Abstract
Understanding the origins and roles of cardiac progenitor cells is important for elucidating the pathogenesis of congenital and acquired heart diseases1,2. Moreover, manipulation of cardiac myocyte progenitors has potential for cell-based repair strategies for various myocardial disorders3. Here we report the identification in mouse of a previously unknown cardiac myocyte lineage that derives from the proepicardial organ. These progenitor cells, which express the T-box transcription factor Tbx18, migrate onto the outer cardiac surface to form the epicardium, and then make a substantial contribution to myocytes in the ventricular septum and the atrial and ventricular walls. Tbx18-expressing cardiac progenitors also give rise to cardiac fibroblasts and coronary smooth muscle cells. The pluripotency of Tbx18 proepicardial cells provides a theoretical framework for applying these progenitors to effect cardiac repair and regeneration.
Emergence of recent data has generated a paradigm shift for our understanding of cardiogenesis, with consequent implications for an understanding of cardiac progenitors and the aetiology of congenital heart disease. It was recognized that cardiac muscle cells derived from precardiac mesoderm subsequently form the primitive heart tube. More recently, the discovery of the secondary or anterior heart field, which contributed cells to the outflow tract and potentially to the right ventricle, suggested the presence of two distinct cardiac lineages4. Subsequent lineage studies based on expression of the LIM homeodomain transcription factor Islet 1 (Isl1), and retrospective clonal analysis in the mouse, have confirmed two cardiac lineages, the first and second lineage, based on their timing of entry into the heart and the timing of their differentiation4–6. Here we report a previously unknown myocardial lineage, derived from Tbx18-expressing epicardial cells, that makes a substantial contribution to the heart.
Previous studies in avian species have demonstrated that the proepicardium and/or epicardium is a source for coronary vascular progenitors and cardiac fibroblasts7–9. During embryogenesis, cells from the proepicardium emigrate to form the epicardium—the epithelial outer lining of the heart. Epicardial cells undergo an epithelial-to-mesenchymal transition and invade the heart, giving rise to vascular endothelial cells, coronary vascular support cells and adventitial fibroblasts7–9. Recently, regeneration of the zebrafish heart has been shown to be associated with re-activation of an early marker of the proepicardium/epicardium, the T-box transcription factor Tbx18 (refs 10, 11). Tbx18-expressing cells appear to cluster at the wound site of zebrafish heart, with concurrent appearance of neo-vasculature during regeneration10.
To facilitate visualization of Tbx18 expression, we generated an nlacZ (nuclear lacZ) knock-in into the endogenous Tbx18 locus in mouse (Supplementary Fig. 3), and found that nlacZ expression mirrored that of the endogenous Tbx18 gene (Fig. 1 and Supplementary Fig. 2). Neither Tbx18 mRNA nor Tbx18:nlacZ expression are observed within the heart up to embryonic day (E)11.5 (Fig. 1).
Figure 1. LacZ expression in Tbx18:nlacZ knock-in mice recapitulates endogenousTbx18 expression.
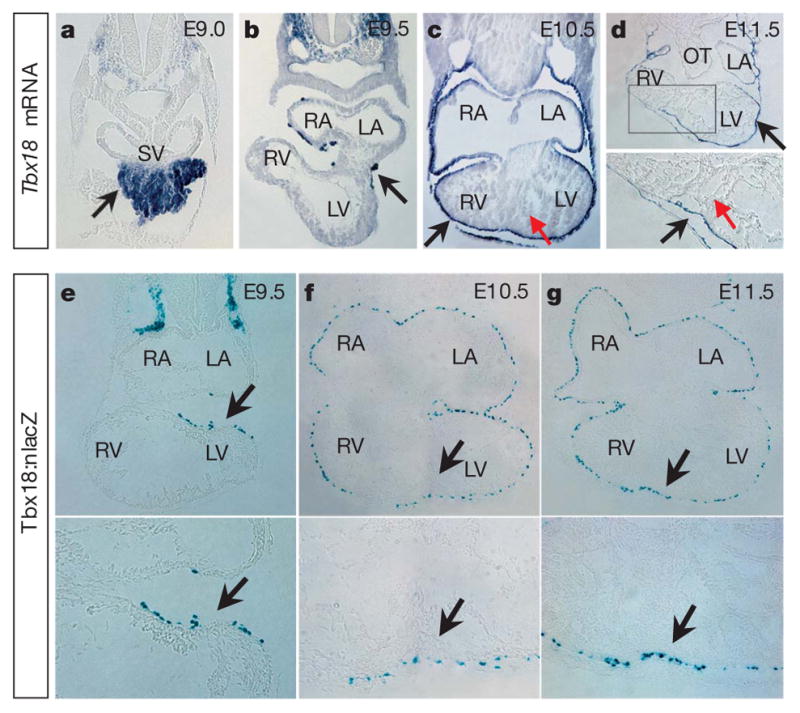
a–d, Section RNA in situ hybridization (ISH) of Tbx18 in mouse embryos (E9.0–E11.5). Tbx18 is expressed very early in the proepicardium (a) and epicardium (b–d). For parallel whole-mount RNA ISH, see Supplementary Fig. 2. e–g, X-gal staining on cardiac sections from Tbx18:nlacZ embryos at E9.5–E11.5. Tbx18:nlacZ cells are detected in the early epicardium at E9.5 (e) and in all epicardial cells covering the heart after E10.5 (f, g, data not shown for E12.5–E13.5). X-gal staining on Tbx18:nlacZ mouse tissues is consistent with Tbx18 mRNA ISH. For the Tbx18:nlacZ targeting strategy, see Supplementary Fig. 3. Black arrows indicate Tbx18 mRNA expression (a–d) and Tbx18:nlacZ cells (e–g); red arrows in c and d indicate that Tbx18 is not expressed in heart between E9.5 and E11.5. For e–g, lower panels are high-magnification views of the upper panels in the heart (black arrow regions). LA/RA, left/right atrium; LV/RV, left/right ventricle; OT, outflow tract; and SV, sinus venosus.
To investigate epicardial lineages in the mouse, we also generated a Cre knock-in into the endogenous Tbx18 locus (Supplementary Fig. 5), and crossed Tbx18:Cre mice with the lineage reporter R26RlacZ (ref. 12) mice. Analysis of lacZ expression in embryos from this cross demonstrated early expression consistent with that of endogenous Tbx18 (ref. 11; Figs 1 and 2 and Supplementary Figs 2 and 7a, b). In contrast to active expression of Tbx18, lineage analysis revealed the presence of Tbx18-derived lineages within the heart by E10.5 in the region of the forming ventricular septum and in scattered regions within both ventricular walls and atria (Fig. 2d–f and Supplementary Fig. 7b). Co-immunostaining with cardiac troponin T (cTnT, also known as cTnnt), cardiac troponin I (cTnI, also known as cTnni), MF20 (a sarcomeric myosin antibody) and the transcription factors Gata4 and Nkx2.5 demonstrated that these Tbx18-derived cells were cardiomyocytes (Fig. 2g–i and Supplementary Fig. 6). Tbx18-lineage-traced, Nkx2.5-positive cells could first be observed within the heart at E9.75 (Supplementary Fig. 6a). Complementary lineage studies using an organ explant culture system were consistent with Tbx18:Cre/R26RlacZ lineage studies. Outer epicardial cells of embryonic hearts (E11–E13) were selectively labelled with 5-(and 6-)carboxyfluorescein diacetate succinimidyl ester (CFSE)13. Hearts harvested at time 0 exhibited specific labelling of the epicardium (Supplementary Fig. 7c). After culture for 18–24 h, fluorescently labelled epicardial cells were observed within the heart, and expressed cTnT (Supplementary Fig. 7d–f).
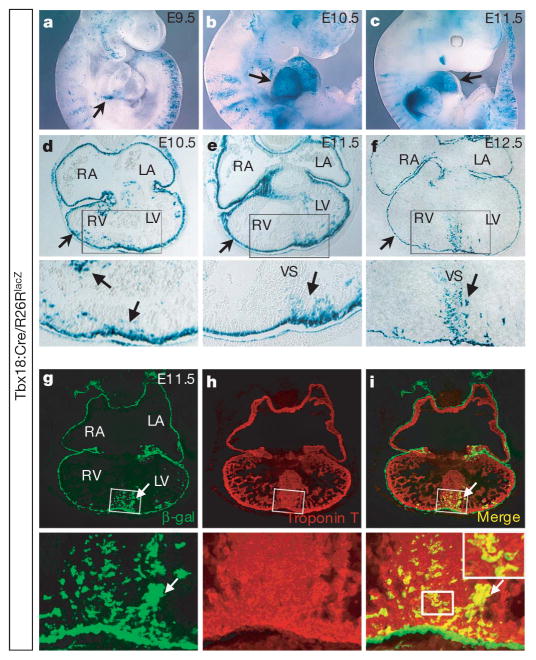
Figure 2. Cells derived fromTbx18-expressing cells are observed within the heart by E10.5, and exhibit a cardiomyocyte identity.
Tbx18:Cre mice were crossed to R26RlacZ indicator mice12 (Tbx18:Cre/R26RlacZ). a–c, X-gal staining on whole-mount embryos (E9.5–E11.5) shows Tbx18-lineage-traced cells in the proepicardium and epicardium (arrows in a–c). d–f, X-gal staining on cryosections from E10.5 to E12.5. Tbx18-lineage-traced cells are observed in the epicardium (arrows in the upper panels of d–f) and within the heart, particularly within the apical region in the developing ventricular septum (arrows in the lower panels of d–f). The lower panels of d–f show high-magnification views of the ventricles in the upper panel micrographs. For Tbx18-lineage-traced cells at E13.5, see Supplementary Fig. 7. g, β-galactosidase antibody staining on Tbx18-lineage-traced tissue at E11.5. h, Cardiac troponin T antibody staining. i, Overlay of g and h revealed that Tbx18-lineage-traced cells in heart were cardiac-troponin-T-positive cells. In g–i, lower panels are high-magnification views of the upper panels in the ventricular septum region. Arrows in d–i indicate Tbx18-lineage-traced cells in the ventricular septum region. LA/RA, left/right atrium; LV/ RV, left/right ventricle; and VS, ventricular septum.
In contrast to earlier stages of heart development, cells actively expressing Tbx18 are present within the heart at E12.5 (ref. 14). We examined sections from Tbx18:nlacZ mice from E10.5 to E17.5 and from neonatal stages using X-gal staining to detect β-galactosidase enzyme, encoded by the lacZ gene. (Fig. 1f, g and Supplementary Fig. 4d–f, data not shown for other stages), and by co-immunostaining with β-galactosidase antibody and cell-type-specific markers including cTnT and Nkx2.5 (myocytes), Pdgfrb (vascular support cells15) and Pecam1 (endothelial cells). Results demonstrated that cells actively expressing Tbx18 within the heart at or after E12.5 were neither myocytes (Nkx2.5-negative, Supplementary Fig. 4a–c, data not shown for other stages) nor endothelial cells (Pecam1-negative, data not shown). Using mice containing a fibroblast-specific promoter from the Col1a2 gene driving expression of a fusion protein in which cre recombinase is fused to a tamoxifen-inducible mutant oestrogen receptor (ER(T))16, and the Tbx18-floxed nlacZ/nGFP (nuclear green fluorescent protein) allele (Supplementary Fig. 3), we demonstrated that some Tbx18-expressing cells were fibroblasts (Supplementary Fig. 4g). Also, a population of Tbx18:nlacZ cells co-expressed Pdgfrb (Supplementary Fig. 4h panels 1–6), a marker of vascular support cells (both pericytes and vascular smooth muscle)15. Tbx18-expressing fibroblasts were not stained with the Pdgfrb antibody (Supplementary Fig. 4i, panels 1–6), demonstrating that both fibroblasts and vascular support cells actively express Tbx18 within the heart. Active expression of Tbx18 was not observed in cardio-myocytes, except within the sinus horns17.
Adult lineage analysis revealed that Tbx18 derivatives were observed in the differentiated smooth muscle of the coronary vessels (Fig. 3a, b, c (insets 2 and 3), h, green or white arrows), including the coronary arteries, as evidenced by co-localization of β-galactosidase antibody with neuropilin-1 (NP-1, also known as Nrp1; Fig. 3i), a specific marker for arteries18. Tbx18 expression was maintained in some coronary vascular smooth muscle cells (Fig. 3j, k). At adult stages, a substantial population of ventricular and atrial myocytes were observed to derive from Tbx18-expressing progenitors (Fig. 3c–f and Supplementary Fig. 8, black or white notched arrows). Cardiomyocytes isolated from Tbx18:Cre/R26RlacZ-lineage-traced hearts exhibited X-gal-positive staining (Fig. 3g). Calcium transient analysis performed on myocytes isolated from Tbx18:Cre/R26REYFP-lineage-traced adult hearts demonstrated no differences between Tbx18-labelled myocytes and their non-labelled counterparts (Supplementary Fig. 9). Tbx18 descendant cells also contributed significantly to the atrioventricular valves (Fig. 3c, inset 4). Co-localization of Tbx18-derived cells with endothelial lineages (as marked by Pecam1) was not observed in either embryonic or at adult stages (Fig. 3l).
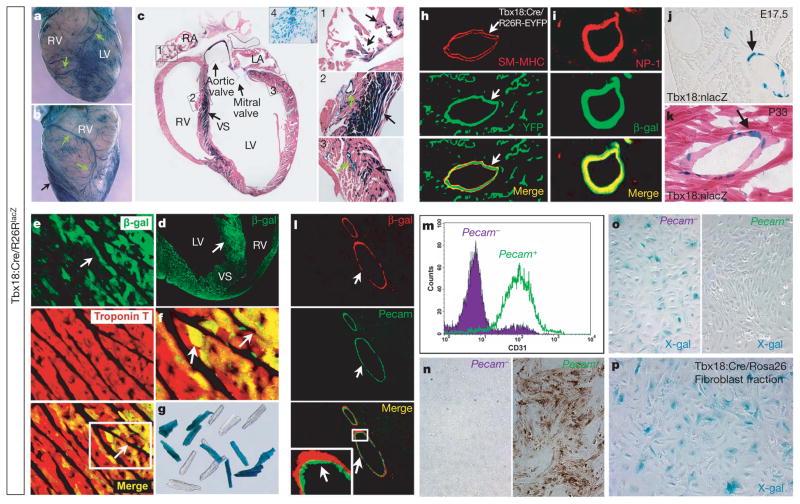
Figure 3. Tbx18 lineage tracing in the adult heart.
a, b, Whole-mount X-gal staining of Tbx18:Cre/R26RlacZ adult mouse heart (6 weeks). Coronary vasculature (green arrows) is derived from Tbx18 lineages. Dense staining in the septum is visible (black arrow) c, X-gal staining of tissues from Tbx18:Cre/R26RlacZ adult mouse heart (6 weeks). Tbx18 can give rise to: cardiomyocytes within atria (arrows in c, inset 1 and Supplementary Fig. 8), the ventricular septum (black arrow in c, inset 2) and the ventricular wall (black arrow in c, inset 3); coronary vascular support cells (green arrows in c, insets 2 and 3); and atrioventricular valves (c, inset 4, high magnification for bicuspid valve). Most Tbx18-lineage-traced cells within the ventricular septum co-stained with cardiac troponin T (arrows in d–f; f is a high magnification of the lower panel of e), demonstrating they are cardiomyocytes. g, A subset of cardiomyocytes isolated from adult Tbx18-lineage-traced hearts exhibited X-gal-positive staining. h, Tbx18 lineages give rise to coronary vascular smooth muscle cells (co-localized with smooth muscle myosin heavy chain). i, Tbx18 lineages give rise to coronary artery smooth muscle cells (co-localized with NP-1, an artery marker). j, k, X-gal staining on Tbx18:nLacZ knock-in mice shows that Tbx18 expression is maintained in some coronary vascular smooth muscle cells from embryonic stages to adulthood. l–o, Tbx18 lineages do not give rise to coronary vascular endothelial cells (not co-localized with Pecam1, l). m, Coronary vascular endothelial cells were isolated from the hearts (green fraction)26 and react with Pecam1 antibody (n, right panel), whereas Pecam1− fraction (purple) cells do not (n, left panel). o, Purified Pecam1+ cells are Xgal− (o, right panel), confirming that Tbx18-expressing lineages do not give rise to coronary vascular endothelial cells. A portion of cells in Pecam1− fraction are Xgal+ (o, left panel). p, Cardiac fibroblasts isolated from Tbx18-lineage-traced hearts25 demonstrated that approximately 30% of these cells derive from Tbx18 lineages. Abbreviations are as in Fig. 2.
Because previous studies have demonstrated that epicardial cells give rise to coronary endothelial cells7–9,19, we verified immunohisto-chemical results by X-gal staining of affinity-purified endothelial cells isolated from Tbx18-lineage-traced hearts (Fig. 3m–o). Purified cardiac endothelial cell fractions did not contain Tbx18-lineage-positive cells (Fig. 3o, right panel). We then investigated whether the pro-epicardium contained endothelial lineages distinct from Tbx18-expressing cells. Immunostaining demonstrated a population of Flk-1 (also known as Kdr) cells within the proepicardium that did not express Tbx18 (Fig. 4a). No Nkx2.5-positive myocardial progenitors were observed within the proepicardium (Fig. 4b). We also isolated fibroblasts from Tbx18-lineage-traced hearts. Consistent with our Col1a2-Cre-ER(T)16 results (Supplementary Fig. 4g), approximately one-third of cardiac fibroblasts expressed β-galactosidase (Fig. 3p).
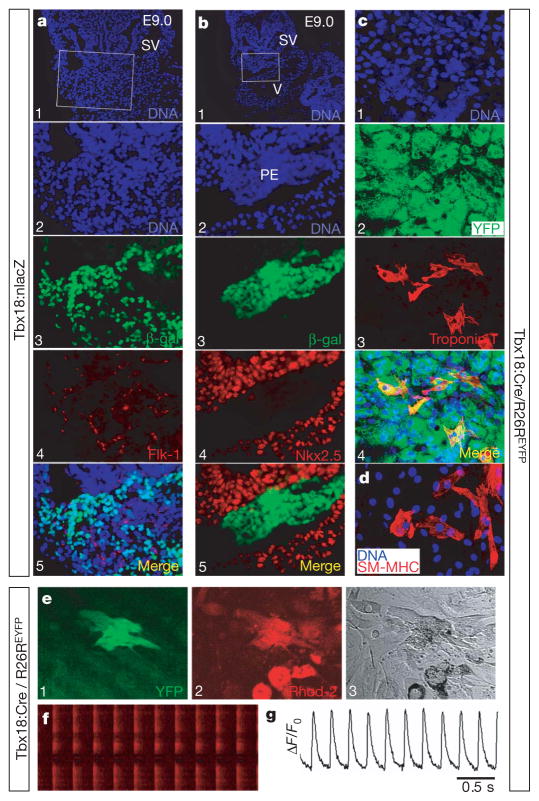
Figure 4. Tbx18-expressing progenitors within proepicardium are distinct, and retain the capacity to differentiate into cardiomyocytes and smooth muscle cells in vitro.
a, b, Tbx18-expressing progenitors within the proepicardium are not co-stained with antibodies to Flk-1 (a) or Nkx2.5 (b) (E9.0, data not shown for E9.5), suggesting that Flk-1 endothelial progenitors within the proepicardium are a distinct population from Tbx18-expressing cells. Tbx18 progenitors with myocyte potential do not yet express Nkx2.5 within the proepicardium, although Tbx18-derived myocytes do express Nkx2.5 within the heart. The second panels in a and b are high-magnification images of the top panels of a and b, respectively. c, Tbx18-lineage-traced proepicardial cells (Tbx18:Cre/R26REYFP) were disassociated and single cells were expanded in proepicardial culture medium. A significant percentage of clones (34%) derived from single proepicardial cells adopt cardiomyocyte fate after incubating in the differentiation medium21 (co-stained with cardiac troponin T, bottom panel of c). d, Every clone (40 out of 40) derived from single Tbx18 proepicardial cells displayed staining for smooth muscle myosin heavy chain (SM-MHC) antibody after incubating in smooth muscle culture medium. e–g, Real-time Ca2+ transients of cells derived from single Tbx18-lineage-traced proepicardial cells after myocytic differentiation. e, Left panel: monitored cell is YFP+, suggesting it derived from the Tbx18:Cre/R26REYFP proepicardium. e, Middle panel: differentiated cells were monitored under an inverted confocal microscope after they were preloaded with Ca2+ indicator Rhod-2-AM. e, Right panel, transmitted light image of the monitored cell. f, Line-scanning images and time course (g) of spontaneous Ca2+ transients obtained from differentiated YFP+ cells. PE, proepicardium; SV, sinus venosus; and V, ventricle.
Isl1-expressing second heart field progenitors contribute cardio-myocytes that ultimately reside in the outflow tract, the right ventricle, the ventricular septum, the left ventricle, the atria and the atrial septum5,20. Tbx18-derived cardiomyocytes contributed significantly to the ventricular septum and atria as well as to small numbers of cells scattered throughout both ventricular walls (Figs 2d–i and 3c–g and Supplementary Figs 7a, b and 8). A comparison of Tbx18:Cre, Isl1:Cre- and Isl1:Cre/Tbx18:Cre-lineage-traced hearts indicated complementarity of these two lineages (Fig. 3c and Supplementary Figs 10 and 11a, b), as demonstrated by quantitative analysis of myocytes isolated from lineage-traced hearts (ventricular septum, left ventricle and right ventricle; Supplementary Table 1). Immuno-staining and RNA in situ hybridization of Tbx18, Isl1 and MLC2a (also known as Myl7) revealed that Tbx18 was not co-expressed with Isl1 and MLC2a during early embryogenesis (Supplementary Figs 2a–e and 11c).
We isolated Tbx18:Cre/R26REYFP-lineage-traced proepicardial cells and cultured them under conditions to favour differentiation into cardiomyocytes or smooth muscle cells21. Immunostaining analyses demonstrated efficient conversion of Tbx18 lineages to myocytes or smooth muscle cells (Supplementary Fig. 12). To evaluate the pluripotency of individual proepicardial cells, single-cell clonal analysis was performed. Out of 336 single proepicardial cells plated on OP9 feeder layers, approximately 37% (124 out of 336) proliferated and formed clones by day seven. Forty clones were picked randomly, each dispersed into two wells, and cultured under conditions to favour either myocyte or smooth muscle cell fates. After five days of culture in differentiation medium, 34% of single-cell clonal derivatives differentiated into cardiomyocytes with an obvious striated cytoarchitecture and expressed cTnT (Fig. 4c). Spontaneous contraction was observed in some wells after four days of culture (Supplementary Fig. 13 and Supplementary Video), and myocyte identity was further confirmed by calcium transients (Fig. 4e–g). Each clone (40 out of 40) cultured with smooth muscle culture medium stained with smooth muscle myosin heavy chain (Fig. 4d). No instances were observed where derivatives of a single clone formed only cardiomyocytes and not smooth muscle cells. This demonstrated that a large proportion of proepicardial cells are pluripotent and can adopt either cardiomyocyte or smooth muscle cell fates.
Adult epicardial cells can be activated to migrate in vitro and adopt vascular cell fates22. Migratory adult epicardial cells from Tbx18:nlacZ mice expressed Tbx18 (Supplementary Fig. 14). To investigate whether postnatal and adult epicardium retains a similar pluripotency to that of the proepicardium, postnatal and adult epicardial cells isolated from Tbx18:Cre/R26REYFP-lineage-traced mice were cultured on OP9 cells, and then subsequently cultured in myocyte- or smooth-muscle-specific differentiation medium. In contrast to results with proepicardial cells, adult epicardial cells did not convert to cTnT-expressing cells under these culture conditions (data not shown). Understanding the underlying causes of this difference in potential will have significant implications for the possible use of epicardial cells for cardiac repair.
Our data are consistent with a model whereby the first cells to enter the heart from the proepicardium/epicardium give rise to myocyte lineages, first observed at approximately E9.75, whereas subsequent Tbx18-positive epicardial lineages that will give rise to vascular support cells and fibroblasts are first observed entering the heart at approximately E12.5. Endothelial lineages within the proepicardium are distinct from those of Tbx18 cells (Supplementary Fig. 1). Tbx18 is not actively expressed within cardiac myocyte derivatives, but is observed in a subset of vascular smooth muscle and cardiac fibro-blasts, in addition to the epicardium (Fig. 3j, k and Supplementary Fig. 4g–i).
Previous studies in avian embryos have failed to demonstrate that proepicardial lineages give rise to cardiomyocytes within the heart, although myocytic potential has been demonstrated in vitro7–9,23. It is unknown whether observed differences reflect distinct experimental approaches or species-specific differences. In mouse, Tbx18-expressing cells of the septum transversum that are in continuity with the sinus venosus contribute to the myocardial sleeve of the latter17. These cells, however, never express Nkx2.5, in contrast to the Tbx18-derived intracardiac myocytes of the ventricles and atria described here. The distinct embryonic origin of a substantial number of cardiomyocytes from an epicardial lineage provides a new perspective on heart development and congenital or adult heart disease affecting these lineages.
METHODS
Whole-mount RNA in situ hybridization and histological analysis
Whole-mount RNA ISH was carried out as described previously24.
Generation of Tbx18:nlacZ/nGFP, Tbx18:Cre and Isl1:Cre knock-in mouse models28
Tbx18 and Isl1 genomic clones were isolated by screening a mouse genomic lambda library (129/sv, Stratagene). Arms of targeting constructs were amplified on phage DNA templates with high-fidelity DNA polymerase (Pfu, Stratagene, catalogue number 600153). For individual targeting strategy, see Supplementary Figs 3, 5 and 10.
X-gal staining
For the whole-mount staining, mouse embryos and tissues were collected from timed pregnant females. Embryos and tisssues were fixed in 4% paraformaldehyde for 15–60 min. After permeabilization (10% Na-deoxycholate, 10% NP40 in PBS), embryos and tissues were stained in X-gal solution (50 mM K-ferricyanide, 50 mM K-ferrocyanide, 200 mM MgCl2, 100 mg ml−1 X-gal in PBS) for 4–12 h and were then post-fixed with 4% para-formaldehyde. For section staining, embryos and tissues were fixed in 4% para-formaldehyde and were then dehydrated in series sucrose solution. X-gal staining was performed on 6–8-μM cryosections with previous additional fixation in 4% paraformaldehyde for 6–8 min.
Fluorescent dye lineage tracing of epicardial cells with a heart explant culture system
Embryos were dissected at E11.5 in PBS. About 20 μl dye solution (30 μg ml−1) was injected into the pericardial cavity (CFSE, Molecular Probes, C1157). The chest expands under the pressure and it was ensured that it did not shrink back, indicating the dye was not able to leak from the cavity. Embryos were then submerged in media (DMEM) and left at room temperature (23 °C) for 15 min in the dark. Hearts were removed and washed in media for 5 min. Some hearts were selected randomly for fixation in 4% paraformaldehyde. The rest were cultured on a 0.4 μm polycarbonate membrane insert with 10% embryonic stem cell qualified fetal bovine serum (FBS) (Gibco, 10439024) supplemented DMEM (Gibco, D5796) for 18–24 h. Hearts were fixed and frozen as described previously for immunohistochemistry.
Isolation of cardiomyocytes, endothelial cells, fibroblasts and epicardial cells from adult mouse hearts
For cardiomyocyte isolation, pure adult cardiomyocytes (6–8 weeks) were isolated according to a previously described method29. For Ca2+ imaging and Tbx18/Isl1 lineage contribution quantification, different parts of the heart (ventricular septum, left ventricular wall and right ventricular wall) were dissected and separated after retrograde collagenase perfusion via the aorta. After an additional collagenase incubation for 10 min at 37 °C, cardiomyocytes were dispersed mechanically29. Pure populations of myocytes were obtained by centrifugation and cell filtration. Quantification was performed on a number of independent hearts for each sample by cell counts of numerous, random, field-of-view micrographs to determine the percentage of total cardiomyocytes expressing the YFP lineage marker. Statistical analyses were performed on the data sets. For isolation of endothelial cells, adult mouse hearts were minced with a razor blade. Endothelial cells from the hearts were isolated according to a previously described method26. Cardiac fibroblasts were isolated and cultured as described previously25. Pre-plating of cells isolated from the heart digest was shortened to 30 min at 37 °C to increase the purity of fibroblasts. For epicardial cell isolation, adult epicardial cells were isolated from explanted hearts by treatment with collagenase at 37 °C for 10 min (prepared as described for cardiomyocyte isolation). The epicardium was then manually extracted as a monolayer sheet.
Immunostaining
Embryos and tissues were fixed immediately in 4% paraformaldehyde for 10–30 min after dissection. Tissues were embedded and cut by cryosectioning (5–10 μm). For sections with endogenous YFP, a post-fixation for 5 min on ice was performed before staining. Cells obtained in culture were washed with warm media and then were fixed for 7 min with 10% formalin. Primary antibodies used in this study were: rabbit polyclonal anti-β-galactosidase (Cappel, product number 55978, 1:200), goat polyclonal anti-β-galactosidase (Biogenesis, 4600–1409, 1:200), rabbit polyclonal anti-smooth muscle myosin heavy chain (Biomedical Technologies Inc., BT562, 1:200), rabbit polyclonal anti-NKX2.5 (Santa Cruz Biotechnology, SC14033, 1:50), rat polyclonal anti-Pecam1 (Pharmingen, 550274, 1:100), mouse monoclonal anti-cardiac troponin T (NeoMarkers, MSZ-295-P, 1:200), mouse monoclonal anti-α-smooth muscle actin (Abcam, ab7817, 1:200), mouse monoclonal anti-α-actinin (sarcomeric) (Sigma-Aldrich, A7811, 1:200), rabbit polyclonal anti-WT1 (Santa Cruz Biotechnology, sc-846, 1:75), goat polyclonal anti-GATA-4 (Santa Cruz Biotechnology, sc-1237, 1:75), mouse polyclonal anti-Isl1 (Developmental Studies Hybridoma Bank, 39.4D5, 1:100). Mouse monoclonal cardiac troponin I was provided by J. Lin (Developmental Studies Hybridoma Bank, 1:100), mouse monoclonal anti-MF20 (1:200) was provided by D. Fischman, and rabbit polyclonal anti-Pdgfrb (1:200) was provided by W. Stallcup. Rabbit polyclonal anti-neuropilin 1 (1:200) was provided by A. L. Kolodkin.
Isolation and differentiation of proepicardial cells
Tbx18-lineage-traced embryos (Tbx18:Cre/R26REYFP, E9.0–E9.5) were dissected from timed pregnant mice and were quickly washed in cold PBS after dissection. A check was made under the epifluorescent microscope that Tbx18-positive cells in the proepicardium have not begun to migrate to the heart. Proepicardial cells were isolated with a pulled-tip glass pipette. The isolated proepicardial cells were cultured on gelatin with a medium designed to induce differentiation into cardiac myocytes or smooth muscle cells21.
Single proepicardial cells preparation
Proepicardial cells collected from seven to ten E9.0–E9.5 embryos (Tbx18:Cre/R26REYFP) were digested with 0.05% trypsin/EDTA (Invitrogen) for 5 min at 37 °C. Single-cell suspension was obtained by pipetting cells in culture medium (85% IMDM (Gibco), 15% FBS (selected batches), 100 μM non-essential amino acids (Gibco), 2 mM sodium pyruvate (Gibco) and 100 μM β-mercaptoethanol (Sigma)). Disassociated cells were plated at 0.5 cells per well onto a 96-well plate with a mitomycin C (Sigma)-inactivated OP9 cell feeder layer (YFP signals were examined to assure single-cell colonies derived from Tbx18 lineages). After seven days of culture, clones proliferated from single cells were dispersed to small clumps with 0.05% trypsin/ EDTA for 30 s. These cells were re-plated onto gelatin-coated 16-well chamber slides with one colony split into two wells and then left in stem cell culture medium for two days. Cells were cultured with a medium designed to induce differentiation into cardiac myocytes or smooth muscle cells21. Immunostaining was performed after five days of culture.
Ca2+ imaging
Cells were loaded with Rhod-2-AM (5 μM, 15 min; Molecular Probes, R1245MP), and imaged with an Olympus Fluoview 1000 inverted confocal microscope with a ×40 oil immersion lens (numerical aperture 1.3). The line-scan imaging mode was used to measure Ca2+ transients of paced cells. The extracellular solution contained 116 mM NaCl, 5.0 mM KCl, 0.8 mM MgSO4, 1.0 mM NaH2PO4, 5.5 mM glucose, 20 mM HEPES and 1.0 mM CaCl2 (pH 7.4). Image processing and data analysis were performed as previously described27.
Supplementary Material
Acknowledgments
We thank A. Kleckner, J. Lam and M. Zamora for critical technical assistance; D. Bader for discussion; P. Soriano and F. Costantini for providing R26RlacZ and R26REYFP indicator mice; H. Kubo for providing OP9 cells; and J. Lin, D. Fischman and A. Kolodkin for providing troponin I, Pdgfrb and neuropilin 1 antibodies, respectively. We are also grateful to B. Gelb for his critical comments and revisions of the manuscript. This work was supported by AHA National Scientist Development Grant to C.-L.C. and NIH1RO1 to S.M.E.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Olson EN. A decade of discoveries in cardiac biology. Nature Med. 2004;10:467–474. doi: 10.1038/nm0504-467. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Murry CE, Field LJ, Menasche P. Cell-based cardiac repair: reflections at the 10-year point. Circulation. 2005;112:3174–3183. doi: 10.1161/CIRCULATIONAHA.105.546218. [DOI] [PubMed] [Google Scholar]
- 4.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nature Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 5.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly R, Evans SM. The secondary/anterior heart field. In: Rosenthal N, Harvey RP, editors. Heart Development and Regeneration. Academic; San Diego: in the press. [Google Scholar]
- 7.Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci USA. 1992;89:9504–9508. doi: 10.1073/pnas.89.20.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with in growth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 9.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res. 1998;82:1043–1052. doi: 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- 10.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 11.Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene Tbx18. Mech Dev. 2001;100:83–86. doi: 10.1016/s0925-4773(00)00494-9. [DOI] [PubMed] [Google Scholar]
- 12.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 13.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 14.Franco D, et al. Left and right ventricular contributions to the formation of the interventricular septum in the mouse heart. Dev Biol. 2006;294:366–375. doi: 10.1016/j.ydbio.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 15.Armulik A, Abramsson A, Betsholtz C. Endothelial/pericyte interactions. Circ Res. 2005;97:512–523. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- 16.Zheng B, Zhang Z, Black CM, de Crombrugghe B, Denton CP. Ligand-dependent genetic recombination in fibroblasts: a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol. 2002;160:1609–1617. doi: 10.1016/S0002-9440(10)61108-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christoffels VM, et al. Formation of the venous pole of the heart from an Nkx2–5-negative precursor population requires Tbx18. Circ Res. 2006;98:1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 18.You LR, et al. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- 19.Perez-Pomares JM, et al. Origin of coronary endothelial cells from epicardial mesothelium in avian embryos. Int J Dev Biol. 2002;46:1005–1013. [PubMed] [Google Scholar]
- 20.Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol. 2005;287:134–145. doi: 10.1016/j.ydbio.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 21.Moretti A, et al. Multipotent embryonic Isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 22.Smart N, et al. Thymosin β4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 23.Kruithof BP, et al. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol. 2006;295:507–522. doi: 10.1016/j.ydbio.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 24.Cai CL, et al. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian SV, Kelm RJ, Polikandriotis JA, Orosz CG, Strauch AR. Reprogramming of vascular smooth muscle α-actin gene expression as an early indicator of dysfunctional remodeling following heart transplant. Cardiovasc Res. 2002;54:539–548. doi: 10.1016/s0008-6363(02)00270-5. [DOI] [PubMed] [Google Scholar]
- 26.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nature Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 27.Ouyang K, et al. Ca2+ sparks and secretion in dorsal root ganglion neurons. Proc Natl Acad Sci USA. 2005;102:12259–12264. doi: 10.1073/pnas.0408494102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjantonakis AK, Dickinson ME, Fraser SE, Papaioannou VE. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nature Rev Genet. 2003;4:613–625. doi: 10.1038/nrg1126. [DOI] [PubMed] [Google Scholar]
- 29.Zhou YY, et al. Culture and adenoviral infection of adult mouse cardiac myocytes: methods for cellular genetic physiology. Am J Physiol Heart Circ Physiol. 2000;279:H429–H436. doi: 10.1152/ajpheart.2000.279.1.H429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.