on an orbital shaker. Secondary antibodies were added at 1:500 in antibody solution and incubated at RT for 2 hours. DAPI solution (2 µg/mL) was added for 5 minutes, then slides were washed as done after primary antibody application, with the final washing using PBS. Coverslips were mounted using DAKO mounting medium (Agilent Pathology Solutions, cat. no. S3023) and allowed to harden overnight. Slides were then stored at 4°C until imaging, or at -20°C for long-term storage.
For long-term slice cultures, slices grown on cell culture inserts were rinsed in PBS, and then fixed in 4% PFA for 2 days at 4°C. The sections were washed 3x10 minutes in PBS, then permeabilized/blocked as performed for cryosections on slides. Primary and secondary antibody incubations were performed overnight at 4°C, followed by washing 3x10 minutes in PBS after each step. The sections were mounted in Vectashield containing DAPI (Vector Labs).
Imaging/Microscopy
Fluorescent cell culture imaging was performed using a Zeiss Axio Vert.A1 widefield microscope and an Axiocam ERc 5s camera (Zeiss, Zeiss GmbH) using Zeiss Plan-Neofluar 2.5x 0.085 and Zeiss LD A-Plan 10x 0.25 Ph1 objectives. Both tdtomato and GFP channels were recorded separately and subsequently pseudocolored and merged using the Fiji package of ImageJ.
Widefield imaging of IHC stainings and long-term slice cultures was performed using an Axio Imager Z2 (Zeiss, Zeiss GmbH) using a Sola SM2 illumination source, (5x 0.15 plan-neofluar, 10x 0.3 plan-neofluar, 20x 0.5 plan-neofluar) objective and a Hamamatsu Orca Flash 4 camera. Filters used were Ex360/40nm Em 445/50nm, Ex480/40nm Em 535/50nm and Ex560/55nm Em 645/75nm.
Confocal imaging was performed using a Zeiss LSM700 AxioImager with a 20x 0.8 plan-apochromat dry objective. Lasers of 405nm (5mW), 488nm (10mW), 555nm (10mW) and 639nm (5mW) together with, corresponding to wavelength, filters SP490, SP555, SP640 and LP490, LP560, LP640 were used for recording. For whole organoid tile scans the XY Scanning Stage and the Zeiss Zen implemented stitching algorithm was used. For colocalization of markers, Z-scans at 20x were performed.
Imaging of GFP stained slides used for quantification, and time-lapse migration were acquired using a Yokogawa W1 spinning disk confocal microscope (VisiScope, Visitron Systems GmbH, Puchheim, Germany) controlled with the Visitron VisiView software and mounted on the Eclipse Ti-E microscope (Nikon, Nikon Instruments BV) using ex 488 laser and em filter 525/50. Measurements were recorded using the 10x 0.45 CFI plan Apo lambda (Nikon, Nikon Instruments BV) objective with the sCMOS camera PCO edge 4.2m. Whole IHC slice imaging for cell counting was performed using the tile scan acquisition function. Tile scans were then stitched using the Grid/Collection stitching plugin64 in Fiji. For time lapse movies, tile scan Z-stacks were recorded. For the time-lapse recordings, we recorded a 300 µm z-stack to account for any drift and curling of the tissue over time. Some sections were not perfectly flat, so a larger z-depth allowed us to account for any unevenness. We set the beginning of the z-stack below the focal plane of the tissue, and then focused through the detectable fluorescent signal (~50 µm) until no more signal was detectable, and set the end of the z-stack. This allowed cells to be followed continuously for 3 days. After stitching as performed for cell counting, a maximum Z-projection was created, and the Z-projection time stacks used for global visualization of cell migration. Cropped regions were created using Fiji and saved as uncompressed AVI files. The AVI files were converted to the mp4 format using Handbrake software to generate the supplemental movies.
For imaging the morphology of GFP+ cells, z-stacks (0.5 µm z-steps) were collected with the VisiScope spinning disk confocal using the CFI plan Apo lambda 40x/0.95 objective with a 2x lens switch. A maximum z-projection was created in Fiji.
For generation of figures and presenting images, the “crop” function, and changing the max/min levels using the “Brightness/Contrast” function of Fiji were used.
Cell density quantification
The number of GFP+ cells migrating into GFP- target regions of cerebral organoid fusions was counted manually using the “Cell Counter” plugin of Fiji. The GFP- area in organoid fusions was outlined with the ROI tool and the area calculated using the “measure” function. The cell counts were divided by the area to determine the density of migrated GFP+ cells. For the migration time course (Figure 2E,F) and organoid fusion mixing (Figure 3B,C) experiments, confocal tile scan images of tissue cryosections were used for quantification. For the long-term slice culture experiment (Figure 6D,E), a wide-field 5x image containing the GFP- region was used for counting.
Marker expression quantification
For determining the percentage of ventricular zone (VZ)-like regions expressing various markers, immunostained organoid fusion cryosections were imaged on a spinning disk confocal microscope (see microscopy section) using a 10x objective to collect single-plane, tile scan images of entire organoids. The VZ-like regions were identified in the DAPI channel as circular regions with a packed arrangement of cells in a radialized pattern. The VZ-regions were marked manually using the “Cell Counter” plugin of Fiji. Then the additional channels containing the signal from various marker immunostainings were overlaid, and the VZ-like regions scored for expression of each marker of interest. Two different sections of each organoid were counted and summed. Then the counts for each organoid were calculated as a percentage of total VZ-regions expressing a given marker. The percentages from 4 different organoids from 2 independent differentiations were averaged.
For determining the percentage of GFP+ cells expressing various markers, immunostained organoid fusion cryosections were imaged on a spinning disk confocal microscope (see microscopy section) using a 20x objective to collect z-stacks (1 µm step size) through the entire 20µm section. For each organoid, 2-3 positions were collected that included migrated GFP+ cells within the dorsal region of an organoid fusion. The number of GFP+ cells expressing various markers was counted manually using the “Cell Counter” plugin of Fiji. The cell counts for each organoid were summed and the percentage of GFP+ cells expressing each marker was calculated. The percentages were averaged across 4 organoids.
Statistics
All graphs and statistical analyses were generated using Prism 7 (GraphPad). When comparing only two groups (Figure 1C, 6E, Supplemental Figure 1) an unpaired two-tailed Student’s t-test was performed, for the remaining data comparing multiple groups a one-way ANOVA with posthoc Tukey’s test was used (Figure 2F, 3C). Samples were tested for normality prior to testing statistical significance. No statistical methods were used to predetermine the sample size. Sample sizes for experiments were estimated based on previous experience with a similar setup that showed significance. Experiments were not randomized and investigator was not blinded.
Supplementary Material
Ventral drug-patterning treatment induces ventral forebrain identity in cerebral organoids. (A-F) Whole-organoid confocal tile scan images of dorsalUnt and ventral organoids (48-55 days old) immunostained for forebrain (FOXG1), dorsal (TBR1, PAX6), or ventral (NKX2-1, DLX2, or GSX2) markers. (G) Quantification of the percentages (mean±SEM) of VZ-like regions expressing each marker: FOXG1 (dorsalUnt 82.5±9.1%, ventral 82.5±4.2%, p>0.999), NKX2-1 (dorsalUnt 0.0±0.0% , ventral 73.1±7.5%, p<0.001), TBR1 (dorsalUnt 95.5±3.6% , ventral 0.0±0.0%, p<0.001), DLX2 (dorsalUnt 4.75±2.3% , ventral 96.8±1.4%, p<0.001), PAX6 (dorsalUnt 76.3±11.2% , ventral 0.0±0.0%, p<0.001), GSX2 (dorsalUnt 6.0±2.0% , ventral 100.0±0.0%, p<0.001). For each marker, 4 organoids were used for counting. Statistical significance was tested using the student’s t-test (df=6). Scale bars are 500µm.
The gross morphology of cerebral organoid fusions changes with age. Whole-organoid confocal tile-scans to visualize the gross structural organization of ventral::dorsalCycA cerebral organoid fusion tissue at different ages. (A) 46 day old and (B) 61 day old organoid fusions contained VZ-like progenitor regions (insets A and B). Older, 80 day old organoid fusions contained less or no VZ-like progenitor regions. Scale bars are 500µm.
Migrating GFP+ cells in organoid fusions are highly non-proliferative. (A) Confocal images showing GFP/Ki67 immunostaining of migrated GFP+ cells in the dorsal region of 46 and 80 day old ventral::dorsalCycA organoid fusion cryosections. Very few GFP+ cells (blues arrows) also express Ki67 (yellow arrows). (B) Quantification of the percentage (mean±SEM) of GFP+ migrated cells expressing Ki67 from 46 day old (1.1±0.2%, 2420 cells counted from n=4 organoids), and 80 day old ventral::dorsalCycA fusions (0.7±0.2%, 3067 cells counted from n=4 organoids). Scale bar is 20µm.
Migrating GFP+ cells in organoid fusions do not express the Cajal Retzius cell marker Reelin (RELN). (A) A confocal image of GFP/RELN immunostaining in the dorsal region of an 80-day old ventral::dorsalCycA organoid fusion cryosection showing that migrated GFP+ cells (arrows) do not express RELN. Scale bar is 20µm.
Migrating GFP+ cells in organoid fusions express immature and mature neuronal markers. (A) A confocal image of GFP/DCX/NeuN immunostaining in the dorsal region of a 58-day old ventral::dorsalCycA organoid fusion cryosection showing that migrating GFP+ cells are DCX+ immature neurons (yellow arrows), and some are mature (DCX+/NeuN+) neurons (blue arrows). (B) A confocal image of GFP/MAP2 immunostaining in the dorsal region of an 80-day old ventral::dorsalCycA organoid fusion cryosection showing that some migrating GFP+ are mature (MAP2+) neurons (yellow arrows). Scale bars are 20µm.
The morphology of GFP+ cells migrating within cerebral organoid fusions. (A-C) Cropped z-projections of 80x spinning disc z-stacks to visualize the morphology of single GFP+ cells that migrated from ventral into dorsal organoid tissue within 80 day old ventral::dorsalCycA cerebral organoid fusions. (A) A GFP+GAD1+ interneuron with a branched morphology. The branches extend in many directions, and the cell body is large and round. (B-C) GFP+/GAD1+ interneurons with a migratory morphology consisting of an elongated cell body as well as branched leading processes and a trailing process. The cell in C has a leading process with 3 branches, and a bifurcated trailing process. Scale bars are 10µm.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. The cell migrates in a single direction. The leading process is branched with the different branches dynamically extending and retracting seemingly independent of one another. The trailing process follows as the cell body moves forward, and multiple times a leading process becomes a trailing process. As the cell moves forward, one leading process is extended while the remaining processes retract. Then the whole migratory dynamic cycle is repeated as the cell progresses forward. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. This movie is an example of a cell exhibiting many changes of direction involving the dynamic extension and retracting of several processes. As the cell body remains static, branches are extended in multiple directions, and then each of the main branches extends additional higher order branches. Finally, a branch is extended in a particular direction followed by the retraction of the other main branch. The cell body is then moved in the direction of the extending branch. The cycle is repeated as the cell decides which direction to migrate. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. This movie shows multiple migrating cells. 1) Initially a cell in the middle of the field of view is migrating upward. The upward process is retracted as a new leading process is extended downward and becomes branched. The cell migration direction is then changed downward. The bifurcated leading process is dynamic such that one process is extended as the other process is retracted. The cell body then moves toward the extended leading process. Prior to nucleokinesis, a swelling is observed moving from the cell body into the proximal portion of the leading process. Then the cell body is moved in parallel to the swelling, and finally the cell body moves into the swelling. 2) A second cell migrates from the left field of view toward the right, changes direction back toward the left, and then again changes direction toward the right, and finally changes once again back toward the left. With each change of direction, the trailing process becomes the leading process. The new leading process is extended toward the direction of travel as the trailing process is retracted. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. This movie shows multiple cells migrating in different directions with extending and retracting processes. Beginning around 45 hours, one cell migrates throughout the entire field of view beginning in the top left corner and traveling toward the bottom right corner. The cell travels rapidly in a constant direction, but at around 70 hours the progress is slowed as the leading process branches. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral::dorsalCycA organoid fusion. This movie shows multiple cells migrating. Around 6 hours a cell can be seen migrating into view from the bottom left corner of the field of view. This cell initially migrates left to right with a branched leading process. At multiple times, 3 branches are observed. As the cell progresses forward, branches are extended in the direction of travel, while other branches are retracted. Around 23 hours the cell changes direction abruptly from moving right to moving left. This involves an extension of a new process toward the left, while the previous leading process oriented to the right is retracted. The cell proceeds to the left, but around 39 hours, the leading process begins turning toward the right. The leading process makes a 180-degree turn, and then extends. The cell body then follows the leading process as the cell migrates rapidly from top to bottom and eventually proceeds out of view in the z-direction. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of ventral-derived GFP+ neurites growing within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. The neurites appear to be axons with an enlarged tuft at the end the processes which resembles that of a growth cone. The processes are highly dynamic, and exhibit extension in single directions, but also abrupt changes in direction. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
Primary Antibodies.
| Species | Antigen | Company | Catalog no. | Dilution |
|---|---|---|---|---|
| Rb | FoxG1 | Abcam | ab18259 | 1:200 |
| Rb | TBR1 | Abcam | ab31940 | 1:300 |
| Rb | PV | Swant | PV27 | 1:1000 |
| Rb | Calbindin | Swant | CD38a D-28K | 1:1000 |
| Ms | Calretinin | Swant | 6B3 | 1:1000 |
| Ms | GAD1/GAD67 | Millipore | MAB5406 | 1:800 |
| Rb | VGAT | Synaptic Systems | 131 013 | 1:500 |
| Chk | GFP | Abcam | ab13970 | 1:1000 |
| Rb | NPY | Abcam | ab30914 | 1:1000 |
| Ms | Relin | Millipore | MAB5366 | 1:200 |
| Rb | SOX6 | Abcam | ab30455 | 1:500 |
| Ms | COUP-TFII | Perseus Proteomics | PP-H7147-00 | 1:300 |
| Gt | Sp8 | Santa Cruz | SC-104661 | 1:300 |
| Rb | VIP | Immunostar | 20077 | 1:750 |
| Gt | DCX | Santa Cruz | SC-8066 | 1:200 |
| Ms | NeuN | Millipore | MAB377 | 1:500 |
| Rat | SOM | Millipore | MAB354 | 1:200 |
| Ms | Nkx2.1 | Dako | M3575 | 1:50 |
| Ms | PAX6 | DSHB | PAX6-s | 1:200 |
| Rb | DsRed/tdtomato | Clontech | 632496 | 1:250 |
| Rb | GSX2 | Millipore | ABN162 | 1:500 |
| Ms | Ki67 | BD | 550609 | 1:100 |
| Ms | HuC/D | Invitrogen | AB21271 | 1:200 |
| Ms | MAP2 | Millipore | MAB3418 | 1:500 |
| Gt | DLX2 | Santa Cruz | SC-18140 | 1:100 |
Secondary Antibodies.
| Host | Recognizes | Fluophore | Company | Catalog no. | Dilution |
|---|---|---|---|---|---|
| Donkey | rabbit | Alexa Fluor 568 | Invitrogen | A10042 | 1:500 |
| Donkey | rabbit | Alexa Fluor 647 | Invitrogen | A31573 | 1:500 |
| Donkey | mouse IgG | Alexa Fluor 568 | Invitrogen | A10036 | 1:500 |
| Donkey | mouse IgG | Alexa Fluor 647 | Invitrogen | A31571 | 1:500 |
| Donkey | chicken | Alexa Fluor 488 | Jackson Immuno | 703-605-155 | 1:500 |
| Donkey | goat | Alexa flour 647 | Invitrogen | A21447 | 1:500 |
| Donkey | rat | Alexa Fluor 647 | Jackson Immuno | 712-605-150 | 1:500 |
| Goat | Ms IgG1 | Alexa Fluor 568 | Invitrogen | A21124 | 1:500 |
| Goat | Ms IgG2b | Alexa Fluor 568 | Invitrogen | A21144 | 1:500 |
qPCR Primers.
| Gene | Primer 1 sequence | Primer 2 sequence |
|---|---|---|
| DLX2 | ACGTCCCTTACTCCGCCAAG | AGTAGATGGTGCGGGGTTTCC |
| FOXG1 | TGGCCCATGTCGCCCTTCCT | GCCGACGTGGTGCCGTTGTA |
| GSX2 | CACCGCCACCACCTACAAC | CAGGAGTTGCGTGCTAGTGA |
| LHX6 | CCGTCTGCAGGCAAGAACAT | GACACACGGAGCACTCGAG |
| NKX2-1 | GCCGTACCAGGACACCATG | ATGTTCTTGCTCACGTCCCC |
| TBP | GGGCACCACTCCACTGTATC | CGAAGTGCAATGGTCTTTAGG |
| TBR1 | CTCAGTTCATCGCCGTCACC | AGCCGGTGTAGATCGTGTCATA |
Acknowledgements
We are grateful to members of the Knoblich laboratory for technical expertise and feedback. We also thank the Molecular Biology Service Facility, notably Harald Scheuch, and the BioOptics facility, notably Tobias Müller and Pawel Pasierbek, as well as all the core facilities of IMBA/IMP for technical support. We also thank the HistoPathology facility of the Vienna Biocenter Core Facilities (VBCF). We also thank E. Hilary Gustafson and Simone Wolfinger for technical assistance and expertise regarding cell culture and 3D-cerebral organoid culture. J.A.B. received funding from an EMBO post-doctoral fellowship. Work in J.A.K.’s laboratory is supported by the Austrian Academy of Sciences, the Austrian Science Fund (Z_153_B09) and an advanced grant from the European Research Council (ERC).
Footnotes
Data Availability:
The authors declare that the data that support the findings of this study are available from the corresponding author upon request.
Author Contribution
J.A.B. and J.A.K. conceived the project and experimental design and wrote the manuscript. J.A.B. performed experiments and analyzed data. D.R. performed experiments and analyzed data under the supervision of J.A.B. and J.A.K. S.B. and J.L.S. performed cell counting of immunostained tissue. J.A.K. directed and supervised the project.
References
- 1.Harris KD, Shepherd GMG. The neocortical circuit: themes and variations. Nat Neurosci. 2015;18:170–181. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marín O, Müller U. Lineage origins of GABAergic versus glutamatergic neurons in the neocortex. Current Opinion in Neurobiology. 2014;26:132–141. doi: 10.1016/j.conb.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessaris N, Magno L, Rubin AN, Oliveira MG. Genetic programs controlling cortical interneuron fate. Current Opinion in Neurobiology. 2014;26:79–87. doi: 10.1016/j.conb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J, Anton ES. Decision making during interneuron migration in the developing cerebral cortex. Trends in Cell Biology. 2014;24:342–351. doi: 10.1016/j.tcb.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marin O, Valiente M, Ge X, Tsai LH. Guiding Neuronal Cell Migrations. Cold Spring Harbor Perspectives in Biology. 2010;2:a001834–a001834. doi: 10.1101/cshperspect.a001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marín O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- 7.Rossignol E. Genetics and function of neocortical GABAergic interneurons in neurodevelopmental disorders. Neural Plasticity. 2011;2011:649325. doi: 10.1155/2011/649325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lancaster MA, Knoblich JA. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science. 2014;345:1247125–1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 9.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariani J, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. CELL. 2015;162:375–390. doi: 10.1016/j.cell.2015.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renner M, et al. Self-organized developmental patterning and differentiation in cerebral organoids. The EMBO Journal. 2017:e201694700–14. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maroof AM, et al. Directed Differentiation and Functional Maturation of Cortical Interneurons from Human Embryonic Stem Cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholas CR, et al. Functional Maturation of hPSC-Derived Forebrain Interneurons Requires an Extended Timeline and Mimics Human Neural Development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, et al. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8:1670–1679. doi: 10.1038/nprot.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paşca AM, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadoshima T, et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proceedings of the National Academy of Sciences. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martynoga Ben, Morrison H, Price DJ, Mason JO. Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Developmental Biology. 2004;283:113–127. doi: 10.1016/j.ydbio.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Xuan S, et al. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 20.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 21.Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(Suppl 1):i82–8. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- 22.Eisenstat DD, et al. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 23.Hernández-Miranda LR, PARNAVELAS JG, Chiara F. Molecules and mechanisms involved in the generation and migration of cortical interneurons. ASN Neuro. 2009;2:e00031–e00031. doi: 10.1042/AN20090053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flames N, et al. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butt SJB, et al. The Temporal and Spatial Origins of Cortical Interneurons Predict Their Physiological Subtype. Neuron. 2004;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 26.Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh-Li HM, et al. Gsh-2, a murine homeobox gene expressed in the developing brain. Mechanisms of Development. 1995;50:177–186. doi: 10.1016/0925-4773(94)00334-j. [DOI] [PubMed] [Google Scholar]
- 29.Georgala PA, Carr CB, Price DJ. The role of Pax6 in forebrain development. Devel Neurobio. 2011;71:690–709. doi: 10.1002/dneu.20895. [DOI] [PubMed] [Google Scholar]
- 30.Vazin T, et al. Efficient derivation of cortical glutamatergic neurons from human pluripotent stem cells: a model system to study neurotoxicity in Alzheimer's disease. Neurobiology of Disease. 2014;62:62–72. doi: 10.1016/j.nbd.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erlander MG, Tillakaratne NJ, Feldblum S, Patel N, Tobin AJ. Two genes encode distinct glutamate decarboxylases. Neuron. 1991;7:91–100. doi: 10.1016/0896-6273(91)90077-d. [DOI] [PubMed] [Google Scholar]
- 32.Meyer G, Goffinet AM, Fairén A. Feature Article: What is a Cajal–Retzius cell? A Reassessment of a Classical Cell Type Based on Recent Observations in the Developing Neocortex. Cereb Cortex. 1999;9:765–775. doi: 10.1093/cercor/9.8.765. [DOI] [PubMed] [Google Scholar]
- 33.Hevner RF, Neogi T, Englund C, Daza RAM, Fink A. Cajal-Retzius cells in the mouse: transcription factors, neurotransmitters, and birthdays suggest a pallial origin. Brain Res Dev Brain Res. 2003;141:39–53. doi: 10.1016/s0165-3806(02)00641-7. [DOI] [PubMed] [Google Scholar]
- 34.Abranches E, et al. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS ONE. 2008;4:e6286–e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown JP, et al. Transient expression of doublecortin during adult neurogenesis. J Comp Neurol. 2003;467:1–10. doi: 10.1002/cne.10874. [DOI] [PubMed] [Google Scholar]
- 36.Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome Biology. 2005;6:204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nóbrega-Pereira S, et al. Postmitotic Nkx2-1 controls the migration of telencephalic interneurons by direct repression of guidance receptors. Neuron. 2008;59:733–745. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markram H, et al. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 39.Fishell G, Rudy B. Mechanisms of inhibition within the telencephalon: "where the wild things are". Annu Rev Neurosci. 2010;34:535–567. doi: 10.1146/annurev-neuro-061010-113717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sultan KT, Brown KN, Shi S-H. Production and organization of neocortical interneurons. Front Cell Neurosci. 2012;7:221–221. doi: 10.3389/fncel.2013.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 42.Hansen DV, et al. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka DH. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133:2167–2176. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka D, Nakaya Y, Yanagawa Y, Obata K, Murakami F. Multimodal tangential migration of neocortical GABAergic neurons independent of GPI-anchored proteins. Development. 2003;130:5803–5813. doi: 10.1242/dev.00825. [DOI] [PubMed] [Google Scholar]
- 45.Britto JM, Johnston LA, Tan S-S. The stochastic search dynamics of interneuron migration. Biophysical Journal. 2009;97:699–709. doi: 10.1016/j.bpj.2009.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ang ESBC, Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martini FJ, et al. Biased selection of leading process branches mediates chemotaxis during tangential neuronal migration. Development. 2008;136:41–50. doi: 10.1242/dev.025502. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka DH, et al. Random Walk Behavior of Migrating Cortical Interneurons in the Marginal Zone: Time-Lapse Analysis in Flat-Mount Cortex. Journal of Neuroscience. 2009;29:1300–1311. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reiner O. LIS1 and DCX: Implications for Brain Development and Human Disease in Relation to Microtubules. Scientifica. 2013;2013:1–17. doi: 10.1155/2013/393975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fatehullah A, Tan SH, Barker N. Organoids as an in vitro model of human development and disease. Nat Cell Biol. 2016;18:246–254. doi: 10.1038/ncb3312. [DOI] [PubMed] [Google Scholar]
- 51.Ranga A, Gjorevski N, Lutolf MP. Drug discovery through stem cell-based organoid models. Adv Drug Deliv Rev. 2014;69–70:19–28. doi: 10.1016/j.addr.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y, et al. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69:61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian L, Bershteyn M, Paredes MF, Kriegstein AR. Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nature Communications. 2017;8:14167. doi: 10.1038/ncomms14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- 55.Jakovcevski I, Mayer N, Zecevic N. Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cerebral Cortex. 2011;21:1771–1782. doi: 10.1093/cercor/bhq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X, Zecevic N. Dorsal Radial Glial Cells Have the Potential to Generate Cortical Interneurons in Human But Not in Mouse Brain. Journal of Neuroscience. 2011;31:2413–2420. doi: 10.1523/JNEUROSCI.5249-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stumm RK, et al. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis DA. Inhibitory neurons in human cortical circuits: substrate for cognitive dysfunction in schizophrenia. Current Opinion in Neurobiology. 2014;26:22–26. doi: 10.1016/j.conb.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakano T, et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 60.Sakaguchi H, et al. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nature Communications. 2014;6:8896–8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muguruma K, Nishiyama A, Kawakami H, Hashimoto K, Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. CellReports. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 62.Jo J, et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Preibisch S, Saalfeld S, Tomancak P. Globally optimal stitching of tiled 3D microscopic image acquisitions. 2009;25:1463–1465. doi: 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ventral drug-patterning treatment induces ventral forebrain identity in cerebral organoids. (A-F) Whole-organoid confocal tile scan images of dorsalUnt and ventral organoids (48-55 days old) immunostained for forebrain (FOXG1), dorsal (TBR1, PAX6), or ventral (NKX2-1, DLX2, or GSX2) markers. (G) Quantification of the percentages (mean±SEM) of VZ-like regions expressing each marker: FOXG1 (dorsalUnt 82.5±9.1%, ventral 82.5±4.2%, p>0.999), NKX2-1 (dorsalUnt 0.0±0.0% , ventral 73.1±7.5%, p<0.001), TBR1 (dorsalUnt 95.5±3.6% , ventral 0.0±0.0%, p<0.001), DLX2 (dorsalUnt 4.75±2.3% , ventral 96.8±1.4%, p<0.001), PAX6 (dorsalUnt 76.3±11.2% , ventral 0.0±0.0%, p<0.001), GSX2 (dorsalUnt 6.0±2.0% , ventral 100.0±0.0%, p<0.001). For each marker, 4 organoids were used for counting. Statistical significance was tested using the student’s t-test (df=6). Scale bars are 500µm.
The gross morphology of cerebral organoid fusions changes with age. Whole-organoid confocal tile-scans to visualize the gross structural organization of ventral::dorsalCycA cerebral organoid fusion tissue at different ages. (A) 46 day old and (B) 61 day old organoid fusions contained VZ-like progenitor regions (insets A and B). Older, 80 day old organoid fusions contained less or no VZ-like progenitor regions. Scale bars are 500µm.
Migrating GFP+ cells in organoid fusions are highly non-proliferative. (A) Confocal images showing GFP/Ki67 immunostaining of migrated GFP+ cells in the dorsal region of 46 and 80 day old ventral::dorsalCycA organoid fusion cryosections. Very few GFP+ cells (blues arrows) also express Ki67 (yellow arrows). (B) Quantification of the percentage (mean±SEM) of GFP+ migrated cells expressing Ki67 from 46 day old (1.1±0.2%, 2420 cells counted from n=4 organoids), and 80 day old ventral::dorsalCycA fusions (0.7±0.2%, 3067 cells counted from n=4 organoids). Scale bar is 20µm.
Migrating GFP+ cells in organoid fusions do not express the Cajal Retzius cell marker Reelin (RELN). (A) A confocal image of GFP/RELN immunostaining in the dorsal region of an 80-day old ventral::dorsalCycA organoid fusion cryosection showing that migrated GFP+ cells (arrows) do not express RELN. Scale bar is 20µm.
Migrating GFP+ cells in organoid fusions express immature and mature neuronal markers. (A) A confocal image of GFP/DCX/NeuN immunostaining in the dorsal region of a 58-day old ventral::dorsalCycA organoid fusion cryosection showing that migrating GFP+ cells are DCX+ immature neurons (yellow arrows), and some are mature (DCX+/NeuN+) neurons (blue arrows). (B) A confocal image of GFP/MAP2 immunostaining in the dorsal region of an 80-day old ventral::dorsalCycA organoid fusion cryosection showing that some migrating GFP+ are mature (MAP2+) neurons (yellow arrows). Scale bars are 20µm.
The morphology of GFP+ cells migrating within cerebral organoid fusions. (A-C) Cropped z-projections of 80x spinning disc z-stacks to visualize the morphology of single GFP+ cells that migrated from ventral into dorsal organoid tissue within 80 day old ventral::dorsalCycA cerebral organoid fusions. (A) A GFP+GAD1+ interneuron with a branched morphology. The branches extend in many directions, and the cell body is large and round. (B-C) GFP+/GAD1+ interneurons with a migratory morphology consisting of an elongated cell body as well as branched leading processes and a trailing process. The cell in C has a leading process with 3 branches, and a bifurcated trailing process. Scale bars are 10µm.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. The cell migrates in a single direction. The leading process is branched with the different branches dynamically extending and retracting seemingly independent of one another. The trailing process follows as the cell body moves forward, and multiple times a leading process becomes a trailing process. As the cell moves forward, one leading process is extended while the remaining processes retract. Then the whole migratory dynamic cycle is repeated as the cell progresses forward. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. This movie is an example of a cell exhibiting many changes of direction involving the dynamic extension and retracting of several processes. As the cell body remains static, branches are extended in multiple directions, and then each of the main branches extends additional higher order branches. Finally, a branch is extended in a particular direction followed by the retraction of the other main branch. The cell body is then moved in the direction of the extending branch. The cycle is repeated as the cell decides which direction to migrate. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. This movie shows multiple migrating cells. 1) Initially a cell in the middle of the field of view is migrating upward. The upward process is retracted as a new leading process is extended downward and becomes branched. The cell migration direction is then changed downward. The bifurcated leading process is dynamic such that one process is extended as the other process is retracted. The cell body then moves toward the extended leading process. Prior to nucleokinesis, a swelling is observed moving from the cell body into the proximal portion of the leading process. Then the cell body is moved in parallel to the swelling, and finally the cell body moves into the swelling. 2) A second cell migrates from the left field of view toward the right, changes direction back toward the left, and then again changes direction toward the right, and finally changes once again back toward the left. With each change of direction, the trailing process becomes the leading process. The new leading process is extended toward the direction of travel as the trailing process is retracted. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. This movie shows multiple cells migrating in different directions with extending and retracting processes. Beginning around 45 hours, one cell migrates throughout the entire field of view beginning in the top left corner and traveling toward the bottom right corner. The cell travels rapidly in a constant direction, but at around 70 hours the progress is slowed as the leading process branches. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of migrating GFP+ cells within the dorsal region of a ventral::dorsalCycA organoid fusion. This movie shows multiple cells migrating. Around 6 hours a cell can be seen migrating into view from the bottom left corner of the field of view. This cell initially migrates left to right with a branched leading process. At multiple times, 3 branches are observed. As the cell progresses forward, branches are extended in the direction of travel, while other branches are retracted. Around 23 hours the cell changes direction abruptly from moving right to moving left. This involves an extension of a new process toward the left, while the previous leading process oriented to the right is retracted. The cell proceeds to the left, but around 39 hours, the leading process begins turning toward the right. The leading process makes a 180-degree turn, and then extends. The cell body then follows the leading process as the cell migrates rapidly from top to bottom and eventually proceeds out of view in the z-direction. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.
A time-lapse movie of ventral-derived GFP+ neurites growing within the dorsal region of a ventral/GFP::dorsalCycA organoid fusion. The neurites appear to be axons with an enlarged tuft at the end the processes which resembles that of a growth cone. The processes are highly dynamic, and exhibit extension in single directions, but also abrupt changes in direction. This recording was from a slice culture of an organoid fusion created fusing a ventral H9 hESC-derived organoid containing a CAG-eGFP-WPRE construct to a dorsalCycA iPSC-derived organoid.