Abstract
Previous studies in the mouse indicated that ARID3A plays a critical role in the first cell fate decision required for generation of trophectoderm (TE). Here, we demonstrate that ARID3A is widely expressed during mouse and human placentation and essential for early embryonic viability. ARID3A localizes to trophoblast giant cells and other trophoblast-derived cell subtypes in the junctional and labyrinth zones of the placenta. Conventional Arid3a knockout embryos suffer restricted intrauterine growth with severe defects in placental structural organization. Arid3a null placentas show aberrant expression of subtype-specific markers as well as significant alteration in cytokines, chemokines and inflammatory response-related genes, including previously established markers of human placentation disorders. BMP4-mediated induction of trophoblast stem (TS)-like cells from human induced pluripotent stem cells results in ARID3A up-regulation and cytoplasmic to nuclear translocation. Overexpression of ARID3A in BMP4-mediated TS-like cells up-regulates TE markers, whereas pluripotency markers are down-regulated. Our results reveal an essential, conserved function for ARID3A in mammalian placental development through regulation of both intrinsic and extrinsic developmental programs.
Introduction
Two sequential cell fate decisions during blastocyst formation establish three distinct cellular lineages—trophectoderm (TE), epiblast, and primitive endoderm (Rossant and Tam, 2009; Zernicka-Goetz, 2004). The first cell fate decision in mice and humans occurs at the 8- to 16-cell stage and leads to segregation of the inner cell mass (ICM), which gives rise to all tissues of the body, and the TE, which is required for implantation into the uterus and formation of the placenta (Niwa, 2007). The placenta is essential for survival of the mammalian embryo, as it transports nutrients, produces hormones, provides structural support within the womb, provides immunological protection, and acts as a physiological buffer between the mother and the fetus (Simister and Story, 1997). Abnormal placental development underlies a wide range of complications during pregnancy, including preeclampsia (PE), miscarriage, and predisposition to chronic disease in adulthood (Roberts et al., 1989; Suzuki, 2008). PE, a pregnancy-specific placental disorder characterized by the development of hypertension during gestation, is a major obstetric problem that contributes substantially to maternal and perinatal morbidity and mortality worldwide (Ananth et al., 2013).
While genome-wide analyses have identified genes deregulated in PE, only a few transcription factors (TFs) have been shown to be associated with normal TE specification and/or human placental differentiation (Hemberger et al., 2010; Martinez-Fierro et al., 2016). Its strong similarity with human placentation makes the mouse an excellent model to elucidate key mechanisms of placental development.
Our previous studies showed that AT-Rich Interactive Domain 3A (ARID3A) is essential for the first cell fate decision (Rhee et al., 2014). We found that overexpression (OE) of ARID3A alone is sufficient for trans-differentiation of embryonic stem (ES) cells to trophoblast stem (TS)-like cells—the in vitro counterpart of the TE layer of the blastocyst. Global expression profiles of ARID3A-OE ES cells and TS cells are highly similar. Arid3a-OE ES cells gain the capacity to incorporate into the TE of developing embryos—an indication of the development of functional TS cells.
To gain insight into the role of Arid3a in placentation, we have carried out further in vivo analyses in the mouse and in vitro analyses in the human. Our data indicate that ARID3A provides an indispensable and conserved function in mammalian placental development and may provide a novel diagnostic marker for PE.
Results
1. Arid3a is highly expressed during mouse and human placentation
We first analyzed published global expression profiling for each stage of mouse embryonic development (Smith et al., 2014). Arid3a expression initiates in the morula, then later, becomes highly expressed in extraembryonic components (Fig. 1A). Because a subset of these extraembryonic components are expressed in the placenta, we compared the levels of Arid3a with other key placental markers, including Gata3, Tfap2c, Hand1, and Id2 in vivo and upon time-course differentiation of TS cells-the in vitro counterpart of the TE (Kidder and Palmer, 2010). As shown in Figure 1A, both Arid3a and these transcripts are highly enriched within extraembryonic components and TE. However, other TE markers such as Cdx2, Eomes, Id2, and Hand1, are down-regulated upon TS differentiation (Fig. 1B). This suggested that Arid3a differs from previously studied TE markers in that it expresses in mouse placentation both in vivo and in vitro.
Figure 1. ARID3A is highly expressed during mammalian placentation.
(A) Global expression profiling of TE markers in early mouse embryonic developmental stages. (B) Expression levels of TE markers upon differentiation of mouse TS cells. (C) Global expression profiling of TE and pluripotency markers in human tissues, including placenta. (D) Absolute expression levels of ARID3A in placenta throughout gestation from 1st to 3rd trimester. Two ARID3A values were two different human ARID3A probes used in human microarray data. Two values were detected from two different regions of sequences within ARID3A coding DNA sequences. (E) Expression levels of TE and pluripotency markers upon BMP4-mediated differentiation of human ES and iPS to TS cells. Differentiation day, d; replication, r.
Similar to the mouse, ARID3A levels in human are the highest in the placenta as compared to all other tissues (Li et al., 2013; Rhee et al., 2014)(Fig. 1C). Analyses of publically available data sets (Mikheev et al., 2008) further revealed that ARID3A is highly expressed throughout placental gestation from the 1st to 3rd trimester (Fig. 1D) as well as in human TS-like cells induced from ES cells by BMP4 (Xu et al., 2002)(Fig. 1E). Unlike BMP4-induced TS-like cells, neither human ES cell-derived endodermal cells (Supplemental Fig. S1A) nor embryoid bodies (Supplemental Fig. S1B) displayed significant induction of human ARID3A.
2. Loss of ARID3A during early mouse gestation results in intrauterine growth restriction and defects in placental development
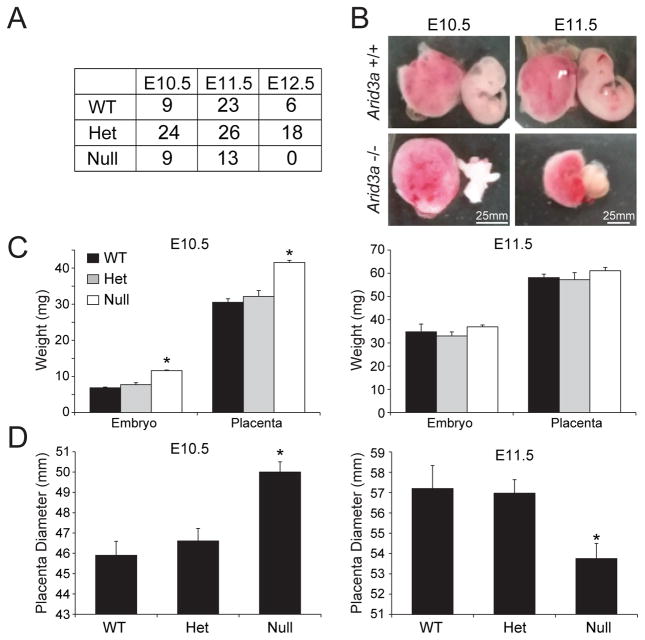
Arid3a−/− null C57BL/6 mice (Webb et al., 2011) were employed to investigate the role of Arid3a in early placental development. Breeding heterozygous Arid3a mice resulted in non-Mendelian ratios from E10.5–E12.5, with no homozygous mutants obtained at E12.5 (Fig. 2A). Since our previous studies (Rhee et al., 2014) detected high ARID3A expression in the TE, we carefully examined the gross anatomy of all E10.5 and E11.5 surviving embryos and placentas from Arid3a mutant heterozygous crosses. Arid3a mutants exhibited a range of phenotypes, from indistinguishable, to small, to paler embryos and placentas—further indications of the vasoconstriction characterized previously (Webb et al., 2011)(Figs. 2B,C).
Figure 2. Arid3a KO in mouse embryos results in defects in placental development.
(A) The numbers of viable placentas and embryos at E10.5, E11.5 and E12.5. Homozygous lethality of Arid3a null embryos was observed primarily at E12.5. (B) Representative examples of Arid3a KO and WT mouse embryos and placentas at E10.5 and E11.5. (C) Weight comparisons among WT, heterozygous and null embryos and placentas. (D) Placental diameter comparisons among WT, heterozygous and null placentas.
E10.5 nulls were frequently observed undergoing absorption (data not shown), and perhaps the adhesive properties underlying this as yet to be determined phenomenon account for their heavier weights relative to E10.5 wild-type (WT) controls (Fig. 2C). Mutant placental diameters at E10.5 also significantly exceeded those of WTs but were significantly smaller at E11.5 (Fig. 2D). One explanation for these data was that Arid3a-deficient placentas might be undergoing inflammatory-mediated swelling at E10.5 followed by apoptotic-mediated atrophy at E11.5 (further addressed below and in Fig. 4H).
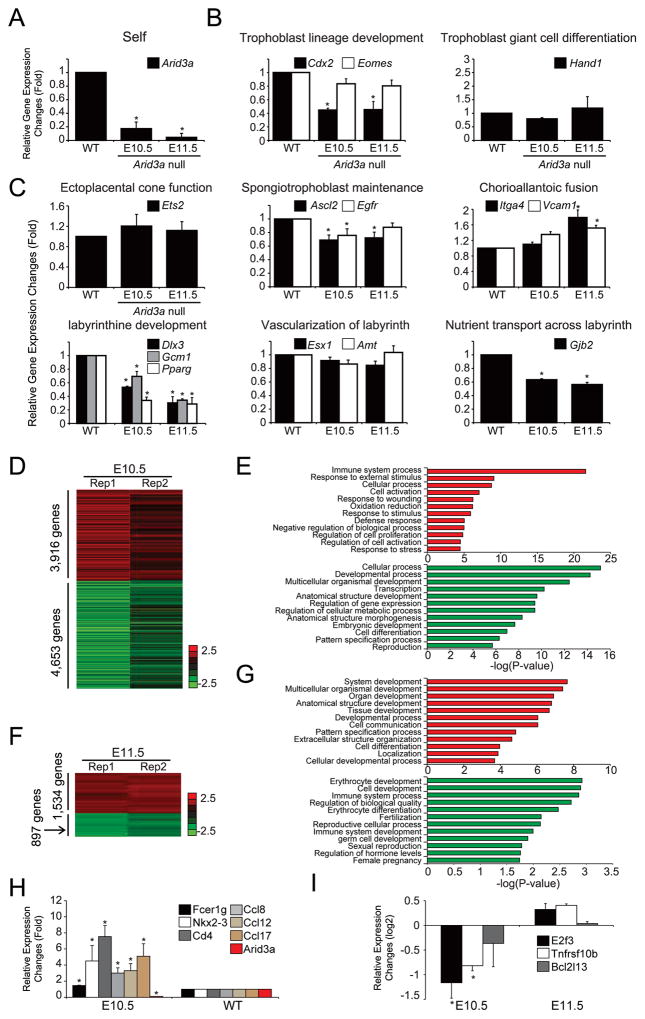
Figure 4. Arid3a KO results in aberrant expression of subtype-specific placental markers.
(A) Expression levels of Arid3a in WT and KO placentas; The E10.5 WTs were used as comparison for E10.5 null and E11.5 WTs for E11.5 null. (B,C) Expression levels of placental lineage markers as determined by RT-qPCR. Transcripts measured are associated with different trophoblast lineages or are required for achieving placental growth milestones. All data are plotted relative to WT levels. Error bars indicate standard deviations of biological triplicates; asterisks indicate significance (minimally) of p≤0.05 of at least 3 independent biological replicates. (D, F) An unsupervised hierarchical clustering of transcript disruption by Arid3a KO normalized to WT using a cutoff threshold of 1.5-fold from expression profiles at E10.5 and E11.5, respectively. (E, G) Significantly enriched GO terms (biological functions) of differentially expressed genes upon KO of Arid3a at E10.5 and E11.5; red, up-regulated and green, down-regulated genes. (H) Alteration of expression associated with innate immunity in Arid3a KO placentas assessed by RT-qPCR. (I) Expression levels of apoptosis mediators altered in Arid3a KO placentas as assessed by RNA-seq.
3. Arid3a mutant placentas have structural defects in placental organization
We first addressed abnormal size and weight mutant phenotypes by hematoxylin and eosin (H&E) staining. As shown in Figure 3, both E10.5 and E11.5 mutant placentas displayed abnormal organization of the junctional zone, disruption of both trophoblast giant cell (TGC) and spongiotrophoblast (SpT) layers, as well as an anomalous labyrinth layer. Consistent with these observations, immunohistochemistry (IHC) at E10.5 and E11.5 revealed that ARID3A protein was predominantly localized within nuclei of TGCs in WTs (Fig. 3B). Additionally, we tested TGC markers at E9.5. Initially, TGCs were present in E9.5 Arid3a null placentas, but then are lost as placentas further developed (Supplemental Fig. S2A). Quantification indicated that the TGC densities within the junctional zone were also significantly reduced in Arid3a KO placentas (Supplemental Fig. S2B).
Figure 3. Arid3a KO in mouse embryos results in defects in placental structural organization.
(A–H) Immunohistochemistry of Arid3a WT and KO sagittal placental cross sections stained with Hematoxyline and Eosin (H&E) and antibodies against: (A) ARID3A; (B) HAND1; (C) PROLIFERN; (D) PLACENTAL LACTOGEN (PL1); (E) ASCL2; (F) CDX2; (G) GCM1 (H). D, Decidua; TGC, Trophoblast giant cells; SpT, Spongiotrophoblasts
Next we examined the expression of HAND1, PROLIFERIN, and PLACENTAL LACTOGEN (PL1), which are required for the differentiation of cellular subtypes expressed in the TGC layer of the junctional zones (Scott et al., 2000)(Figs. 3C–E). While HAND1 expression was evident in E10.5 WT placentas, mutants showed significantly reduced staining around the junctional zones (Fig. 3C). Levels of PROLIFERIN and PL1 were highly disrupted in null placentas (Figs. 3D,E). Consistent with disruption of the SpT layer, expression levels of SpT markers such as ASCL2 and CDX2, were significantly reduced at E11.5, as confirmed by RT-qPCR (Figs. 3F,G;Figs. 4B,C). These results indicate that Arid3a is required for the structural organization of various layers of the placenta.
4. Aberrant expression of subtype-specific markers in Arid3a null placentas
We further examined expression levels of key genes previously shown to be vital to placental development (Rossant and Cross, 2001). Upon KO of Arid3a (Fig. 4A), genes required for trophoblast lineage development, SpT maintenance, labyrinthine development, and nutrient transport across the labyrinth pathway were highly down-regulated (Figs. 4B,C). Conversely, key markers of ectoplacental cone function and chorioallantonic fusion were unchanged (Fig. 4C).
To confirm and extend the RT-qPCR results, we generated expression profiles of Arid3a KO and WT placentas by RNA-seq. We identified approximately 8,000 and 2,000 differentially expressed genes at E10.5 and E11.5, respectively (cut-off threshold ≥1.5-fold; Figs. 4D,F). Differentially up-regulated genes at E10.5 were strongly enriched in gene ontology (GO) terms associated with immune system-related processes, including the response to external stimuli and defense response (Fig. 4E). Conversely, genes down-regulated upon KO were enriched in general metabolic and structural processes, such as anatomical structure and pattern specification (Fig. 4E).
5. Activation of the inflammatory response in Arid3a null placentas
Further inspection of our global expression profiles (Figs. 4E,G) revealed that ARID3A is a critical regulator of innate immunity responses in the placenta. As shown in Figures 4H,I, and in S3A, KO of Arid3a at E10.5 led to up-regulation of transcripts encoding inflammatory chemokines (eg, Ccl12, Ccl17, Ccl8, Ccl5), cytokines (eg, Il1a, Lta, Ltb, Mif), and several other inflammatory mediators (eg, Fcer1g, Nkx2–3, and Cd4), whereas at E11.5, transcripts encoding pro-apoptotic factors (eg, E2f3, Tnfrsf10b, and Bcl2l13) were up-regulated. These results are consistent with the data of Figure 2D and our recent analyses of Arid3a KO hematopoiesis (Kim et al., 2016), suggesting that E10.5 Arid3a-deficient placentas are undergoing inflammatory-mediated swelling followed at E11.5 by apoptotic-mediated atrophy.
We also noted that several angiogenic factors and their receptors, including the soluble Fms-like tyrosine kinase (sFlt-1) and placental growth factor (Plgf) are significantly deregulated in Arid3a KO placentas at E10.5 but return close to normal levels at E11.5 (Fig. 4F;Supplemental Fig. S3B). Excessive inflammation and angiogenic imbalance often underlies symptoms associated with PE (Perez-Sepulveda et al., 2014); readdressed in Discussion).
6. Loss or gain of Arid3a disrupts expression levels of key TS and TGC differentiation markers
We first examined the effect of Arid3a loss on inflammatory chemokine expression following shRNA-based KD of Arid3a. We obtained > 70% KD efficiency at the mRNA level (Fig. 5A). Arid3a-deficient TS cells revealed deregulation of several inflammatory chemokines (e.g., Ccl8, Ccl12, Ccl17) as well as upregulation of several inflammatory mediators (e.g., Nkx2–3, and Cd4) (Fig. 5A). Next, we assessed expression levels of key markers of TE development. As shown in Figures 5B and 5C, well established markers (e.g., Cdx2, Hand1, Gata3, Eomes, and Tcfap2c) were down- or up-regulated upon Arid3a KD or OE, respectively.
Figure 5. Loss or gain of Arid3a disrupts expression levels of key TE markers in mouse TS cells.
(A) Inflammatory chemokine markers are up-regulated following Arid3a KD in mouse TS cells as measured by RT-qPCR. Asterisks indicate significance (minimally) of p≤0.05 of at least 3 independent biological replicate; error bars indicate standard deviation of biological triplicates. (B,C) Expression levels of key markers of TE development are down- or up-regulated upon Arid3a KD (B) or OE (C) in TS cells, respectively. (D) In vitro differentiation of Arid3a-deficient TS cells perturbs gene expression corresponding to essential layers of the placenta as compared to differentiated WT TS cells.
Finally, to better understand the consequences of Arid3a loss on further differentiation to mature trophoblastic lineages, we performed in vitro differentiation of Arid3a-deficient TS cells as previously described (Tanaka et al., 1998)(Fig. 5D). We observed marked deregulation of Hand1, which is essential for TGC differentiation (Scott et al., 2000), Cdx2, an early and essential marker of TE polarity and integrity of the TE epithelium (Strumpf et al., 2005), Gcm1, which promotes differentiation of underlying cytotrophoblast cells into the outer syncytiotrophoblast layer (Bainbridge et al., 2012) and Ascl2, which is required for the maintenance of TGC precursors (Guillemot et al., 1994). Also significantly deregulated was the labyrinth marker, Pparγ, which promotes labyrinthine trophoblast differentiation via Gcm1-regulation (Fournier et al., 2008).
These results suggest that Arid3a gain or lost leads to significant changes in TS cell inflammation, development and differentiation, particularly within the TGC lineage, whose numbers and disorganization are apparent by IHC (Fig. 3;Supplemental Fig. S2B).
7. Human ARID3A levels are induced by BMP4 and ARID3A OE leads to down-regulation of pluripotency and up-regulation of TE-associated gene expression
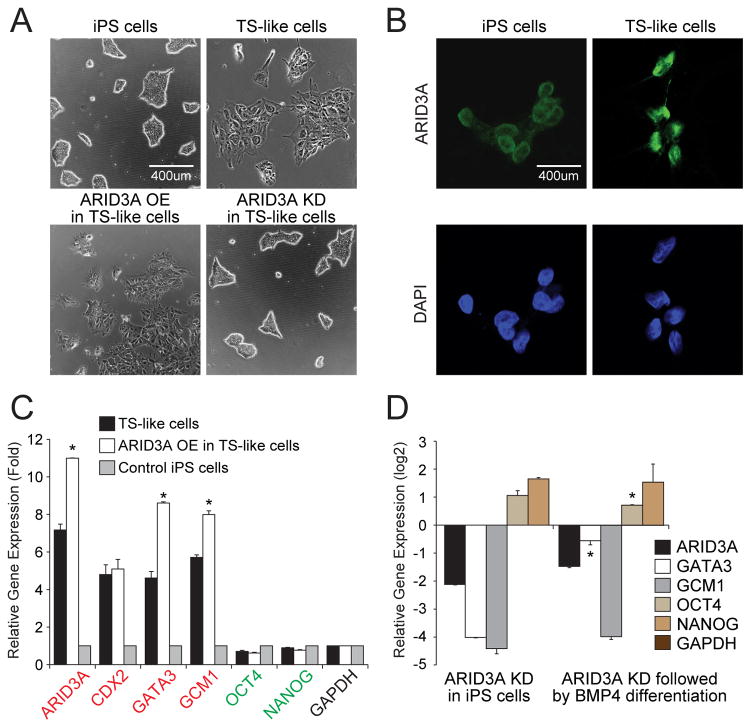
Our meta-analyses of the data of Li et al (2013) indicated that ARID3A is highly expressed in the developing human placenta and in TS-like cells (Figs. 1C,E). As an initial approach to circumvent the ethical limitations of studying human placental function in vivo, we generated human “TS-like” cells (Xu et al., 2002) by addition of BMP4 to human iPS cells in feeder-conditioned medium (FCM) cultures. This treatment, while controversial (further addressed in Discussion), is generally acknowledged to induce TE differentiation.
BMP4 induction led to both increased ARID3A levels as well as cytoplasmic to nuclear translocation of ARID3A (Figs. 6A,B). Transient transfection of ARID3A into these TS-like cells increased its levels ~8-fold (without alteration of the differentiated morphology induced by BMP4/FCM) and up-regulated the TE-markers, GATA3 and GCM1 (Figs. 6A,C). Conversely, shRNA-mediated ARID3A KD de-repressed transcription of pluripotency genes (OCT4 and NANOG) while repressing transcription of TE genes (Fig. 6D). This may provide a mechanism by which ARID3A loss delays differentiation of human ES cells to TS-like cells.
Figure 6. human ARID3A loss or gain disrupts expression levels of key TE markers in human TS-like cells.
(A) BMP4-mediated human TS-like cells with ARID3A OE show differentiated morphology. ARID3A KD in TS-like induces a rounded morphology, indicative of a pluripotent state indistinguishable from human iPS cells. (B) Immunofluorescence images of control iPS and TS-like cells stained with directly conjugated anti-ARID3A (green; Alexa 594) and DAPI (blue). (C) Trophoblast lineage markers (red) and pluripotency factors (green) are up-regulated following ARID3A OE as measured by RT-qPCR. Error bars indicate standard deviation of biological triplicates. (D) Trophoblast lineage markers and pluripotency factors are down-regulated following KD of ARID3A. Asterisks indicate significance (minimally) of p≤0.05 of at least 3 independent biological replicates as compared to non-differentiated cells.
Discussion
Here we show that ARID3A plays a vital role in mouse placentation by contributing to the structural integrity of the placenta as well as by mediating communication among critical effectors, such as cells of the innate immune system. Our initial in vitro investigation in the human model further supports a requirement for ARID3A in initiation of ES to TS-like cell conversion through modulation of underlying transcriptional programs. Taken with our previous mechanistic observations (Rhee et al., 2014), our results identify ARID3A as an indispensable component of placental commitment, hemostasis and immune tolerance.
Anchoring the conceptus to the uterus is a critical placental function. In mice, TGCs perform this role, and the bHLH TF, Hand1, is critical (Riley et al., 1998; Scott et al., 2000). Hand1 expression was not altered by loss of Arid3a. Instead, the most striking consequence of Arid3a deficiency was displayed morphologically. While specific sub-lineages can be delineated, they are highly disorganized with reduced cellularity (Fig. 3). That along with the expression data of Figure 4 supports an interpretation in which both layers and genes underlying their formation are deregulated in Arid 3a nulls. Addressing this observation in vitro by employing isolated cell cultures would contribute significantly, but it would not fully resolve the in vivo defects. That will require loss of function analyses of specific sub-lineage-restricted master transcription factors. Additional important questions to be resolved by future studies are the mechanisms by which Arid3a maintains this structural integrity and the precise consequences of Arid3a loss on TGC transcriptional programs.
Global RNA expression profiling confirmed that ARID3A function extends beyond the “first cell fate decision” (Rhee et al., 2014) to additional cellular processes vital to placental development, including placental structure, metabolism, immune tolerance and angiogenesis. Innate immune cells, typically placental NK, switch from a tolerogenic, anti-inflammatory phenotype to a cytotoxic, pro-inflammatory phenotype upon the sensing of pathogens or endogenous danger signals (Perez-Sepulveda et al., 2014). In our case, we suggest the trigger was loss of Arid3a. As cytotoxic effectors, innate immune cells create a state of inflammation, via cytokine and chemokine release, and placental ischemia, through reduced angiogenesis and increased vasoconstriction—the phenotype observed at E10.5 in Arid3a−/− placentas. We recently published a detailed analysis (Kim et al., 2016) of the inflammatory consequences of ARID3A loss on embryonic hematopoiesis. In those studies, conducted at the same embryonic time points as analyzed here, we observed inflammatory consequences of a prototypical TH1 response—most notably, a significant increase in IFNα. However, the immune profiles of our null placentas (Figs. 4E–H) displayed no significant overlap with the INFα target genes of Kim et al. (2016) nor with other targets deregulated in the null maternal embryo. This indicates that a large component of the observed placental inflammation is intrinsically derived. ARID3A is also required at later stages of embryogenesis for normal erythroid lineage differentiation and hematopoietic stem cell production (Kim et al., 2016; Webb et al., 2011).
The in vivo authenticity of the previously characterized mesoderm inducer, BMP4, in human TS induction has been challenged (Bernardo et al., 2011; Ezashi et al., 2012; Li and Parast, 2014). However, more recently and well after the BMP4 debate arose, a number of groups have confirmed that the BMP4 system can induce TS-like cell phenotype. For example, Kurek et al (2015) showed conclusively that BMP-alone targets are required and sufficient for mesoderm induction, whereas trophoblast induction is WNT dependent, suggesting that exclusive differentiation toward either lineage is possible in BMP4 cultures. Yabe et al (2016) found that, when BMP4 is used in combination with ACTIVIN and FGF2 signaling inhibitors, trophoblast differentiation was significantly more efficient and synchronous. Taken in this context, we feel that our approach—to determine the effect of ARID3A loss or gain in producing in vitro “TS-like” cells from human ES/iPS cells—is rational.
We observed that, as in the mouse (Rhee et al., 2014), Arid3a overexpression under BMP4-mediated culture conditions did convert human iPS to a more TS-like phenotype via downregulation of pluripotency genes and upregulation of trophoblast genes (Figs. 6C,D). However, not all our findings in mouse were conserved in the human. For example, in mouse blastocysts and placenta (Rhee et al., 2014), ARID3A activates CDX2, a transcription factor required for the initiation of TS commitment (Dietrich and Hiiragi, 2007). However, in the BMP4-induction hES cell system, ARID3A loss had no effect on CDX2 expression. This is consistent with observations that CDX2 expression is maximized in the non-trophoblast (mesoderm-derived) component of the human chorion (Bernardo et al., 2011; Niakan and Eggan, 2013). A further difference was that the reciprocal CDX2-OCT4 expression patterns established in the mouse are not conserved in the human (Hay et al., 2004). The human embryo shows a lag in trophoblast lineage segregation, with a period of time during which OCT4 and CDX2 are co-expressed in the TE (Niakan and Eggan, 2013). Nonetheless, we observed ARID3A-mediated repression of human OCT4, even though its human regulatory elements have diverged to a point such that they are not repressed when enforced into mouse TE (Molineris et al., 2011). While its specific role in human placentation remains to be elucidated, our findings establish that ARID3A is instrumental to trophoblast lineage determination.
Preeclampsia (PE) is a major cause of pregnancy-associated morbidity and mortality, affecting 2–5% of pregnancies worldwide after 20 weeks of gestation (Ananth et al., 2013). Normal placental function depends on trophoblastic invasion of the maternal decidua, myometrium, and blood vessels (Li and Parast, 2014). Fms-like tyrosine kinase 1 (sFLT-1) and placental growth factor (PlGF) are key among angiogenic and anti-angiogenic and mediators implicated in PE pathology (Levine et al., 2006). A recent study (Zeisler et al., 2016) demonstrated that the ratio of sFLT-1 to PlGF is elevated in the blood of pregnant women prior to the onset of PE. As shown in Supplemental Figure S3, Arid3a activates both sFLT1 and PIGF, as well as the additional inflammatory agents, ENDOGEN, and VEGFR-2. Additional Arid3a targets associated with PE include PPARγ and GCM1 (Chen et al., 2004; Waite et al., 2000). Our meta-analyses of placental PE data sets (Bilban et al., 2009; Founds et al., 2011) identified ARID3A and ELF5 as the only two deregulated transcription factors associated with human TE specification and/or differentiation.
In summary, ARID3A is vital not only for maintaining proper intrauterine growth but also for maintaining a properly balanced immune system during pregnancy. Our data indicate that, as suggested by previous TS cell studies in vitro (Rhee et al., 2014), ARID3A is required for normal murine trophoblast development in vivo. However, it is important to note that while numerous pathways are deregulated, the etiology of the initiating placental defect in the mouse remains to be determined. It is unclear and the focus of future studies as to whether the primary defect is a direct consequence of the Arid3a null placenta, is secondary to a failing embryo/fetus or is derived from a combination of these events. Finally, we suggest that the use of ARID3A as a biomarker may provide a diagnostic, noninvasive predictive molecule to identify mothers at risk for defective deep placentation syndromes.
Materials and Methods
E10.5 or E11.5 staged Arid3a−/− mouse embryos (Webb et al., 2011) were obtained using timed pregnancies of C57BL/6 females. Placentas were isolated under a dissecting microscope and either stained with H&E or stained by IHC for various markers prior to sectioning. Whole embryos were dissociated to single cells for RNA isolation (RNeasy Mini Kit; Qiagen) and then subjected to RNA-qPCR (SYBR PerfeCta SYBR Green) or RNA-seq. Human iPS cells (Yamanaka retrovirus reprogrammed hiPS cells, ATCC) were maintained in mTeSR1 media (Stemcell technologies), cultured in BMP4/FCM (for TS-like cell generation), and then detached and plated on supplemented matrigel. ARID3A OE and KD lentiviral vector construction and propagation, transfection conditions, placental dissection as well as details of additional method are in provided in Supplementary information. Data sets analyzed in our meta-analyses (Fig. 1) were obtained from Gene Expression Omnibus (GEO): GSE18507, GSE49354, and GSE9984. Sequencing data are submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE79638.
Supplementary Material
Highlights.
ARID3A is highly expressed during mammalian placentation
Arid3a KO placentas have structural defects in placental organization
Arid3a KO placentas results in aberrant expression of subtype-specific placental genes
ARID3A loss or gain disrupts key TE-associated gene expression in human TS-like cells
Acknowledgments
We thank Dr. Richard Finnell for help with human pluripotent cells; Nancy Otto for help with placental IHC; Luis Coletta for RNA sample processing and NGS performed at the NGS core of the MD Anderson. This work was supported by awards from the NIH (R01GM112722) to JK and from the NIH (R01CA31534), CPRIT (RP120348 and RP120459) and the Marie Betzner Morrow Centennial Endowment to HOT.
References
- Ananth CV, Keyes KM, Wapner RJ. Pre-eclampsia rates in the United States, 1980–2010: age-period-cohort analysis. Bmj. 2013;347:f6564. doi: 10.1136/bmj.f6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge SA, Minhas A, Whiteley KJ, Qu D, Sled JG, Kingdom JC, Adamson SL. Effects of reduced Gcm1 expression on trophoblast morphology, fetoplacental vascularity, and pregnancy outcomes in mice. Hypertension. 2012;59:732–739. doi: 10.1161/HYPERTENSIONAHA.111.183939. [DOI] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell stem cell. 2011;9:144–155. doi: 10.1016/j.stem.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilban M, Haslinger P, Prast J, Klinglmuller F, Woelfel T, Haider S, Sachs A, Otterbein LE, Desoye G, Hiden U, et al. Identification of novel trophoblast invasion-related genes: heme oxygenase-1 controls motility via peroxisome proliferator-activated receptor gamma. Endocrinology. 2009;150:1000–1013. doi: 10.1210/en.2008-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Chen CY, Yang YC, Su TH, Chen H. Decreased placental GCM1 (glial cells missing) gene expression in pre-eclampsia. Placenta. 2004;25:413–421. doi: 10.1016/j.placenta.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Dietrich JE, Hiiragi T. Stochastic patterning in the mouse pre-implantation embryo. Development. 2007;134:4219–4231. doi: 10.1242/dev.003798. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Telugu BP, Roberts RM. Model systems for studying trophoblast differentiation from human pluripotent stem cells. Cell and tissue research. 2012;349:809–824. doi: 10.1007/s00441-012-1371-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founds SA, Terhorst LA, Conrad KP, Hogge WA, Jeyabalan A, Conley YP. Gene expression in first trimester preeclampsia placenta. Biological research for nursing. 2011;13:134–139. doi: 10.1177/1099800410385448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier T, Therond P, Handschuh K, Tsatsaris V, Evain-Brion D. PPARgamma and early human placental development. Current medicinal chemistry. 2008;15:3011–3024. doi: 10.2174/092986708786848677. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- Hemberger M, Udayashankar R, Tesar P, Moore H, Burton GJ. ELF5-enforced transcriptional networks define an epigenetically regulated trophoblast stem cell compartment in the human placenta. Human molecular genetics. 2010;19:2456–2467. doi: 10.1093/hmg/ddq128. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome research. 2010;20:458–472. doi: 10.1101/gr.101469.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PG, Canver MC, Rhee C, Ross SJ, Harriss JV, Tu HC, Orkin SH, Tucker HO, Daley GQ. Interferon-alpha signaling promotes embryonic HSC maturation. Blood. 2016 doi: 10.1182/blood-2016-01-689281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RJ, Lam C, Qian C, Yu KF, Maynard SE, Sachs BP, Sibai BM, Epstein FH, Romero R, Thadhani R, et al. Soluble endoglin and other circulating antiangiogenic factors in preeclampsia. The New England journal of medicine. 2006;355:992–1005. doi: 10.1056/NEJMoa055352. [DOI] [PubMed] [Google Scholar]
- Li Y, Moretto-Zita M, Soncin F, Wakeland A, Wolfe L, Leon-Garcia S, Pandian R, Pizzo D, Cui L, Nazor K, et al. BMP4-directed trophoblast differentiation of human embryonic stem cells is mediated through a DeltaNp63+ cytotrophoblast stem cell state. Development. 2013;140:3965–3976. doi: 10.1242/dev.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Parast MM. BMP4 regulation of human trophoblast development. The International journal of developmental biology. 2014;58:239–246. doi: 10.1387/ijdb.130341mp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Fierro ML, Reyes-Oliva EA, Cabral-Pacheco GA, Garza-Veloz I, Aceves-Medina MC, Luevano M, Barbosa-Cisneros OY, Galvan-Valencia M, Yahuaca-Mendoza P, Delgado-Enciso I, et al. MYC-induced nuclear antigen (MINA) and preeclampsia. Hypertension in pregnancy. 2016:1–13. doi: 10.3109/10641955.2015.1130833. [DOI] [PubMed] [Google Scholar]
- Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, Unadkat JD. Profiling gene expression in human placentae of different gestational ages: an OPRU Network and UW SCOR Study. Reproductive sciences. 2008;15:866–877. doi: 10.1177/1933719108322425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineris I, Grassi E, Ala U, Di Cunto F, Provero P. Evolution of promoter affinity for transcription factors in the human lineage. Molecular biology and evolution. 2011;28:2173–2183. doi: 10.1093/molbev/msr027. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Developmental biology. 2013;375:54–64. doi: 10.1016/j.ydbio.2012.12.008. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Perez-Sepulveda A, Torres MJ, Khoury M, Illanes SE. Innate immune system and preeclampsia. Frontiers in immunology. 2014;5:244. doi: 10.3389/fimmu.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee C, Lee BK, Beck S, Anjum A, Cook KR, Popowski M, Tucker HO, Kim J. Arid3a is essential to execution of the first cell fate decision via direct embryonic and extraembryonic transcriptional regulation. Genes & development. 2014;28:2219–2232. doi: 10.1101/gad.247163.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nature genetics. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Roberts JM, Taylor RN, Musci TJ, Rodgers GM, Hubel CA, McLaughlin MK. Preeclampsia: an endothelial cell disorder. American journal of obstetrics and gynecology. 1989;161:1200–1204. doi: 10.1016/0002-9378(89)90665-0. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nature reviews Genetics. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. doi: 10.1242/dev.017178. [DOI] [PubMed] [Google Scholar]
- Scott IC, Anson-Cartwright L, Riley P, Reda D, Cross JC. The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Molecular and cellular biology. 2000;20:530–541. doi: 10.1128/mcb.20.2.530-541.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister NE, Story CM. Human placental Fc receptors and the transmission of antibodies from mother to fetus. Journal of reproductive immunology. 1997;37:1–23. doi: 10.1016/s0165-0378(97)00068-5. [DOI] [PubMed] [Google Scholar]
- Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Berghout J, Campbell J, Corbani LE, Forthofer KL, Frost PJ, et al. The mouse Gene Expression Database (GXD): 2014 update. Nucleic acids research. 2014;42:D818–824. doi: 10.1093/nar/gkt954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Suzuki S. Clinical significance of pregnancies with circumvallate placenta. The journal of obstetrics and gynaecology research. 2008;34:51–54. doi: 10.1111/j.1447-0756.2007.00682.x. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- Waite LL, Person EC, Zhou Y, Lim KH, Scanlan TS, Taylor RN. Placental peroxisome proliferator-activated receptor-gamma is up-regulated by pregnancy serum. The Journal of clinical endocrinology and metabolism. 2000;85:3808–3814. doi: 10.1210/jcem.85.10.6847. [DOI] [PubMed] [Google Scholar]
- Webb CF, Bryant J, Popowski M, Allred L, Kim D, Harriss J, Schmidt C, Miner CA, Rose K, Cheng HL, et al. The ARID family transcription factor bright is required for both hematopoietic stem cell and B lineage development. Molecular and cellular biology. 2011;31:1041–1053. doi: 10.1128/MCB.01448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nature biotechnology. 2002;20:1261–1264. doi: 10.1038/nbt761. [DOI] [PubMed] [Google Scholar]
- Zeisler H, Hund M, Verlohren S. The sFlt-1:PlGF Ratio in Women with Suspected Preeclampsia. The New England journal of medicine. 2016;374:1785–1786. doi: 10.1056/NEJMc1602338. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. First cell fate decisions and spatial patterning in the early mouse embryo. Seminars in cell & developmental biology. 2004;15:563–572. doi: 10.1016/j.semcdb.2004.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.