Abstract
Helicobacter pylori is the most common bacterial infection worldwide, and virtually all infected persons develop co-existing gastritis. H. pylori is able to send and receive signals from the gastric mucosa, which enables both host and microbe to engage in a dynamic equilibrium. In order to persist within the human host, H. pylori has adopted dichotomous strategies to both induce inflammation as a means of liberating nutrients while simultaneously tempering the immune response to augment its survival. Toll-like receptors (TLRs) and Nod proteins are innate immune receptors that are present in epithelial cells and represent the first line of defense against pathogens. To ensure persistence, H. pylori manipulates TLR-mediated defenses using strategies that include rendering its LPS and flagellin to be non-stimulatory to TLR4 and TLR5, respectively; translocating peptidoglycan into host cells to induce NOD1-mediated anti-inflammatory responses; and translocating DNA into host cells to induce TLR9 activation.
1. Introduction
Helicobacter pylori is a Gram-negative pathogen that infects the stomachs of human hosts; greater than fifty percent of the world’s population is colonized by this pathogen, thus making it the most common bacterial infection worldwide. Although most persons infected with H. pylori remain asymptomatic, a minority of patients can develop gastric or duodenal ulcers (10–15%) or gastric adenocarcinoma (1–3%) (Peek and Blaser 2002; Amieva and Peek 2016). However, a mere one percent of half the world’s population represents a prodigious number of individuals that are plagued by cancer of the stomach, which is why gastric cancer is currently the third leading cause of cancer-related death worldwide and why the World Health Organization has classified H. pylori as a class I carcinogen (Carneiro 2014; Ferlay et al. 2015).
With only 1% of infected individuals progressing toward gastric cancer, H. pylori-induced disease is the exception rather than the rule. Carcinogenic capabilities of H. pylori are mediated by a triumvirate of factors including bacterial virulence constituents, host genetics, and environmental influences, and there is increasing evidence that a dis-symbiosis of these factors mediates disease progression. In order to survive, H. pylori must initiate colonization as well as liberate nutrients from the host; however, a consequence of this is a phenotype that is pro-inflammatory and thus not sustainable for prolonged infection. Therefore, to be successful, H. pylori must develop and maintain strategies to both liberate nutrients and also avoid the host immune response.
2. Toll-like Receptor Modulation by H. pylori
2.1 Toll-like Receptors
Toll-like receptors (TLRs) belong to a class of transmembrane receptors known as pattern recognition receptors (PRRs) that detect pathogen-associated molecular patterns (PAMPs) (Peek et al. 2010; Castano-Rodriguez et al. 2014; Pachathundikandi et al. 2015). PAMPs can originate from a wide array of molecules such as lipids, nucleic acids, and specific proteins that can be derived from organisms of bacterial, viral, or fungal origin. So far there are 10 identified human TLRs, however, not all are manipulated by H. pylori; TLR2 as a homo- or hetero-dimer with TLR1 or TLR6 (lipoteichoic acid, NapA, Hsp60), TLR4 (lipopolysaccharide), TLR5 (flagella), TLR9 (hypo-methylated DNA), and TLR10 hetero-dimerized with TLR2 (hypothesized to detect lipopolysaccharides) have been shown to interact with H. pylori (Peek et al. 2010; Kim et al. 2013; Nagashima et al. 2015; Koch et al. 2015; Pachathundikandi and Backert 2016). TLRs are an essential component of the innate immune system and are expressed both on cell surfaces and intracellularly. They are localized to a wide array of cell types including macrophages and dendritic cells, as well as non-immune cells including gastric epithelial cells. Upon microbial ligand binding to a leucine-rich repeat (LRR) ectodomain, TLRs dimerize and adaptor molecules such as MyD88 or TRIF complex with the intra-cytoplasmic Toll/IL-1 receptor (TIR) domain (Motshwene et al. 2009). Activation of TLRs induces signaling cascades that eventually lead to the transcription of both pro- and anti-inflammatory cytokines, as well as type I interferons. Despite extensive research into the mechanisms of TLR activation, their exact role in H. pylori infection remains controversial.
2.2 TLR4/H. pylori Lipopolysaccharide Interactions
H. pylori lipopolysaccharide (LPS) is a highly specialized structure that is uniquely adapted to maintain persistence within the gastric niche. This is primarily accomplished through decoration of the H. pylori LPS O-antigen with Lewis antigen, which is the outermost domain of the LPS molecule. Lewis antigens mimic host Lewis antigens that are expressed on the apical surface epithelium and within the glands of both the antrum and corpus (Kobayashi et al. 1993; Nogueira et al. 2004). Through this molecular mimicry, H. pylori can evade immune detection, but this heightens the risk of eliciting autoimmune responses (Monteiro 2001). Additionally, H. pylori harbors unique modifications to the lipid A core domain (Perez-Perez et al. 1995; Hajjar et al. 2002; Cullen et al. 2011). The lipid A core is the inner most domain of LPS, referred to as endotoxin, and is the ligand for the TLR4-MD2 immune complex. Compared to other bacterial LPS moieties such as Escherichia coli, H. pylori LPS has ~ 1000 fold less endotoxicity (King et al. 2009; Li et al. 2016), and this reduction is attributed to 3 major modifications to the lipid A core. The first modification is a hypoacylation pattern where H. pylori is tetra-acylated compared to hexa- or penta-acylated chains. Secondly, hypoacy-lated fatty acids have longer carbon chain lengths (2 18-carbon and 2 16-carbon chains) compared to the optimal chain lengths required for robust TLR4 activation. Lastly, H. pylori LPS is hypo-phosphorylated, an adaptation that renders it less susceptible to destruction by cationic antimicrobial peptides (CAMPs) (Cullen et al. 2011; Li et al. 2016). The role of TLR4 in immune activation is controversial. Monoclonal anti-TLR4 antibodies in the presence of H. pylori-epithelial cell co-cultures failed to block IL-8 secretion (Su et al. 2003) and H. pylori infected HEK293 cells transfected with TLR4 failed to induce NF-κB activation (Smith et al. 2003b). However, H. pylori can up-regulate TLR4 expression in gastric epithelial cell lines, and it has been shown that H. pylori may up-regulate TLR4 for adherence to the epithelial cell surface (Su et al. 2003). In contrast, immune recognition of H. pylori has also been shown to be TLR4 independent (Backhed et al. 2003; Ishihara et al. 2004), and H. pylori LPS may actually antagonize TLR4 (Lepper et al. 2005).
2.3 TLR5/H. pylori Flagellin Interactions
The natural ligand of TLR5 is flagellin, specifically the highly conserved N-terminus of the D1 domain (Smith et al. 2003a). Since H. pylori is a flagellated bacterium, and TLR5 is expressed in the gastric epithelium, this pathogen should be capable of inducing TLR5-mediated pro-inflammatory signaling cascades. However, H. pylori flagellin is not recognized by TLR5 (Lee et al. 2003; Gewirtz et al. 2004; Andersen-Nissen et al. 2005), due to a mutation in the conserved domain of FlaA (Andersen-Nissen et al. 2005). This mutation, which occurs in the D0-D1 domain between amino acids 89–96, renders H. pylori flagella inert for TLR5 activation, and when these amino acids are substituted into the corresponding region of the Salmonella enterica serovar Typhimurium FliC, its ability to activate TLR5 is similarly lost (Andersen-Nissen et al. 2005). Taken together, these data suggest an important role for H. pylori FlaA in maintaining persistence within the gastric niche by dampening the innate immune response.
3 TLR9 Functions in the Innate Immune Response
DNA is the fundamental molecule of nearly all living organisms and consequently is normally sequestered within eukaryotic nuclear envelopes, bacterial cell walls, or viral capsids. During the course of infection, DNA can be actively released or released as a result of degradation from invading microbes or damaged host cells. TLR9 is an endosome-bound, transmembrane receptor that detects these aberrant DNAs and orchestrates a cognate immune response (Hemmi et al. 2000). TLR9 expression is most abundant in dendritic cells (DCs), B cells, macrophages, and other antigen presenting cells (APCs); however, it is also expressed in epithelial cells. In plasmacytoid dendritic cells (pDCs) and B cells, TLR9 activation classically leads to the release of pro-inflammatory cytokines and type I interferons, while epithelial responses are less well defined. TLR9 was originally thought to discern pathogenic DNAs based upon the presence of hypo-methylated CpG DNA motifs (which are rare in eukaryotic genomes); however, accumulating evidence suggests that TLR9 can also recognize DNA in a sequence-independent manner via structural components such as the saccharide backbone (Haas et al. 2008; Wagner 2008; Ohto et al. 2015). Therefore, in an attempt to prevent the recognition of “self” DNA, evolutionary pressure has relegated TLR9 to endosomal sequestration (Barton et al. 2006). The consequences of TLR9 surface expression were characterized in a study in which TLR9 transmembrane expressing mutant mice were generated by exchanging the localization signal of TLR9 with that of TLR4. TLR9 transmembrane mutant mice died from systemic inflammation and anemia within 4 weeks (Mouchess et al. 2011). Since TLR9 is constrained to the endosome, cells of the immune system must internalize pathogens or pathogenic DNA before it can be detected by TLR9. Most immune cells accomplish this task through either receptor-mediated endocytosis in response to scavenger receptor binding, phagocytosis of complement-mediated opsonized material, or Fc receptor-mediated uptake of antibody opsonized material or a combination of these processes (Brencicova and Diebold 2013). In the case of “self” DNA detected from apoptotic cells, select marginal zone B (MZB) and B1a B cells can internalize chromatin complexes and induce a TLR9-mediated immunoregulatory response through IL-10 to counteract the pro-inflammatory signaling induced by DNA-antibody complexes internalized by rheumatoid factor expressing B cells (Miles et al. 2012). The TLR9-mediated immune response, therefore, is a highly complex response that is not only dependent on cellular localization both between cell types and within cells, but is also dependent upon the ligand and its origin.
3.1 TLR9 Regulation and Signaling
Takeshita et al. have extensively studied tlr9 gene regulation in both humans and mice. The authors found that tlr9 is regulated by 4 cis promoter elements: CRE, 5′-PU Box, 3′-PU box, and C/EBP. The transcription factors that interact with these cis promoter elements include CREB1 (CRE site), Ets2, Elf1, Elk1 (5′PU site), and C/EBPa (CEBP site); the authors could not identify the protein that bound the 3′-PU box. Additionally, this group found that CREB and members of the C/EBP family directly transactivate the tlr9 promoter while the Ets family members Ets2, Elk1, and Elf1 enhance trans activity. Furthermore, the authors posit that CREB1, Ets2, Elf1, Elk1, and C/EBPα physically interact with each other and with CBP/p300 to attain maximal transcription. To garner a deeper comprehension of tlr9 gene regulation in response to stimuli such as CpG DNA, the authors challenged cells transfected with a luciferase reporter linked to the entire tlr9 promoter. Challenge with CpG DNA significantly reduced tlr9 promoter activity which resulted from an interaction with the pro-inflammatory transcription factors NF-κB, c-Jun, and AP-1. The authors also found that the Ets family member Spi1 acted as a tlr9 gene repressor but could not identify if this protein acted directly or indirectly to inhibit transcription (Takeshita et al. 2004). Taken together, these data can provide valuable insight into cell type-specific tlr9 gene regulation and could lead to more targeted strategies for the use of CpG DNAs as therapeutics.
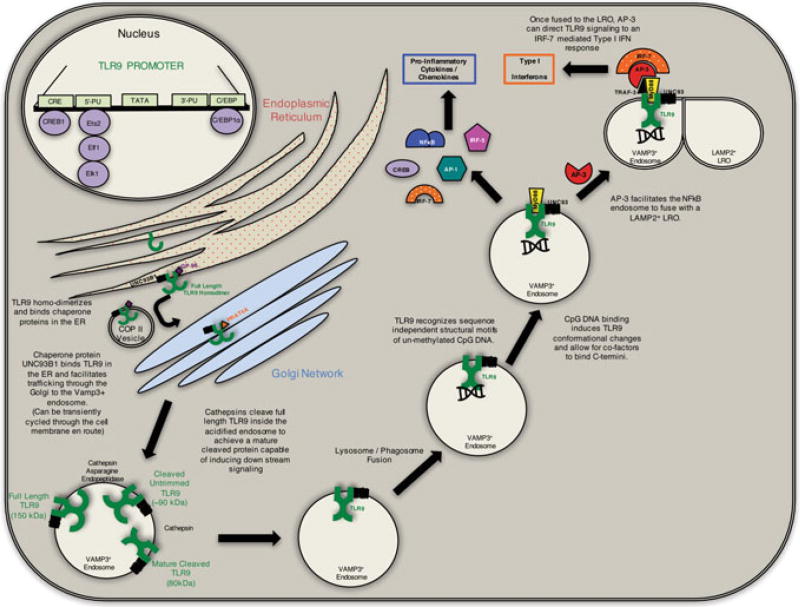
Posttranscriptional regulation of TLR9 begins in the endoplasmic reticulum (ER) where it is synthesized as a full-length TLR9 (FL-TLR9) protein under the supervision of chaperone proteins GP96 and PRAT4A, which ensure proper protein folding and function. Inside the ER, FL-TLR9 homo-dimerizes and binds the chaperone protein UNC93B1 (Kim et al. 2008). UNC93B1 is a multipass transmembrane protein that is integral in TLR9 trafficking. UNC93B1 facilitates the transport of FL-TLR9 to the Golgi via a COP-II vesicle (Lee et al. 2013). Once in the Golgi, FL-TLR9 bound UNC93B1 complexes with adaptor-protein 2 and is either secreted in a VAMP3+ endosome or may be transiently cycled through the plasma membrane before being compartmentalized into an endosome (Lee et al. 2013). Inside the acidified endosome, FL-TLR9 (~150 kDa) is cleaved by cathepsin and asparagine endopetidases to form a cleaved, untrimmed TLR9 protein (~90 kDa). This version of TLR9 is cleaved once more by cathepsin to form the mature, cleaved form of TLR9 that is capable of binding DNA (Ewald et al. 2008; Park et al. 2008; Ewald et al. 2011). Granulin, HMGB1, and LL-37 have all been implicated as a DNA binding proteins that may facilitate DNA delivery to TLR9 (Lande et al. 2007; Tian et al. 2007; Yanai et al. 2009; Park et al. 2011). Although DNA can bind un-cleaved TLR9, downstream signaling cannot occur until after proteolysis (Ewald et al. 2008). CpG DNA binding to mature, cleaved TLR9 within the endo-lysosome induces a conformational change that enables the interaction between the cytoplasmic TIR domains of the homodimer (Owyang et al. 2012). These dimerized TIR domains recruit the adaptor molecule MyD88 to form the myddosome (Latz et al. 2004). Once localized to an endosome, TLR9 trafficking dictates the type of downstream response (Honda et al. 2005; Guiducci et al. 2006). For example, ligand binding within the VAMP3+ endosome can yield pro-inflammatory responses mediated by transcription factors such as NF-κB, AP-1, or CREB (Latz et al. 2004; Leifer et al. 2004; Kim et al. 2008). However, adaptor-protein 3 can bind the myddosome and facilitate endosomal fusion with a LAMP2+ lysosome-related organelle (LRO), and together with TRAF-3, facilitate an IRF-7-mediated, type I interferon response to CpG DNA (Sasai et al. 2010) (Fig. 1).
Fig. 1.
TLR9 regulation and signaling: TLR9 expression is regulated by cis and trans elements including the CRE site, 5′-PU box, 3′-PU box, and a C/EBP site. These sites bind the transcription factors CREB1, Ets2, Elf1, Elk1, and C/EBP1α. Full-length TLR9 (FL-TLR9) is then synthesized in the ER as a homodimer with the aid of chaperone proteins UNC93B1 and GP96. The full-length homodimer is then shuttled to the Golgi via a COPII vesicle where it binds the chaperone protein PRAT4a to facilitate trafficking. The fully synthesized FL-TLR9 is then incorporated into a VAMP3+ endosome where cathepsin and asparagine endopeptidases cleave the receptor to form the mature, cleaved form. This mature form of TLR9 is then free to induce downstream signaling cascades upon ligand binding. Cellular localization and interactions with adaptor proteins can further differentiate the type of signaling response upon binding its cognate ligand
3.2 Duality of TLR9-Mediated Responses
Not all TLR activation promotes pro-inflammatory signaling cascades, and there have been numerous investigations into how TLR9 may promote both pro- and anti-inflammatory responses in the gut. These studies were particularly focused on how dysregulation of TLR9 signaling could lead to the development of inflammatory bowel disease or colitis (Rachmilewitz et al. 2004; Katakura et al. 2005; Lee et al. 2006; Luther et al. 2011; Hotte et al. 2012; Jounai et al. 2012; O’Hara et al. 2012; Owyang et al. 2012).
pDCs perpetually monitor the microbiota and present microbial antigens to immune cells within the lamina propria. However, healthy individuals do not develop overt inflammatory responses to commensals while they can develop inflammatory responses to pathogens. In a study designed to elucidate the mechanism of how TLR9 may contribute to the maintenance of intestinal homeostasis, the authors found that TLR9 deficiency alone was not sufficient to alter the general composition and signaling capacity of the intestinal immune system (Hofmann et al. 2014). However, when the authors investigated specific sub-populations of immune cells from wild-type and TLR9 knockout mice, they found significant functional changes. These were related to differences in the regulation of the transcription factors NF-κB and CREB from both sets of mice. While both transcription factors compete for binding with CBP, the NF-κB-CBP complex predominantly regulates pro-inflammatory gene transcription while the CREB-CBP complex is critical for the regulation of the anti-inflammatory cytokine IL-10. In TLR9 deficient mice, myeloid cells in the lamina propria displayed a significant bias toward NF-κB activity, while intestinal T cell populations displayed a lack of counter-regulatory mechanisms to control the inflammatory response (perhaps due to lack of APC derived IL-10). Furthermore, when CD4+ T cells from Tlr9−/− mice were transplanted into wild-type mice, recipient mice developed severe colitis similar to mice that received naive splenic T cells (Hofmann et al. 2014). These findings were corroborated by another study that revealed that the colitogenic potential of transplanted splenic T cells from germ-free mice could be reversed by exclusively exposing them to synthetic CpG oligodeoxynucleotides (ODNs) treatment (Bleich et al. 2009) The anti-inflammatory responses induced by TLR9 in the gut may be due to the ability of this receptor to regulate the secretion of type I interferon (Katakura et al. 2005; Mancuso et al. 2007; Otani et al. 2012; Steinhagen et al. 2012). TLR9 is localized in many different cell types; however, it is the most abundant in pDCs, a class of immune cell that readily secrete type I interferons (IFNs). Studies have shown that TLR9 signaling activated by probiotic DNA binding can inhibit colitis through the induction of type I IFNs (Katakura et al. 2005). The same study also suggested that the probiotic DNAs were absorbed in the small intestine and could have a systemic anti-inflammatory response that manifests in reduced severity of colitis (Katakura et al. 2005). Taken together, these data suggest that the physiological interaction of TLR9 with bacterial CpG DNA is essential for the maintenance of intestinal homeostasis by inducing counter-regulatory, anti-inflammatory mechanisms.
While pDCs selectively sample the gut lumen and activate adaptive immune responses, epithelial cells are continuously exposed to the luminal microbiota and their products. TLR9 has also been shown to modulate both pro- and anti-inflammatory signaling in intestinal epithelial cells. Like gastric epithelial cells, intestinal epithelial cells express TLR9 in both the apical and basolateral compartments (Schmausser et al. 2004; Lee et al. 2006). When polarized intestinal epithelial cells were challenged with CpG DNA at either the apical or basolateral surface, only basolateral challenge resulted in NF-κB activation and IL-8 secretion. Apical stimulation failed to induce NF-κB activation due to an inability to degrade β-TrCP ubiquitinated proteins (such as p105/NF-κB1). Consequently, when the genes induced by apical versus basolateral stimulation were examined, less than half of them were shared. Although apical stimulation did not result in a canonical NF-κB mediated anti-microbial response, it did induce targets of the Wnt pathway with anti-microbial activity, particularly cryptidins. Additionally, apical TLR9 stimulation promotes immune tolerance and can inhibit pro-inflammatory IL-8 secretion resultant from basolateral challenge with TLR9 agonists, as well as TLR2, TLR3, or TLR5 agonists. Basolateral TLR9 activation also resulted in a tolerogenic response when challenged repeatedly. Compared to the polarized intestinal epithelial cells, non-polarized cells induce a pro-inflammatory phenotype similar to that of basolaterally challenged cells (Lee et al. 2006). This suggests that cell polarization can modify the signaling pathway of TLR9 in the intestinal epithelium. These studies highlight the importance of TLR9 in maintaining gut homeostasis and provide insight into possible mechanisms of how TLR9 deficiency or site-specific activation may contribute to IBD or colitis.
How TLR9 regulates its response to DNA remains controversial; some studies posit that the micro-environment may regulate the type of response (Hiramatsu et al. 2013), while others suggest that the response is regulated by signal localization (either apically or basolaterally) (Lee et al. 2006). Another hypothesis suggests that the response is sequence-/structure-dependent since some commensal DNAs have been shown to be anti-inflammatory compared to pro-inflammatory pathogenic DNAs (Jijon et al. 2004; Rachmilewitz et al. 2004; Rakoff-Nahoum et al. 2004; Ghadimi et al. 2010). Despite its mode of action, TLR9 signaling has been shown to be of great importance in maintaining gut homeostasis and has vast potential as a therapeutic target in diseases where the inflammatory response is dysregulated.
4 DNA Translocation Strategies
Bacterial T4SSs are responsible for the mobilization of macromolecules including monomeric proteins, multimeric toxins, and DNA-protein complexes across the envelopes of Gram-negative or Gram-positive bacteria (Cascales and Christie 2003; Alvarez-Martinez and Christie 2009; Fronzes et al. 2009a; Backert et al. 2015). T4SSs can be categorized into three groups: conjugation systems, effector translocation systems, and DNA release/uptake systems (Alvarez-Martinez and Christie 2009). Although there is extensive versatility in bacterial T4SSs, the most common and well-studied subset of bacterial T4SSs is the conjugative system. The conjugative T4SSs are of significant medical relevance due to their capacity for widespread transmission of antibiotic resistance genes or virulence genes through plasmid or chromosomal DNA transfer (Cascales et al. 2013). Conjugative bacterial T4SS are not only capable of translocating DNA out of donor cells, but also they can take up DNA from the extracellular milieu. Additionally, organisms can use the DNA release/uptake T4SSs to release DNA substrates into the extracellular milieu in a contact independent manner (Cascales and Christie 2003). DNA transfer events between bacterial species or into the extracellular milieu are very common; however, transkingdom DNA transfer events are rare. To date, there are only a few bacterial species capable of translocating DNA into a eukaryotic host including: Agrobacterium tumefaciens, E. coli (Waters 2001) (Fernandez-Gonzalez et al. 2011), Bartonella henselae (Schroder et al. 2011), and most recently H. pylori (Varga et al. 2016).
4.1 DNA Translocation Through the T4SS
A. tumefaciens uses a VirB/D4 conjugation system to deliver oncogenic DNA (T-DNA) in conjunction with effector chaperone proteins into susceptible plant cells, resulting in crown gall disease. This bacterial T4SS has served as the canonical conjugative secretion system and has been studied most extensively. The A. tumefaciens T4SS is composed of 12 proteins, VirB1-11, and VirD4 (Schroder and Lanka 2005). Most bacterial T4SS, including the H. pylori cag T4SS and the comB T4SS, share some homology in sequence or function to the VirB/D4 proteins of this archetypal secretion system (Frick-Cheng et al. 2016).
Conjugation can be categorized into three distinct biochemical reactions. First, DNA transfer and replication (Dtr) proteins bind to its cognate origin of transfer (oriT) sequences and initiate the processing of DNA for transfer (de la Cruz et al. 2010; Zechner et al. 2012) (Wong et al. 2012). Second, the Dtr-oriT complex (relaxosome) binds to the type IV coupling protein (T4CP) (Gomis-Ruth et al. 2002). Lastly, the T4CP delivers the DNA substrate to the transenvelope channel composed of mating pair formation (mpf) proteins that shuttle DNA across the cell membrane (Schroder and Lanka 2005; Wallden et al. 2010). The process of DNA transfer is energized by the ATPases VirD4, VirB11, and VirB4 components (Christie and Cascales 2005; Schroder and Lanka 2005; Fronzes et al. 2009a). In Gram-negative bacteria such as A. tumefaciens or E. coli, the central hub protein, VirB10, spans both the inner and outer membranes and is decorated by structural proteins VirB7 and VirB9 in a 1:1:1 ratio to form a 14-fold symmetrical outer membrane pore which termed the core complex (Fronzes et al. 2009b; Low et al. 2014). This core complex serves as the structural scaffold for the biogenesis of the outer membrane pore and the pilus (composed mostly of VirB2 and few VirB5 proteins) (Schroder and Lanka 2005; Fronzes et al. 2009a; Low et al. 2014). The channel gate opens as a result of structural changes of VirB10 in response to ATP utilization by VirD4 and VirB11, as well as DNA substrate docking (Cascales et al. 2013). Once the channel is open, the DNA substrate is able to move to the bacterial cell surface (Cascales et al. 2013). The pilus and core complex have been well characterized to date; however, the structure of the interacting proteins outside of the core complex, termed the inner membrane complex (IMC), are less well defined. In the E. coli R388 encoded T4SS, the core complex is connected to the IMC via a central stalk region (Low et al. 2014). At the distal end of the stalk, there are two barrel-shaped densities that display C2 symmetries composed mostly of VirB4 (Low et al. 2014). The structure of the E. coli R388 system is suggestive of a two-step DNA transfer process in which VirD4 pumps the relaxosome-DNA substrate into the periplasm or by an IMC-mediated passage through the inner membrane. Once in the periplasm, the DNA substrate can then be secreted via the core complex (Low et al. 2014).
4.2 The H. pylori cag Pathogenicity Island (PAI)
The cagPAI is a 40kb gene locus that encodes proteins required to assemble a type IV secretion system (T4SS). The H. pylori cag T4SS retains significant homology to the archetypal T4SS of A. tumefaciens and has been shown to translocate the effector molecules CagA and peptidoglycan into host cells and has recently been shown to translocate DNA as well (Odenbreit et al. 2000; Viala et al. 2004; Varga et al. 2016).
The primary effector of the cag T4SS is CagA, a bacterial oncoprotein that is injected into host cells upon bacterial attachment. Following translocation into host epithelial cells, CagA can either be phosphorylated by Src and Abl kinases at specific EPIYA amino acid motifs at the 3′ terminus or can remain un-phosphorylated (Mueller et al. 2012). Phosphorylated CagA activates multiple cellular phosphatases which can subsequently lead to morphological changes and induce responses similar to those of growth factor stimulation (Selbach et al. 2009). Similarly, unmodified CagA has been shown to induce mitogenic and pro-inflammatory responses as well as weaken cell-cell junctions to induce a loss of cellular polarity (Amieva et al. 2003; Bagnoli et al. 2005; Suzuki et al. 2005; Murata-Kamiya et al. 2007; Saadat et al. 2007; Franco et al. 2008).
4.3 Structure of the H. pylori Cag T4SS
In contrast to the conjugative T4SS of A. tumefaciens or E. coli R388 which can translocate DNA substrates into eukaryotic hosts (Chilton et al. 1977; Waters 2001), the H. pylori T4SS has been categorized as an effector/translocator T4SS because until recently, only CagA and peptidoglycan have been shown to be translocated (Odenbreit et al. 2000; Viala et al. 2004). Compared to the conjugative T4SS, relatively little is known about the structure of effector translocation T4SSs (Frick-Cheng et al. 2016). The H. pylori cag pathogenicity island is composed of 18 genes required for T4SS-dependent phenotypes (Fischer et al. 2001). While some of these genes share sequence relatedness to other bacterial species, the sequence conservation is quite low, and 9 of the 18 genes are fundamentally unique to H. pylori and do not display any homology to other bacterial proteins (Frick-Cheng et al. 2016). Recent studies have shown that the H. pylori cag T4SS core complex is composed of proteins unrelated to components of T4SS in other bacteria, and similarly, the architecture of this T4SS is vastly different from those of other T4SS (Frick-Cheng et al. 2016). The cag core complex is 41 nm in diameter and is composed of 5 cag-encoded proteins (Cag3, CagT, CagM, CagX, and CagY) (Frick-Cheng et al. 2016). In contrast, the A. tumefaciens core complex has a diameter of only 20 nm and is composed of 3 proteins, VirB7, VirB9, and VirB10 (Chandran et al. 2009; Fronzes et al. 2009b). Of note, CagX and CagY share some homology to VirB9 and VirB10, respectively, and both core complexes share a 14-fold symmetry (Frick-Cheng et al. 2016). The H. pylori cag island also encodes for a T4SS pilus; however, its composition and structure are not well defined. Upon host cell contact, multiple cag-encoded structural proteins have been reported to localize to the cag pilus, as well as the effector protein CagA (Kwok et al. 2007; Backert et al. 2008; Jimenez-Soto et al. 2009; Barrozo et al. 2013). Importantly, H. pylori cag mutants that fail to form pili are also incapable of translocating CagA, suggesting an important role for pili in T4SS function (Kwok et al. 2007; Jimenez-Soto et al. 2009; Shaffer et al. 2011; Johnson et al. 2014). Furthermore, the H. pylori cag-encoded pilus protein CagL has been proposed to interact with beta-1 integrin receptors on the host cell surface to facilitate effector molecule translocation (Conradi et al. 2012; Barden et al. 2013). In contrast, A. tumefaciens does not require biogenesis of the vir T4SS or host receptor contact to deliver its oncogenic T-DNA effector into plant cells (Cascales and Christie 2004; Jakubowski et al. 2009; Garza and Christie 2013). Taken together, these data suggest that the H. pylori cag T4SS is highly unique, and this notion is further evidenced by the fact that H. pylori cag T4SS is the only known T4SS to date that is capable of translocating a protein substrate (CagA), non-proteinaceous substrate (peptidogly-can), and a DNA substrate (Christie and Cascales 2005; Varga et al. 2016).
4.4 H. pylori T4SS ComB
The H. pylori comB T4SS is primarily involved in natural competence and is functionally independent of the cag T4SS (Israel et al. 2000). Generally, competence systems share some homology with proteins involved in the T4SS pilus formation or type II secretion systems (Chen and Dubnau 2004). Indeed, the H. pylori comB system is composed of many conserved T4SS components, and its proteins have thus been named after their orthologues in the A. tumefaciens VirB/D4 system (Karnholz et al. 2006). The comB system shares a similar gene cluster organization of the VirB/D4 system and only lacks the VirB1, VirB5, and VirB11 components (Fernandez-Gonzalez and Backert 2014). ComB2 and ComB3 are orthologous to VirB2 and VirB3 in secondary structure despite having only 27.2% or 19.5% sequence homology, respectively (Karnholz et al. 2006). Although comB systems are nearly ubiquitous in H. pylori strains, they are unique from other DNA uptake T4SSs in that: they incorporate DNA in a pilus-independent manner, their DNA uptake efficiency varies between strains, and their DNA uptake is saturable, sensitive to length, symmetry, and the strandedness of the DNA substrate (Hofreuter et al. 1998; Levine et al. 2007). The VirB2 protein, and by association the ComB2 protein, is a subunit of the surface-exposed pilus structure of the VirB/D4 system. One study has suggested that the ComB2 protein may be a component of an incomplete “stump structure” in the cell envelope required for DNA uptake (Schroder and Lanka 2005). Unlike the VirB2 component, the functions of VirB3 homologues are unknown except that they are essential for T4SS function (Karnholz et al. 2006). The H. pylori ComB DNA uptake system is energized by ComB4, an orthologous protein to VirB4 (Hofreuter et al. 2001). The remaining components of the H. pylori ComB system are the ComB6–10 proteins that together form the core complex of the import channel. ComB6 is a polytopic membrane protein essential for T4SS function (Karnholz et al. 2006), while ComB7 is a non-essential membrane-attached protein that stabilizes the central channel structure (Hofreuter et al. 2003). ComB8 is a membrane-associated protein that contains a periplasmic N-terminus, ComB9 is a periplasmic protein anchored to the membrane, and lastly, ComB10 is also mostly periplasmic with its N-terminus anchored to the cytoplasmic membrane (Hofreuter et al. 2003). Based on its homology to the archetypal T4SS of A. tumefaciens, a two-step DNA uptake model has been proposed (Stingl et al. 2010). The ComB T4SS facilitates double-stranded DNA uptake into the periplasm, and a second, ComEC system is then utilized to incorporate the periplasmic DNA across the cytoplasmic membrane and into the host cell (Stingl et al. 2010; Kruger et al. 2016).
4.5 tfs3 and tfs4 Secretion Systems
In addition to the comB and cag systems, a novel 16.3 kb gene cluster was recently discovered in one of three plasticity regions of the H. pylori genome (Kersulyte et al. 2003). First described in H. pylori strain PeCan18B, this gene cluster houses 16 ORFs, 7 of which share homology to the virB/D4 operon (Kersulyte et al. 2003). Based on these data, this gene cluster is speculated to represent a third T4SS involved in conjugative DNA transfer and has thus been named tfs3. Furthermore, this gene cluster has a lower G + C content compared to the rest of the H. pylori genome, and it has been suggested that tfs3 was acquired by a horizontal gene transfer event from a different bacterial species (Kersulyte et al. 2003). When 94 clinical H. pylori isolates were screened for the presence of tfs3, 19% of the strains contained either the full-length genes or truncated genes (Kersulyte et al. 2003). Upon further investigation into the tfs3, significant sequence variation and number of genes were present among different isolates, and subsequently re-classified the Tfs3 into two sub-groups based upon their homology, termed tfs3a and tfs3b (Yamaoka 2010). However, tfs3a and tfs3b were clearly distinct, and tfs3b was renamed to tfs4, in reference to a fourth T4SS (Fischer et al. 2010; Fernandez-Gonzalez and Backert 2014).
The tfs4 gene cluster was first observed and subsequently characterized in H. pylori strain P12 (Fischer et al. 2010). Similar to tfs3, tfs4 was found in another plasticity zone of the H. pylori genome and contained enough orthologues of the A. tumefaciens virB/D4 genes to constitute a complete T4SS (Fischer et al. 2010). The tfs4 cluster contained the disease marker dupA, a virB4 homolog that has been affiliated with increased risk for duodenal ulcer (Lu et al. 2005) and a TraG/VirD4 homolog which encodes a type IV coupling protein independent of the established VirD4 protein in the cag T4SS (Selbach et al. 2002). In vitro experiments have shown that tfs4 encodes for proteins that can excise the gene cluster and facilitate its horizontal transfer to recipient cells via the Tfs4 T4SS (Fischer et al. 2010). Additional studies have shown that H. pylori can translocate genomic DNA to another bacterial species, Campylobacter jejuni, in a unidirectional manner on agar plates (Oyarzabal et al. 2007). This transfer was independent of the comB, cag, and tfs3 genes in the donor (Oyarzabal et al. 2007), and thus leaves the open possibility that tfs4 genes could facilitate genomic (gDNA) transfer events (Fernandez-Gonzalez and Backert 2014).
5 H. pylori/TLR9 Interactions
Studies investigating the role of TLR9 in gastric cancer suggest that it is up-regulated in cancer tissue and may play a role in cancer progression (Schmausser et al. 2004, 2005; Wang et al. 2014). Similarly, TLR9 SNPs have been implicated in increasing gastric cancer risk (Hold et al. 2009; Wang et al. 2013; Trejo-de la et al. 2015). Given the highly established relationship between H. pylori and gastric cancer, we and others have sought to determine the effects of H. pylori on TLR9-mediated responses and its possible implications in gastric carcinogenesis.
5.1 TLR9-Mediated Responses to H. pylori
In the human gastric niche, TLR9 expression is primarily localized to the apical surface epithelium; however, H. pylori-induced chronic active gastritis alters the expression pattern to a basolateral only pattern (Schmausser et al. 2004). As mentioned previously, in polarized intestinal epithelial cells, TLR9 activation in the basolateral compartment has been linked to pro-inflammatory signaling; however, the effects of TLR9 localization in gastric epithelial cells have not been elucidated (Schmausser et al. 2005; Lee et al. 2006). Previous studies have shown that H. pylori can induce gastric epithelial cell expression of Cox2 in a TLR2-/TLR9-dependent manner. In this study, the authors show that TLR2/TLR9 signaling in gastric epithelial cells induce MAPKs and subsequently allow transcription factors binding to both the CRE and AP-1 sites within the Cox2 promoter. As a result of Cox2 expression, prostaglandin E2 is released and promotes gastric cancer cell invasion and angiogenesis (Chang et al. 2005). The oncogenic potential of H. pylori-TLR9 interactions was also exemplified by a study in which purified H. pylori DNA induced invasion of gastric epithelial cells in vitro, an effect that could be partially reduced with the endosomal inhibitor chloroquine (Kauppila et al. 2013). In murine models of H. pylori infection, Rad et al. have demonstrated that TLR9 detects H. pylori in vivo and induces pro-inflammatory responses (Rad et al. 2009). These studies are complicated by more recent investigations in which TLR9 was shown to promote anti-inflammatory signaling during the acute phase of H. pylori infection, which was mediated by type I IFNs (Otani et al. 2012). Moreover, purified H. pylori DNAs have been shown to alleviate DSS-induced colitis in mouse models (Luther et al. 2011; Owyang et al. 2012). Collectively, these data highlight the dichotomous role of TLR9 during H. pylori infection. H. pylori may utilize TLR9 signaling to dampen the inflammatory response during the acute phase to establish infection; however, in an inflammatory micro-environment in which cells have lost their polarity, TLR9 may execute pro-inflammatory cascades and further exacerbate the progression toward gastric cancer.
Although both murine and human TLR9 recognize CpG DNA motifs, caution must be taken in translating findings from mouse studies into human disease. TLR9 receptors from mice and humans recognize different DNA ligands, and mouse TLR9 is expressed in a broader number of cell types compared to humans. Human TLR9 is expressed in epithelial cells, B cells, neutrophils, and pDCs whereas murine TLR9 is expressed in those same cell types and additionally in myeloid DCs, as well as macrophages (Bauer et al. 2001; Rehli 2002). These data suggest that the DNA sequences or structures that activate murine TLR9 may not elicit the same responses in humans while the more abundant localization of TLR9 in mouse species suggests that it may be exposed to more and varied ligands. Together, the differences in DNA sequence recognition and cellular distribution may account for the contrasting observations between human and murine responses to TLR9 activation and must be considered in translating into human studies or contemplating possible therapeutic targets.
5.2 H. pylori Activates TLR9 via the T4SS
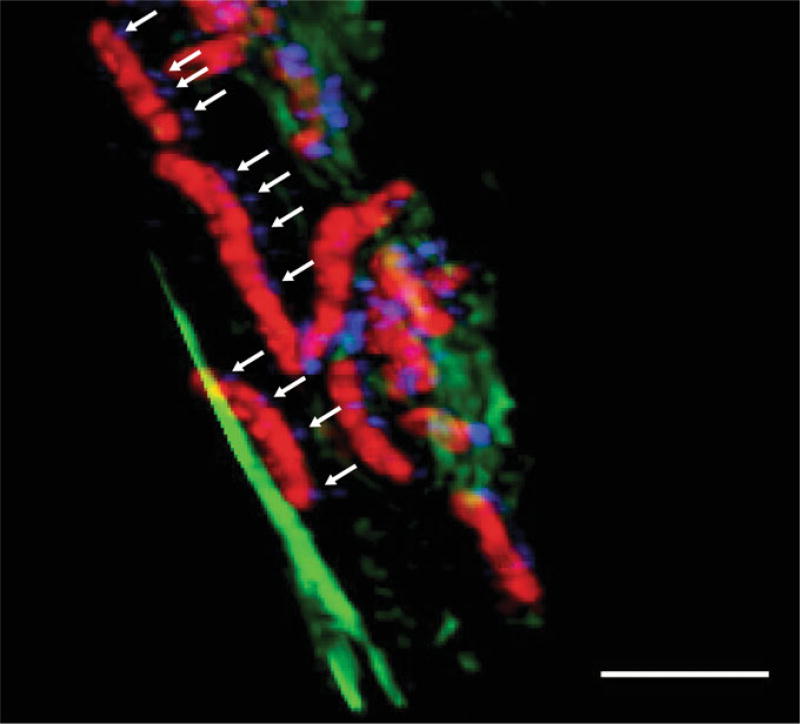
As studies of H. pylori-TLR9 interactions have generated mixed results, we sought to determine how H. pylori could engage this endosome-bound innate immune receptor (Varga et al. 2016). Since H. pylori primarily interact with the epithelial cell surface, we focused our studies on possible modalities of DNA transfer into host epithelial cells. Through the use of a TLR9 reporter assay, we found that H. pylori strains could activate TLR9 in vitro, and that the activation was independent of the ComB, Tfs3, and Tfs4 secretion systems. Once these known DNA transfer T4SSs had been eliminated as possible candidates for TLR9 activation, we shifted our focus to the cag PAI. We tested H. pylori cag isogenic mutants and found that an intact, functional cag T4SS was required for TLR9 activation. Since some bacterial T4SSs, like that of Neisseria gonorrhoeae use the T4SS to translocate DNA into the extracellular milieu (Hamilton et al. 2005), we also tested H. pylori conditioned media for its capacity to activate TLR9. Bacterial cell free, H. pylori conditioned media had no effect on TLR9 activation at concentrations ranging up to 30%. Lastly, although rare, H. pylori have the capacity to invade host epithelial cells. Thus, we performed gentamicin protection assays to determine whether the cag isogenic mutants could be endocytosed efficiently compared to wild-type H. pylori as a means to explain the differences observed in TLR9 activation. We found that there were no significant differences in the degree of endocytosis of wild-type versus cag mutant bacteria in our reporter cell line. Together, these data suggest that TLR9 activation is mediated directly through the cag T4SS. To confirm and validate these results, we cultured H. pylori in media enriched with the DNA nucleoside thymidine analog, bromodeoxyuridine (BrdU). In this manner, the microbial DNA can be easily discerned from host cell DNA. We challenged the labeled bacteria with AGS gastric epithelial cells with labeled H. pylori, and through structured illumination confocal microscopy, we were able to directly observe H. pylori DNA within host cells (Fig. 2), a result that was further validated by flow cytometry.
Fig. 2.
H. pylori DNA translocation: Confocal micrograph demonstrating H. pylori DNA translocation in the presence of host epithelial cells. H. pylori is shown in red, DNA in blue, and actin filaments in green. Arrows indicate areas of DNA exiting H. pylori. Scale bar, 2µm. Image acquired by Drs. Carrie Shaffer and Maria Hadjifrangiskou
The ability of the cag T4SS to translocate DNA is a unique finding, but not without precedent as both A. tumefaciens and B. henselae can also translocate plasmid DNA into eukaryotic hosts. However, the H. pylori system is different from the two aforementioned species because we observed TLR9 activation with H. pylori strains that were devoid of plasmid DNA, and the H. pylori cag T4SS is the first known T4SS that can translocate different classes of substrates including protein (CagA), non-protein (peptidoglycan), and DNA. We therefore wanted to next examine whether H. pylori cag T4SS-mediated DNA translocation mirrored that of A. tumefaciens (Cascales and Christie 2004; Jakubowski et al. 2009; Garza and Christie 2013). Similar to Agrobacterium, we found that DNA translocation required de novo protein synthesis, and that DNA was both protected and exposed to the extracellular milieu during transport.
To expand our findings into the natural niche of H. pylori, TLR9 expression and localization were examined in human gastric biopsies obtained from patients residing within either a high-risk or low-risk region of gastric cancer. Both populations share a similar incidence of H. pylori infection (90%); however, patients residing within the low-risk region displayed significantly less TLR9 expression compared to those within the high cancer-risk region. Moreover, when H. pylori strains isolated from patients in the low-risk region were tested for their ability to activate TLR9 in our in vitro reporter assay, they induced significantly less activation compared to strains isolated from the high-risk region.
6 Conclusions and Outlook
H. pylori has co-evolved with its human host over the course of millennia, and its prolonged colonization can be attributed to its ability to modulate the host immune system. Upon first encounter, H. pylori contacts the gastric epithelium and receptors of the innate immune system. Thus, it is critical that H. pylori harbors the ability to evade these innate immune responses to establish infection and persist for the lifetime of the host. H. pylori has adopted mechanisms to either hide or modulate its potentially immunogenic components such as altering its LPS to avoid TLR4 detection or harboring amino acid substitutions in its flagella to subvert the TLR5 immune recognition. In addition to those established mechanisms, we have recently shown that H. pylori can also translocate its gDNA through the cag T4SS to activate TLR9. Due to the dual roles that TLR9 plays in facilitating both pro- and anti-inflammatory responses, H. pylori’s ability to engage this receptor suggests yet another way in which this pathogen can manipulate the host response to maintain its persistence within the gastric niche.
Acknowledgments
We would like to acknowledge Drs. Carrie Shaffer and Maria Hadjifrangiskou for providing the confocal micrograph.
Contributor Information
Matthew Gordon Varga, Department of Cancer Biology, Vanderbilt University School of Medicine, Nashville, TN, USA.
Richard M. Peek, Department of Cancer Biology, Vanderbilt University School of Medicine, Nashville, TN, USA Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN, USA.
References
- Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev MMBR. 2009;73(4):775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva M, Peek RM., Jr Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150(1):64–78. doi: 10.1053/j.gastro.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300(5624):1430–1434. doi: 10.1126/science.1081919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, Aderem A. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci USA. 2005;102(26):9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Fronzes R, Waksman G. VirB2 and VirB5 proteins: specialized adhesins in bacterial type-IV secretion systems? Trends Microbiol. 2008;16(9):409–413. doi: 10.1016/j.tim.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Backert S, Tegtmeyer N, Fischer W. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol. 2015;10(6):955–965. doi: 10.2217/fmb.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, Rokbi B, Torstensson E, Zhao Y, Nilsson C, Seguin D, Normark S, Buchan AM, Richter-Dahlfors A. Gastric mucosal recognition of Helicobacter pylori is independent of Toll-like receptor 4. J Infect Dis. 2003;187(5):829–836. doi: 10.1086/367896. [DOI] [PubMed] [Google Scholar]
- Bagnoli F, Buti L, Tompkins L, Covacci A, Amieva MR. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad Sci USA. 2005;102(45):16339–16344. doi: 10.1073/pnas.0502598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barden S, Lange S, Tegtmeyer N, Conradi J, Sewald N, Backert S, Niemann HH. A helical RGD motif promoting cell adhesion: crystal structures of the Helicobacter pylori type IV secretion system pilus protein CagL. Structure. 2013;21(11):1931–1941. doi: 10.1016/j.str.2013.08.018. [DOI] [PubMed] [Google Scholar]
- Barrozo RM, Cooke CL, Hansen LM, Lam AM, Gaddy JA, Johnson EM, Cariaga TA, Suarez G, Peek RM, Jr, Cover TL, Solnick JV. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9(2):e1003189. doi: 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton GM, Kagan JC, Medzhitov R. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat Immunol. 2006;7(1):49–56. doi: 10.1038/ni1280. [DOI] [PubMed] [Google Scholar]
- Bauer S, Kirschning CJ, Hacker H, Redecke V, Hausmann S, Akira S, Wagner H, Lipford GB. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci USA. 2001;98(16):9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich A, Janus LM, Smoczek A, Westendorf AM, Strauch U, Mahler M, Hedrich HJ, Fichtner-Feigl S, Scholmerich J, Falk W, Hofmann C, Obermeier F. CpG motifs of bacterial DNA exert protective effects in mouse models of IBD by antigen-independent tolerance induction. Gastroenterology. 2009;136(1):278–287. doi: 10.1053/j.gastro.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Brencicova E, Diebold SS. Nucleic acids and endosomal pattern recognition: how to tell friend from foe? Frontiers Cell Infect Microbiol. 2013;3:37. doi: 10.3389/fcimb.2013.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro F. World cancer report 2014. WHO Press, International Agency for Research on Cancer; 2014. [Google Scholar]
- Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1(2):137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304(5674):1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E, Atmakuri K, Sarkar MK, Christie PJ. DNA substrate-induced activation of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol. 2013;195(11):2691–2704. doi: 10.1128/JB.00114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Rodriguez N, Kaakoush NO, Mitchell HM. Pattern-recognition receptors and gastric cancer. Front Immunol. 2014;5:336. doi: 10.3389/fimmu.2014.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462(7276):1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Wu MS, Lin JT, Chen CC. Helicobacter pylori-induced invasion and angiogenesis of gastric cells is mediated by cyclooxygenase-2 induction through TLR2/TLR9 and promoter regulation. J Immunol. 2005;175(12):8242–8252. doi: 10.4049/jimmunol.175.12.8242. [DOI] [PubMed] [Google Scholar]
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW. Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell. 1977;11(2):263–271. doi: 10.1016/0092-8674(77)90043-5. [DOI] [PubMed] [Google Scholar]
- Christie PJ, Cascales E. Structural and dynamic properties of bacterial type IV secretion systems (review) Mol Membr Biol. 2005;22(1–2):51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradi J, Tegtmeyer N, Wozna M, Wissbrock M, Michalek C, Gagell C, Cover TL, Frank R, Sewald N, Backert S. An RGD helper sequence in CagL of Helicobacter pylori assists in interactions with integrins and injection of CagA. Front Cell Infect Microbiol. 2012;2:70. doi: 10.3389/fcimb.2012.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7(12):e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34(1):18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456(7222):658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopep-tidase. J Exp Med. 2011;208(4):643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez E, Backert S. DNA transfer in the gastric pathogen Helicobacter pylori. J Gastroenterol. 2014;49(4):594–604. doi: 10.1007/s00535-014-0938-y. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez E, de Paz HD, Alperi A, Agundez L, Faustmann M, Sangari FJ, Dehio C, Llosa M. Transfer of R388 derivatives by a pathogenesis-associated type IV secretion system into both bacteria and human cells. J Bacteriol. 2011;193(22):6257–6265. doi: 10.1128/JB.05905-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42(5):1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- Fischer W, Windhager L, Rohrer S, Zeiller M, Karnholz A, Hoffmann R, Zimmer R, Haas R. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38(18):6089–6101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AT, Johnston E, Krishna U, Yamaoka Y, Israel DA, Nagy TA, Wroblewski LE, Piazuelo MB, Correa P, Peek RM., Jr Regulation of gastric carcinogenesis by Helicobacter pylori virulence factors. Cancer Res. 2008;68(2):379–387. doi: 10.1158/0008-5472.CAN-07-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL. Molecular and structural analysis of the Helicobacter pylori cag type IV secretion system core complex. mBio. 2016;7(1):e02001–e02015. doi: 10.1128/mBio.02001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Christie PJ, Waksman G. The structural biology of type IV secretion systems. Nat Rev Microbiol. 2009a;7(10):703–714. doi: 10.1038/nrmicro2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fronzes R, Schafer E, Wang L, Saibil HR, Orlova EV, Waksman G. Structure of a type IV secretion system core complex. Science. 2009b;323(5911):266–268. doi: 10.1126/science.1166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza I, Christie PJ. A putative transmembrane leucine zipper of Agrobacterium VirB10 is essential for T-pilus biogenesis but not type IV secretion. J Bacteriol. 2013;195(13):3022–3034. doi: 10.1128/JB.00287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewirtz AT, Yu Y, Krishna US, Israel DA, Lyons SL, Peek RM., Jr Helicobacter pylori flagellin evades toll-like receptor 5-mediated innate immunity. J Infect Dis. 2004;189(10):1914–1920. doi: 10.1086/386289. [DOI] [PubMed] [Google Scholar]
- Ghadimi D, Vrese M, Heller KJ, Schrezenmeir J. Effect of natural commensal-origin DNA on toll-like receptor 9 (TLR9) signaling cascade, chemokine IL-8 expression, and barrier integrity of polarized intestinal epithelial cells. Inflamm Bowel Dis. 2010;16(3):410–427. doi: 10.1002/ibd.21057. [DOI] [PubMed] [Google Scholar]
- Gomis-Ruth FX, de la Cruz F, Coll M. Structure and role of coupling proteins in conjugal DNA transfer. Res Microbiol. 2002;153(4):199–204. doi: 10.1016/s0923-2508(02)01313-x. [DOI] [PubMed] [Google Scholar]
- Guiducci C, Ott G, Chan JH, Damon E, Calacsan C, Matray T, Lee KD, Coffman RL, Barrat FJ. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J Exp Med. 2006;203(8):1999–2008. doi: 10.1084/jem.20060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T, Metzger J, Schmitz F, Heit A, Muller T, Latz E, Wagner H. The DNA sugar backbone 2′ deoxyribose determines toll-like receptor 9 activation. Immunity. 2008;28(3):315–323. doi: 10.1016/j.immuni.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Hajjar AM, Ernst RK, Tsai JH, Wilson CB, Miller SI. Human Toll-like receptor 4 recognizes host-specific LPS modifications. Nat Immunol. 2002;3(4):354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55(6):1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hiramatsu Y, Satho T, Irie K, Shiimura S, Okuno T, Sharmin T, Uyeda S, Fukumitsu Y, Nakashima Y, Miake F, Kashige N. Differences in TLR9-dependent inhibitory effects of H(2)O(2)-induced IL-8 secretion and NF-kappa B/I kappa B-alpha system activation by genomic DNA from five Lactobacillus species. Microbes Infect/Institut Pasteur. 2013;15(2):96–104. doi: 10.1016/j.micinf.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Hofmann C, Dunger N, Doser K, Lippert E, Siller S, Edinger M, Falk W, Obermeier F. Physiologic TLR9-CpG-DNA interaction is essential for the homeostasis of the intestinal immune system. Inflamm Bowel Dis. 2014;20(1):136–143. doi: 10.1097/01.MIB.0000436276.19755.c1. [DOI] [PubMed] [Google Scholar]
- Hofreuter D, Odenbreit S, Henke G, Haas R. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol Microbiol. 1998;28(5):1027–1038. doi: 10.1046/j.1365-2958.1998.00879.x. [DOI] [PubMed] [Google Scholar]
- Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41(2):379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- Hofreuter D, Karnholz A, Haas R. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int J Med Microbiol: IJMM. 2003;293(2–3):153–165. doi: 10.1078/1438-4221-00258. [DOI] [PubMed] [Google Scholar]
- Hold GL, Rabkin CS, Gammon MD, Berry SH, Smith MG, Lissowska J, Risch HA, Chow WH, Mowat NA, Vaughan TL, El-Omar EM. CD14-159C/T and TLR9-1237T/C polymorphisms are not associated with gastric cancer risk in Caucasian populations. Eur J Cancer Prev Official J Eur Cancer Prev Organ. 2009;18(2):117–119. doi: 10.1097/CEJ.0b013e3283101292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hotte NS, Salim SY, Tso RH, Albert EJ, Bach P, Walker J, Dieleman LA, Fedorak RN, Madsen KL. Patients with inflammatory bowel disease exhibit dysregulated responses to microbial DNA. PLoS ONE. 2012;7(5):e37932. doi: 10.1371/journal.pone.0037932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara S, Rumi MA, Kadowaki Y, Ortega-Cava CF, Yuki T, Yoshino N, Miyaoka Y, Kazumori H, Ishimura N, Amano Y, Kinoshita Y. Essential role of MD-2 in TLR4-dependent signaling during Helicobacter pylori-associated gastritis. J Immunol. 2004;173(2):1406–1416. doi: 10.4049/jimmunol.173.2.1406. [DOI] [PubMed] [Google Scholar]
- Israel DA, Lou AS, Blaser MJ. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol Lett. 2000;186(2):275–280. doi: 10.1111/j.1574-6968.2000.tb09117.x. [DOI] [PubMed] [Google Scholar]
- Jakubowski SJ, Kerr JE, Garza I, Krishnamoorthy V, Bayliss R, Waksman G, Christie PJ. Agrobacterium VirB10 domain requirements for type IV secretion and T-pilus biogenesis. Mol Microbiol. 2009;71(3):779–794. doi: 10.1111/j.1365-2958.2008.06565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijon H, Backer J, Diaz H, Yeung H, Thiel D, McKaigney C, De Simone C, Madsen K. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology. 2004;126(5):1358–1373. doi: 10.1053/j.gastro.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5(12):e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun. 2014;82(8):3457–3470. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai K, Ikado K, Sugimura T, Ano Y, Braun J, Fujiwara D. Spherical lactic acid bacteria activate plasmacytoid dendritic cells immunomodulatory function via TLR9-dependent crosstalk with myeloid dendritic cells. PLoS ONE. 2012;7(4):e32588. doi: 10.1371/journal.pone.0032588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnholz A, Hoefler C, Odenbreit S, Fischer W, Hofreuter D, Haas R. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J Bacteriol. 2006;188(3):882–893. doi: 10.1128/JB.188.3.882-893.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakura K, Lee J, Rachmilewitz D, Li G, Eckmann L, Raz E. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J Clin Investig. 2005;115(3):695–702. doi: 10.1172/JCI22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppila JH, Karttunen TJ, Saarnio J, Nyberg P, Salo T, Graves DE, Lehenkari PP, Selander KS. Short DNA sequences and bacterial DNA induce esophageal, gastric, and colorectal cancer cell invasion. APMIS: Acta Pathologica, Microbiologica, et Immunologica Scandinavica. 2013;121(6):511–522. doi: 10.1111/apm.12016. [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Velapatino B, Mukhopadhyay AK, Cahuayme L, Bussalleu A, Combe J, Gilman RH, Berg DE. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol. 2003;185(13):3764–3772. doi: 10.1128/JB.185.13.3764-3772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452(7184):234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Park JH, Franchi L, Backert S, Núñez G. The Cag pathogenicity island and interaction between TLR2/NOD2 and NLRP3 regulate IL-1β production in Helicobacter pylori infected dendritic cells. Eur J Immunol. 2013;43(10):2650–2658. doi: 10.1002/eji.201243281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JD, Kocincova D, Westman EL, Lam JS. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate immunity. 2009;15(5):261–312. doi: 10.1177/1753425909106436. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Sakamoto J, Kito T, Yamamura Y, Koshikawa T, Fujita M, Watanabe T, Nakazato H. Lewis blood group-related antigen expression in normal gastric epithelium, intestinal metaplasia, gastric adenoma, and gastric carcinoma. Am J Gastroenterol. 1993;88(6):919–924. [PubMed] [Google Scholar]
- Koch KN, Hartung ML, Urban S, Kyburz A, Bahlmann AS, Lind J, Backert S, Taube C, Müller A. Helicobacter urease-induced activation of the TLR2/NLRP3/IL-18 axis protects against asthma. J Clin Invest. 2015;125(8):3297–3302. doi: 10.1172/JCI79337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger NJ, Knuver MT, Zawilak-Pawlik A, Appel B, Stingl K. Genetic diversity as consequence of a microaerobic and neutrophilic lifestyle. PLoS Pathog. 2016;12(5):e1005626. doi: 10.1371/journal.ppat.1005626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449(7164):862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- la Trejo-de OA, Torres J, Sanchez-Zauco N, Perez-Rodriguez M, Camorlinga-Ponce M, Flores-Luna L, Lazcano-Ponce E, Maldonado-Bernal C. Polymorphisms in TLR9 but not in TLR5 increase the risk for duodenal ulcer and alter cytokine expression in the gastric mucosa. Innate Immun. 2015;21(7):706–713. doi: 10.1177/1753425915587130. [DOI] [PubMed] [Google Scholar]
- Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449(7162):564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5(2):190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- Lee SK, Stack A, Katzowitsch E, Aizawa SI, Suerbaum S, Josenhans C. Helicobacter pylori flagellins have very low intrinsic activity to stimulate human gastric epithelial cells via TLR5. Microbes Infect/Institut Pasteur. 2003;5(15):1345–1356. doi: 10.1016/j.micinf.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Lee J, Mo JH, Katakura K, Alkalay I, Rucker AN, Liu YT, Lee HK, Shen C, Cojocaru G, Shenouda S, Kagnoff M, Eckmann L, Ben-Neriah Y, Raz E. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006;8(12):1327–1336. doi: 10.1038/ncb1500. [DOI] [PubMed] [Google Scholar]
- Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, Schekman R, Barton GM. UNC93B1 mediates differential trafficking of endosomal TLRs. eLife. 2013;2:e00291. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173(2):1179–1183. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepper PM, Triantafilou M, Schumann C, Schneider EM, Triantafilou K. Lipopolysaccharides from Helicobacter pylori can act as antagonists for Toll-like receptor 4. Cell Microbiol. 2005;7(4):519–528. doi: 10.1111/j.1462-5822.2005.00482.x. [DOI] [PubMed] [Google Scholar]
- Levine SM, Lin EA, Emara W, Kang J, DiBenedetto M, Ando T, Falush D, Blaser MJ. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J (Official Publication of the Federation of American Societies for Experimental Biology) 2007;21(13):3458–3467. doi: 10.1096/fj.07-8501com. [DOI] [PubMed] [Google Scholar]
- Li H, Liao T, Debowski AW, Tang H, Nilsson HO, Stubbs KA, Marshall BJ, Benghezal M. Lipopolysaccharide structure and biosynthesis in Helicobacter pylori. Helicobacter. 2016 doi: 10.1111/hel.12301. [DOI] [PubMed] [Google Scholar]
- Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. Structure of a type IV secretion system. Nature. 2014;508(7497):550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128(4):833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther J, Owyang SY, Takeuchi T, Cole TS, Zhang M, Liu M, Erb-Downward J, Rubenstein JH, Chen CC, Pierzchala AV, Paul JA, Kao JY. Helicobacter pylori DNA decreases pro-inflammatory cytokine production by dendritic cells and attenuates dextran sodium sulphate-induced colitis. Gut. 2011;60(11):1479–1486. doi: 10.1136/gut.2010.220087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Midiri A, Biondo C, Beninati C, Zummo S, Galbo R, Tomasello F, Gambuzza M, Macri G, Ruggeri A, Leanderson T, Teti G. Type I IFN signaling is crucial for host resistance against different species of pathogenic bacteria. J Immunol. 2007;178(5):3126–3133. doi: 10.4049/jimmunol.178.5.3126. [DOI] [PubMed] [Google Scholar]
- Miles K, Heaney J, Sibinska Z, Salter D, Savill J, Gray D, Gray M. A tolerogenic role for Toll-like receptor 9 is revealed by B cell interaction with DNA complexes expressed on apoptotic cells. Proc Natl Acad Sci USA. 2012;109(3):887–892. doi: 10.1073/pnas.1109173109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro MA. Helicobacter pylori: a wolf in sheep’s clothing: the glycotype families of Helicobacter pylori lipopolysaccharides expressing histo-blood groups: structure, biosynthesis, and role in pathogenesis. Adv Carbohydr Chem Biochem. 2001;57:99–158. doi: 10.1016/s0065-2318(01)57016-x. [DOI] [PubMed] [Google Scholar]
- Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284(37):25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchess ML, Arpaia N, Souza G, Barbalat R, Ewald SE, Lau L, Barton GM. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35(5):721–732. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Tegtmeyer N, Brandt S, Yamaoka Y, De Poire E, Sgouras D, Wessler S, Torres J, Smolka A, Backert S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J Clin Investig. 2012;122(4):1553–1566. doi: 10.1172/JCI61143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Kamiya N, Kurashima Y, Teishikata Y, Yamahashi Y, Saito Y, Higashi H, Aburatani H, Akiyama T, Peek RM, Jr, Azuma T, Hatakeyama M. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26(32):4617–4626. doi: 10.1038/sj.onc.1210251. [DOI] [PubMed] [Google Scholar]
- Nagashima H, Iwatani S, Cruz M, Jimenez Abreu JA, Uchida T, Mahachai V, Vilaichone RK, Graham DY, Yamaoka Y. Toll-like receptor 10 in Helicobacter pylori infection. J Infect Dis. 2015;212(10):1666–1676. doi: 10.1093/infdis/jiv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira AM, Marques T, Soares PC, David L, Reis CA, Serpa J, Queiroz DM, Rocha GA, Rocha AC. Lewis antigen expression in gastric mucosa of children: relationship with Helicobacter pylori infection. J Pediatr Gastroenterol Nutr. 2004;38(1):85–91. doi: 10.1097/00005176-200401000-00019. [DOI] [PubMed] [Google Scholar]
- Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287(5457):1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- O’Hara JR, Feener TD, Fischer CD, Buret AG. Campylobacter jejuni disrupts protective Toll-like receptor 9 signaling in colonic epithelial cells and increases the severity of dextran sulfate sodium-induced colitis in mice. Infect Immun. 2012;80(4):1563–1571. doi: 10.1128/IAI.06066-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, Uchiyama S, Miyake K, Shimizu T. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520(7549):702–705. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- Otani K, Tanigawa T, Watanabe T, Nadatani Y, Sogawa M, Yamagami H, Shiba M, Watanabe K, Tominaga K, Fujiwara Y, Arakawa T. Toll-like receptor 9 signaling has anti-inflammatory effects on the early phase of Helicobacter pylori-induced gastritis. Biochem Biophys Res Commun. 2012;426(3):342–349. doi: 10.1016/j.bbrc.2012.08.080. [DOI] [PubMed] [Google Scholar]
- Owyang SY, Luther J, Owyang CC, Zhang M, Kao JY. Helicobacter pylori DNA’s anti-inflammatory effect on experimental colitis. Gut Microbes. 2012;3(2):168–171. doi: 10.4161/gmic.19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzabal OA, Rad R, Backert S. Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J Clin Microbiol. 2007;45(2):402–408. doi: 10.1128/JCM.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachathundikandi SK, Backert S. Differential expression of interleukin 1beta during Helicobacter pylori infection of Toll-like receptor 2 (TLR2)- and TLR10-expressing HEK293 cell lines. J Infect Dis. 2016;214(1):166–167. doi: 10.1093/infdis/jiw154. [DOI] [PubMed] [Google Scholar]
- Pachathundikandi SK, Lind J, Tegtmeyer N, El-Omar EM, Backert S. Interplay of the gastric pathogen Helicobacter pylori with toll-like receptors. BioMed Res Int. 2015;2015:192420. doi: 10.1155/2015/192420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9(12):1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B, Buti L, Lee S, Matsuwaki T, Spooner E, Brinkmann MM, Nishihara M, Ploegh HL. Granulin is a soluble cofactor for toll-like receptor 9 signaling. Immunity. 2011;34(4):505–513. doi: 10.1016/j.immuni.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Peek RM, Jr, Fiske C, Wilson KT. Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol Rev. 2010;90(3):831–858. doi: 10.1152/physrev.00039.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez GI, Shepherd VL, Morrow JD, Blaser MJ. Activation of human THP-1 cells and rat bone marrow-derived macrophages by Helicobacter pylori lipopolysaccharide. Infect Immun. 1995;63(4):1183–1187. doi: 10.1128/iai.63.4.1183-1187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilewitz D, Katakura K, Karmeli F, Hayashi T, Reinus C, Rudensky B, Akira S, Takeda K, Lee J, Takabayashi K, Raz E. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126(2):520–528. doi: 10.1053/j.gastro.2003.11.019. [DOI] [PubMed] [Google Scholar]
- Rad R, Ballhorn W, Voland P, Eisenacher K, Mages J, Rad L, Ferstl R, Lang R, Wagner H, Schmid RM, Bauer S, Prinz C, Kirschning CJ, Krug A. Extracellular and intracellular pattern recognition receptors cooperate in the recognition of Helicobacter pylori. Gastroenterology. 2009;136(7):2247–2257. doi: 10.1053/j.gastro.2009.02.066. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Rehli M. Of mice and men: species variations of Toll-like receptor expression. Trends Immunol. 2002;23(8):375–378. doi: 10.1016/s1471-4906(02)02259-7. [DOI] [PubMed] [Google Scholar]
- Saadat I, Higashi H, Obuse C, Umeda M, Murata-Kamiya N, Saito Y, Lu H, Ohnishi N, Azuma T, Suzuki A, Ohno S, Hatakeyama M. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447(7142):330–333. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329(5998):1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmausser B, Andrulis M, Endrich S, Lee SK, Josenhans C, Muller-Hermelink HK, Eck M. Expression and subcellular distribution of toll-like receptors TLR4, TLR5 and TLR9 on the gastric epithelium in Helicobacter pylori infection. Clin Exp Immunol. 2004;136(3):521–526. doi: 10.1111/j.1365-2249.2004.02464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmausser B, Andrulis M, Endrich S, Muller-Hermelink HK, Eck M. Toll-like receptors TLR4, TLR5 and TLR9 on gastric carcinoma cells: an implication for interaction with Helicobacter pylori. Int J Med Microbiol: IJMM. 2005;295(3):179–185. doi: 10.1016/j.ijmm.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Schroder G, Lanka E. The mating pair formation system of conjugative plasmids-A versatile secretion machinery for transfer of proteins and DNA. Plasmid. 2005;54(1):1–25. doi: 10.1016/j.plasmid.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Schroder G, Schuelein R, Quebatte M, Dehio C. Conjugative DNA transfer into human cells by the VirB/VirD4 type IV secretion system of the bacterial pathogen Bartonella henselae. Proc Natl Acad Sci USA. 2011;108(35):14643–14648. doi: 10.1073/pnas.1019074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect Immun. 2002;70(2):665–671. doi: 10.1128/iai.70.2.665-671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Paul FE, Brandt S, Guye P, Daumke O, Backert S, Dehio C, Mann M. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 2009;5(4):397–403. doi: 10.1016/j.chom.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011;7(9):e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SLR, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003a;4(12):1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- Smith MF, Jr, Mitchell A, Li G, Ding S, Fitzmaurice AM, Ryan K, Crowe S, Goldberg JB. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-kappa B activation and chemokine expression by epithelial cells. J Biol Chem. 2003b;278(35):32552–32560. doi: 10.1074/jbc.M305536200. [DOI] [PubMed] [Google Scholar]
- Steinhagen F, Meyer C, Tross D, Gursel M, Maeda T, Klaschik S, Klinman DM. Activation of type I interferon-dependent genes characterizes the “core response” induced by CpG DNA. J Leukoc Biol. 2012;92(4):775–785. doi: 10.1189/jlb.1011522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl K, Muller S, Scheidgen-Kleyboldt G, Clausen M, Maier B. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA. 2010;107(3):1184–1189. doi: 10.1073/pnas.0909955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Ceponis PJ, Lebel S, Huynh H, Sherman PM. Helicobacter pylori activates Toll-like receptor 4 expression in gastrointestinal epithelial cells. Infect Immun. 2003;71(6):3496–3502. doi: 10.1128/IAI.71.6.3496-3502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Mimuro H, Suzuki T, Park M, Yamamoto T, Sasakawa C. Interaction of CagA with Crk plays an important role in Helicobacter pylori-induced loss of gastric epithelial cell adhesion. J Exp Med. 2005;202(9):1235–1247. doi: 10.1084/jem.20051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita F, Suzuki K, Sasaki S, Ishii N, Klinman DM, Ishii KJ. Transcriptional regulation of the human TLR9 gene. J Immunol. 2004;173(4):2552–2561. doi: 10.4049/jimmunol.173.4.2552. [DOI] [PubMed] [Google Scholar]
- Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8(5):487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- Varga MG, Shaffer CL, Sierra JC, Suarez G, Piazuelo MB, Whitaker ME, Romero-Gallo J, Krishna US, Delgado A, Gomez MA, Good JAD, Almqvist F, Skaar EP, Correa P, Wilson KT, Hadjifrangiskou M, Peek RM. Pathogenic Helicobacter pylori strains translocate DNA and activate TLR9 via the cancer-associated cag type IV secretion system. Oncogene. 2016 doi: 10.1038/onc.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5(11):1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Wagner H. The sweetness of the DNA backbone drives Toll-like receptor 9. Curr Opin Immunol. 2008;20(4):396–400. doi: 10.1016/j.coi.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Wallden K, Rivera-Calzada A, Waksman G. Type IV secretion systems: versatility and diversity in function. Cell Microbiol. 2010;12(9):1203–1212. doi: 10.1111/j.1462-5822.2010.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xue L, Yang Y, Xu L, Zhang G. TLR9 promoter polymorphism is associated with both an increased susceptibility to gastric carcinoma and poor prognosis. PLoS ONE. 2013;8(6):e65731. doi: 10.1371/journal.pone.0065731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TR, Peng JC, Qiao YQ, Zhu MM, Zhao D, Shen J, Ran ZH. Helicobacter pylori regulates TLR4 and TLR9 during gastric carcinogenesis. Int J Clin Exp Pathol. 2014;7(10):6950–6955. [PMC free article] [PubMed] [Google Scholar]
- Waters VL. Conjugation between bacterial and mammalian cells. Nat Genet. 2001;29(4):375–376. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- Wong JJ, Lu J, Glover JN. Relaxosome function and conjugation regulation in F-like plasmids—a structural biology perspective. Mol Microbiol. 2012;85(4):602–617. doi: 10.1111/j.1365-2958.2012.08131.x. [DOI] [PubMed] [Google Scholar]
- Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nature Rev Gastroenterol Hepatol. 2010;7(11):629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, Savitsky D, Ronfani L, Akira S, Bianchi ME, Honda K, Tamura T, Kodama T, Taniguchi T. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature. 2009;462(7269):99–103. doi: 10.1038/nature08512. [DOI] [PubMed] [Google Scholar]
- Zechner EL, Lang S, Schildbach JF. Assembly and mechanisms of bacterial type IV secretion machines. Philos Trans R Soc Lond B Biol Sci. 2012;367(1592):1073–1087. doi: 10.1098/rstb.2011.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]