Abstract
Caspases of the non-canonical inflammasome (caspases-4/5/11) directly bind endotoxin (LOS/LPS) and can be activated in the absence of any co-factors. Models of LPS-induced caspase activation have postulated that 1:1 binding of endotoxin monomers to caspase trigger caspase oligomerization and activation, analogous to that established for endotoxin-induced activation of MD-2/TLR4. However, using metabolically radiolabeled LOS and LPS, we now show high affinity and selective binding of caspase-4 to high molecular-mass aggregates of purified endotoxin and to endotoxin-rich outer membrane vesicles without formation of 1:1 endotoxin:caspase complexes. Our findings thus demonstrate markedly different endotoxin recognition properties of caspase-4 from that of MD-2/TLR4 and strongly suggest that activation of caspase-4 (and presumably -5 and -11) are mediated by interactions with activating endotoxin-rich membrane interfaces rather than by endotoxin monomers.
Keywords: Caspase-4, Caspase-11, non-canonical inflammasome, lipopolysaccharide, outer membrane vesicle
Introduction
LOS and LPS are unique and highly abundant glycolipids present in the outer leaflet of the outer membrane of most Gram-negative bacteria. After extraction and purification, these molecules spontaneously aggregate even at sub-nM concentrations and form LPS-rich interfaces reflecting their amphipathic properties. A remarkable feature of these molecules is their potent activation of innate immunity, due in substantial part to their ability to activate TLR4 at pM concentrations.1 This potency and specificity requires a complex multi-step set of ordered protein-LPS interactions that are initiated by proteins (e.g., LPS-binding protein, LBP) that interact with and alter LPS-rich interfaces leading to extraction and delivery of LPS monomers (to CD14 and then to MD-2) as needed for presentation and activation of TLR4.
Activation of TLR4 signaling is initiated at the cell surface of TLR4-expressing cells and potentially within endosomal compartments after internalization of activated MD-2/TLR4.2,3 More recently, an additional cellular mechanism capable of detecting intracellular LPS via a cytosolic sensor and causing pyroptotic cell death – the non-canonical inflammasome – was recognized.4–7 Importantly, this non-canonical inflammasome can be activated by direct binding of an inflammatory caspase (murine caspase-11; human caspase-4 and -5, 60% and 54% sequence identity to caspase-11, respectively) to LPS, in contrast to canonical inflammasomes which require accessory proteins to generate a complex that efficiently activates caspase-1.8 Limited studies of LPS-caspase-4/5/11 interactions have been performed, but the physical state of the LPS used was not rigorously analyzed.8,9 Many models of LPS-induced caspase activation have presumed a 1:1 interaction between protein and LPS monomers, but without direct testing of that concept. Based on studies of protein-LPS interactions leading to TLR4 activation and the behavior of isolated LPS, it seems more likely that caspase initially interacts with LPS-rich interfaces and is activated in situ (i.e., on the interface) or after extraction of an endotoxin monomer.
To begin to directly test these hypotheses, we have in this study assessed the physical state of LPS recognized by caspase-4 during initial interactions. Specifically, we have characterized quantitatively and qualitatively human caspase-4-LPS binding and demonstrate an apparently preferential interaction of caspase-4 with LPS presented as part of LPS-rich interfaces, both in isolated form and as part of spontaneously shed outer membrane vesicles (OMVs).
Materials and methods
Proteins and bacteria
Purified recombinant His/3XFLAG-tagged human caspase-4 (C258A) was expressed in sf21 insect cells, and was a gift from Dr Feng Shao (National Institute of Biological Sciences, Beijing 102206, China). This point mutation renders the protein catalytically inactive and minimizes autoproteolysis. rBPI21 and LBP were gifts from Xoma (Berkley, CA, USA), and CD14 was a gift from Amgen (Thousand Oaks, CA, USA). rBPI21 is a recombinant derivative of the amino-terminal portion of BPI that possesses the LPS recognition properties of holo-BPI.10 BN and BN2pL strains of Escherichia coli were generous gifts of M Stephen Trent (University of Georgia, Athens, GA).11 Unlabeled Re, Rc, and Ra E. coli LPS and E. coli lipid A were obtained from Enzo Life Sciences (Farmingdale, NY). [1, 2-14C]- Or [3H]-acetic acid sodium salt was purchased from Moravek Biochemicals Inc. (Brea, CA, USA). [14C]-Oleic acid was from Perkin Elmer (Waltham, MA). Chromatographic matrices (Superdex 200, Sephracryl 400, and Ni2+ FF-sepharose) were purchased from GE Healthcare (Piscataway, NJ, USA). Dulbecco’s phosphate buffered saline without calcium and magnesium and RPMI 1640 were purchased from Cellgro (Manassas, VA, USA). Lysostaphin and fatty acid standards [12:0, 14:0, 16:0, 12:0(3-OH), 14:0(3-OH)] were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Metabolic labeling and isolation of LOS and LPS aggregates and OMVs
[3H]-LOS was isolated from an aceE mutant of Neisseria meningitidis serogroup B after metabolic labeling as described previously.12,13 Size-exclusion chromatography of LOS aggregates was performed in PBS without divalent cations as previously described.14 Recovery of [3H]-LOS was ≥ 70%. Conditioned medium of metabolically labeled meningococcal cultures was fractionated as previously described to recover radiolabeled OMVs.15 The LOS content of the recovered OMVs was determined based on the known specific radioactivity of the metabolically labeled LOS and the fraction of the total OMV radioactivity that was due to radiolabeled LOS.15 Purified LOS aggregates were used to produce and purify monomeric [3H]-LOS:sCD14 complexes as previously described.1,16 E. coli were grown shaking at 37°C in the presence of 10 µci/ml [1,2-14C]-acetic acid sodium for 7 h and washed twice in PBS. Lipid A from E. coli was isolated as described.17 The specific radioactivity (counts per minute (cpm)/ng) of lipid A was estimated by preparing unlabeled lipid A in parallel and subjecting unlabeled material to LC-MS quantitation of lipid A-specific 14:0(3-OH). The specific radioactivity of 14:0(3-OH) was used to derive the specific radioactivity of the other fatty acyl constituents of lipid A and of lipid A.
Generation of metabolically labeled staphylococcal membrane protoplasts
Radiolabeled membrane protoplasts were generated from Staphylococcus aureus SA113 metabolically labeled with [14C]-oleic acid as previously described.18 The sedimented protoplasts were re-suspended in RPMI supplemented with 30% raffinose after confirmation of absence of any remaining viable bacteria by plating on chocolate agar. Approximately 90% of incorporated [14C]-oleate is in phospholipids.18 Thus, recovery of bacterial cpm in protoplasts was used to estimate recovery of bacterial phospholipids in the protoplasts and phospholipid content (ca. 1 µg/108 bacteria).
Ni2+-Sepharose co-capture assay
Radiolabeled LOS (or lipid A) and His-tagged caspase-4 were incubated in PBS without divalent cations for 30 min at 37°C in siliconized 1.5 ml tubes. Complexes were captured by addition of washed Ni2+ FF-Sepharose beads and incubation on a rotor-mixer for 45 min at room temperature. Samples were brought to 1 ml with PBS, pelleted and washed twice with PBS. The amount of [3H]-LOS in the supernatant, each of the washes and the bead pellet was measured by liquid scintillation spectroscopy. For experiments comparing binding of caspase-4 to protoplasts and to OMVs, incubations were conducted in RPMI supplemented with 30% raffinose. The percent of [3H]-LOS/OMV/protoplast captured equaled [(radioactivity in pellet)/(total radioactivity recovered)]*100. Competition assays were conducted by addition of increasing concentrations of rBPI21 or of unlabeled E. coli LPS at the same time as caspase-4 and [3H]-LOS. Caspase-4-dependent LOS capture was determined by subtracting the % of cpm captured in the caspase-4 negative controls from the % of cpm captured in samples with caspase-4 added.
Isolation and characterization of co-captured radiolabeled LOS aggregates and OMVs
[3H]-LOS aggregates or OMVs were incubated with 100 ng caspase-4 for 30 min at 37°C and subjected to Ni2+-Sepharose co-capture as described above. Captured material was eluted from Ni2+-Sepharose beads using 20 mM phosphate, 0.5 M NaCl, 0.5 M imidazole (pH 7.5). For fatty acid analysis, eluted samples were treated with 4 N HCl, then 4 N NaOH at 100°C followed by neutralization and Bligh-Dyer extraction.15 Samples containing recovered [3H]-free fatty acids were applied to a 5μ Grace Prevail organic acid column (10 mm × 250 mm) (Waters, Inc. Milford, MA, USA) as described previously.19 Recoveries of cpm exceeded 85%. Data are plotted as percent of the height of the maximum radiolabeled peak.
Statistical analysis and data presentation
Indicated statistical tests were performed using Prism 6 (GraphPad Software, San Diego, CA, USA). Graphs were compiled and annotated with OmniGraffle Pro 6 (Omni Group, Seattle, WA, USA).
Results
Size-exclusion chromatography analysis of purified metabolically labeled LOS aggregates
To better define the molecular and physical determinants and products of human caspase-4 interactions with endotoxin, we began with studies of aggregates of metabolically radiolabeled LOS (LOS-agg) derived from N. meningitidis. These aggregates have been extensively used in the past for studies of endotoxin interaction with several different human proteins that regulate delivery and/or activation of TLR4 by extracted monomers of endotoxin.1,2,14–16 Moreover, the availability of an aceE mutant of the meningococci permits metabolic labeling with [14C]-acetate to a specific radioactivity of purified [14C]-LOS of up to 3,000 cpm/pmol and with [3H]-acetate up to 25,000 cpm/pmol. In both radiolabeled forms, every LOS molecule is uniformly and selectively radiolabeled within the six fatty acyl chains of LOS.15 These properties of the radiolabeled LOS have greatly facilitated measurements of the physical size of complexes containing LOS and the stoichiometry and affinity of interactions with specific endotoxin binding proteins.1,13,14,20,21
We first analyzed our purified [14C]-LOS-agg by size-exclusion chromatography in Sephacryl S-400. As shown in Figure 1A, nearly all applied [14C]-LOS was recovered as a single major peak that eluted in the void volume, indicative of aggregates of LOS of large size roughly corresponding to polypeptides of Mr ≥ 8 × 106.
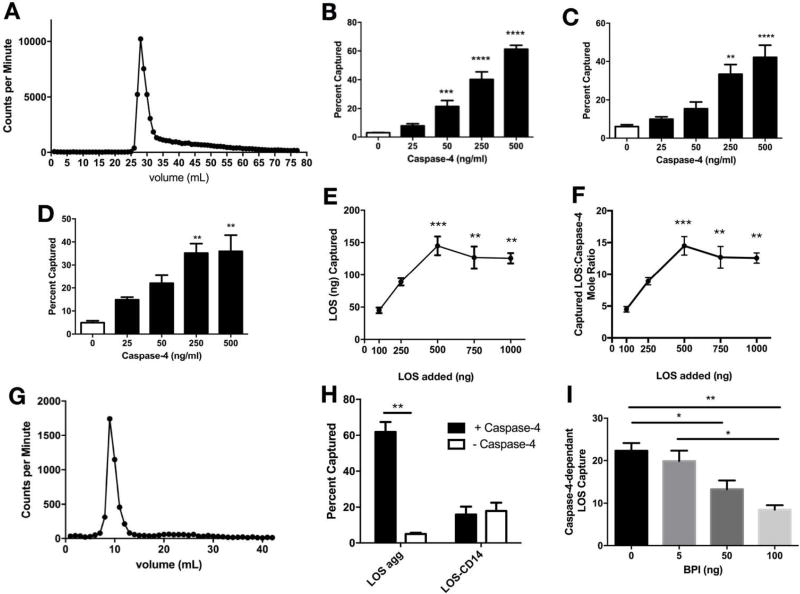
Figure 1.
Selective binding of human caspase-4 to aggregates of purified meningococcal LOS. A) LOS aggregates were incubated for 30 min at 37°C in PBS, then analyzed by size-exclusion Sephacryl S400 chromatography. Elution of [3H]-LOS was monitored by presence of radioactivity (counts per min) in eluted fractions. Capture of [3H]-LOS aggregates after incubation of 500 (B), 50 (C), and 5 (D) ng/ml LOS with increasing amounts of caspase. ** P < 0.01, *** P < 0.001, **** P < 0.0001 vs. no caspase present (1-way ANOVA and Dunnet’s post-test) E) Capture of LOS following incubation of increasing amounts of [3H]-LOS with 100 ng caspase 4 in 0.2 ml PBS, expressed as ng LOS captured (E) or the mol ratio of captured LOS/caspase-4 (F). ** P < 0.01, *** P < 0.001 (1-way ANOVA and Dunnet’s post-test vs 100 ng loaded). G) Superdex 200 chromatography of LOS:caspase-4 complexes that were eluted from Ni2+-Sepharose beads. H) [3H]-LOS aggregates (10 ng) or [3H]-LOS /sCD14 complexes (each containing 10 ng LOS) were incubated with 100 ng caspase-4 in 0.2 ml PBS for 30 min at 37°C followed by nickel capture. ** P < 0.01, paired T-test. I) Dose-dependent inhibition by rBPI21 of caspase (100 ng)-dependent capture of LOS aggregates (10 ng) in 0.2 ml of PBS. * P < 0.05, ** P < 0.01, one-way ANOVA and Tukey’s post-test. Results shown represent the mean ± SEM of 3 or more determinations.
Dose-dependent interactions of [3H]-LOS-agg with His6-rcaspase-4
Binding experiments were carried out in PBS, which is compatible with assays that depend on co-capture of LOS to activated Ni2+-beads to monitor LOS interactions with His-tagged recombinant caspase-4. These assays were initially carried out with 500 ng/ml LOS (100 nM; Figure 1B) and subsequently with 50 ng/ml LOS (10 nM; Figure 1C) and 5 ng/ml LOS (1 nM; Figure 1D) and increasing concentrations of caspase-4. Recombinant catalytically inactive (C258A) caspase-4 was used to assay the initial interaction of caspase-4 with endotoxin preceding caspase activation (autoproteolysis) and avoid the potential complicating effects of caspase fragmentation on detection of His-tag-dependent caspase-endotoxin complexes. Similar caspase dose-dependent co-capture of LOS was observed at each concentration of LOS tested (Figures 1B–D), indicating nM (or higher) affinity of caspase-4 for LOS-agg. At each LOS dose tested, maximal co-capture of LOS was observed at 100 ng caspase-4 per assay (0.2 ml), corresponding to approximately 10 nM caspase-4.
Molar ratio of LOS:caspase-4 complexes
Early models of endotoxin-murine caspase-11 or human caspase-4 complexes have postulated formation of monomeric endotoxin:caspase complexes8,22 as has been documented for complexes of endotoxin with CD14, MD-2, or MD-2/TLR4.1,13,21,23 The avid binding of caspase-4 to aggregates of LOS having ≥ 1000 molecules of LOS per aggregate suggested physical and molecular determinants of endotoxin interactions with caspase-4 that were markedly different from that of endotoxin interactions with MD-2 and MD-2/TLR4. However, it was conceivable that while the initial interacting form of endotoxin with caspase-4 is a large endotoxin-rich supramolecular structure, the final product is a monomeric complex like that of MD-2 and MD-2/TLR4. We addressed this possibility by: i) measuring the apparent mol ratio of LOS to caspase-4 in co-captured complexes at maximal caspase-4-dependent co-capture of LOS; and ii) assessing by size-exclusion chromatography the size of the LOS:caspase-4 complex (final product).
Figure 1E shows LOS dose-dependent co-capture following incubation with 100 ng caspase-4. Maximum LOS co-capture corresponded to an apparent mol ratio of ~14 mol LOS/mol caspase-4 (Figure 1F), using a molecular mass for LOS of 5 kDa and of 50 kDa for recombinant caspase-4. Moreover, following elution of the co-captured LOS:caspase-4 complex by imidazole and subsequent size-exclusion chromatography, the eluted radiolabeled material was recovered in the void volume (corresponding to globular proteins of Mr ≥ 6 × 105, Figure 1G), consistent with aggregates of LOS containing bound caspase-4 and not oligomers of monomeric LOS:caspase-4 complexes.
Caspase-4 does not bind to LOS:sCD14 or extract LOS from LOS:sCD14
The experiments above demonstrate the capacity of caspase-4 to bind to endotoxin-rich supramolecular interfaces formed by aggregates of purified endotoxin. However, these findings do not exclude the possibility that caspase can bind to both supramolecular interfacial and monomeric forms of endotoxin. Because of the amphipathic properties of endotoxin, endotoxin monomers do not exist stably in isolated form in aqueous environments but rather as a complex with a (host) protein that can extract an endotoxin monomer from an interface (e.g., CD14) or receive an endotoxin monomer that is bound to another protein molecule (e.g., MD-2 and MD-2/TLR4 from endotoxinm:CD14 and LOSm:albumin).1,14,21,24 To test whether caspase-4 shows that interactive ability as well, we measured by co-capture assay interaction of [14C]-LOS:sCD14 with His-tagged caspase-4. As shown in Figure 1H, caspase-4 neither binds to nor extracts LOS from LOS:sCD14.
rBPI21 inhibits binding of caspase-4 to LOS
Other mammalian LPS-binding proteins have been described [e.g., bactericidal/permeability-increasing protein (BPI)] that bind preferentially and with high affinity (low nM) to aggregates of LPS or to LPS in the outer membrane of intact bacteria or OMVs.10,25,26 To determine if BPI and caspase-4 share overlapping or otherwise mutually incompatible binding sites within endotoxin-rich interfaces, we tested the effect of increasing concentrations of rBPI21 on caspase-4-dependent co-capture of radiolabeled LOS. Figure 1I demonstrates dose-dependent inhibition by rBPI21 of caspase-4-LOS aggregate interactions. Approximately 50% inhibition was observed at a 1:1 mol ratio of caspase-4 and rBPI21, consistent with a high affinity interaction of caspase-4 with LOS aggregates.
Binding of human caspase-4 to purified meningococcal OMVs
LPS is an integral component of most Gram-negative bacterial outer membranes and hence requires extraction with organic solvents and selective precipitation/fractionation to be recovered in purified form.17 Therefore, it is extremely unlikely that caspase will be exposed under natural conditions to isolated LPS rather than to membranes containing LPS or soluble protein complexes to which LPS is bound. The former could include OMVs that are shed by Gram-negative bacteria.15,27 Thus, we tested, by co-capture assay, the ability of caspase-4 to interact with purified metabolically labeled LOS-rich meningococcal OMVs.15
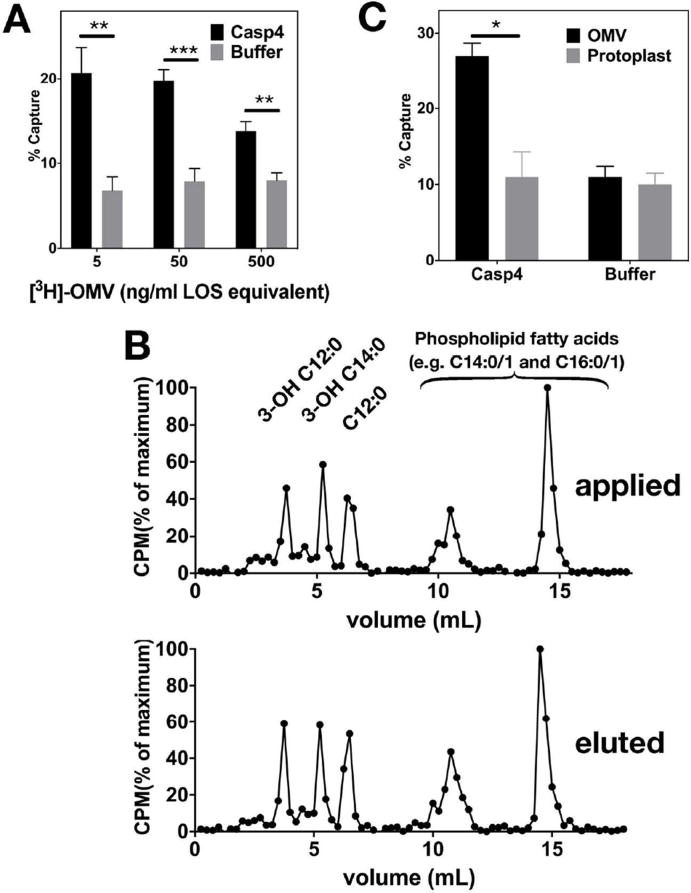
Figure 2A shows dose- and caspase-4-dependent capture of [3H]-OMVs. At an input of OMVs of only 3 ng/ml LOS (corresponding to LOS content of 3 × 105 meningococci/ml), caspase-4-specific capture of OMVs could be demonstrated. Elution of co-captured radiolabeled material followed by fatty acid analysis revealed that the eluted (Figure 2B, lower panel) material had the same fatty acyl composition as that of the OMVs that were added to and incubated with caspase-4 (Figure 2B, upper panel). Note that the fatty acyl composition of the OMVs includes substantial amounts of longer-chain non-hydroxylated fatty acids (14:0/1 and 16:0/1) not present in LOS, reflecting the presence of outer membrane-derived LOS and phospholipids in the purified OMVs.15 The compositional analysis of the recovered co-captured material indicates that interaction of caspase-4 with membranes containing LOS at the exposed interface is not followed by extraction of individual LOS molecules from the LOS-rich outer leaflet of the OMV.
Figure 2.
Binding of human caspase-4 to purified meningococcal OMVs, but not to staphylococcal protoplasts. A) Increasing amounts of OMVs, quantified by LOS content, were incubated with or without 200 ng/ml of caspase-4 in PBS for 30 min at 37°C. ** P < 0.01, *** P < 0.001 (unpaired T-tests). B) Captured OMVs were eluted from beads and the fatty acid composition of starting material (top panel) and captured OMVs (lower panel) was determined by HPLC (see methods). Identities of peaks were determined using purified commercial and laboratory-derived standards. Results are representative of two experiments. C) OMVs containing ca. 100 ng LOS and 50 ng phospholipids and protoplasts containing ca. 100 ng phospholipids were incubated with 100 ng of caspase-4 in 0.2 ml of RPMI 1640 supplemented with 30% raffinose for 30 min at 37°C and then captured with nickel beads. * P < 0.05 (unpaired T-test). Results shown represent the mean ± SEM, n = 2.
Caspase-4 does not bind to cell wall-depleted membrane protoplasts of S. aureus
To test if the interaction of caspase-4 with meningococcal OMVs was selective for membranes containing endotoxin (LOS), we repeated co-capture experiments with OMVs and, for comparison, membrane protoplasts derived from S. aureus. These membranes are highly sensitive to added secretory phospholipases A2 that require binding and penetration of the outer leaflet of the membrane to produce phospholipid hydrolysis.18 In contrast to the ability of caspase-4 to bind to LOS-rich OMVs, no caspase-dependent co-capture of the radiolabeled membrane protoplasts was observed (Figure 2C). These experiments suggest that the capacity of caspase-4 to bind to biological membranes is selective for those membranes containing exposed endotoxin.
Caspase-4 binding to other LPS and lipid A species
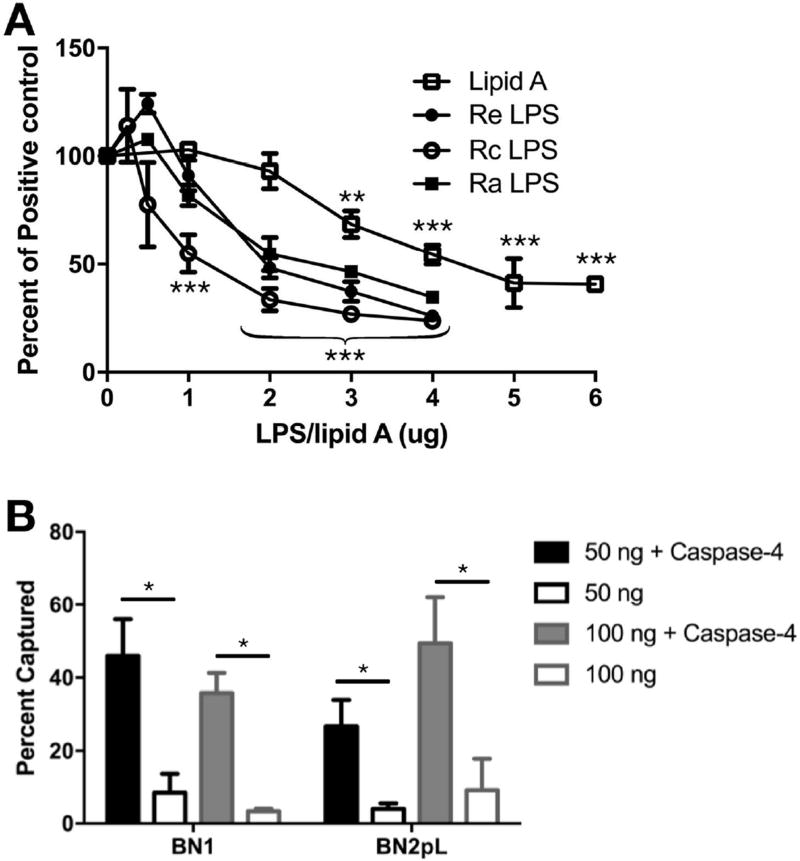
To extend our analyses to other endotoxin species, we compared the ability of E. coli lipid A and LPS of increasing core polysaccharide chain length (Ra > Rc > Re chemotype) to inhibit (by competition) caspase-4 binding and co-capture of radiolabeled LOS. Figure 3A shows similar dose-dependent effects of all three LPS chemotypes indicating no major effect of changes in the length of core polysaccharide in E. coli LPS on its interactions with caspase-4. Somewhat higher concentrations of lipid A were required to achieve comparable inhibition.
Figure 3.
Caspase-4 binds to E. coli lipid A and various chemotypes of LPS. A) Increasing amounts of unlabeled LPS or lipid A were incubated with 10 ng [3H]-LOS and 100 ng caspase-4 followed by Ni2+-Sepharose capture. Results are expressed as percent of [3H]-LOS captured relative to positive control (no LPS added). Statistical significance of differences vs. no LPS control, ** P < 0.01, *** P < 0.001, n = 3. B) 50 ng or 100 ng of [14C]-BN1 (hex-acyl LPS) or [14C]-BN2pL (tetra-acyl LPS) were incubated with 100 ng capsase-4 and subjected to capture. * P < 0.05, 1-way ANOVA and Tukey’s post-test, n = 3. Results shown represent the mean ± SEM.
Preliminary findings of others have suggested that alterations of lipid A structure that reduce the potency of lipid A and LPS as MD-2/TLR4 agonists also reduce activation of murine caspase-11 and human caspase-4.5,8 To test if differences in acylation of E. coli lipid A affect binding to caspase-4, as judged by the co-capture assay, we isolated lipid A from two metabolically labeled strains of E. coli that had been engineered via mutagenesis to generate relatively homogeneous and distinct lipid A structures.11 Both strains produce lipid A with two phosphates, but strain BN1 produces a typical (potently bioactive) hexa-acylated lipid A whereas BN2pL produces tetra-acyl lipid A with markedly reduced MD-2/TLR4 agonist potency.11 Similar caspase-dependent co-capture of radiolabeled lipid A from both strains was observed (Figure 3B).
Discussion
Using metabolically radiolabeled meningococcal LOS and E. coli LPS (lipid A), we have demonstrated high affinity (nM) binding of recombinant enzymatically inactive caspase-4 to endotoxin when these purified endotoxin species are presented as high Mr supramolecular aggregates. Caspase binding was also observed to endotoxin-rich meningococcal OMVs but not to cell wall-depleted cytoplasmic membrane protoplasts derived from Gram-positive bacteria (S. aureus), suggesting a selective affinity of caspase-4 for the Gram-negative bacterial outer membrane and perhaps other membrane interfaces that become rich in endotoxin (see below; Figure 4). In contrast, no interaction of caspase-4 was detected when endotoxin (LOS) was presented as a monomeric complex with sCD14. Taken together, these findings indicate markedly different endotoxin recognition properties of human caspase-4 (and, presumably, murine caspase-11) in comparison to MD-2/TLR4, which requires presentation of endotoxin as a monomeric complex with CD14 for most efficient endotoxin binding and displays little or no reactivity with aggregates of purified endotoxin.
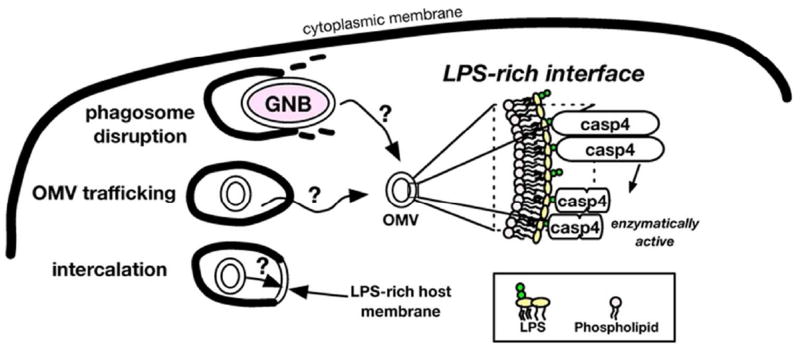
Figure 4.
Proposed role for LPS-rich interfaces in binding and activation of caspase-4. We propose that an LPS-rich interface provides a supramolecular platform on which caspase-4 molecules can assemble in the appropriate proximity and/or orientation to induce activation and autoproteolysis of caspase-4. LPS-rich interfaces could be exposed to the cytoplasm via escape or disruption of phagosomes containing viable Gram-negative bacteria (GNB) and any shed OMVs,42 uptake and trafficking of extracellular OMVs out of early endosomes,41 or intercalation of LPS into host membranes and topological rearrangement.43
The use of metabolically labeled purified LOS (aggregates) and OMVs together with His-tagged caspase-4 and Ni2+-Sepharose made possible sensitive and quantitative assay of caspase-LOS (OMV) interactions, including the apparent stoichiometry of these interactions. The high mol ratio of LOS bound to caspase-4 (Figure 1F), the high Mr of eluted LOS-caspase complexes (Figure 1G), and the unmodified lipid composition of OMVs recovered after co-capture by His6-caspase-4 (Figure 2B) are all compatible with accumulation of caspase-4 on endotoxin-rich supramolecular structures, without accompanying extraction of endotoxin monomers and formation of monomeric endotoxin/caspase-4 complexes. We used a catalytically-inactive caspase-4 mutant as a probe for the primary binding interactions between endotoxin and caspase-4 to preclude endotoxin-induced autoproteolysis and possible separation of the endotoxin-binding domain of caspase-4 from the N-terminal polyhistidine-tag. Although this experimental model eliminated the possibility of testing how caspases-4/5/11 are activated by LPS, the characteristics of the initial caspase-4-LPS interactions that were revealed by this approach suggest novel hypotheses that can be tested in the future.
For canonical inflammasomes and pyroptosomes, incorporation of caspases into protein complexes orients them in sufficient proximity to initiate activating proteolytic events.28,29 Given the ability of caspases-4/5/11 to be activated by LPS without co-factors, we hypothesize that LPS-rich interfaces bring caspases 4/5/11 into sufficient proximity to initiate proteolytic events. The recognition that caspase-4 binds to high molecular mass aggregates of endotoxin may help explain the altered migratory and elution properties of caspase-4 following incubation with activating endotoxins. Thus, the increase in size of caspase-4-containing complexes observed by Shi et al.8 under non-denaturing conditions may reflect the binding of multiple molecules of caspase to large (>> 400 kDa) LPS aggregates rather than, as suggested, the assembly of oligomeric protein complexes.
Our findings of similar caspase-4 binding to bis-phosphoryl hexa-acyl and tetra-acyl lipid A are consistent with prior observations by Shi et al.8 who compared caspase-4 binding to hexa-acyl lipid A vs. tetra-acyl lipid IVA. Unlike hexa-acyl lipid A, lipid IVA failed to cause pyroptotic cell death,8 implying that binding and induced caspase activation represent distinct events with different structural requirements. With respect to the initial binding interactions, it has become apparent that the amino-terminal CARD domain of caspases-4/5/11 is indispensable. Specifically, the importance of basic residues in the CARD domain,8 suggests that negative charges (e.g., phosphates) on lipid A/LPS are important targets. This structural feature is reminiscent of the importance to binding of basic residues in other proteins that interact with LPS-rich interfaces: e.g. LBP,30 BPI,31 Limulus factor C,32 and Limulus anti-LPS factor (LALF).33 The higher concentration of lipid A (vs. Re, Rc, and Ra LPS) required to inhibit binding of caspase-4 to [3H]-LOS (Figure 4A) may reflect differences in dispersion of lipid A relative to LPS in aqueous solution. Alternatively, it may reflect an added contribution of acidic sugars (3-deoxy-d-manno-oct-2-ulosonic acid, Kdo) and substituted phosphates in the inner core region of LPS to initial electrostatic interactions of LPS with caspases-4/5/11. We did not observe binding of caspase-4 to membrane protoplasts derived from S. aureus (Figure 2C), despite the fact that their most abundant constituent is an anionic phospholipid, phosphatidylglycerol.17 This result implies that net negative charge alone on a membrane surface is insufficient for caspase-4 binding and that other structural and compositional properties of the LPS-rich membrane are likely important.
Hexa- and tetra-acylated LPS bind with similar affinity to MD-2/TLR4 but differ markedly in their ability to induce TLR4 activation (hexaacylated >> tetraacylated).20,34 However, whereas the lower TLR4 agonist potency of under-acylated LPS has been clearly linked to altered properties of monomeric LPS/MD-2 and LPS/MD-2/TLR4 complexes,20,30–34 the absence of detectable formation of monomeric endotoxin/caspase-4 complexes suggests a different means by which variables in lipid A acylation regulate caspase-4 activation. Based on our findings, we predict that the reduced caspase activating properties of under-acylated LPS reflect instead effects of LPS acylation on interfacial properties of LPS-rich surfaces. Indeed, biophysical studies have demonstrated that variables in the chemistry and geometry of lipid A are correlated with differences in the physical and functional properties of LPS-containing membranes.37–39 Of note, factor C of Limulus amebocytes, a pro-coagulant protease that exists in zymogen form until contact with LPS and does not require cofactors for activation,8,32,40 also shows similar structure-activity relationships as MD-2/TLR4 as defined by variables in LPS molecular structure but is most efficiently activated by aggregates of endotoxin or OMVs (Hoppe K et al., manuscript in preparation). As noted above, we did not use catalytically active caspase-4 in order to minimize autoproteolysis and thereby permit accurate assessments of initial binding interactions. Future studies should address the extent to which caspase-4 auto-processing takes place on LPS-rich interfaces and if so, the extent to which caspase-4 fragments remain associated with this supramolecular interface.
In summary, we have presented evidence that endotoxin recognition by caspase-4 is mediated by protein interactions with LPS-rich membrane interfaces. Such interfaces may function to concentrate and activate non-canonical inflammasome caspases when sufficient density and/or appropriate alignment of these proteins are achieved. Better understanding the mechanistic basis for endotoxin recognition will require studies to define the precise LPS structural features (molecular and supramolecular) that confer caspase-4/5/11 binding and/or activation. Our demonstration that caspase-4 can bind directly to meningococcal OMVs is particularly important given recent evidence that OMVs from a variety of Gram-negative bacteria can activate caspase-11-dependent pyroptosis in murine cells.41 Thus, LPS-rich interfaces may be presented to cytoplasmic caspases either by shedding of OMVs from intracellular (cytosolic) bacteria, uptake and release of extracellular OMVs from early endosomes, or perhaps by uptake and fusion of extracellular endotoxin-rich complexes with host cell membranes that provide an alternative membrane interface accessible to cytosolic caspases (Figure 4, schema).
References
- 1.Gioannini TL, Teghanemt A, Zhang D, et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc Natl Acad Sci U S A. 2004;101:4186–4191. doi: 10.1073/pnas.0306906101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 3.Kagan JC, Su T, Horng T, et al. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008 doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aachoui Y, Leaf IA, Hagar JA, et al. Caspase-11 protects against bacteria that escape the vacuole. Science. 2013;339:975–978. doi: 10.1126/science.1230751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hagar JA, Powell DA, Aachoui Y, et al. Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science. 2013;341:1250–1253. doi: 10.1126/science.1240988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayagaki N, Warming S, Lamkanfi M, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 7.Broz P, Ruby T, Belhocine K, et al. Caspase-11 increases susceptibility to Salmonella infection in the absence of caspase-1. Nature. 2012;490:288–291. doi: 10.1038/nature11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi J, Zhao Y, Wang Y, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 9.Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 10.Weiss J. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem Soc Trans. 2003;31:785–790. doi: 10.1042/bst0310785. [DOI] [PubMed] [Google Scholar]
- 11.Needham BD, Carroll SM, Giles DK, et al. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc Natl Acad Sci U S A. 2013;110:1464–1469. doi: 10.1073/pnas.1218080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giardina PC, Gioannini T, Buscher BA, et al. Construction of acetate auxotrophs of Neisseria meningitidis to study host-meningococcal endotoxin interactions. J Biol Chem. 2001;276:5883–5891. doi: 10.1074/jbc.M009273200. [DOI] [PubMed] [Google Scholar]
- 13.Prohinar P, Re F, Widstrom R, et al. Specific high affinity interactions of monomeric endotoxin.protein complexes with Toll-like receptor 4 ectodomain. J Biol Chem. 2007;282:1010–1017. doi: 10.1074/jbc.M609400200. [DOI] [PubMed] [Google Scholar]
- 14.Esparza GA, Teghanemt A, Zhang D, et al. Endotoxin•albumin complexes transfer endotoxin monomers to MD-2 resulting in activation of TLR4. Innate Immun. 2012;18:478–491. doi: 10.1177/1753425911422723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Post DM, Zhang D, Eastvold JS, et al. Biochemical and functional characterization of membrane blebs purified from Neisseria meningitidis serogroup B. J Biol Chem. 2005;280:38383–38394. doi: 10.1074/jbc.M508063200. [DOI] [PubMed] [Google Scholar]
- 16.Teghanemt A, Prohinar P, Gioannini TL, et al. Transfer of monomeric endotoxin from MD-2 to CD14: characterization and functional consequences. J Biol Chem. 2007;282:36250–36256. doi: 10.1074/jbc.M705995200. [DOI] [PubMed] [Google Scholar]
- 17.Hankins JV, Madsen JA, Needham BD, et al. The outer membrane of Gram-negative bacteria: lipid A isolation and characterization. Methods Mol Biol. 2013;966:239–258. doi: 10.1007/978-1-62703-245-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koprivnjak T, Weidenmaier C, Peschel A, et al. Wall teichoic acid deficiency in Staphylococcus aureus confers selective resistance to mammalian group IIA phospholipase A(2) and human beta-defensin 3. Infect Immun. 2008;76:2169–2176. doi: 10.1128/IAI.01705-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker JH, Kaufman JW, Zhang DS, et al. Metabolic labeling to characterize the overall composition of Francisella lipid A and LPS grown in broth and in human phagocytes. Innate Immun. 2014;20:88–103. doi: 10.1177/1753425913485308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gioannini TL, Teghanemt A, Zhang D, et al. Purified monomeric ligand.MD-2 complexes reveal molecular and structural requirements for activation and antagonism of TLR4 by Gram-negative bacterial endotoxins. Immunol Res. 2014;59:3–11. doi: 10.1007/s12026-014-8543-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gioannini TL, Teghanemt A, Zhang D, et al. Monomeric endotoxin:protein complexes are essential for TLR4-dependent cell activation. J Endotoxin Res. 2005;11:117–123. doi: 10.1179/096805105X35198. [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Zhao Y, Shao F. Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr Opin Immunol. 2015;32C:78–83. doi: 10.1016/j.coi.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Yu L, Phillips RL, Zhang D, et al. NMR studies of hexaacylated endotoxin bound to wild-type and F126A mutant MD-2 and MD-2·TLR4 ectodomain complexes. J Biol Chem. 2012;287:16346–16355. doi: 10.1074/jbc.M112.343467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teghanemt A, Widstrom RL, Gioannini TL, et al. Isolation of monomeric and dimeric secreted MD-2. Endotoxin.sCD14 and Toll-like receptor 4 ectodomain selectively react with the monomeric form of secreted MD-2. J Biol Chem. 2008;283:21881–21889. doi: 10.1074/jbc.M800672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gioannini TL, Teghanemt A, Zarember KA, et al. Regulation of interactions of endotoxin with host cells. Journal of Endotoxin Research. 2003;9:401–408. doi: 10.1179/096805103225002773. [DOI] [PubMed] [Google Scholar]
- 26.Iovine N, Eastvold J, Elsbach P, et al. The carboxyl-terminal domain of closely related endotoxin-binding proteins determines the target of protein-lipopolysaccharide complexes. J Biol Chem. 2002;277:7970–7978. doi: 10.1074/jbc.M109622200. [DOI] [PubMed] [Google Scholar]
- 27.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kagan JC, Magupalli VG, Wu H. SMOCs: supramolecular organizing centres that control innate immunity. Nat Rev Immunol. 2014;14:821–826. doi: 10.1038/nri3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandes-Alnemri T, Wu J, Yu JW, et al. The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14:1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamping N, Hoess A, Yu B, et al. Effects of site-directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of lipopolysaccharide (LPS)-binding protein on binding and transfer of LPS and subsequent immune cell activation. J Immunol. 1996;157:4648–4656. [PubMed] [Google Scholar]
- 31.Beamer LJ, Carroll SF, Eisenberg D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koshiba T, Hashii T, Kawabata S. A structural perspective on the interaction between lipopolysaccharide and factor C, a receptor involved in recognition of Gram-negative bacteria. J Biol Chem. 2007;282:3962–3967. doi: 10.1074/jbc.M609198200. [DOI] [PubMed] [Google Scholar]
- 33.Hoess A, Watson S, Siber GR, et al. Crystal structure of an endotoxin-neutralizing protein from the horseshoe crab, Limulus anti-LPS factor, at 1.5 A resolution. EMBO J. 1993;12:3351–3356. doi: 10.1002/j.1460-2075.1993.tb06008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teghanemt A, Zhang D, Levis EN, et al. Molecular basis of reduced potency of underacylated endotoxins. J Immunol. 2005;175:4669–4676. doi: 10.4049/jimmunol.175.7.4669. [DOI] [PubMed] [Google Scholar]
- 35.Park BS, Song DH, Kim HM, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009 doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 36.Kim HM, Park BS, Kim JI, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Snyder S, Kim D, McIntosh TJ. Lipopolysaccharide bilayer structure: effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry. 1999;38:10758–10767. doi: 10.1021/bi990867d. [DOI] [PubMed] [Google Scholar]
- 38.Brandenburg K, Seydel U. Investigation into the fluidity of lipopolysaccharide and free lipid A membrane systems by Fourier-transform infrared spectroscopy and differential scanning calorimetry. Eur J Biochem. 1990;191:229–236. doi: 10.1111/j.1432-1033.1990.tb19114.x. [DOI] [PubMed] [Google Scholar]
- 39.Richter W, Vogel V, Howe J, et al. Morphology, size distribution, and aggregate structure of lipopolysaccharide and lipid A dispersions from enterobacterial origin. Innate Immun. 2011;17:427–438. doi: 10.1177/1753425910372434. [DOI] [PubMed] [Google Scholar]
- 40.Ding JL, Ho B. Endotoxin detection--from Limulus amebocyte lysate to recombinant factor C. Subcell Biochem. 2010;53:187–208. doi: 10.1007/978-90-481-9078-2_9. [DOI] [PubMed] [Google Scholar]
- 41.Vanaja SK, Russo AJ, Behl B, et al. Bacterial Outer Membrane Vesicles Mediate Cytosolic Localization of LPS and Caspase-11 Activation. Cell. 2016;165:1106–1119. doi: 10.1016/j.cell.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pilla DM, Hagar JA, Haldar AK, et al. Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1321700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schromm AB, Brandenburg K, Rietschel ET, et al. Lipopolysaccharide-binding protein mediates CD14-independent intercalation of lipopolysaccharide into phospholipid membranes. FEBS Lett. 1996;399:267–271. doi: 10.1016/s0014-5793(96)01338-5. [DOI] [PubMed] [Google Scholar]