Abstract
A central challenge in evolutionary biology concerns the mechanisms by which complex adaptations arise. Such adaptations depend on the fixation of multiple, highly specific mutations, where intermediate stages of evolution seemingly provide little or no benefit. It is generally assumed that the establishment of complex adaptations is very slow in nature, as evolution of such traits demands special population genetic or environmental circumstances. However, blueprints of complex adaptations in molecular systems are pervasive, indicating that they can readily evolve. We discuss the prospects and limitations of non-adaptive scenarios, which assume multiple neutral or deleterious steps in the evolution of complex adaptations. Next, we examine how complex adaptations can evolve by natural selection in changing environment. Finally, we argue that molecular ’springboards’, such as phenotypic heterogeneity and promiscuous interactions facilitate this process by providing access to new adaptive paths.
Introduction
What are the limits to perfection in nature1? Certain molecular traits may be difficult to evolve not because they are made impossible by laws of physics and chemistry, but because multiple mutations must be fixed together to provide a benefit (Figure 1). In this work, such traits will be referred to as complex adaptations. Darwin himself was highly aware of this problem2. He stated that “if it could be demonstrated that any complex organ existed, which could not possibly have been formed by numerous, successive, slight modifications, my theory would absolutely break down”. Complex adaptations highlight a persistent problem in evolutionary discussions. Many analyses focus on the adaptive nature of the end result of evolution only. However, what looks like an optimal organism design may not be attainable by a series of adaptive mutations.
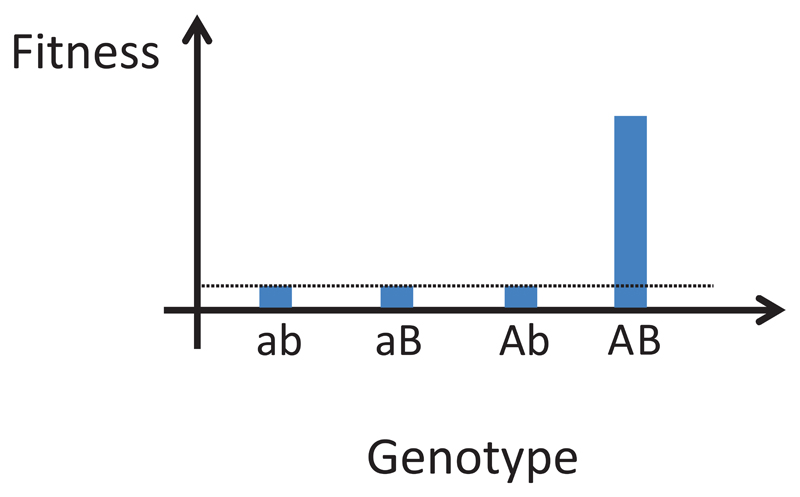
Figure 1. Concept of complex adaptations.
Two mutations (a → A and b → B) have to occur simultaneously to provide a fitness advantage. Note that the individual mutations depicted here are neutral, but they could also be deleterious.
The problem of complex adaptation relates to another central issue in evolutionary biology. Since the early 1920s, Ronald Fisher pioneered the view that adaptation is by and large a hill-climbing process: it proceeds through progressive accumulation of beneficial mutations3. By contrast, Sewall Wright proposed that fixation of conditionally deleterious mutations acts as stepping-stones by providing access to evolutionary routes that are otherwise inaccessible4. This issue has practical implications. For example, directed protein evolution typically employs random mutagenesis coupled with strong functional selection5. Such protocols generally fail to discover new enzymatic activities that demand the accumulation of many neutral or slightly deleterious mutations. Computer scientists face similar problems: standard evolutionary algorithms have a tendency to quickly converge on a local solution, and hence frequently fail to identify more promising regions of the parameter space6.
There are two fundamental problems with reconstructing the evolutionary history of complex adaptations: shortage of intermediate forms in extant organisms and improbable fossilization (‘missing links’). This review focuses on complex adaptations in cellular systems, where these problems are especially severe. Recent advances in experimental evolution, deep-scan mutational assays and systems biology allowed studying the problem of complex adaptation in a rigorous manner. These studies unequivocally demonstrated that complex adaptations in molecular systems are pervasive and can readily evolve in the laboratory.
It is worth emphasizing what this review is not about. We do not discuss the concept of facilitated variation7,8, modularity9 or the evolution of biological complexity per se, as these topics are not directly related and have been reviewed elsewhere. We do not explicitly review key innovations, defined as phenotypic traits that allow subsequent evolutionary radiation of a taxonomic group10. Evolution of key innovations could be limited by physicochemical constraints, environmental conditions or shortage of mutational combinations required for functional change. Therefore, the notions of key innovations and complex adaptation are different from each other.
We start by providing a classification scheme with examples, followed by prospects and limitations of current theories on how and why complex adaptations arise. We introduce conceptual (Box 1) and methodological advances (Box 2), and end the paper by discussing future directions and predictions that have so far remained untested.
Box 1. Complex adaptations – theoretical considerations.
By definition, complex adaptations are phenotypic traits requiring multiple, specific mutations to yield a functional advantage. Despite substantial efforts, the population genetic mechanisms driving complex adaptations are unclear99,100. In a nutshell, the paradox is as follows. As mutational events are rare, complex adaptations are unlikely to occur in small populations: the waiting time for the rise of multiple specific mutations would be very long. This would suggest that the evolution of complex adaptations is facilitated in large populations. The issue is more complicated though. If the intermediate mutational steps towards complex adaptation are individually deleterious, they will be purged in large populations. As the fixation of deleterious mutations is exceedingly unlikely with growing population size, shift from one adaptive peak to another through weakly deleterious intermediates can only occur when genetic drift prevails, i.e. in small populations.
How can the seeming paradox be resolved? First, the intermediate mutation may confer a benefit under alternative environmental or genetic conditions (see main text). Alternatively, we may need to abandon the idea of sequential fixation of intermediate mutations. Although individual deleterious intermediate-stage mutations have negligible chance for fixation in large populations, there is a steady input of such mutations. Indeed, segregating deleterious mutations are common in natural populations of yeast101 and human102,103. Such a stable reservoir of non-adaptive mutations is poised for the rise of a second mutation that is adaptive in the specific genetic background, leading to simultaneous fixation of the two mutations. As the size of the reservoir increases with growing population size, the time required for this process declines sharply as population size increases88,104,105. Thus, selection alone can offer a solution to escape local fitness peaks in natural populations. One may argue that this theory is unlikely to work in the case of three or more non-adaptive intermediate steps, as the simultaneous rise of multiple mutations in a single genotype is exceedingly unlikely. Although this criticism may hold when the intermediate steps are deleterious, the effect is partly offset by the elevated number of paths toward the final adaptation when the intermediate states are neutral105.
Evolutionary escape from local fitness peaks has been a central problem for over 85 years now99. Unfortunately, the lack of consensus on the relative roles played by mutation, recombination and random genetic drift hinders empirical tests. The role of recombination is particularly controversial: it can either facilitate the escape from local fitness peaks by combining mutations from different individuals or hinder it by breaking up adaptive combinations106. As a result, only low recombination rates can speed up crossing fitness valleys, while high rates are predicted to be disadvantageous106.
Rates of recombination, mutation and the importance of genetic drift vary enormously across evolutionary lineages, but do they influence which evolutionary pathways are realized in nature? For instance, it is currently unknown whether multicellular eukaryotic species with relatively low population size and high mutation rate have greater or smaller acquisition of complex adaptations on a per generation basis105. As a further complication, the answer likely depends on the molecular basis of adaptation, e.g. the number of sites involved and whether the intermediate states are deleterious or neutral.
Finally, most theoretical considerations have focused on the rate of traversing a single specific mutational path, which is expected to be low when neutral or deleterious mutations are involved. However, in realistic fitness landscapes there could be multiple different possible mutational paths to the same genotype, and there might be several possible beneficial genotypes that are phenotypically equivalent. As a result, evolution may follow trajectories that involve neutral or deleterious intermediates even in the presence of directly uphill trajectories106. Addressing this issue demands quantification of the frequencies of these different modes of mutational trajectories on realistic fitness landscapes.
Box 2. Methods to study complex adaptations.
Population genetics has a long tradition of studying the problem of complex adaptations4. Theoretical studies indicate that the time to establishment of complex adaptations depends on population size, mutation rate, recombination, and the magnitude of the selective disadvantage of intermediate-state alleles (Box 1). Specific examples of complex adaptations can be studied by phylogenetic analysis, molecular laboratory experiments and computational systems biology.
Functionally related genes which are jointly needed for a cellular function are not expected to be accessible gradually, and therefore they should arise and be lost together on the same branch of the phylogenetic tree. Comparative genomics has provided ample evidence for such co-evolving gene clusters, especially in the case of macromolecular protein complexes107 and linear metabolic pathway28. Other complex traits are assumed to be accessible only in a defined order49,53, as certain mutations pave the way for subsequent adaptive changes.
Molecular studies in the laboratory unequivocally demonstrated that complex adaptations in molecular systems are pervasive. Three main types of analysis have been deployed: mutational analysis coupled with exploration of the adaptive landscape, reconstruction of ancestral molecules, and laboratory experimental evolution. Deep-scan mutational scanning showed that in spite of the prevalence of harmful mutations, mutational effects vary due to epistatic interactions with other mutations108. Reconstruction and functional analysis of ancestral molecules showed that adaptive evolution frequently demands prior fixation of other, functionally silent mutations89,109. Directed protein evolution studies investigated the impact of neutral exploration of the genotype space on acquisition of new enzymatic functions (see main text for details).
Finally, systems biology models offer a new angle to study the underlying molecular processes of complex adaptations that involve multiple gene products110. By providing a mapping between genotype and phenotype, molecular network models provide valuable insights into the exploration of the adaptive landscape, with the ultimate goal of predicting which particular evolutionary trajectories are realized, while others are not. For instance, using genome-scale metabolic modelling, recent works tested the evolutionary mechanisms whereby complex bacterial metabolic innovations arise43,110. In the main text, we demonstrate the utility of these methodological developments on testing theories of complex adaptations.
Classification of complex adaptations
Complex adaptations can occur within a single gene (intramolecular, Figure 2). One form involves mutually dependent mutations that stabilize local elements in a protein structure and simultaneously promote new functions11. Here, researchers face “the chicken or the egg” dilemma: mutations in the enzymatic active site are typically destabilizing, whereas structural mutations elsewhere in the protein provide no obvious change in enzymatic function but mitigate the fitness cost of the former. It is not trivial which of the two types of mutations come first. Another common form of intramolecular complex adaptations concerns the establishment of pairwise interactions between interdependent sites within a single RNA or protein molecule. Well-known examples include the origin of stem-loop interactions in tRNA12 and disulfide bonds in protein molecules13. As the disruption of such highly specific interactions often compromises stability or function, explaining their evolution as a stepwise process is challenging.
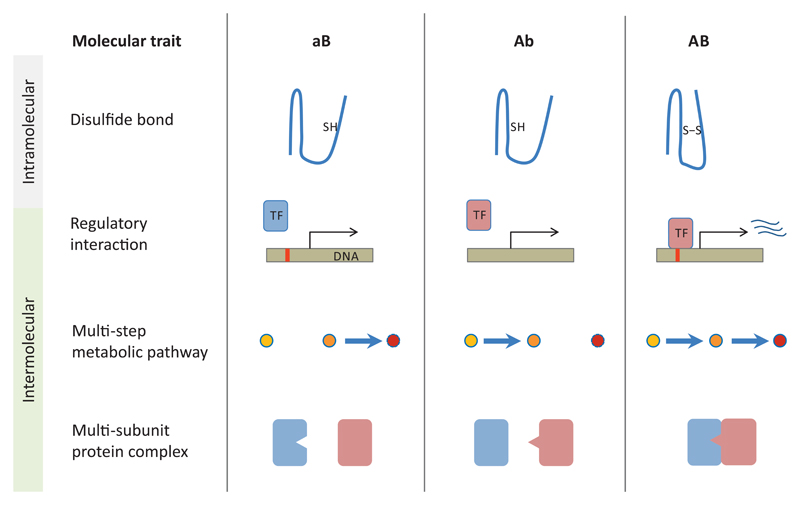
Figure 2. Main classes and examples of complex adaptations in molecular systems.
Establishment of a new disulfide bond (S-S) from two adjacent sulfhydryl groups (-SH) within the same protein molecule represents an example of intramolecular complex adaptation. The origin of new transcription factor – DNA binding site interactions, multi-step metabolic pathways and multi-subunit complexes all qualify as intermolecular complex adaptations requiring specific mutations in multiple genes.
Intermolecular complex adaptations involve genetic changes at multiple loci, and belong to three main classes (Figure 2). The most elementary form entails the establishment of new interactions between two macromolecules, such as seen in transcription factor - binding site interactions, two-component signal transduction systems, and more generally, protein-protein binding interfaces. Although complementarity is essential for maintenance of function, these systems frequently show substantial divergence across species14. The main issue is how new interaction specificities arise without deleterious cross-talks or non-functional intermediate stages. The second class involves the origin of multi-gene molecular pathways that demand the coordinated operation of multiple gene products. A prime example is metabolic pathways that typically work through series of enzymatic steps to produce a desired end-product15. As the intermediate steps do not necessarily produce useful metabolites, explaining the stepwise evolution of such pathways demands careful consideration. The third class of complex adaptations concerns the origin of macromolecular cellular structures, such as bacterial flagella16,17, eukaryotic centrioles and ion channels. Such structures require the establishment of both novel specific interaction events and the coordinated action of multiple proteins. Although precursors of these complex traits are widespread across species, deciphering step-by-step evolutionary histories have proved to be generally difficult. Multimeric protein complexes are generally assembled from subunits derived from gene duplication16,18, but the evolutionary forces driving their diversification remain largely unknown.
Leaps in the genotype space
In his New Essays on Human Understanding, Leibniz proposed the principle of continuity in nature. This maxim says that nature does not make jumps (“natura non facit saltum”): objects and properties in nature change gradually, rather than suddenly. Darwin was also very explicit about this issue2. He stated that natural selection proceeds by the accumulation of numerous slight favorable steps, all of which have individually small effects on organisms’ performance. Similarly, modern scholars generally state that evolution proceeds in small discrete steps in the genotype space, where steps are limited to immediate mutational neighbors (but see refs 19,20). The justification of this assumption is that the rise of double mutants is usually negligible, and mutations with large effects tend to have deleterious side consequences. We do not discuss the validity of these assumptions in detail, but rather focus on some notable exceptions.
Several mutational mechanisms initiate major phenotypic transitions. First, a sizeable fraction of nucleotide substitutions occur simultaneously within short stretches of DNA21 and may therefore affect multiple interacting sites within a protein22. Moreover, structural alterations such as deletions and insertions induce point mutations in nearby regions23,24. Second, as recombination events can reorganize protein modules, proteins or entire cellular networks, they can lead to evolutionary novelties inaccessible by point mutations only25,26. Third, and on a related note, horizontal gene transfer, a major source of evolutionary novelties in bacteria, often results in the simultaneous acquisition of multiple functionally related genes27,28. Fourth, large-scale duplications and aneuploidy events simultaneously increase the dosage of multiple genes, and thereby produce large phenotypic leaps29. A case study on adaptive evolution in yeast demonstrated that extra copies of a chromosome increased fitness by simultaneous overexpression of two specific regulatory genes30. Remarkably, overexpressing any of the two genes individually provided no fitness advantage30. The frequency and mechanisms by which gross chromosomal mutations aid the crossing of suboptimal intermediate states remain an important open question for future studies.
Non-adaptive origins of complex adaptations
How do complex adaptations arise in multiple steps? One theory suggests that they involve mutations that are individually neutral or even deleterious, but together provide a fitness advantage31–33. Non-adaptive mutations may accumulate by genetic drift first, and thereby prepare the ground for later mutations that confer new, adaptive traits. Alternatively, multiple mutations may be fixed simultaneously in large populations (see Figure 3A and Box 1 for more details).
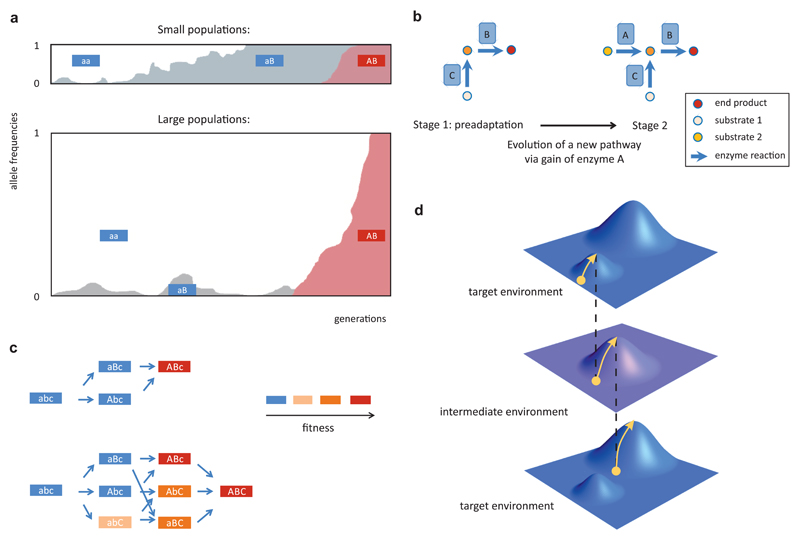
Figure 3. Evolutionary mechanisms of establishing complex adaptations.
A) Non-adaptive origin of complex adaptations can occur through sequential fixation of mutations in small populations where the intermediate mutation first goes to fixation through genetic drift (upper plot). Alternatively, in large populations a beneficial second mutation can arise in a descendant of the intermediate mutation and they fix simultaneously without the population ever going through a pure intermediate state (lower plot). Allele frequency plots depict the dynamics of the neutral intermediate mutation and the beneficial second mutation. B) Simplified metabolic network scheme in which enzyme functions A and B are asymmetrically dependent on each other as a consequence of preadaptation. In the first evolutionary stage, the network can metabolize substrate 1 and this serves as a preadaptation to utilize substrate 2 via the acquisition of enzyme A. Because in the second stage both enzymes A and C produce the same intermediate metabolite, the activity of downstream enzyme B does not exclusively depend on A. In contrast, A can only be active under steady state conditions when B is active, hence their functional dependence is asymmetric. C) Evolution of complex adaptations via adaptive by-passes in extra genotype dimensions. The figure depicts a simplified scenario where evolution from a low-fitness genotype (ab) to a high-fitness one (AB) involves neutral intermediary steps, but a mutation at a third locus (c → C) opens new uphill trajectories where all intermediate steps are beneficial. Fitness of genotypes is represented by a color scale. D) Evolution in alternating environments promotes escape from local fitness optima through series of purely adaptive walks. The horizontal plane represents genotype space, the vertical axis represents fitness, and arrows indicate uphill evolutionary trajectories. Environmental change alters the fitness landscape in such a way that a fitness valley in the target environment becomes a fitness peak in the intermediate environment hence facilitating valley crossing.
The best evidence for the theory comes from laboratory evolution studies on catalytic RNAs34 and proteins5,32,35. Several works demonstrated that neutral mutations promote the evolution of new protein functions in the laboratory35,36. Moreover, large populations of RNA enzymes with accumulated neutral variation adapted more rapidly to cleave a novel RNA substrate than a population without such genetic variation34.
Non-adaptive evolution may also contribute to the initial establishment of altered multi-protein complexes37,38 and DNA regulatory interactions39 prior to adaptive diversification. For example, epistasis across the molecular interface of a transcription factor and the binding site allows the evolution of novel binding specificities to arise by neutral evolution39. Similarly, a new subunit in a fungal proton pump arose by duplication followed by complementary degeneration of binding interfaces in the two copies. During this process both copies became obligate components of the complex without conferring any new function40.
Despite its apparent success, the theory faces several problems. First, when high-fitness genotypes are well-isolated from each other and sparse on the adaptive landscape, molecules subjected to genetic drift will fail to escape the local fitness peak. Indeed, the protocol of neutral drift followed by positive selection has met with mixed success in certain studies41,42. For instance, drift produced only protein variants and positions of mutation that had also been described in conventional directed evolution experiments based on positive selection only42.
Second, laboratory studies of genetic drift in protein and RNA molecules typically applied exceedingly high mutation rates32,34,35. Whether neutral mutations promote the evolution of new molecular functions under more realistic population genetic settings remains to be tested.
Third, the role of non-adaptive forces is questionable in the origin of multi-step pathways. Most notably, it has been proposed that “additions of individual reactions to a metabolic network will not change the phenotype until a second added reaction connects the first one to an already existing pathway”33. There is no direct empirical support for this scenario in bacteria, which are especially prolific in producing metabolic innovations43. Genes under no selection are rapidly inactivated and subsequently lost in free-living bacteria, not least because there is a pervasive mutational bias towards deletions of genomic segments44,45.
Finally, multiple non-adaptive steps may render the establishment of new complex traits exceedingly unlikely. The process is expected to be much faster with the availability of adaptive bypasses (see below). Given these considerations, it is important to consider scenarios that assume stepwise evolution of complex adaptations by natural selection.
Preadaptations and indirect evolutionary paths
As Darwin himself elaborated, many complex traits evolved from earlier traits that had served different functions2. Examples for molecular preadaptations are abundant in the literature. For instance, several components of the bacterial flagellum share homologous proteins with the type III secretion system (TTSS), indicating that one evolved from the other17. Another example is the citrate acid cycle: it most likely originated by assembling chemical steps previously functioning in amino acid biosynthesis46,47. Similarly, studies on digital organisms revealed that populations often evolve complex features by building on simpler functions that had evolved earlier48. Accordingly, early rising mutations serve as stepping-stones in the evolution of complex traits.
The theory has three main predictions. First, asymmetric relationships between proteins or cellular subsystems should be common: the function of one protein (A) depends on another protein (B), but the function of protein B does not depend on A. The best examples come from metabolism28,49. When multiple enzymes produce the same metabolite, enzymes in the converging reaction (A) depend on the metabolite flux through B, but not vice versa (Figure 3B). Such asymmetry is reflected in gene expression, gene essentiality and most importantly in the evolution of gene content49. Similarly, the functional relationships between regulators and target proteins are also frequently asymmetric: the target may exert its function without the regulator in specific contexts, but not necessarily vice versa. Indeed, transcription factors in bacteria are typically less conserved than their target genes and evolve independently of them50. More generally, large-scale genetic interaction screens uncovered a plethora of asymmetric functional relationships between gene pairs where mutation in one gene modifies the effect of mutation in the other51.
Second, as a consequence of asymmetrical protein relationships, evolution should proceed via a defined order43,49. In agreement with the expectations, enzymes that serve as stepping-stones towards multi-step adaptations were gained on an earlier branch of the phylogenetic tree as compared to enzymes that provide benefit only in the presence of the partner enzymes43. More generally, genes preexisting in an organism should influence the functionality of a horizontally acquired gene product if it operates on an ancestral pathway52. A recent large-scale analysis of bacterial genomes provided general support for this idea53. It showed that specific molecular functions tend to be gained sequentially by horizontal gene transfer53, suggesting that bacterial evolution is governed by functional assembly patterns. Other studies aimed to reconstruct adaptive landscapes in a single protein by functional analysis of mutational combinations54. It appears that evolutionary novelties by horizontal gene transfer and point mutations are profoundly constrained by epistasis, and therefore frequently occur via a defined order.
Third, evolutionary traps can be circumvented by “extra-dimensional bypasses” (Figure 3C) Accordingly55, the necessity of non-adaptive steps reflects limited dimensionality of the adaptive landscapes considered, and therefore is more apparent than real (Box 1). Indeed, most molecular evolution studies are confined to the immediate neighbourhood around the wild-type sequence and are limited in scope. A recent study investigated this problem systematically55. It confirmed that many direct evolutionary paths are indeed blocked by pairwise epistatic interactions (i.e. populations along these paths must proceed via one or more non-adaptive steps). By exhaustive analysis of 160,000 mutational variants of a single protein, the authors show that such evolutionary traps can be circumvented by indirect paths through gain and subsequent loss of mutations55,56. These results indicate that higher-order epistasis is critically important for understanding evolutionary processes57.
Dynamic environment
The dynamic environment scenario heavily relies on the notion of pre-adaptation43,58,59. It claims that stepping-stone mutations fixed earlier are beneficial in specific environments only, and therefore complex adaptation is accelerated in varying environments (Figure 3D). Computer simulations of genetic circuits and RNA molecules60 and verbal arguments on early expansion of molecular pathways61 reached similar conclusions.
The scenario is attractive for two reasons. Environmental change is ubiquitous, and epistasis changes qualitatively across environments62–64. Mutations neutral or even deleterious in one environment may turn into beneficial in another environmental context. Therefore, environmental change may facilitate evolution by exposing populations to new adaptive routes. Recent studies provide direct empirical support for the theory. The examples include the evolution of new protein functions, transcription factor - DNA binding site interfaces and metabolic pathways.
Protein evolution was examined by analyzing single amino acid mutants in an enzyme under selection for a wild-type function and for a new function65,66. Interestingly, resistance to a new antibiotic emerged from mutations that are neutral at low levels to another drug but deleterious at high levels; thus the capacity to evolve a new function also depends on the strength of selection65,66. It appears that fluctuating environments select for enzymes with especially high activities66. Another study explicitly demonstrated that alteration of the adaptive landscape by environmental change permits exploration of novel regions of the sequence space that are otherwise selected against, and this process leads to superior phenotypes67.
The lac operon in Escherichia coli tells a similar story68. In a fixed environment, the evolution of transcription factor and binding sites is trapped at a local solution, as beneficial mutational combinations cannot be reached by natural selection only. However, in a fluctuating environment, many of them become accessible via adaptive steps. Such escapes from adaptive stasis readily occurred because numerous mutations had opposite effects in the two environments and therefore environmental changes opened up new possibilities for beneficial mutations68.
Finally, it has been proposed that temporally varying nutrient conditions selects for new enzymatic reactions that, as a by-product, serve as stepping-stones towards the establishment of complex metabolic pathways43. Three complementary approaches provided support for the theory. First, computational analysis of bacterial metabolic networks revealed that new complex pathways can evolve via the acquisition of single biochemical reactions that confer a benefit under specific environmental conditions43. Second, by reconstructing the evolutionary history of gene gains in bacteria, it was shown that complex metabolic pathways are indeed established in a defined order as predicted by the dynamic environment model43. Third, laboratory evolution studies showed that adaptation to one carbon source promotes utilization of other nutrients43,69.
Together, these studies indicate that complex traits can emerge in complex environments without the need to invoke neutral exploration of genotype space, a view that is in sharp contrast with non-adaptive scenarios of complex adaptations. This conclusion has important ramifications for those studying the design principles of complex molecular pathways and those aiming to create industrially useful microbes. First, deciphering the adaptive value of molecular pathways might often require studying their operation under multiple environmental conditions. Second, evolutionary engineering of microbes to obtain desired phenotypes may demand complex, temporally varying selection pressure in the laboratory.
Although the dynamic environment model is a promising alternative to non-adaptive scenarios, it nevertheless faces several conceptual challenges. Most notably, it is unclear why mutations adaptive in one transient environment are not selected against and lost upon environmental change. A major barrier to complex adaptations may be the absence of a relevant series of environmental conditions, rather than the shortage of relevant mutations in the population. Answering this question demands exploring the network of evolutionary trade-offs across environments in the laboratory70.
Molecular springboards
Finally, we suggest three molecular mechanisms that potentiate the evolution of complex adaptations (Figure 4). The common feature of these mechanisms is that they do not confer new molecular function. Rather, they provide access to many new evolutionary paths or reduce the number of mutations required for phenotypic change.
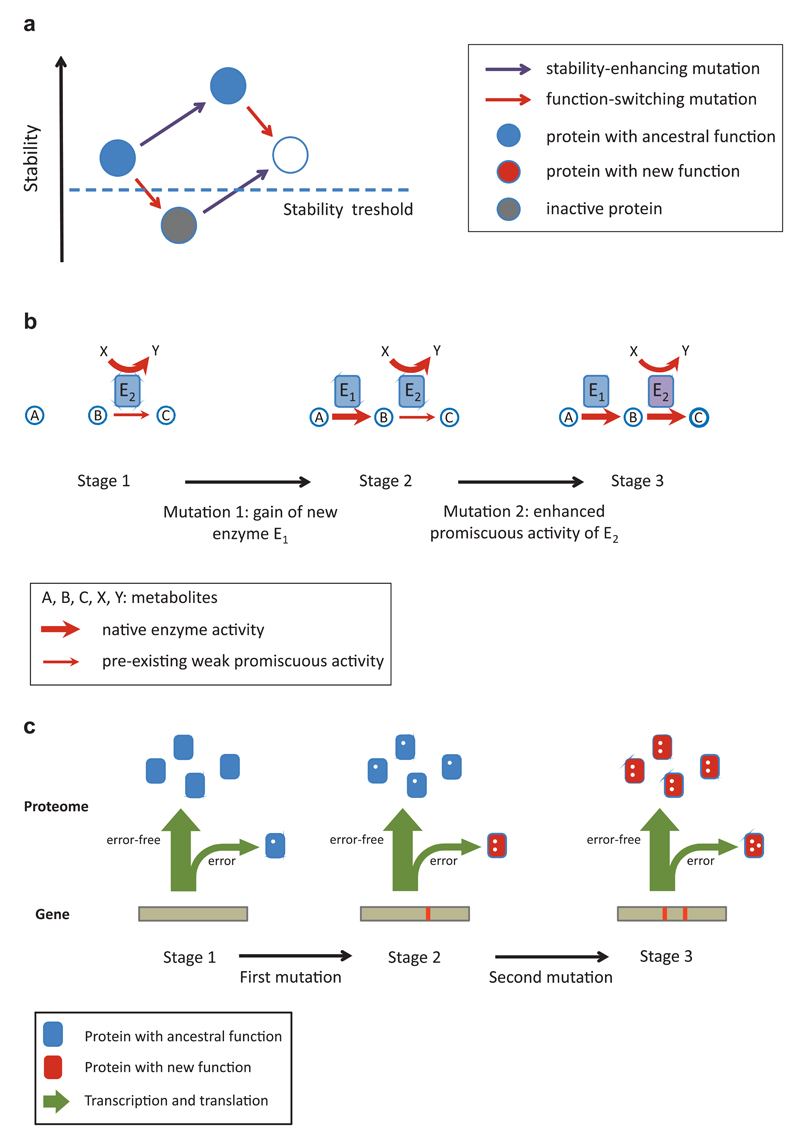
Figure 4. Molecular springboards of complex adaptations.
Molecular mechanisms that potentiate the establishment of complex adaptations by eliminating fitness valleys (A) or opening up more direct mutational paths (B-C). A) Permissive mutations that increase protein stability allow the fixation of function-altering mutations that would otherwise inactivate the protein. Note that the stability-enhancing mutation might not have any fitness effect. B) The presence of low-level enzyme side activities facilitates the adaptive evolution of multi-step metabolic pathways. A two-step pathway with metabolites A-C is depicted where the second metabolic step can be weakly catalyzed by the promiscuous side activity of enzyme E2 (stage 1). Note that E2 has a primary enzymatic activity outside of the pathway of interest. Gain of an enzyme (E1) catalyzing the first reaction immediately confers a fitness advantage as it allows the operation of the pathway, albeit with low activity (stage 2). A second mutation enhances the side activity of E2 and thereby results in a fully functional pathway (stage 3). C) Phenotypic mutations allow selection for intermediate mutations that would otherwise be neutral. The figure depicts a situation where two mutations are required for a novel protein function. Owing to transcriptional and translational errors, a small fraction of the proteome already possesses one of the mutations in a non-heritable form (stage 1). A genotype carrying the other mutation thus has a selective advantage as some of its proteins will carry out the new function (stage 2). A later adaptive genetic mutation provides the full fitness benefit by converting all protein molecules within the cell from the ancestral into the new function (stage 3). Mutations / errors are depicted by white dots.
Gatekeepers of protein stability
A central problem in protein evolution is that mutations that cause functional change simultaneously compromise protein stability, solubility or folding rate. Therefore, the evolution of new molecular functions frequently demands prior fixation of mutations that elevate robustness to such perturbations11. Such permissive mutations can act within a gene or across many genes. Permissive mutations within the same protein alter global biophysical properties of the protein, and thereby mitigate the pleiotropic side effects of function-altering mutations (Figure 4A). They have been observed in the laboratory35,71 and in nature72 as well. Chaperones are often implicated in buffering harmful structure-altering mutations, and thereby facilitate accumulation of neutral mutations and molecular adaptation across the genome73,74. Indeed, overexpression of molecular chaperones mitigates the constraints on protein stability, and thereby improves the specificity and activity of the evolved enzymes75. In principle, both forms of stability-enhancing mechanisms allow the accumulation of a variety of function-changing mutations and thereby have the potential to open new evolutionary paths.
Promiscuous molecular interactions
Enzymatic side reactions, spurious transcription factor – DNA interactions, promiscuous protein-protein interactions arise as inevitable byproducts of infidelity in molecular recognition. They appear to be prevalent in metabolic, protein-protein interactions and transcriptional networks alike76–78. While promiscuous interactions are often weak and physiologically irrelevant, their specificity and strength can be enhanced by further mutations79,80. Given that they exhibit sufficient initial activities, such fortuitous interactions may provide a selective environment for the further emergence of entirely novel molecular interactions and hence facilitate the establishment of multi-step pathways via adaptive walks (Figure 4B). Recent evolution of a novel metabolic pathway for the degradation of a toxic xenobiotic provides indirect evidence for this process81. In addition, promiscuous interactions that initially arose as by-products of evolution are also known to have contributed to the stepwise evolution of specific hormone-receptor interactions82 and have been implicated in the formation of novel regulatory binding sites83. A particularly interesting corollary of the presence of promiscuous interactions concerns the co-evolution of residues participating in protein-protein interactions80. It is generally assumed that a disrupting mutation in one protein drives the selection of a compensatory mutation in its partner during evolution. Alternatively, interacting proteins can coevolve through the generation of promiscuous variants, which serve as mutational intermediates that preserve the ability of the two proteins for functional interaction80.
Phenotypic heterogeneity
Infidelity of biochemical processes pervades multiple levels from single molecules to pathways and cell. It has long been suggested that the resulting phenotypic heterogeneity can be a source of evolutionary adaptations84. A specific model concerns transcriptional and translational infidelity85, and claims that errors speed up the evolution of complex adaptation by allowing selection for intermediate mutations which would otherwise be neutral (Figure 4C). In a nutshell, the model proposes that when two mutations are needed for a novel phenotype, the second “mutation” is first delivered by errors during transcription and translation. Therefore, the single mutant allele will be selectively advantageous, even though it does not yet encode the complete trait. Thus, phenotypic errors allow protein sequences to “look ahead” for a more direct path to a complex trait. The theory is appealing, since translational errors are a thousand-fold more frequent than mutations, thus generating protein variants from non-mutant genes. However, it is important to keep in mind that an elevated translational error rate has a substantial fitness cost, not least because it promotes protein aggregation86,87.The role of mistranslation in protein evolution has recently been studied in the laboratory. The authors showed that phenotypic mutations paved the path to what later, after gene duplication, became newly compartmentalized enzymes84. Thus, gene duplication followed rather than initiated the divergence of this new trait.
Future directions
Complex adaptations occur at several organizational levels. They are abundant in single proteins, protein-protein binding interfaces, metabolic pathways and in transcription factor - binding site interactions. In this review, we argued that despite the apparent functional differences, the governing evolutionary mechanisms in these systems could be similar. Perhaps the most important issue for future studies is to test non-adaptive scenarios and the impact of environmental changes on the origin of complex adaptations in nature. This ambitious goal demands discriminating predictions. Population genetic models indicate that when the population size is large, sequential fixation of non-adaptive intermediate mutations in the population becomes exceedingly unlikely, regardless of the future benefit they may confer in combination with other alleles (Box 1). Accordingly, in large populations, complex adaptations most likely evolve either through the simultaneous fixation of mutational combinations88 or through the sequential fixation of intermediate mutations that confer an advantage in another environmental or genetic context. These two scenarios can be discriminated by the phylogenetic and molecular reconstruction of intermediate steps of evolution and by testing the phenotypic properties of these variants. Pioneering evolutionary studies embarked on this challenge, and showed that permissive mutations in a viral protein had been fixed ahead of the mutation that confers a novel resistance phenotype89. Given the large size of virus populations, this result appears to be inconsistent with the non-adaptive model, and rather suggests that the intermediate mutations themselves were driven by selection.
Are permissive mutations generally subject to neutral or adaptive evolution? As permissive mutations in proteins often alter thermal stability, solubility or folding rate90, they could potentially confer a fitness advantage in specific environments, such as increased temperature or ionizing radiation90. Recent laboratory studies unequivocally demonstrated that antagonistic pleiotropy is prevalent: mutations deleterious in one environment are beneficial in another91. This raises the possibility that mutations with antagonistic effects, rather than neutral mutations contribute to the evolution of complex adaptations. A case study showed that this is indeed a realistic possibility67. Systematic testing of this hypothesis demands exploring mutational effects and epistasis across environments.
We expect conceptual breakthroughs through innovative application of novel molecular techniques. Deep mutational scanning92 and novel genome engineering approaches93,94 offer unprecedented resolutions to map the genotype – fitness landscapes of single proteins and multi-gene subsytems alike. Studying mutational trajectories around adaptive peaks in multiple environments will give insights into the frequency by which varying environments escape adaptive stasis. Moreover, realistic fitness landscapes are highly multidimensional and may therefore contain adaptive by-passes in extra dimensions55. Experimental mapping of high-dimensional landscapes will provide information about the prevalence and dimensionality of such indirect paths.
These considerations have practical implications. Current practices of structure-guided protein engineering focus on mutating active sites to attain novel enzymatic activities95. However, such mutations frequently result in complete loss of function or destabilized protein structure96. Addition of permissive mutations distributed through the protein structure is a prerequisite for new functional specificities. Hence, function-altering mutations should be combined with structurally non-obvious allosteric mutations (see ref 97 for details). We anticipate that evolutionary engineering to obtain a desired function could be facilitated by temporally varying the selection regime. Finally, we note that in computer science, standard genetic algorithms have a tendency to quickly converge to a local solution, and hence frequently fail to identify more promising regions of the search space6. Application of dynamically changing ‘environments’ offers a natural strategy to maintain the diversity required to explore the adaptive surface98.
Acknowledgements
We thank Dan Tawfik for his valuable comments on an earlier version of this paper. This work was supported by the 'Lendület' Programme of the Hungarian Academy of Sciences (BP and CP), The Wellcome Trust (CP and BP), H202-ERC-2014-CoG (CP), GINOP-2.3.2-15-2016-00014 (EVOMER, CP and BP) and GINOP-2.3.2-15-2016-00020 (MolMedEx TUMORDNS, CP).
Footnotes
Author contributions statements
All authors contributed to writing and revising the manuscript.
Competing financial interests statement
The authors declare no competing financial interests.
References
- 1.Smith JM, Burian R, Kauffman S, Alberch P, et al. Developmental constraints and evolution: a perspective from the Mountain Lake conference on development and evolution. Quarterly Review of Biology. 1985:265–287. [Google Scholar]
- 2.Darwin C. The origin of species. London: Murray; 1859. [Google Scholar]
- 3.Fisher RA. The genetical theory of natural selection: a complete variorum edition. Oxford University Press; 1930. [Google Scholar]
- 4.Wright S. Surfaces of selective value revisited. The American Naturalist. 1988;131:115–123. [Google Scholar]
- 5.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banzhaf W, Beslon G, Christensen S, Foster JA, et al. Guidelines: From artificial evolution to computational evolution: a research agenda. Nat Rev Genet. 2006;7:729–735. doi: 10.1038/nrg1921. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner M, Gerhart J. Evolvability. Proc Natl Acad Sci U S A. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parter M, Kashtan N, Alon U. Facilitated variation: how evolution learns from past environments to generalize to new environments. PLoS Comput Biol. 2008;4:e1000206. doi: 10.1371/journal.pcbi.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner GP, Altenberg L. Perspective: complex adaptations and the evolution of evolvability. Evolution. 1996:967–976. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 10.Givnish TJ, Sytsma KJ. Molecular evolution and adaptive radiation. Cambridge University Press; 2000. [DOI] [PubMed] [Google Scholar]
- 11.Harms MJ, Thornton JW. Evolutionary biochemistry: revealing the historical and physical causes of protein properties. Nature Reviews Genetics. 2013;14:559. doi: 10.1038/nrg3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meer MV, Kondrashov AS, Artzy-Randrup Y, Kondrashov FA. Compensatory evolution in mitochondrial tRNAs navigates valleys of low fitness. Nature. 2010;464:279–282. doi: 10.1038/nature08691. [DOI] [PubMed] [Google Scholar]
- 13.Ivankov DN, Finkelstein AV, Kondrashov FA. A structural perspective of compensatory evolution. Curr Opin Struct Biol. 2014;26C:104–112. doi: 10.1016/j.sbi.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch M, Hagner K. Evolutionary meandering of intermolecular interactions along the drift barrier. Proceedings of the National Academy of Sciences. 2015;112:E30–E38. doi: 10.1073/pnas.1421641112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada T, Bork P. Evolution of biomolecular networks: lessons from metabolic and protein interactions. Nat Rev Mol Cell Biol. 2009;10:791–803. doi: 10.1038/nrm2787. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Ochman H. Stepwise formation of the bacterial flagellar system. Proc Natl Acad Sci U S A. 2007;104:7116–7121. doi: 10.1073/pnas.0700266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pallen MJ, Matzke NJ. From The Origin of Species to the origin of bacterial flagella. Nature Reviews Microbiology. 2006;4:784–790. doi: 10.1038/nrmicro1493. [DOI] [PubMed] [Google Scholar]
- 18.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome biology. 2007;8:1. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dietrich MR. From hopeful monsters to homeotic effects: Richard Goldschmidt's integration of development, evolution, and genetics. American Zoologist. 2000;40:738–747. [Google Scholar]
- 20.Theißen G. Saltational evolution: hopeful monsters are here to stay. Theory in Biosciences. 2009;128:43–51. doi: 10.1007/s12064-009-0058-z. [DOI] [PubMed] [Google Scholar]
- 21.Schrider D, Hourmozdi J, Hahn M. Pervasive Multinucleotide Mutational Events in Eukaryotes. Current Biology. 2011;21:1051–1054. doi: 10.1016/j.cub.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nik-Zainal S, Alexandrov L, Wedge D, Van Loo P, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De S, Babu MM. A time-invariant principle of genome evolution. Proceedings of the National Academy of Sciences. 2010;107:200914454. doi: 10.1073/pnas.0914454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–85. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peisajovich SG, Garbarino JE, Wei P, Lim WA. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science. 2010;328:368–372. doi: 10.1126/science.1182376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YX, Perry K, Vinci VA, Powell K, et al. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- 27.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 28.Pál C, Papp B, Lercher MJ. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 29.Sheltzer JM, Amon A. The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends in Genetics. 2011;27:446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rancati G, Pavelka N, Fleharty B, Noll A, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch M. The evolution of genetic networks by non-adaptive processes. Nature Reviews Genetics. 2007;8:803–813. doi: 10.1038/nrg2192. [DOI] [PubMed] [Google Scholar]
- 32.Amitai G, Gupta RD, Tawfik DS. Latent evolutionary potentials under the neutral mutational drift of an enzyme. HFSP J. 2007;1:67–78. doi: 10.2976/1.2739115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wagner A. The origins of evolutionary innovations: a theory of transformative change in living systems. Oxford University Press; 2011. [Google Scholar]
- 34.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474:92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- 35.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. Journal of molecular biology. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Bloom JD, Romero PA, Lu Z, Arnold FH. Neutral genetic drift can alter promiscuous protein functions, potentially aiding functional evolution. Biology Direct. 2007;2:17. doi: 10.1186/1745-6150-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science. 2010;330:920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 38.Lynch M. The evolution of multimeric protein assemblages. Mol Biol Evol. 2012;29:1353–1366. doi: 10.1093/molbev/msr300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson DW, McKeown AN, Thornton JW. Intermolecular epistasis shaped the function and evolution of an ancient transcription factor and its DNA binding sites. Elife. 2015;4:e07864. doi: 10.7554/eLife.07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finnigan GC, Hanson-Smith V, Stevens TH, Thornton JW. Evolution of increased complexity in a molecular machine. Nature. 2012;481:360–364. doi: 10.1038/nature10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrie KL, Joyce GF. Limits of neutral drift: Lessons from the in vitro evolution of two ribozymes. Journal of molecular evolution. 2014;79:75–90. doi: 10.1007/s00239-014-9642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith WS, Hale JR, Neylon C. Applying neutral drift to the directed molecular evolution of a β-glucuronidase into a β-galactosidase: Two different evolutionary pathways lead to the same variant. BMC Research Notes. 2011;4:138. doi: 10.1186/1756-0500-4-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szappanos B, Fritzemeier J, Csörgő B, Lázár V, et al. Adaptive evolution of complex innovations through stepwise metabolic niche expansion. Nature Communications. 2016;7:11607. doi: 10.1038/ncomms11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends in Genetics. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 45.Kuo CH, Ochman H. Deletional bias across the three domains of life. Genome Biol Evol. 2009;1:145–152. doi: 10.1093/gbe/evp016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meléndez-Hevia E, Waddell TG, Cascante M. The puzzle of the Krebs citric acid cycle: Assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. Journal of Molecular Evolution. 1996;43:293–303. doi: 10.1007/BF02338838. [DOI] [PubMed] [Google Scholar]
- 47.Huynen MA, Dandekar T, Bork P. Variation and evolution of the citric-acid cycle: a genomic perspective. Trends in microbiology. 1999;7:281–291. doi: 10.1016/s0966-842x(99)01539-5. [DOI] [PubMed] [Google Scholar]
- 48.Lenski RE, Ofria C, Pennock RT, Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- 49.Notebaart RA, Kensche PR, Huynen MA, Dutilh BE. Asymmetric relationships between proteins shape genome evolution. Genome biology. 2009;10:1. doi: 10.1186/gb-2009-10-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J Mol Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 51.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen HD, Jewett MW, Groisman EA. Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits. PLOS Genet. 2011;7:e1002184. doi: 10.1371/journal.pgen.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Press MO, Queitsch C, Borenstein E. Evolutionary assembly patterns of prokaryotic genomes. Genome Res. 2016;26:826–833. doi: 10.1101/gr.200097.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 55.Wu NC, Dai L, Olson CA, Lloyd-Smith JO, Sun R. Adaptation in protein fitness landscapes is facilitated by indirect paths. Elife. 2016;5 doi: 10.7554/eLife.16965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palmer AC, Toprak E, Baym M, Kim S, et al. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nature Communications. 2015;6:7385. doi: 10.1038/ncomms8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinreich DM, Lan Y, Wylie CS, Heckendorn RB. Should evolutionary geneticists worry about higher-order epistasis? Curr Opin Genet Dev. 2013;23:700–707. doi: 10.1016/j.gde.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mustonen V, Lässig M. From fitness landscapes to seascapes: non-equilibrium dynamics of selection and adaptation. Trends in genetics. 2009;25:111–119. doi: 10.1016/j.tig.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 59.Wagner A. The White-Knight Hypothesis, or Does the Environment Limit Innovations? Trends in Ecology & Evolution. 2016 doi: 10.1016/j.tree.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Kashtan N, Noor E, Alon U. Varying environments can speed up evolution. Proceedings of the National Academy of Sciences. 2007;104:13711–13716. doi: 10.1073/pnas.0611630104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horowitz NH. On the Evolution of Biochemical Syntheses. Proc Natl Acad Sci U S A. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bell G. Fluctuating selection: the perpetual renewal of adaptation in variable environments. Philos Trans R Soc Lond B Biol Sci. 2010;365:87–97. doi: 10.1098/rstb.2009.0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayden EJ, Wagner A. Environmental change exposes beneficial epistatic interactions in a catalytic RNA. Proceedings of the Royal Society of London B: Biological Sciences. 2012;279:3418–3425. doi: 10.1098/rspb.2012.0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taute KM, Gude S, Nghe P, Tans SJ. Evolutionary constraints in variable environments, from proteins to networks. Trends in Genetics. 2014;30:192–198. doi: 10.1016/j.tig.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Bershtein S, Tawfik DS. Ohno's model revisited: measuring the frequency of potentially adaptive mutations under various mutational drifts. Mol Biol Evol. 2008;25:2311–2318. doi: 10.1093/molbev/msn174. [DOI] [PubMed] [Google Scholar]
- 66.Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 67.Steinberg B, Ostermeier M. Environmental changes bridge evolutionary valleys. Science Advances. 2016;2:e1500921–e1500921. doi: 10.1126/sciadv.1500921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Vos MG, Dawid A, Sunderlikova V, Tans SJ. Breaking evolutionary constraint with a tradeoff ratchet. Proc Natl Acad Sci U S A. 2015;112:14906–14911. doi: 10.1073/pnas.1510282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proceedings of the National Academy of Sciences. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lázár V, Pal Singh G, Spohn R, Nagy I, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proceedings of the National Academy of Sciences. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong LI, Suchard MA, Bloom JD. Stability-mediated epistasis constrains the evolution of an influenza protein. Elife. 2013;2:e00631. doi: 10.7554/eLife.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bogumil D, Dagan T. Cumulative impact of chaperone-mediated folding on genome evolution. Biochemistry. 2012;51:9941–9953. doi: 10.1021/bi3013643. [DOI] [PubMed] [Google Scholar]
- 74.Fares MA, Moya A, Barrio E. GroEL and the maintenance of bacterial endosymbiosis. TRENDS in Genetics. 2004;20:413–416. doi: 10.1016/j.tig.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 75.Tokuriki N, Tawfik DS. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature. 2009;459:668–673. doi: 10.1038/nature08009. [DOI] [PubMed] [Google Scholar]
- 76.Copley SD. An evolutionary biochemist's perspective on promiscuity. Trends in Biochemical Sciences. 2015;40:72–78. doi: 10.1016/j.tibs.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Notebaart RA, Szappanos B, Kintses B, Pál F et al. Network-level architecture and the evolutionary potential of underground metabolism. Proc Natl Acad Sci U S A. 2014;111:11762–11767. doi: 10.1073/pnas.1406102111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levy ED, Landry CR, Michnick SW. How perfect can protein interactomes be? Sci Signal. 2009;2 doi: 10.1126/scisignal.260pe11. pe11. [DOI] [PubMed] [Google Scholar]
- 79.Aharoni A, Gaidukov L, Khersonsky O, Gould SMQ, et al. The'evolvability'of promiscuous protein functions. Nature genetics. 2004;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 80.Aakre CD, Herrou J, Phung TN, Perchuk BS, et al. Evolving New Protein-Protein Interaction Specificity through Promiscuous Intermediates. Cell. 2015;163:594–606. doi: 10.1016/j.cell.2015.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Copley SD. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: the patchwork approach. Trends Biochem Sci. 2000;25:261–265. doi: 10.1016/s0968-0004(00)01562-0. [DOI] [PubMed] [Google Scholar]
- 82.Bridgham JT, Carroll SM, Thornton JW. Evolution of Hormone-Receptor Complexity by Molecular Exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 83.Nourmohammad A, Lässig M. Formation of regulatory modules by local sequence duplication. PLoS Comput Biol. 2011;7:e1002167. doi: 10.1371/journal.pcbi.1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yanagida H, Gispan A, Kadouri N, Rozen S, et al. The Evolutionary Potential of Phenotypic Mutations. PLoS Genet. 2015;11:e1005445. doi: 10.1371/journal.pgen.1005445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E. The look-ahead effect of phenotypic mutations. Biol Direct. 2008;3:18. doi: 10.1186/1745-6150-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalapis D, Bezerra AR, Farkas Z, Horvath P et al. Evolution of Robustness to Protein Mistranslation by Accelerated Protein Turnover. PLoS Biol. 2015;13:e1002291. doi: 10.1371/journal.pbio.1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pouplana LR, Santos MA, Zhu JH, Farabaugh PJ, Javid B. Protein mistranslation: friend or foe? Trends Biochem Sci. 2014;39:355–362. doi: 10.1016/j.tibs.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 88.Weinreich DM, Chao L. Rapid Evolutionary Escape by Large Populations from Local Fitness Peaks Is Likely in Nature. Evolution. 2005;59:1175–1182. [PubMed] [Google Scholar]
- 89.Bloom JD, Gong LI, Baltimore D. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Trudeau DL, Kaltenbach M, Tawfik DS. On the Potential Origins of the High Stability of Reconstructed Ancestral Proteins. Molecular Biology and Evolution. 2016 doi: 10.1093/molbev/msw138. [DOI] [PubMed] [Google Scholar]
- 91.Qian W, Ma D, Xiao C, Wang Z, Zhang J. The genomic landscape and evolutionary resolution of antagonistic pleiotropy in yeast. Cell Rep. 2012;2:1399–1410. doi: 10.1016/j.celrep.2012.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fowler DM, Fields S. Deep mutational scanning: a new style of protein science. Nat Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nyerges Á, Csörgő B, Nagy I, Bálint B et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc Natl Acad Sci U S A. 2016;113:2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lutz S. Beyond directed evolutionsemi-rational protein engineering and design. Current opinion in biotechnology. 2010;21:734–743. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Current Opinion in Structural Biology. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 97.Goldsmith M, Tawfik DS. Enzyme engineering by targeted libraries. Methods Enzymol. 2013;523:257–283. doi: 10.1016/B978-0-12-394292-0.00012-6. [DOI] [PubMed] [Google Scholar]
- 98.O’Neill M, Vanneschi L, Gustafson S, Banzhaf W. Open issues in genetic programming. Genetic Programming and Evolvable Machines. 2010;11:339–363. [Google Scholar]
- 99.Coyne JA, Barton NH, Turelli M. Perspective: a critique of Sewall Wright's shifting balance theory of evolution. Evolution. 1997:643–671. doi: 10.1111/j.1558-5646.1997.tb03650.x. [DOI] [PubMed] [Google Scholar]
- 100.Orr HA. The genetic theory of adaptation: a brief history. Nat Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 101.Doniger SW, Kim HS, Swain D, Corcuera D, et al. A catalog of neutral and deleterious polymorphism in yeast. PLoS genetics. 2008;4:e1000183. doi: 10.1371/journal.pgen.1000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henn BM, Botigué LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nature Reviews Genetics. 2015;16:333–343. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.MacArthur DG, Balasubramanian S, Frankish A, Huang N, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lynch M, Abegg A. The rate of establishment of complex adaptations. Mol Biol Evol. 2010;27:1404–1414. doi: 10.1093/molbev/msq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lynch M. Scaling expectations for the time to establishment of complex adaptations. Proc Natl Acad Sci U S A. 2010;107:16577–16582. doi: 10.1073/pnas.1010836107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weissman DB, Feldman MW, Fisher DS. The rate of fitness-valley crossing in sexual populations. Genetics. 2010;186:1389–1410. doi: 10.1534/genetics.110.123240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ramani AK, Marcotte EM. Exploiting the co-evolution of interacting proteins to discover interaction specificity. Journal of molecular biology. 2003;327:273–284. doi: 10.1016/s0022-2836(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 108.Podgornaia AI, Laub MT. Pervasive degeneracy and epistasis in a protein-protein interface. Science. 2015;347:673–677. doi: 10.1126/science.1257360. [DOI] [PubMed] [Google Scholar]
- 109.Bridgham JT, Ortlund EA, Thornton JW. An epistatic ratchet constrains the direction of glucocorticoid receptor evolution. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Papp B, Notebaart RA, Pál C. Systems-biology approaches for predicting genomic evolution. Nat Rev Genet. 2011;12:591–602. doi: 10.1038/nrg3033. [DOI] [PubMed] [Google Scholar]