Silver nanoparticles inhibit nitrification but enhance nitrous oxide production by nitrifying organisms.
Abstract
Silver nanoparticles (AgNPs) are the most common materials in nanotechnology-based consumer products globally. Because of the wide application of AgNPs, their potential environmental impact is currently a highly topical focus of concern. Nitrification is one of the processes in the nitrogen cycle most susceptible to AgNPs but the specific effects of AgNPs on nitrification in aquatic environments are not well understood. We report the influence of AgNPs on nitrification and associated nitrous oxide (N2O) production in estuarine sediments. AgNPs inhibited nitrification rates, which decreased exponentially with increasing AgNP concentrations. The response of nitrifier N2O production to AgNPs exhibited low-dose stimulation (<534, 1476, and 2473 μg liter−1 for 10-, 30-, and 100-nm AgNPs, respectively) and high-dose inhibition (hormesis effect). Compared with controls, N2O production could be enhanced by >100% at low doses of AgNPs. This result was confirmed by metatranscriptome studies showing up-regulation of nitric oxide reductase (norQ) gene expression in the low-dose treatment. Isotopomer analysis revealed that hydroxylamine oxidation was the main N2O production pathway, and its contribution to N2O emission was enhanced when exposed to low-dose AgNPs. This study highlights the molecular underpinnings of the effects of AgNPs on nitrification activity and demonstrates that the release of AgNPs into the environment should be controlled because they interfere with nitrifying communities and stimulate N2O emission.
INTRODUCTION
Nanotechnology has advanced rapidly over the past few decades (1, 2). Large quantities of nanomaterials are being discharged into the environment because of the production and widespread application of nanotechnology-based consumer products. These nanomaterials trigger potential human health and eco-environmental problems (3). Nanoparticles with high surface/volume ratios and increased catalytic properties are more reactive than their bulk counterparts and therefore could be more toxic. Nanoparticles can also serve as carriers for dangerous contaminants and can thus promote their translocation in the environment. Silver nanoparticles (AgNPs) are commonly used nanomaterials because of their broad-spectrum antimicrobial activity (4). They are used in products such as laundry additives, personal care products, children’s toys, textiles, and medical products, which when combined account for approximately 25% of all nanotechnology-enhanced products (4, 5). In general, AgNPs are immobilized with capping agents, which are usually negatively charged species or relatively large, hydrophilic polymers, such as citrate, dextran, gum arabic, polyethylene glycol, and polyvinylpyrrolidone (PVP) (6). Among these, PVP is an attractive and commonly used capping reagent that can stabilize and protect AgNPs by steric stabilization (7). Stabilized AgNPs can be exposed to the environment through a variety of routes including during their manufacture, use, recycling, and disposal (8). Recently, the rapid increase in AgNP usage has raised extensive public concern regarding potential environmental effects (9), especially the unintended impacts of AgNPs on microbial ecosystems and associated ecological risks.
The global production of AgNPs ranges from 5.5 to 550 metric tons per year (10). However, annual production is predicted to increase to 1216 metric tons by 2020 (5). The environmental concentration of AgNPs in surface waters is estimated to be approximately 0.04 to 0.32 μg liter−1, with a predicted sedimentation rate of approximately 2 to 14 mg kg−1 a−1 (11). By 2020, concentrations of AgNPs in surface water and sediment may reach approximately 0.2 to 1.8 μg liter−1 and 87.8 to 614.4 mg kg−1, respectively (5). AgNPs have been shown to readily undergo sulfidation and transform into highly insoluble Ag2S during their transport, especially in sewage disposal systems (12–15). However, part of the released AgNPs cannot be fully transformed into Ag2S before they exert toxicity in the environment, depending on particle size, aggregation level, and concentration of the released AgNPs (16). Notably, silver exists in the natural environment in various species, including Ag2S, AgCl, Ag+, and particulate silver (8). It has been demonstrated that AgNPs could be substantially reformed from Ag2S in the presence of Fe(III) under sunlit conditions in natural aquatic environments (17). In addition, Ag+ could be photochemically reduced to AgNPs by dissolved organic matter (DOM) in environmental waters exposed to sunlight (8). AgCl has also been demonstrated to transform into AgNPs under irradiation (18). Because Fe(III) and DOM are ubiquitous in natural aquatic ecosystems, the process of reformation of AgNPs from Ag2S and Ag+ might also be ubiquitous, and thus increase the persistence of AgNPs in natural aquatic environments, such as rivers and estuaries (19, 20). Therefore, with the constant increase of AgNP usage, the risk associated with AgNPs in aquatic environments will also increase, as will the need for corresponding ecotoxicological assessments. The toxic effects of AgNPs have been suggested to stem mainly from dissolution and release of silver ions (Ag+), reactive oxygen species (ROS) generation, endocrine disruption, and cell injury and inflammation (5, 21–23). Although toxicity mechanisms are being explored, little is known about the effects of AgNPs on nitrification rates and the associated production of N2O, which is a greenhouse gas, in aquatic environments. Nitrification is a critical process for the balance of reduced and oxidized nitrogen pools in the environment, linking mineralization to nitrogen loss pathways of denitrification and anaerobic ammonium oxidation (anammox) (24–26). The slow growth of nitrifiers and their high sensitivity to environmental perturbations often result in cell growth inhibition by toxicants, including AgNPs (4). Nitrification is an important pathway for N2O production in the environment (27, 28). During nitrification, N2O can be produced either as a by-product of hydroxylamine (NH2OH) oxidation (NH2OH→NOH→N2O) or as an end product or intermediate product of nitrite (NO2−) reduction via nitrifier denitrification (29, 30). N2O has a >300-fold stronger effect on climate warming than carbon dioxide (CO2) and can react with ozone in the stratosphere (30, 31). Stress from low oxygen can induce nitrifier N2O production, and we hypothesize that stress from AgNP exposure can also exacerbate nitrifier N2O production.
Estuaries, especially large-river deltaic systems (for example, the Yangtze Estuary), are dynamic regions where rivers, lands, and oceans interact, and they are among the most productive ecosystems (32). Estuarine ecosystems have long been disturbed by human activities due to their ability to support large human populations (32). In addition to receiving large quantities of AgNPs (19, 20), estuaries have also suffered from a substantial loading of anthropogenic nitrogen (33), which has threatened their overall quality and function (34). Hence, a deeper understanding of how AgNPs affect key nitrogen transformations in estuarine environments is required. Here, we examine the toxicity of PVP-coated AgNPs on the activity of nitrifying communities from the Yangtze estuarine sediments and the effects of AgNPs on associated N2O production during nitrification. Isotopomer analyses were conducted to identify the pathway of N2O production affected most by AgNPs. Metatranscriptomes were analyzed to elucidate the metabolic response of nitrifying organisms by tracking the expression of AgNP-responsive genes. This research provides a new view of the AgNP toxicity mechanism affecting nitrification in estuarine environments.
RESULTS AND DISCUSSION
Effect of AgNPs on nitrification rate
The toxicity of AgNPs to the activity of nitrifying communities from the intertidal sediments of the Yangtze Estuary was examined via a sediment slurry incubation experiment (fig. S1). Three different sizes (10, 30, and 100 nm) of PVP-coated AgNPs were chosen. The toxicity of Ag+ (in the form of AgNO3) was also investigated to compare its toxicity with that of AgNPs. Within 30 hours of acute exposure, the nitrification inhibition level increased gradually for the first 12 hours (fig. S2) and then remained relatively stable, probably due to nearly saturated silver toxicity. Thus, dose-response curves were constructed by plotting the measured nitrification inhibition level against AgNP exposure concentrations over 12 hours (Fig. 1). Nitrification inhibition increased exponentially as the AgNP dose increased (P < 0.01). On the basis of the dose-response curves, we determined AgNP concentrations that produced 10% (EC10 or effective concentration) and 50% (EC50) reductions in the nitrification rate relative to the control. The toxicity of AgNPs to nitrification activity was size-dependent, with EC10 values of 32, 145, and 502 μg liter−1 and EC50 values of 421, 1775, and 6020 μg liter−1 for the 10-, 30-, and 100-nm AgNPs, respectively (Fig. 1). The increased toxicity of the smaller AgNPs was perhaps caused by increased contact between cell membranes and the surface of the smaller particles or indirectly caused by the increased dissolution of Ag+ from the smaller AgNPs due to their greater surface area–to–volume ratio (35).
Fig. 1. Percentage reduction of nitrification rate in AgNP or Ag+ treatments compared to the no-silver control (incubation time = 12 hours; n = 3).
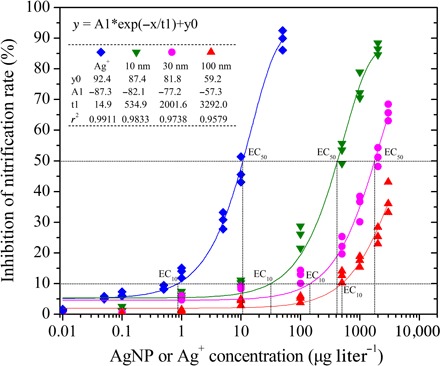
EC10 and EC50 represent the concentrations that produced a 10 or 50% reduction in nitrification rate relative to the control, respectively. Nonlinear fitted curves (ExpDec1) and equations are given (P < 0.01).
The mechanisms by which AgNPs exert their toxicity are debatable (36). Recently, the direct particle-specific effect of AgNPs was ruled out because no toxicity was observed when they were synthesized and tested under anaerobic conditions where Ag+ release was precluded (36). AgNP morphological properties—such as particle size, shape, and capping agent—might still be indirect effectors influencing Ag dissolution (22, 36). This point is confirmed in this study, wherein AgNP toxicity to nitrification correlated significantly with dissolved Ag+ concentration (P < 0.01) (fig. S3 and table S1). Dissolved Ag+ exerted toxicity via interaction with the thiol groups of the cysteine residues in respiratory chain enzymes, such as NADH (reduced form of NAD+) dehydrogenase, which decoupled respiration and adenosine 5′-triphosphate (ATP) synthesis (22, 37). In addition, Ag+ may bind with transport proteins, leading to proton leakage and proton motive force collapse (37, 38).
AgNO3 (as a source of dissolved Ag+) exhibited a greater inhibitory effect on nitrification than AgNPs, with EC10 and EC50 values of 1 and 11 μg liter−1, respectively (Fig. 1). However, the Ag+ dissolved from AgNPs was more toxic than the same concentration of the Ag+ present as AgNO3 (Fig. 1 and fig. S3). This result may indicate that AgNPs deliver Ag+ more effectively to nitrifiers because they are less susceptible to binding and reduced bioavailability by common natural ligands, such as chloride, sulfide, thiosulfate, and dissolved organic carbon, compared with AgNO3 (36, 39). During the AgNP inhibition experiment, no NO2− accumulation (always below the detection limit) was detected in the sediment slurries, supporting the idea that AgNPs inhibit ammonia (NH3) oxidation more than NO2− oxidation (4). The PVP capping agent (less than 0.5% coating on AgNPs by weight) may contribute to the toxicity of AgNPs. However, on the basis of the independent inhibition experiment conducted for PVP, less than a 1.5% reduction in the nitrification rate, accounting for only 0 to 5% of the AgNP toxicity, resulted from exposure to PVP (0.05 to 15 μg liter−1) for 12 hours (fig. S4).
Effect of AgNPs on N2O production
The increasing release and atmospheric accumulation of the powerful greenhouse gas N2O have caused concern during this era of rapid environmental change (27, 34). N2O production during nitrification was affected by AgNP exposure (figs. S5 and S6). For 10-nm AgNPs, although concentrations below 0.1 μg liter−1 apparently did not affect the N2O emission (Student’s t test, P > 0.05), those between 1 and 10 μg liter−1 stimulated N2O emission by up to 17.6% during the 30-hour incubation (Student’s t test, P < 0.05). When exposed to higher concentrations (100 to 500 μg liter−1), an initial inhibition (up to 23.5%) of N2O emission occurred (Student’s t test, P < 0.05) but switched to stimulation (up to 60.8%) after 12 hours of incubation (Student’s t test, P < 0.05). However, when concentrations were increased to 1000 to 2000 μg liter−1, inhibition of up to 90.0% was always observed (Student’s t test, P < 0.01). Similar results were recorded for the effects of increasing dosages of 30- and 100-nm AgNPs and Ag+ (provided as AgNO3) on nitrifier N2O production (fig. S6). Thus, nitrifying communities respond rapidly to AgNPs or Ag+ exposure, probably by regulating gene transcription and adjusting metabolic pathways. One important response was the regulation of the by-product N2O emission.
Dose-response curves were constructed between Ag exposure concentration and N2O emission over 12 hours (Fig. 2). Although nitrification rates decreased exponentially as the AgNP or Ag+ dose increased (P < 0.01) (Fig. 1), the dose-response relationship between Ag concentration and N2O emission showed both low-dose stimulation and high-dose inhibition [that is, hormesis (40)] (Fig. 2 and fig. S7). N2O emission increased with the increasing Ag concentrations, reaching maximum increases of 71.8, 70.9, 97.2, and 125.3% under Ag concentrations of 5.2 μg liter−1 for Ag+, and 257, 713, and 1173 μg liter−1 for 10-, 30-, and 100-nm AgNPs, respectively, based on the constructed curves (Fig. 2). The stimulation of N2O production due to Ag addition might be a stress response, as is observed under low-O2 conditions (41). However, with increasing Ag concentrations, the degree to which N2O emission was stimulated was initially reduced, after which the emission of N2O became inhibited compared with the no-silver control, showing that the damage from Ag to nitrifying cells increased until it was strong enough to counteract the stimulated N2O production. This observed hermetic effect of AgNPs on N2O production during nitrification has not been reported previously. This finding is globally significant as environmental inputs of AgNPs have been increasing exponentially due to their rising usage and disposal levels worldwide (20, 42, 43).
Fig. 2. Effect of AgNPs or Ag+ on N2O emission during nitrification (incubation time = 12 hours).
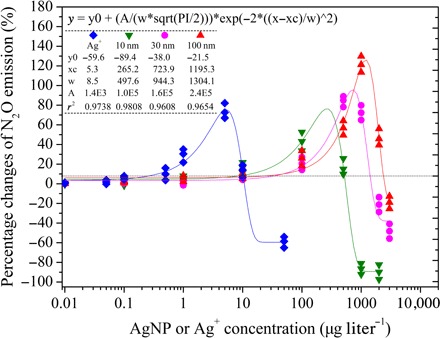
Data show the percentage changes of N2O emission in the AgNP or Ag+ treatments compared to the no-silver control (n = 3). Nonlinear fitted curves (Gauss) and equations are given (P < 0.01).
Although low dissolved oxygen (DO) conditions (<3 mg liter−1) enhance N2O emissions, mainly through the nitrifier denitrification (41), the DO concentration in this study ranged from 6.3 to 9.1 mg liter−1 (fig. S8). The DO concentration was significantly lower in the no-silver control (6.3 to 6.5 mg liter−1) than in the AgNP-treated samples where N2O production was greatly enhanced (Student’s t test, P < 0.01). Therefore, the simulated N2O emission due to the AgNP treatments was likely not caused by DO stress. High NO2− concentrations can promote nitrifier denitrification and N2O production (44). However, NO2− concentrations remained below the detection limit during the incubation, indicating that NO2− stress did not enhance N2O production. The possibility that the PVP capping agent might contribute to increased N2O emission was excluded, as the impact of PVP (0.05 to 15 μg liter−1) on N2O emission remained at only 0.1 to 1.2% of inhibition (fig. S4).
Identification of key N2O production pathways
As shown here, increasing AgNP concentrations in estuarine environments can enhance N2O emissions. However, the mechanisms by which AgNPs stimulate N2O production are still unclear. N2O isotopomer analysis is a powerful tool to distinguish whether N2O originates from NH2OH oxidation or NO2− reduction (30). It is based on the intramolecular distribution of 15N in the central position (14N15N16O) and the end position (15N14N16O) of asymmetric N2O molecules (45). The 15N-site preference (SP) is defined as the difference in the bulk nitrogen isotope ratios of N2O between δ15Nα and δ15Nβ, where 15Nα and 15Nβ represent the 15N/14N ratios at the central (α) and end (β) sites of the nitrogen atoms, respectively (45). Because N2O produced through NH2OH oxidation and NO2− reduction have different SP values [33‰ (per mil) for NH2OH oxidation and 0‰ for NO2− reduction (46, 47)], analyzing the SP enables the identification of the sources of N2O produced during these two respective processes.
The isotopomer analysis showed that SP in the no-silver control was 29.3‰ at 12 hours, indicating that approximately 89% of the released N2O was produced via NH2OH oxidation, whereas only approximately 11% was attributed to NO2− reduction (Fig. 3, A and B), assuming that each process is linearly proportional to SP (47). When exposed to AgNPs (100 μg liter−1, 10 nm; 500 μg liter−1, 30 nm; or 1000 μg liter−1, 100 nm), under which N2O production was enhanced by 43.0, 84.1, and 121.5%, the SP values increased to 30.0, 31.5, and 35.6‰, respectively, at 12 hours (Student’s t test, P < 0.05) (Fig. 3A). In addition, SP increased to 32.2‰ when exposed to Ag+ (5 μg liter−1) (Student’s t test, P < 0.01), exhibiting a 73.9% N2O enhancement. Under low-dose AgNP exposure, δ15Nα increased, whereas δ15Nβ simultaneously decreased (Fig. 3, C and D), thus resulting in higher SP. The increased SP indicated that low-dose AgNPs stimulated NH2OH oxidation, which contributed to more than 90% of N2O emission or even became the sole pathway, whereas the contribution of NO2− reduction was minor or nil (Fig. 3B).
Fig. 3. Identification of key N2O production pathways.
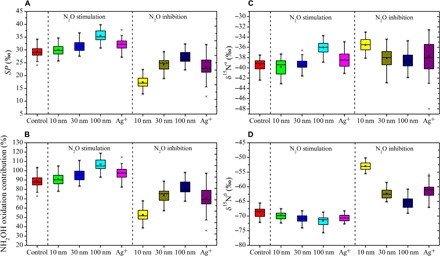
(A) SP values. (B) Contribution of NH2OH oxidation pathway to N2O emission. (C) Isotopomer ratios at the central site of N2O. (D) Isotopomer ratios at the end site of N2O. Horizontal lines indicate the median, five-point stars show the mean, asterisks indicate outlier, the boxes give the 25th and 75th percentiles, and whiskers show range from the 5th to 95th percentile. Control group represents the incubation without silver. The 10 nm, 30 nm, 100 nm, and Ag+ in the “N2O stimulation” area represent the incubations with AgNPs (100 μg liter−1, 10 nm; 500 μg liter−1, 30 nm; and 1000 μg liter−1, 100 nm) and Ag+ (5 μg liter−1), wherein 43.0, 84.1, 121.5, and 73.9% of N2O emission enhancement were detected, respectively. The 10 nm, 30 nm, 100 nm, and Ag+ in the “N2O inhibition” area represent the incubations with AgNPs (2000 μg liter−1, 10 nm; 3000 μg liter−1, 30 nm; and 3000 μg liter−1, 100 nm) and Ag+ (500 μg liter−1), wherein 89.9, 48.3, 19.0, and 59.3% of N2O emission inhibition were detected, respectively. The incubation time was 12 hours.
The inhibition mechanism of high AgNP concentrations on N2O production during nitrification was investigated. When exposed to AgNPs (2000 μg liter−1 10 nm, 3000 μg liter−1 30 nm, and 100 nm) or Ag+ (50 μg liter−1), wherein N2O production decreased by 89.9, 48.3, 19.0, and 59.2%, respectively, SP decreased to 17.4, 24.2, 26.9, and 23.3‰, respectively, at 12 hours (Student’s t test, P < 0.01) (Fig. 3A). These results imply that, although NH2OH oxidation remained the main contributor of N2O, its contribution was reduced under high-dose Ag exposure, which was reduced to approximately 53% when the N2O emission was inhibited by 89.9% (Fig. 3B). Therefore, the regulation of N2O production by AgNPs during nitrification occurred via affecting NH2OH oxidation, whereas the production of N2O via NO2− reduction was minor and insensitive to AgNP exposure. Previously, N2O reduction to N2 was reported to increase SP, and the effect of NO2− reduction might be underestimated when N2O reduction is intense (48). However, no significant increase in the δ18O of N2O was detected during the incubation (it remained approximately 20‰, Student’s t test, P > 0.05), showing that N2O reduction to N2 was not important (47). This study is the first to explore and distinguish the effect of AgNPs on different N2O production pathways of nitrifying communities.
Although NO2− reduction was not the main contributor to N2O production in this study, N2O isotopomer analysis does not distinguish the relative contributions of nitrifier denitrification and heterotrophic denitrification (30). When the headspace O2 concentration is between 0.5 and 3% (v/v), at least 50% of the total N2O production generally derives from the NH3 oxidation pathway rather than heterotrophic denitrification (41). In addition, when the dissolved O2 concentration is ca. 0.06 mg liter−1, the N2O production by denitrification is completely inhibited (49). Therefore, considering that the dissolved O2 concentration remained above 6.3 mg liter−1, NO2− reduction via heterotrophic denitrifiers is likely to be minor (fig. S8). This conclusion was supported further by the metatranscriptomic analyses (see below).
Transcriptional response to AgNP exposure
AgNP toxicity may cause intracellular metabolic disturbances. To explore the impacts of AgNPs on nitrogen metabolic pathways in nitrifying communities, we conducted metatranscriptomic analyses. Nitrifying communities are important in the global nitrogen cycle, but they account for only a small fraction of the complex microbial populations in estuarine and coastal environments (50–52). The complexity of sediment communities complicates exploring toxic AgNP stress mechanisms and monitoring the transcriptional response of nitrifying populations to AgNP exposure. Therefore, a nitrifying bioreactor was set up with fresh sediments from intertidal flats of the Yangtze Estuary to enrich nitrifiers. The continuously operated bioreactor achieved a steady state after approximately half a month, characterized by complete NH4+ removal (fig. S9A) and the dominance of nitrifying populations (>45%; figs. S9B and S10). Nitrifying enrichment is a representative culture to investigate the transcriptional response of nitrifiers in estuarine environment to AgNP exposure, as the in situ nitrifying populations were all enriched herein (fig. S10). Metatranscriptomes were then obtained from the AgNP (500 μg liter−1 30 nm)–treated samples (12 hours of incubation), in which a 25% nitrification inhibition (Student’s t test, P < 0.01) and a hormetic stimulation (80%) on N2O production (Student’s t test, P < 0.01) were observed. Metatranscriptome studies showed that community compositions were not significantly affected by AgNP exposure during the 12-hour incubation period, and nitrifiers accounted for 51.7 and 47.7% of the total organisms in the AgNP-treated and no-silver control samples, respectively (fig. S11).
Metatranscriptomic analyses indicated that the expression of known nitrification-associated genes varied between the AgNP-exposure and control groups (Fig. 4, A and B). The expression of the potentially active subunit of the ammonia monooxygenase gene (amoA) was down-regulated by 1.4-fold when exposed to 500 μg liter−1 of 30-nm AgNPs for 12 hours, consistent with the nitrification inhibition (25%). Real-time quantitative polymerase chain reaction (qPCR) based on RNA samples also showed that amoA expression was down-regulated under AgNP exposure (1.5-fold; Student’s t test, P < 0.01) (table S3). In addition, expression of amoB [recently suggested as a catalytic subunit (53)] decreased by 1.2-fold (Student’s t test, P < 0.05), whereas the transcript level of amoC remained unchanged (Student’s t test, P > 0.05), after exposure to AgNPs (Fig. 4, A and B, and table S3). Posttranscriptional disturbance might occur, because ammonia monooxygenase (amo) uses a copper metal center. Dissolved Ag+ from AgNPs may interfere with enzyme activity by substituting copper with Ag+ or inducing the production of chelators, both of which could deactivate amo (22, 54). The amo transcripts of the recently discovered comammox bacteria were also discovered (fig. S13), which exhibit up to 99% similarity with the amoABC sequences of comammox Nitrospira (55, 56). However, these genes only showed less than 80% similarity with those of known AOB (fig. S13), and their expression accounted for approximately 45 and 60% of amo transcripts in the AgNP treatment and no-silver control samples, respectively. These results suggest that comammox bacteria are present, possibly playing an important role in NH3 oxidation in estuarine sediments.
Fig. 4. Response of nitrifying communities to AgNP exposure.
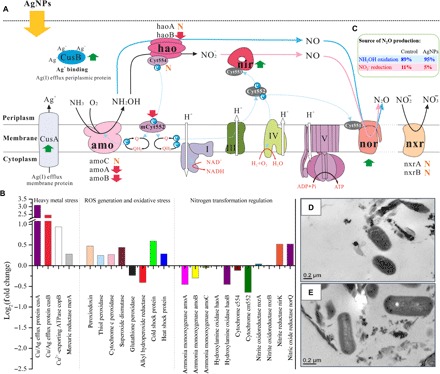
(A) Schematic model depicting the effects of AgNPs on the expression of gene families involved in nitrification. N2O can be produced through NO2− reduction (the bold pink arrows) or incomplete NH2OH oxidation (the bold blue arrows). Upward green arrows indicate that the gene expressions were up-regulated when exposed to AgNPs, the downward red arrows indicate that the gene expressions were down-regulated, and “N” denotes that gene expression was not affected by AgNP exposure. CusA, Cu(I)/Ag(I) efflux system membrane protein cusA; CusB, Cu(I)/Ag(I) efflux system periplasmic protein cusB; amo, ammonia monooxygenase; hao, hydroxylamine oxidase; Cyt554, cytochrome c554; mCyt552, cytochrome cm552; nir, nitrite reductase (NO-forming) nirK; nor, nitric oxide reductase norQ; nxr, nitrite oxidoreductase; Cyt551, cytochrome c551; Cyt552, cytochrome c552; Cyt553, cytochrome c553; Q, ubiquinone; QH2, ubiquinol. The roman numbers refer to the enzyme complex I (NADH-ubiquinone reductase), complex III (ubiquinol-cytochrome c reductase), complex IV (cytochrome c oxidase), and complex V (F-type ATPase) in the respiratory chain (related gene expression regulations by AgNPs are shown in fig. S12). Colored proteins were detected in the cDNA libraries, whereas those in dark gray were not identified but are included in the model of electron transport for reference (74). Dotted blue arrows show the movement of electrons, and white arrows show movement of protons. The membrane was broken by dotted line, as nitrite oxidation did not often occur in the same organism with ammonia oxidation, with the exception of recently discovered comammox Nitrospira (55, 56). (B) Fold change (FC) of transcripts encoding proteins involved in heavy metal stress response, oxidative stress release, and the nitrogen transformation process of nitrifying organisms when exposed to 30-nm AgNP (500 μg liter−1) for 12 hours. FC in relative gene expression was calculated by comparing AgNP-treated samples to the no-silver control. Gene expression levels were calculated on the basis of FPKM. (C) Contribution of different pathways to N2O emission in the no-silver control and the 30-nm AgNP (500 μg liter−1) treatment. (D) TEM image of the no-silver control at 12 hours. (E) TEM image of the 30-nm AgNP (500 μg liter−1) treatment at 12 hours. No apparent physical damage to the cell surface was observed.
AgNPs exhibited variable impacts on the expression of genes in the hydroxylamine oxidase (hao) gene cluster (haoAB-cycAB) (Fig. 4, A and B). The transcript level of haoA (encoding hao) and cycA (encoding electron transfer protein cytochrome c554) remained unchanged (Student’s t test, P > 0.05), whereas expressions of haoB (encoding the putative membrane anchor protein haoB) and cycB (encoding quinone reducing cytochrome cm552) were down-regulated by 1.4- and 1.6-fold (Student’s t test, P < 0.05), respectively (Fig. 4, A and B, and table S3). The cytochrome c554 is believed to function as the physiological electron acceptor of hao, and then, electrons from cytochrome c554 may traverse the membrane-anchored tetraheme cytochrome cm552 to ubiquinone and then to amo, during which ATP is synthesized (Fig. 4A) (57). Cytochrome c554 can also function as an enzyme with significant nitric oxide (NO) reductase activity (57). However, this function was not confirmed at the transcriptional level, as cycA expression remained unchanged (Student’s t test, P > 0.05) when N2O production was significantly enhanced (75.1%). The expressions of nitrite oxidoreductase genes nxrA and nxrB were not affected by 30-nm AgNP (500 μg liter−1) exposure (Fig. 4, A and B), consistent with the qPCR results (Student’s t test, P > 0.05) (table S3).
When exposed to AgNPs, the expression of the nitrite reductase (NO-forming, nirK) gene in nitrifiers was up-regulated by 1.4-fold (Student’s t test, P < 0.05) (Fig. 4, A and B, and table S3), showing that nitrifier denitrification might be enhanced. However, isotopomer analysis showed that the N2O production via NO2− reduction and the contribution of NO2− reduction to total N2O production were reduced under 30-nm AgNP (500 μg liter−1) exposure (Fig. 4C). It is probable that, under AgNP exposure, NO formed from NO2− reduction was released into the surrounding environment, thus being oxidized rather than reduced to N2O (58, 59). In addition, 94 and 92% of nitrite reductase transcripts were affiliated with nitrifiers in the AgNP treatment and the no-silver control samples, respectively, further suggesting that NO2− reduction via heterotrophic denitrifiers was negligible. Transcription of nitrifying bacterial nitric oxide reductase (norQ) increased 1.4-fold in the AgNP treatment, consistent with N2O production, although the nitric oxide reductase norB and norC genes were not observed. qPCR confirmed that the norQ expression was significantly up-regulated under AgNP exposure (Student’s t test, P < 0.01) (table S3). Alternative enzymes might have also contributed to nitrifier N2O production, such as cytochrome c′-β (encoded by cytS) (60) and cytochrome P460 (61), but expressions of these genes were not observed in this study.
Transcripts encoding Cu(I)/Ag(I) efflux membrane protein (cusA), Cu(I)/Ag(I) efflux periplasmic protein (cusB), and Cu2+-exporting adenosine triphosphatase (ATPase) (copB) were up-regulated by 8.6-, 5.5-, and 1.9-fold, respectively, under AgNP exposure (Fig. 4, A and B). Ag and Cu have similar coordination chemistries and can be treated interchangeably (62). Thus, the up-regulation of these genes (Student’s t test, P < 0.01; table S3) indicated an urgent demand for transporting Ag+, as no extra Cu was added, from the cytoplasm to the exterior of the nitrifying cells. This result also suggests that nitrifiers might have been invaded by AgNPs or released Ag+, although no apparent physical cell damage was observed via transmission electron microscopy (TEM) (Fig. 4, D and E). Transcripts encoding mercuric reductase [merA, converting toxic heavy metal ions into their relatively inert elemental forms (63)] were up-regulated by 1.2-fold in the AgNP treatment (Student’s t test, P < 0.05) (Fig. 4B and table S3). These results imply that the nitrifying cells were under AgNP stress, and the enzymes discussed above might help them cope with this stress.
Transcriptional response to oxidative stress was detected (Fig. 4B). Transcripts encoding superoxide dismutase (SOD2, Fe-Mn family), peroxiredoxin (BCP), thiol peroxidase (TPX), and cytochrome c peroxidase (CCP), all of which convert superoxides and hydroperoxides into innocuous products, were up-regulated by 1.4-, 1.4-, 1.2-, and 1.2-fold, respectively. These genes were up-regulated (Student’s t test, P < 0.05; table S3) to detect and defend the cell from oxidative damage. Thus, we speculate that ROS generation and subsequent oxidative damage to nitrifying cells occurred during the AgNP treatment. Transcripts encoding heat shock protein and cold shock protein were up-regulated by 1.2- and 1.5-fold, respectively, when exposed to AgNPs (Fig. 4B). These proteins were first described in relation to temperature shock (64), but the observed up-regulation of these genes (Student’s t test, P < 0.05; table S3) under AgNP exposure suggests that they might be commonly expressed to protect the organism from stress. In contrast, transcripts of genes encoding glutathione peroxidase (GPX) and alkyl hydroperoxide reductase (ahpC) were both down-regulated by approximately 1.3-fold (Student’s t test, P < 0.05) (Fig. 4B and table S3), indicating that some catalase gene expressions might also be inhibited by AgNP exposure.
CONCLUSIONS
In conclusion, our findings suggest that AgNPs potentially have the capability to reduce nitrification rates while enhancing N2O production. Size-dependent impacts of AgNPs on nitrification and the associated N2O production were observed, and our analyses suggest that the toxicity was due to the Ag+ released from AgNPs. The toxicity data indicate that considerable hormesis occurred after low-dose AgNP exposure, which stimulated N2O production up to twofold. The isotopomer analysis suggests that N2O production was mainly from NH2OH oxidation, which was enhanced by exposure to low doses of AgNPs. The metatranscriptional data illustrate that AgNPs affected the gene expression of nitrifying communities, particularly those involved in the stress response and nitrogen metabolism. Consistent with the stimulation of N2O production, the expression of the nitric oxide reductase norQ gene was up-regulated, as were many stress-response genes. Although the estimated concentrations of AgNPs released into the environment are lower than those used here, our work contributes to a mechanistic understanding of AgNP toxicity to nitrification. Our study reveals that further increases in AgNP concentrations in the environment could disrupt a critical link in the nitrogen cycle and could increase the production of a potent greenhouse gas (N2O). Future work is needed to explore the chronic effect of low doses of AgNPs on nitrification. In addition, future work could expand this study to different salinity conditions, as the changing salinity in estuarine environments has an important influence on the aggregation and dissolution kinetics of AgNPs as well as the ammonia-oxidizer community structure. Last but not least, we should pay attention to the environmental risk of the sulfidation process of AgNPs and focus on the activation of Ag2S driven by estuarine environmental conditions and associated toxic effects on nitrification.
MATERIALS AND METHODS
Sample collection and AgNP preparation
Sediment samples and overlying tidal water were collected from intertidal flats of the Yangtze Estuary. Briefly, surface sediments (0 to 5 cm) were collected using PVC tubes from six to eight plots and then stored in sterile plastic bags. Overlying tidal water was collected at the respective sampling sites with a sterile plastic carboy. After collection, the sediment and overlying water samples were transported to the laboratory on ice within 2 hours. Upon arriving at the laboratory, the sediment samples were homogenized, and the overlying tidal water was filtered through cross-flow ultrafiltration to remove microorganisms and nanoparticles (65) and stored at 4°C. Nitrate (NO3−) and ammonium (NH4+) concentrations and the salinity of the overlying water at the sampling site were approximately 80 μM, 10 μM, and 2‰, respectively, whereas the sediment characteristics were as follows: NOx− (NO2− plus NO3−), ca. 0.6 μmol g−1; NH4+, ca. 1.1 μmol g−1; organic carbon, ca. 0.8%; and total nitrogen, ca. 0.07% (66).
Three different sizes (10, 30, and 100 nm) of AgNPs coated with PVP were purchased from Coldstone Tech. The average particle diameters were confirmed by TEM inspection (fig. S14). The amount of PVP coating in the AgNPs was less than 0.5% by weight. PVP is a hydrophilic, neutral, and high–molecular weight polymer that protects nanoparticles by steric stabilization. Here, PVP-coated AgNPs were chosen, because they are commonly used. Stock solutions of AgNPs were prepared immediately before the experiment by dispersing AgNPs in Milli-Q water at a known concentration. In addition, Ag+ was prepared as silver nitrate (AgNO3) to compare its toxicity with that of AgNPs.
Nitrification inhibition
The concentrations of AgNPs and Ag+ used in the present study were selected on the basis of a range-finding test. Three sizes of AgNPs (10, 30, and 100 nm) and dissolved Ag+ were tested. AgNP stock solutions were added to the overlying water (filtered overlying tidal water with a final concentration of 300 μM NH4+) to final Ag concentrations of 0, 0.01, 0.1, 1, 10, 100, 500, 1000, 2000, and 3000 μg liter−1 for two sizes of AgNPs, 30 and 100 nm. Because of the higher toxicity of 10-nm AgNPs, the 3000 μg liter−1 treatment was excluded. For the most toxic Ag+, a further reduced range of 0 to 50 μg liter−1 was used (0, 0.01, 0.05, 0.1, 0.5, 1, 5, 10, and 50 μg liter−1).
Sediment slurries were made by mixing 1 part wet sediment with 30 parts of the prepared silver-containing or silver-free overlying tidal water (67). Then, 10 ml of the slurry was immediately transferred into 120-ml gas-tight glass vials. A final concentration of 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS, pH adjusted to 7.8) was added to maintain the solution pH at approximately 7.8 during NH3 oxidation (68). The vials were placed on an orbital shaker at 180 rpm and incubated in the dark at room temperature (20°C). At each sampling time (0, 1, 2, 3, 5, 8, 12, 24, and 30 hours), three replicates of each treatment were sacrificed, and 2 ml of the headspace gas was extracted using a gas-tight syringe for N2O analysis. The DO and pH of the suspensions were measured. One part of the suspension (5 ml) in each vial was filtered (0.2 μm, Waterman) immediately for NO3− and NO2− measurement, whereas the other part was for total Ag and dissolved Ag+ determination. Before the beginning of the experiment, all vials were heat-sterilized by autoclaving at 121°C and 15 psi for 20 min. Nitrification rates were expressed as the changes in NO3− concentrations with time, because NO2− was below the detection limit (0.1 μM). The inhibition of the nitrification rate was expressed as the percentage reduction in the nitrification rate in the AgNP or Ag+ treatments compared to the no-silver control. The effects of AgNPs and Ag+ on N2O emission were calculated on the basis of the percentage changes of N2O concentration (in the headspace of the glass vials) in the Ag treatments relative to the no-silver control.
Inhibition experiments for PVP were conducted independently on subsamples to rule out toxicity from AgNP coating. A concentration gradient of 0, 0.05, 0.1, 0.5, 1, 5, 10, and 15 μg liter−1 PVP was chosen on the basis of the amount of PVP on the AgNPs (less than 0.5% by weight). After 12 hours of incubation, the N2O in the headspace, DO, pH, NO3−, and NO2− of the suspension were determined.
Stable isotope analysis
To identify the pathway of N2O production, N2O isotopomer ratios (δ15Nα, δ15Nβ, and δ18O) were measured during the nitrification inhibition experiment. Briefly, sediment slurries with AgNPs (100 μg liter−1, 10 nm; 500 μg liter−1, 30 nm; 1000 μg liter−1, 100 nm; 2000 μg liter−1, 10 nm; and 3000 μg liter−1, 30 and 100 nm), Ag+ (5 μg liter−1), and Ag+ (50 μg liter−1) or no-silver control were prepared and immediately transferred into three replicate 120-ml gas-tight glass vials, as described above. These vials were placed on an orbital shaker at 180 rpm and incubated in the dark at room temperature (20°C) for 12 hours. N2O isotopomer ratios of the headspace gas were then determined using an Isotopic N2O Analyzer (Los Gatos Research), with a precision of <1‰. 15N-site preference (SP) is defined using the following equation
| (1) |
where 15Nα and 15Nβ represent the 15N/14N ratios at the central and end sites of the nitrogen atoms, respectively. Characteristic SP values of 33‰ for NH2OH oxidation and 0‰ for NO2− reduction, which were estimated in pure cultures (46, 47), were used to estimate the contribution of each process, assuming that each process is linearly proportional to the SP value using the following equations (47)
| (2) |
| (3) |
Nitrifying bioreactor setup and operation
A continuous-flow membrane nitrifying bioreactor with a working volume of 4.0 liter was set up with approximately 50 g of fresh sediment as inoculum from the Yangtze Estuary. Filter-sterilized and nanoparticle-free overlying tidal water, with an addition of NH4+ to a final concentration of 3 mM, was supplied at a flow rate of approximately 2 liters day−1. The temperature and pH were maintained at 20°C (room temperature) and 7.8, respectively, with a water bath and a 1 M KHCO3 solution. The DO concentration was maintained at approximately 8.0 mg liter−1 by flushing continuously with air. The reactor was stirred at 250 rpm with two stirrers installed at the bottom and above the inlet airflow. The reactor tank and other related materials were heat-sterilized by autoclaving at 121°C and 15 psi for 20 min, whereas the reagent solutions were filter-sterilized through filters with pore size of 0.2 μm (Waterman) before use. Liquid samples (10 ml) were collected from the effluent of the reactor every day and filtered immediately using filters with pore size of 0.2 μm (Waterman). Filtrates were stored at −20°C for the measurement of NH4+, NO3−, and NO2−. Triplicate sediment slurries (5 ml) were harvested from the reactor every day and pelleted by centrifugation for 5 min at 20,000g. Pellets were immediately used for total DNA extraction, and subsequent qPCR and pyrosequencing to detect the enrichment level of nitrifying prokaryotes in the bioreactor (Supplementary Materials and Methods).
When the nitrifying bioreactor achieved a steady state (figs. S9 and S10), samples were taken from the bioreactor for the nitrification inhibition test. Briefly, nitrifying-enriched sediments were harvested from the reactor by centrifugation (20,000g, 5 min). The precipitate was washed three times using 40 mM KH2PO4 buffer (pH 7.8) and resuspended in culture medium. Then, 100 μl of the suspension was transferred immediately into 120-ml gas-tight glass vials in which 10 ml of culture medium was prepared with AgNPs or no-silver control. Inhibition of nitrification rate and effects of AgNPs on N2O emission based on the nitrifying-enriched sediments were determined as described above.
Nucleic acid extraction
The total RNA was extracted from the triplicate no-silver controls and AgNP-treated nitrifying enrichments using the EZNA Soil RNA kit (Omega Bio-tek, Norcross) and purified by removing residual genomic DNA with the Turbo DNA-free kit (Ambion). Contamination of DNA was ruled out using PCR based on primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 909R (5′-CCCCGYCAATTCMTTTRAGT-3′) (69). The concentration, purity, and RNA integrity number of the RNA were determined using a Nanodrop2000 (Thermo) and Agilent2100 (Agilent). Ribosomal RNA (rRNA) was removed using a Ribo-Zero Magnetic Kit (Epicentre), thus resulting in qualified mRNA samples. One part of the mRNA was used for metatranscriptome sequencing, whereas the other part was used for qPCR analyses.
Metatranscriptomic analysis
Metatranscriptomes were obtained from no-silver controls and AgNP-treated (500 μg liter−1 of 30-nm AgNPs) nitrifying enrichments after the 12-hour incubation (table S2), in which a 25% nitrification inhibition (Student’s t test, P < 0.01) and a hormetic stimulation (80%) on N2O production (Student’s t test, P < 0.01) were observed. Triplicate mRNA samples were pooled and used for library construction with the TruSeq RNA Sample Prep Kit (Illumina) and sequenced on an Illumina HiSeq4000 platform (70).
The quality of the raw reads was visualized using FastQC and cleaned and trimmed using SeqPrep (https://github.com/jstjohn/SeqPrep). Reads that were shorter than 50 base pairs (bp), contained ambiguous (N) bases, and were of low quality (below 20) were discarded using Sickle (https://github.com/najoshi/sickle). rRNA reads were further screened using SortMeRNA (http://bioinfo.lifl.fr/RNA/sortmerna/). The trinity de novo assembly pipeline was used to assemble the high-quality paired-end mRNA reads (71, 72). Resulting sequences were filtered to remove contigs of less than 300 bp in length. TransGeneScan (http://sourceforge.net/projects/transgenescan/) was used to predict open reading frames. Transcripts were assigned to taxonomic affiliations by binning to the best hit (BLASTP, e value ≤ 10−5) in the nr database. The potential function was assigned on the basis of the best homology (BLASTP, e value ≤ 10−5) to proteins within the Kyoto Encyclopedia of Genes and Genomes database. Sequences with hits to proteins in eukaryotic organisms and viruses were identified and removed before statistical analysis. Expression levels of transcripts were calculated with RNA-Seq by Expectation-Maximization (http://deweylab.github.io/RSEM/) based on the fragments per kilobase of transcript per million mapped reads (FPKM) (73). The FC in the relative gene expression was calculated by comparing the AgNP-treated samples to no-silver control samples.
Quantitative polymerase chain reaction
Corroboration of gene expression in metatranscriptomic method was performed by qPCR with cDNA as template on an ABI 7500 Sequence Detection System (Applied Biosystems). The cDNA was obtained from the triplicate mRNA using Superscript Double-Stranded cDNA Synthesis kit (Invitrogen) and a random hexamer primer. Before qPCR, all cDNA samples were normalized to a concentration of 10 ng μl−1. The quantitative standard for each gene was constructed as described previously (67). The qPCRs were performed in triplicate with corresponding primers as given in Supplementary Materials and Methods and table S4.
Membrane integrity testing
Nitrifying enriched samples were harvested and pelletized for thin-section TEM imaging after the nitrification inhibition experiment (22). Briefly, biomass from no-silver controls and AgNP treatments was harvested by centrifugation at 700g for 5 min. The obtained flocs were fixed at 4°C for 12 hours in a solution of 2.5% glutaraldehyde, 0.1 M phosphate buffer (pH 7.0), and ultrapure water. The flocs were washed three times with phosphate buffer (0.1 M, pH 7.0) and fixed with 1% osmic acid for 2 hours and then washed three times with phosphate buffer cycle. Samples were dehydrated in an increasing ethanol series (15 min each in 30, 50, 70, 80, 95, and 100% ethanol) at room temperature and in 100% acetone for 20 min at 4°C. Subsequently, they received penetrating treatment with epoxy resin and acetone at a volume ratio of 1:1 and 3:1 for 1 and 3 hours, respectively, at room temperature. Samples were then embedded in pure epoxy resin for 12 hours at 70°C. Ultramicrotome-cut thin sections of the resin sample and slices were dyed for 5 to 10 min with lead citrate and a solution of uranyl acetate saturated in 50% ethanol, and inspected at 60 kV with a FEI Tecnai G2 Twin TEM. Over 10 images were taken for each sample after surveying a large amount of each TEM grid.
Analytical methods
Total Ag and dissolved Ag+ were quantified using an Agilent 7700 inductively coupled plasma mass spectrometer with a detection limit of 0.03 μg liter−1. In brief, total Ag was measured by adding concentrated trace metal–grade nitric acid (HNO3) at 70°C overnight to dissolve AgNPs and then diluted with 2% HNO3 followed by filtration with a 0.2-μm membrane (Waterman) to remove impurities (4). To measure the dissolved Ag+, we placed samples on an Amicon Ultra-15 Centrifugal Filter Unit (3 kDa, Millipore) and centrifuged them at 4000g for 45 min. Dissolved Ag+ concentration was quantified in the filtrate after a dilution to fit the appropriate measurement range with 2% HNO3 (4). NH4+, NO3−, and NO2− were spectrophotometrically determined using a nutrient analyzer (SAN plus, Skalar Analytical B.V.), with detection limits of 0.5 μM for NH4+ and 0.1 μM for NO2− and NO3−. Headspace N2O was quantified using gas chromatography (GC-2014, Shimadzu) with a detection limit of 0.1 ppb (parts per billion). DO concentration and pH in the incubation slurries were determined using an OXY Meter S/N 4164 with an oxygen needle sensor (Unisense) and a Mettler-Toledo pH Meter, respectively.
Statistical analysis
The unpaired, two-tailed Student’s t test was applied using SPSS (version 16.0) to identify the statistical differences among differently treated groups. Pearson correlation analysis was also conducted to explore any underlying correlations. Results were considered significant when P < 0.05.
Acknowledgments
We thank W. S. Gardner, M. J. McCarthy, and M. A. H. J. van Kessel for discussion and revision on the manuscript. Funding: This work was supported by the Chinese National Key Programs for Fundamental Research and Development (2016YFA0600904) and the National Natural Science Foundations of China (41671463, 41130525, 41322002, 41601530, and 41271114). Author contributions: Y.Z., L.H., and M.L. designed the research. Y.Z., L.H., G.Y., C.Y., H.Z., and X.L. performed the research. Y.Z., L.H., D.G., X.L., J.G., R.W., and C.L. analyzed the data. Y.Z., L.H., M.L., and S.E.N. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. All sequence reads are available in the National Center for Biotechnology Information’s Sequence Read Archive Database under the accession number SRP087705.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1603229/DC1
Supplementary Materials and Methods
fig. S1. Nitrate (NO3−) concentrations in the glass vials during the 30-hour incubation period.
fig. S2. Percentage reduction in nitrification rate in AgNP or Ag+ treatments compared to no-silver control during the 30-hour incubation period.
fig. S3. Pearson correlation between the dissolved Ag+ concentrations released from AgNPs and the percentage reduction in nitrification rate at the end of the 12-hour exposure period.
fig. S4. Percentage effects of PVP coating on nitrification activity and N2O production compared to the control group (incubation time = 12 hours, n = 3).
fig. S5. N2O concentrations in the headspace of the glass vials during the 30-hour incubation period.
fig. S6. Effects of AgNPs and Ag+ on N2O emission during nitrification in the 30-hour incubation period.
fig. S7. Effects of the dissolved Ag+ released from AgNPs on N2O emission during nitrification (incubation time = 12 hours).
fig. S8. DO concentration at the end of the incubation period (incubation time = 12 hours).
fig. S9. Bioreactor performance.
fig. S10. Community compositions of the nitrifying bioreactor during the incubation period and the original sediment samples collected from the intertidal flat of the Yangtze Estuary based on 16S rRNA gene sequencing.
fig. S11. Community compositions of the nitrifying organisms in the no-silver control and the AgNP (500 μg liter−1 of 30 nm AgNPs) treatment at the end of the 12-hour incubation, based on the metatranscriptome sequencing.
fig. S12. FC of transcripts encoding the enzymes complex I (NADH-ubiquinone reductase), complex III (ubiquinol-cytochrome c reductase), complex IV (cytochrome c oxidase), and complex V (F-type ATPase) in the respiratory chain of the nitrifying organisms under 30-nm AgNP (500 μg liter−1) exposure for 12 hours.
fig. S13. Neighbor-joining phylogenetic tree of ammonia monooxygenase based on protein sequences.
fig. S14. TEM imaging of AgNPs used in this study.
table S1. Dissolved Ag+ concentration released from AgNPs after a 12-hour exposure period in the nitrification inhibition experiment.
table S2. Sequencing read statistics of the metatranscriptomic libraries.
table S3. Expression of genes encoding proteins involved in nitrogen transformation, heavy metal stress response, and oxidative stress release of nitrifying organisms in the no-silver control and 30-nm AgNP (500 μg liter−1) treatment based on qPCR method (incubation time = 12 hours).
table S4. Primers and qPCR protocols used in this study.
REFERENCES AND NOTES
- 1.Maynard A. D., Aitken R. J., Butz T., Colvin V., Donaldson K., Oberdörster G., Philbert M. A., Ryan J., Seaton A., Stone V., Tinkle S. S., Tran L., Walker N. J., Warheit D. B., Safe handling of nanotechnology. Nature 444, 267–269 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Gao Y., Luo Z., He N., Wang M. K., Metallic nanoparticle production and consumption in China between 2000 and 2010 and associative aquatic environmental risk assessment. J. Nanopart. Res. 15, 1681–1690 (2013). [Google Scholar]
- 3.Wiesner M. R., Lowry G. V., Alvarez P., Dionysiou D., Biswas P., Assessing the risks of manufactured nanomaterials. Environ. Sci. Technol. 40, 4336–4345 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Yang Y., Li M., Michels C., Moreira-Soares H., Alvarez P. J. J., Differential sensitivity of nitrifying bacteria to silver nanoparticles in activated sludge. Environ. Toxicol. Chem. 33, 2234–2239 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Massarsky A., Trudeau V. L., Moon T. W., Predicting the environmental impact of nanosilver. Environ. Toxicol. Phar. 38, 861–873 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Huynh K. A., Chen K. L., Aggregation kinetics of citrate and polyvinylpyrrolidone coated silver nanoparticles in monovalent and divalent electrolyte solutions. Environ. Sci. Technol. 45, 5564–5571 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malynych S., Luzinov I., Chumanov G., Poly(vinyl pyridine) as a universal surface modifier for immobilization of nanoparticles. J. Phys. Chem. B 106, 1280–1285 (2002). [Google Scholar]
- 8.Yin Y., Liu J., Jiang G., Sunlight-induced reduction of ionic Ag and Au to metallic nanoparticles by dissolved organic matter. ACS Nano 6, 7910–7919 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Nowack B., Krug H., Height M., 120 years of nanosilver history: Implications for policy makers. Environ. Sci. Technol. 45, 1177–1183 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Piccinno F., Gottschalk F., Seeger S., Nowack B., Industrial production quantities and uses of ten engineered nanomaterials in Europe and the world. J. Nanopart. Res. 14, 1109–1119 (2012). [Google Scholar]
- 11.Blaser S. A., Scheringer M., MacLeod M., Hungerbühler K., Estimation of cumulative aquatic exposure and risk due to silver: Contribution of nano-functionalized plastics and textiles. Sci. Total Environ. 390, 396–409 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Lombi E., Donner E., Taheri S., Tavakkoli E., Jämting A. K., McClure S., Naidu R., Miller B. W., Scheckel K. G., Vasilev K., Transformation of four silver/silver chloride nanoparticles during anaerobic treatment of wastewater and post-processing of sewage sludge. Environ. Pollut. 176, 193–197 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Ma R., Levard C., Judy J. D., Unrine J. M., Durenkamp M., Martin B., Jefferson B., Lowry G. V., Fate of zinc oxide and silver nanoparticles in a pilot wastewater treatment plant and in processed biosolids. Environ. Sci. Technol. 48, 104−112 (2014). [DOI] [PubMed] [Google Scholar]
- 14.T. R. Kuech, R. J. Hamers, J. A. Pedersen, Chemical transformation of metal, metal oxide, and metal chalcogenide nanoparticles in the environment, in Engineered Nanoparticles and the Environment: Biophysicochemical Processes and Toxicity, B. Xing, C. D. Vecitis, N. Senesi, Eds. (Wiley, 2016), pp. 261–291. [Google Scholar]
- 15.Wang P., Menzies N. W., Dennis P. G., Guo J., Forstner C., Sekine R., Lombi E., Kappen P., Bertsch P. M., Kopittke P. M., Silver nanoparticles entering soils via the wastewater−sludge−soil pathway pose low risk to plants but elevated Cl concentrations increase Ag bioavailability. Environ. Sci. Technol. 50, 8274−8281 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Kaegi R., Voegelin A., Ort C., Sinnet B., Thalmann B., Krismer J., Hagendorfer H., Elumelu M., Mueller E., Fate and transformation of silver nanoparticles in urban wastewater systems. Water Res. 47, 3866–3877 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Li L., Zhou Q., Geng F., Wang Y., Jiang G., Formation of nanosilver from silver sulfide nanoparticles in natural waters by photoinduced Fe(II, III) redox cycling. Environ. Sci. Technol. 50, 13342−13350 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wang G., Nishio T., Sato M., Ishikawa A., Nambara K., Nagakawa K., Matsuo Y., Niikura K., Ijiro K., Inspiration from chemical photography: Accelerated photoconversion of AgCl to functional silver nanoparticles mediated by DNA. Chem. Commun. 47, 9426–9428 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Chinnapongse S. L., MacCuspie R. I., Hackley V. A., Persistence of singly dispersed silver nanoparticles in natural freshwater, synthetic seawater, and simulated estuarine waters. Sci. Total Environ. 409, 2443–2450 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Gallon C., Flegal A., Sources, fluxes, and biogeochemical cycling of sliver in the oceans. Rev. Environ. Contam. Toxicol. 235, 27–48 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Choi O., Deng K. K., Kim N.-J., Ross L. Jr.. Surampalli R. Y., Hu Z., The inhibitory effects of silver nanoparticles, silver ions, and silver chloride colloids on microbial growth. Water Res. 42, 3066–3074 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Arnaout C. L., Gunsch C. K., Impacts of silver nanoparticle coating on the nitrification potential of Nitrosomonas europaea. Environ. Sci. Technol. 46, 5387−5395 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Molleman B., Hiemstra T., Surface structure of silver nanoparticles as a model for understanding the oxidative dissolution of silver ions. Langmuir 31, 13361–13372 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Prosser J. I., Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30, 125–181 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Kuypers M. M. M., Sliekers A. O., Lavik G., Schmid M., Jørgensen B. B., Kuenen J. G., Damsté J. S. S., Strous M., Jetten M. S. M., Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422, 608–611 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R., Stahl D. A., Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461, 976–979 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Moënne-Loccoz P., Fee J. A., Catalyzing NO to N2O in the nitrogen cycle. Science 330, 1632–1633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoro A. E., Buchwald C., McIlvin M. R., Casciotti K. L., Isotopic signature of N2O produced by marine ammonia-oxidizing Archaea. Science 333, 1282–1285 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Law Y., Ni B.-J., Lant P., Yuan Z. G., N2O production rate of an enriched ammonia-oxidising bacteria culture exponentially correlates to its ammonia oxidation rate. Water Res. 46, 3409–3419 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Ishii S., Song Y., Rathnayake L., Tumendelger A., Satoh H., Toyoda S., Yoshida N., Okabe S., Identification of key nitrous oxide production pathways in aerobic partial nitrifying granules. Environ. Microbiol. 16, 3168–3180 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Ravishankara A. R., Daniel J. S., Portmann R. W., Nitrous oxide (N2O): The dominant ozone-depleting substance emitted in the 21st century. Science 326, 123–125 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Bianchi T. S., Allison M. A., Large-river delta-front estuaries as natural “recorders” of global environmental change. Proc. Natl. Acad. Sci. U.S.A. 106, 8085–8092 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seitzinger S. P., Mayorga E., Bouwman A. F., Kroeze C., Beusen A. H. W., Billen G., van Drecht G., Dumont E., Fekete B. M., Garnier J., Harrison J. A., Global river nutrient export: A scenario analysis of past and future trends. Glob. Biogeochem. Cycles 24, GB0A08 (2010). [Google Scholar]
- 34.Canfield D. E., Glazer A. N., Falkowski P. G., The evolution and future of earth’s nitrogen cycle. Science 330, 192–196 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Radniecki T. S., Stankus D. P., Neigh A., Nason J. A., Semprini L., Influence of liberated silver from silver nanoparticles on nitrification inhibition of Nitrosomonas europaea. Chemosphere 85, 43–49 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Xiu Z.-m., Zhang Q.-b., Puppala H. L., Colvin V. L., Alvarez P. J. J., Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 12, 4271−4275 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Holt K. B., Bard A. J., Interaction of silver(I) ions with the respiratory chain of Escherichia coli: An electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag+. Biochemistry 44, 13214–13223 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Lok C., Ho C., Chen R., He Q., Yu W., Sun H., Tam P., Chiu J., Che C., Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J. Proteome Res. 5, 916–924 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Xiu Z.-M., Ma J., Alvarez P. J. J., Differential effect of common ligands and molecular oxygen on antimicrobial activity of silver nanoparticles versus silver ions. Environ. Sci. Technol. 45, 9003−9008 (2011). [DOI] [PubMed] [Google Scholar]
- 40.Stebbing A. R. D., Hormesis—The stimulation of growth by low levels of inhibitors. Sci. Total Environ. 22, 213−234 (1982). [DOI] [PubMed] [Google Scholar]
- 41.Zhu X., Burger M., Doane T. A., Horwath W. R., Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc. Natl. Acad. Sci. U.S.A. 110, 6328–6333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mueller N. C., Nowack B., Exposure modeling of engineered nanoparticles in the environment. Environ. Sci. Technol. 42, 4447–4453 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Lu D., Liu Q., Zhang T., Cai Y., Yin Y., Jiang G., Stable silver isotope fractionation in the natural transformation process of silver nanoparticles. Nat. Nanotechnol. 11, 682–686 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Chandran K., Stein L. Y., Klotz M. G., van Loosdrecht M. C. M., Nitrous oxide production by lithotrophic ammonia-oxidizing bacteria and implications for engineered nitrogen-removal systems. Biochem. Soc. Trans. 39, 1832–1837 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Toyoda S., Yoshida N., Determination of nitrogen isotopomers of nitrous oxide on a modified isotope ratio mass spectrometer. Anal. Chem. 71, 4711–4718 (1999). [Google Scholar]
- 46.Sutka R. L., Ostrom N. E., Ostrom P. H., Breznak J. A., Gandhi H., Pitt A. J., Li F., Distinguishing nitrous oxide production from nitrification and denitrification on the basis of isotopomer abundances. Appl. Environ. Microbiol. 72, 638–644 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rathnayake R. M. L. D., Song Y., Tumendelger A., Oshiki M., Ishii S., Satoh H., Toyoda S., Yoshida N., Okabe S., Source identification of nitrous oxide on autotrophic partial nitrification in a granular sludge reactor. Water Res. 47, 7078–7086 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Ostrom N. E., Pitt A., Sutka R., Ostrom P. H., Grandy A., Huizinga K. M., Robertson G. P., Isotopologue effects during N2O reduction in soils and in pure cultures of denitrifiers. J. Geophys. Res. 112, G02005 (2007). [Google Scholar]
- 49.Dalsgaard T., Stewart F. J., Thamdrup B., de Brabandere L., Revsbech N. P., Ulloa O., Canfield D. E., DeLong E. F., Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off Northern Chile. mBio 5, e01966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dang H., Li J., Chen R., Wang L., Guo L., Zhang Z., Klotz M. G., Diversity, abundance, and spatial distribution of sediment ammonia-oxidizing Betaproteobacteria in response to environmental gradients and coastal eutrophication in Jiaozhou Bay, China. Appl. Environ. Microbiol. 76, 4691–4702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu A. Y., Hou L. Y., Yu C.-P., Biogeography of planktonic and benthic archaeal communities in a subtropical eutrophic estuary of China. Microb. Ecol. 70, 322–335 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Chen L., Sun R., Dai T., Tian J., Zheng W., Wen D., Population and diversity of ammonia-oxidizing archaea and bacteria in a pollutants’ receiving area in Hangzhou Bay. Appl. Microbiol. Biotechnol. 100, 6035–6045 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Lawton T. J., Ham J., Sun T., Rosenzweig A. C., Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily. Proteins 82, 2263–2267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thurman R. B., Gerba C. P., Bitton G., The molecular mechanisms of copper and silver ion disinfection of bacteria and viruses. Crit. Rev. Environ. Control 18, 295−315 (1989). [Google Scholar]
- 55.van Kessel M. A. H. J., Speth D. R., Albertsen M., Nielsen P. H., Op den Camp H. J. M., Kartal B., Jetten M. S. M., Lücker S., Complete nitrification by a single microorganism. Nature 528, 555–559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daims H., Lebedeva E. V., Pjevac P., Han P., Herbold C., Albertsen M., Jehmlich N., Palatinszky M., Vierheilig J., Bulaev A., Kirkegaard R. H., von Bergen M., Rattei T., Bendinger B., Nielsen P. H., Wagner M., Complete nitrification by Nitrospira bacteria. Nature 528, 504–509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Upadhyay A. K., Hooper A. B., Hendrich M. P., NO reductase activity of the tetraheme cytochrome C554 of Nitrosomonas europaea. J. Am. Chem. Soc. 128, 4330–4337 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skiba U., Fowler D., Smith K. A., Nitric oxide emissions from agricultural soils in temperate and tropical climates: Sources, controls and mitigation options. Nutr. Cycl. Agroecosys. 48, 139–153 (1997). [Google Scholar]
- 59.Loick N., Dixon E. R., Abalos D., Vallejo A., Matthews G. P., McGeough K. L., Well R., Watson C. J., Laughlin R. J., Cardenas L. M., Denitrification as a source of nitric oxide emissions from incubated soil cores from a UK grassland soil. Soil Biol. Biochem. 95, 1–7 (2016). [Google Scholar]
- 60.Stein L., Surveying N2O-producing pathways in bacteria. Methods Enzymol. 486, 131–152 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Caranto J. D., Vilbert A. C., Lancaster K. M., Nitrosomonas europaea cytochrome P460 is a direct link between nitrification and nitrous oxide emission. Proc. Natl. Acad. Sci. U.S.A. 113, 14704–14709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Solioz M., Role of proteolysis in copper homoeostasis. Biochem. Soc. Trans. 30, 688–691 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Fox B., Walsh C. T., Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J. Biol. Chem. 257, 2498–2503 (1982). [PubMed] [Google Scholar]
- 64.Ritossa F., A new puffing pattern induced by temperature shock and DNP in drosophila. Experientia 18, 571–573 (1962). [Google Scholar]
- 65.Maskaoui K., Zhou J. L., Colloids as a sink for certain pharmaceuticals in the aquatic environment. Environ. Sci. Pollut. Res. 17, 898–907 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Zheng Y., Hou L., Liu M., Liu Z., Li X., Lin X., Yin G., Gao J., Yu C., Wang R., Jiang X., Tidal pumping facilitates dissimilatory nitrate reduction in intertidal marshes. Sci. Rep. 6, 21338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng Y., Hou L., Newell S., Liu M., Zhou J., Zhao H., You L., Cheng X., Community dynamics and activity of ammonia-oxidizing prokaryotes in intertidal sediments of the Yangtze Estuary. Appl. Environ. Microbiol. 80, 408–419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi O., Hu Z., Size dependent and reactive oxygen species related nanosilver toxicity to nitrifying bacteria. Environ. Sci. Technol. 42, 4583–4588 (2008). [DOI] [PubMed] [Google Scholar]
- 69.Tamaki H., Wright C. L., Li X., Lin Q., Hwang C., Wang S., Thimmapuram J., Kamagata Y., Liu W.-T., Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLOS ONE 6, e25263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai Y., Zheng Y., Bodelier P. L. E., Conrad R., Jia Z., Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat. Commun. 7, 11728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frischkorn K. R., Harke M. J., Gobler C. J., Dyhrman S. T., De novo assembly of Aureococcus anophagefferens transcriptomes reveals diverse responses to the low nutrient and low light conditions present during blooms. Front. Microbiol. 5, 375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A., Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mellbye B. L., Giguere A., Chaplen F., Bottomley P. J., Sayavedra-Soto L. A., Steady-state growth under inorganic carbon limitation conditions increases energy consumption for maintenance and enhances nitrous oxide production in Nitrosomonas europaea. Appl. Environ. Microbiol. 82, 3310–3318 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chain P., Lamerdin J., Larimer F., Regala W., Lao V., Land M., Hauser L., Hooper A., Klotz M., Norton J., Sayavedra-Soto L., Arciero D., Hommes N., Whittaker M., Arp D., Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185, 2759–2773 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li X., Rui J., Mao Y., Yannarell A., Mackie R., Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar. Soil Biol. Biochem. 68, 392–401 (2014). [Google Scholar]
- 76.Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., Huttley G. A., Kelley S. T., Knights D., Koenig J. E., Ley R. E., Lozupone C. A., McDonald D., Muegge B. D., Pirrung M., Reeder J., Sevinsky J. R., Turnbaugh P. J., Walters W. A., Widmann J., Yatsunenko T., Zaneveld J., Knight R., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R., UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., Brown C. T., Porras-Alfaro A., Kuske C. R., Tiedje J. M., Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muyzer G., de Waal E. C., Uitterlinden A. G., Profiling of complexmicrobial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Röling W. F. M., Milner M. G., Jones D. M., Lee K., Daniel F., Swannell R. J. P., Head I. M., Robust hydrocarbon degradation and dynamics of bacterial communities during nutrient-enhanced oil spill bioremediation. Appl. Environ. Microbiol. 68, 5537–5548 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rotthauwe J.-H., Witzel K.-P., Liesack W., The ammonia monooxygenase structural gene amoA as a functional marker: Molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B., Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 102, 14683–14688 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feng G. F., Sun W., Zhang F. L., Karthik L., Li Z. Y., Inhabitancy of active Nitrosopumilus-like ammoniaoxidizing archaea and Nitrospira nitrite-oxidizing bacteria in the sponge Theonella swinhoei. Sci. Rep. 6, 24966 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/8/e1603229/DC1
Supplementary Materials and Methods
fig. S1. Nitrate (NO3−) concentrations in the glass vials during the 30-hour incubation period.
fig. S2. Percentage reduction in nitrification rate in AgNP or Ag+ treatments compared to no-silver control during the 30-hour incubation period.
fig. S3. Pearson correlation between the dissolved Ag+ concentrations released from AgNPs and the percentage reduction in nitrification rate at the end of the 12-hour exposure period.
fig. S4. Percentage effects of PVP coating on nitrification activity and N2O production compared to the control group (incubation time = 12 hours, n = 3).
fig. S5. N2O concentrations in the headspace of the glass vials during the 30-hour incubation period.
fig. S6. Effects of AgNPs and Ag+ on N2O emission during nitrification in the 30-hour incubation period.
fig. S7. Effects of the dissolved Ag+ released from AgNPs on N2O emission during nitrification (incubation time = 12 hours).
fig. S8. DO concentration at the end of the incubation period (incubation time = 12 hours).
fig. S9. Bioreactor performance.
fig. S10. Community compositions of the nitrifying bioreactor during the incubation period and the original sediment samples collected from the intertidal flat of the Yangtze Estuary based on 16S rRNA gene sequencing.
fig. S11. Community compositions of the nitrifying organisms in the no-silver control and the AgNP (500 μg liter−1 of 30 nm AgNPs) treatment at the end of the 12-hour incubation, based on the metatranscriptome sequencing.
fig. S12. FC of transcripts encoding the enzymes complex I (NADH-ubiquinone reductase), complex III (ubiquinol-cytochrome c reductase), complex IV (cytochrome c oxidase), and complex V (F-type ATPase) in the respiratory chain of the nitrifying organisms under 30-nm AgNP (500 μg liter−1) exposure for 12 hours.
fig. S13. Neighbor-joining phylogenetic tree of ammonia monooxygenase based on protein sequences.
fig. S14. TEM imaging of AgNPs used in this study.
table S1. Dissolved Ag+ concentration released from AgNPs after a 12-hour exposure period in the nitrification inhibition experiment.
table S2. Sequencing read statistics of the metatranscriptomic libraries.
table S3. Expression of genes encoding proteins involved in nitrogen transformation, heavy metal stress response, and oxidative stress release of nitrifying organisms in the no-silver control and 30-nm AgNP (500 μg liter−1) treatment based on qPCR method (incubation time = 12 hours).
table S4. Primers and qPCR protocols used in this study.


