Abstract
The sinoatrial node (SAN) is the dominant pacemaker of the heart. Abnormalities in SAN formation and function can cause sinus arrhythmia, including sick sinus syndrome and sudden death. A better understanding of genes and signaling pathways that regulate SAN development and function is essential to develop more effective treatment to sinus arrhythmia, including biological pacemakers. In this review, we briefly summarize the key processes of SAN morphogenesis during development, and focus on the transcriptional network that drives SAN development.
Keywords: Sinus node development, Pacemaker, Sinus node dysfunction, Transcriptional regulation, Cardiac progenitors, Heart field
Introduction
The sinoatrial node (SAN) is the dominant pacemaker of the heart, located at the junction of the superior vena cava and right atria, that regulates heart beat frequency [1]. Abnormalities in SAN formation and function cause sinus arrhythmia and sudden death. Human sinus node dysfunction (SND), or sick sinus syndrome, is a group of heart rhythm disorders, characterized by an alternating occurrence of sinus bradycardia, sinoatrial (SA) arrest/block, or bradycardia-tachycardia syndrome, due to abnormal sinus pacemaking and conduction, and is the most common reason for pacemaker implantation [2]. SND may be seen at any age, but occurs most frequently in the elderly. Rather than being a single clinical entity, SND is often a result of other heart disorders such as coronary artery disease, atrial fibrillation, cardiac fibrosis, heart failure, diabetes and aging. The pathophysiology of SND is unclear, but involves complex structural and electrical remodeling, and changes in gene expression both in the SAN and surrounding atrial myocardium that eventually perturb pacemaking and impulse conduction [2–4]. Despite great successes in pacemaker implantation for the treatment of SND, limitations to the electronic pacemaker exist. These include limited response to autonomic regulation or increased physical activity, and the fixed size of the electronic device that does not meet the need for growth in pediatric patients. Biological pacemakers have the potential to address these limitations, but their development requires increased understanding of molecular mechanisms underlying SAN development and function.
Highly regulated expression of ion channels and associated proteins, intracellular calcium handling proteins and Connexins in the SAN contribute to proper SAN pacemaking and conduction. Misexpression of a number of these genes results in electrical remodeling and has been implicated in the pathogenesis of SND. However, it remains largely unknown as to how expression of these critical pacemaker channels and related genes are regulated and maintained. The Popeye domain containing (POPDC) genes (Popdc1–3), encoding a family of cAMP effector proteins, are abundantly expressed in heart and skeletal muscle. Ablation of Popdc1 or Popdc2 results in bradycardia, frequent sinus pauses and hypoplastic SAN tail, and the phenotype is stress induced- and age-dependent, closely mimicking human SND [5, 6]. A familial missense variant in POPDC1 (S201F) has been identified that causes cardiac arrhythmia and muscular dystrophy due to impaired membrane trafficking of both Popdc1 and Popdc2 [7]. A number of transcription factors, including Tbx18, Tbx3, Shox2, Isl1, and Tbx5, are required for the SAN development and function. However, the role of these transcription factors in maintenance of postnatal and adult SAN gene expression, cellular identity and functional homeostasis remain largely unknown [8]. Here, we will focus on recent progress in our understanding of progenitor populations that give rise to the SAN during embryonic development, and the transcriptional networks that regulate pacemaker lineage formation. Electrophysiological regulation of pacemaker activity has been the subject of recent reviews [3, 4], and will not be covered here.
SAN structure
The mature SAN is composed of heterogeneous cell types, including myocardial pacemaker cells and a substantial number of non-pacemaker cells, including transitional cells, fibroblasts and endothelial cells. Pacemaker cells are typically packed into clusters that are surrounded by fibroblasts and variable extracellular matrix (fibrous tissues) [4, 9]. Interaction and coupling between different SAN cell types, and surrounding atrial cardiomyocytes, is not fully understood. It is clear, however, that fibrotic shielding and graded expression of low and high conductance Connexins (Cx45 vs Cx40/Cx43) is critical for productive pacemaking, conduction and protection of pacemaker cells from hyperpolarization by surrounding atrial cells [9, 10]. Changes in expression of Connexins and increased fibrosis within the SAN region are observed with aging and associated with SND [9].
The SAN is a multi-compartment structure, composed of a head/center and tail/periphery region, and several specialized conduction pathways (Fig. 1). In the adult, the SAN head/center, densely packed with clusters of pacemaker cells, is the leading pacemaker region, although the leading pacemaker region can shift within the SAN in response to various stimuli [11, 12]. Tbx18 null mouse embryos that are missing the SAN head exhibit normal heart rhythm, suggesting that the SAN tail can compensate for the loss of SAN head, and function as a pacemaker during embryonic stages [13]. Whether the SAN head is essential for adult pacemaking, however, remains unclear. In contrast, ablation of Shox2 within the SAN tail results in severe SND in adult mice, highlighting an essential role of this part of the SAN in both pacemaking and conduction in the adult [14, 15]. The SAN periphery/tail is mixed with both pacemaker and atrial myocardial cells and has several specialized conduction pathways, including the SA conduction pathways (SACPs), chiefly composed of transitional cells of the SAN and intranodal atrial strands (ast), that project to both the peripheral and central SAN [9, 16–18] (Fig. 1). SACPs interdigitated with intranodal atrial strands are thought to be preferential conduction pathways for electrical impulses from the SAN to atrial myocardium [9, 10, 16]. Disruption of these pathways due to fibrosis and miscoupling of SAN cells and atrial myocardium may cause source-sink imbalance, reentry and exit block [10]. The transitional cells have been defined as cells with a phenotype that is intermediate between that of pacemaker cells and atrial myocytes, both morphologically and functionally, and in terms of the number of myofibrils and mitochondria they contain [19, 20]. The molecular and cellular identity of transitional cells and atrial strands are unknown, they may share features of both SAN pacemaker cells and surrounding atrial myocardium [15].
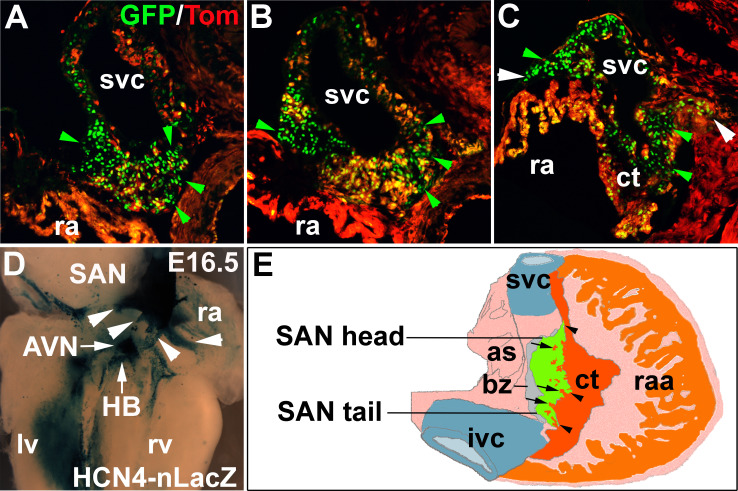
Fig. 1.
SAN structure. a–c Sections across SAN head, center and tail of Hcn4-nGFP, Nkx2–5-Cre and tdTomato triple positive mice showing pacemaker cell clusters of variable sizes (HCN4-GFP+, green arrowhead) intermingled with atrial strands (ast) (tomato+, Nkx2–5-Cre+) and potential sinus-atrial conduction paths (SACPs) (d, white arrowhead). d Wholemount Xgal staining of HCN4-nLacZ heart (endocardial view) showing cardiac conduction system, including SAN with potential SACPs (arrowhead). e A schematic diagram of SAN and the right atria (dorsal view), highlighting the interdigitation of SAN and surrounding atria working myocardium (ct), atria strands (arrow), potential SACPs (arrowhead) (as atrial septum, AVN atrioventricular node, bz block zone, ct crista terminalis, HB his-bundle, ivc inferior vena cava, lv left ventricle, ra right atria, raa right atrial appendage, rv right ventricle, SAN sinoatrial node, svc superior vena cava)
Contributions of heart field progenitors to the SAN
Over the last decade or so, a new paradigm for heart development has emerged,which demonstrates that distinct progenitors of the first and second heart fields, based on their timing of differentiation, give rise to the heart [21–23]. After formation of the early heart tube from the first heart field (FHF), the progenitors of the second heart field (SHF) migrate into the heart from both anterior (arterial) and posterior (venous) poles, contributing to the right ventricle (RV), outflow tract (OFT), and a majority of cells within the left and right atria [24]. Intriguingly, DiI labeling in chick embryos at stage 8 mapped specified SAN progenitors to a discrete region of the right lateral plate mesoderm that is posterior to a known marker of the second heart field, Isl1, and found that canonical Wnt signaling contributes to pacemaker lineage specification [25]. Given that Isl1–Cre lineages give rise to most atrial lineages, including the SAN [24, 26, 27], these observations suggest that the “tertiary heart field” in chick embryos, that contains pacemaker progenitors, as well as progenitors of atria and atrioventricular myocardium, activates expression of Isl1 as it migrates into the heart. It will be of future interest to identify markers that identify this tertiary heart field, and investigate its relevance to heart development in other organisms.
The pacemaker and left–right asymmetry
Located at the right sinoatrial junction, the SAN is by its nature an asymmetric structure. In mouse embryos, left–right identity of the inflow region is specified around the six-somite stage (E8–8.5), correlating with asymmetric expression of the homeodomain transcription factor, Pitx2c, within the left heart field [28–30]. Lineage studies using Dil labeling in chick or mouse embryos revealed that progenitors of the right and left posterior SHF do not mix, and contribute to the right or left atria or sinus venosus (SV), respectively [25, 31, 32].
During mouse and chick development, the first heartbeat is recorded in a left-dominant fashion in the inflow tract (IFT) as early as E8.0–8.5 in mouse or rat, stage 10 in chick [33–38]. These early pacemaker cells will not contribute to the definitive SAN, but rather to the atrioventricular region [25, 39]. By heart looping stages, pacemaking activity is localized to the right inflow, juxtaposed to forming atria [9, 17–22]. These cells will go on to form the SAN which develops at the right sinoatrial junction. The first morphologically discernible SAN is formed at E10.5–11.5, which further proliferates and becomes mature at E13.5 [35, 40].
Expression of the hyperpolarization-activated cyclic-nucleotide gated ion channel, HCN4, marks and contributes to SAN activity during development and in the adult [27, 41–43]. Intriguingly, initial expression of Hcn4 occurs at the cardiac crescent stage in mice (E7.5), and Hcn4 is expressed more highly on the left than on the right [41]. At E8.0, Hcn4 is expressed in a stripe across the junction of the forming atrium and sinus venosus, but is more highly expressed on the left side of the sinus venosus, correlating with an early left-dominant pacemaker. At E8.5, Hcn4 becomes more highly expressed on the right side, at the junction between the atrium and the sinus venosus, in the cells that will go on to form the mature SAN.
Patterning of posterior heart field progenitors, including the SAN
Progenitors of the posterior SHF will contribute to myocardium of the atria, the SV, pulmonary vein (PV), and the SAN. Sublineage segregation of progenitors that give rise to these distinct structures appears to occur early during development, as marked by distinct expression of specific transcription factors, including those of the homeodomain family, including Isl1, Nkx2–5, Shox2, and Pitx2c, and others of the T-box (Tbx) family, including Tbx18, Tbx3, and Tbx5.
After initial formation of the primitive SV from the first differentiated Isl1+/Nkx2.5+ posterior SHF progenitors before E9.5, a caudal-lateral subset of the SHF marked by Tbx18+, Isl1+ low, but Nkx2–5−, starts to emerge at E8.5–9.5 and contributes to myocardium of the sinus horn. Among Tbx18+ sinus horn mesenchyme progenitors, a posterior-most subset of progenitors coexpressing Tbx18 and Isl1 will contribute to SAN formation [13, 31, 44]. In contrast, PV myocardium is derived from a distinct progenitor population of the dorsal mesocardium that is Isl1+/Nkx2.5+/Tbx3+, but Tbx18− [31, 45, 46]. Retrospective clonal analysis using an α-cardiac actin-nlaacZ line demonstrated a clonal relationship between the PV and progenitors of the left SV/coronary sinus [47]. Consistent with this, a recent lineage study with Shox2-Cre demonstrated that PV myocardium shares a common lineage origin with the left SV [14, 15].
The SAN is a multi-compartment structure, composed of a head embedded in the right superior vena cava, and a tail extending into the crista terminalis [18]. Lineage studies with HCN4-CreERT2 revealed that the SAN head (Nkx2–5−) and tail/periphery (Nkx2–5+) may be derived from distinct progenitor populations. Tamoxifen inductions of HCN4-CreERT2 from E7 to 7.5, harvesting at E16.5, selectively labeled a few cells in the SAN tail [27, 43]. Tamoxifen inductions of HCN4-CreERT2 at E8.5 or later resulted in robust labeling of the SAN head [27, 43], consistent with emergence of SAN progenitors at around the six-sormite stages (~E8.5) in the right caudal-most SV [13, 44]. Lineage studies with Tbx18-Cre revealed that both the SAN head and tail are labeled, although Tbx18 is not actively expressed in the SAN tail [13]. Tbx18+ mesenchyme, isolated from the lateral side of the inflow tract of Tbx18-GFP embryos at E9.5 and cultured for 96 h, expresses HCN4 and the myocardial marker MF20, but not Nkx2.5, consistent with having potential to give rise to the SAN head, but not the tail [31].
At early embryonic stages, myocardium of the SV and PV exhibit hybrid phenotypes of atrial working cardiomyocytes and pacemaker cells, expressing atrial myocyte genes (Nkx2–5), as well as pacemaker genes (Hcn4), and marked by a conduction system marker, CCS-LacZ [48]. Later, the majority of SV myocardium loses expression of pacemaker genes as it incorporates into the atrioventricular canal, right atria, right superior vena cava (right), PV (left), and coronary sinus (left, in human) [39, 49]. However, under certain pathological conditions such as ischemia, these cells may re-express a pacemaker phenotype. Thus, adult arrhythmias often map to embryonic pacemaker region-derived myocardium [50, 51].
Transcriptional regulation of SAN development
Formation of the SAN is tightly regulated by a network of transcription factors that display a dynamic and distinct expression pattern in the SAN and surrounding atrial working myocardium. These transcription factors play critical roles both in specification and differentiation of pacemaker cells, and in maintaining pacemaker identity and function (Fig. 2a). Integration of results from a large number of studies has resulted in a cross-repressive model for pacemaker development has emerged (Fig. 2b). Cardiac progenitor factors expressed broadly in the heart, including Tbx5, Nkx2–5 and GATA4, establish a working myocardial cell fate, whereas factors confined to the SAN function as activators to promoter pacemaker cell fate, and/or as repressors to prevent working myocardial cell fate. For example, in cells with increased expression of Nkx2–5 and Tbx5, SAN genes such Shox2 and Tbx3 are repressed to promote a working myocardial cell fate. On the other hand, in cells with sustained expression of Isl1, Shox2 and Tbx3, working myocardial-enriched genes, including Nkx2–5, are repressed, and SAN specific genes are activated. Below we will further discuss the expression and roles of several key transcription factors during SAN development.
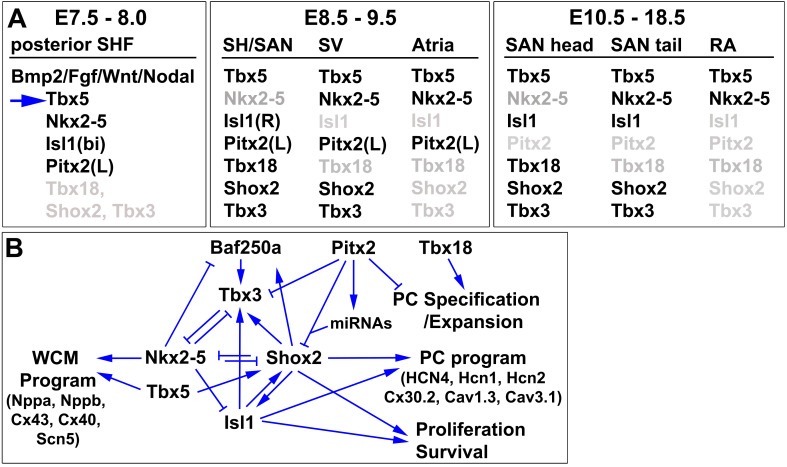
Fig. 2.
Expression of the key SAN transcription factors during inflow tract development and the regulatory network controlling SAN morphogenesis. a Dynamic expression of key transcription factors (Tbx5, Nkx2-5, Isl1, Pitx2, Tbx18, Tbx3, Shox2) during development in the posterior second heart field (SHF), sinus venosus (SV), sinus horn (SH) and sinoatrial node (SAN), and surrounding atrial working myocardium. b A cross-repressive regulatory network that controls pacemaker cell (PC) and atrial working cardiomyocyte (WCM) development
Tbx18
Early in development, Tbx18 marks proepicardium and epicardium, sinus horn myocardium, and SAN. Tbx18 expression is initiated at around E8.25 in mesenchymal cells of the IFT that caudally abut Nkx2–5+ SV myocardium. A subset of Tbx18+ mesenchymal progenitors coexpressing Isl1 at the caudal-lateral most border will give rise to myocardium of the sinus horn and the SAN [13, 30, 31, 44]. At E8.5, coexpression of Tbx18 and Isl1 appears to be bilateral, with the left domain of SV mesenchyme additionally coexpressing Pitx2c [30]. After E8.5, the Tbx18+/Isl1+ area becomes confined to the right lateral side likely due to Isl1 downregulation on the left side [30, 31, 44, 52]. Soon after sinus horn myocardium starts to differentiate, from E9.5 to E10.5, and the first morphologically discernible SAN, coexpressing Tbx18, Isl1 and Tbx3, becomes visible in right sinus horn myocardium [13, 30, 31, 44]. Tbx18–Cre lineage studies revealed that the entire SAN is derived from Tbx18–Cre lineages, although Tbx18 is not actively expressed in the SAN tail/periphery [13], suggesting a transient Tbx18 expression in cells that contribute to SAN tail. Tbx18 expression is gradually downregulated in SAN head and becomes undetectable by birth [53]. However, using a Tbx18-GFP knock-in mouse line, we found that Tbx18-GFP is actively expressed in a small population of adult SAN cells, a subpopulation of which co-express Isl1 (YS, unpublished data).
Tbx18 is required for formation of the sinus horn and SAN head. Ablation of Tbx18 results in failure to form the sinus horn, and a loss of the SAN head. However, the SAN tail of Tbx18 mutants is formed correctly and embryos exhibit normal sinus rhythm [13, 44]. No changes in cell death, proliferation, or SAN gene expression (Tbx3, Isl1, Shox2), and no ectopic expression of atrial chamber genes (Cx40, Nppa, Nkx2–5) are observed in the residual SAN tail of Tbx18 mutants. These observations suggest a specific requirement for Tbx18 in specification and recruitment of the SAN head, and therefore, delineate distinct regulatory mechanisms for the formation of SAN head and tail. Furthermore, expression of genes critical for inflow tract (IFT) morphogenesis, including Gata4, Gata6, Raldh2, Coup-TFII, and Tbx5, is not altered in Tbx18 mutants, suggesting that Tbx18 acts downstream or in an independent molecular pathway [44]. Lineage studies have demonstrated that expression of Tbx18 and Nkx2–5 is mutually exclusive in the IFT. However, in contrast to cross-repressive mechanisms observed for Nkx2–5/Shox2 and Nkx2–5/Tbx3 in establishing the SAN-atrial boundary (see below), neither Tbx18 nor Nkx2–5 were derepressed in Nkx2–5 or Tbx18 mutants, respectively, suggesting that upstream factors and other mechanisms are responsible for formation of the border between the sinus horns and atria [13, 15, 44].
Tbx18 overexpression by adenoviral transduction of neonatal rat ventricular myocytes in vitro, and in adult guinea pig ventricular myocytes in vivo, induces pacemaker-like cells that resemble native SAN cells in their morphology, electrophysiology, gene expression and epigenetic features (H3K4me3 and H3K27me3 modification in promoters of selected candidates, including Cx43, Kir2.1, Actc2 and Hcn4) [53]. Overexpression of TBX18 in ventricle of adult pigs with complete heart block can reprogram ventricular cardiomyocytes to pacemaker cells that function as a de novo pacemaker [54]. This reprogramming by TBX18 is durable and independent of continuous TBX18 expression [54]. Although effective pacing by TBX18 overexpression is transient, this is the most successful example of a single transcription factor inducing adult cardiomyocyte reprogramming in a clinically relevant system. Surprisingly, transgenic overexpression of Tbx18 in embryonic mouse heart is not sufficient to induce an SAN program or SAN differentiation of chamber cardiomyocytes [55], suggesting the presence or absence of (an) unidentified factor(s) in cardiomyocytes that is blocks or is required for reprogramming of chamber myocytes to SAN phenotype in this system. Molecular pathways downstream of Tbx18 in SAN formation during development remain unknown [53, 56].
Shox2
The homeodomain transcription factor Shox2 is expressed from, as early as E8.5, first appearing in the IFT region of developing heart, and later including SV myocardium, SAN and venous valves, and PV [57, 58]. Shox2 is required for IFT development, and plays a critical role in pacemaker cell differentiation, and in defining the boundary between SAN and working cardiomyocytes of the atrium. Shox2 acts mainly as a transcriptional repressor in IFT development by repressing Nkx2–5 and the working myocardial gene program, thus allowing activation of the pacemaker gene program (Tbx3, Isl1, Hcn4). Shox2 null mice die between E11.5 and E17.5, and exhibit bradycardia, hypoplastic SAN and SV, owing to decreased proliferation [57, 58]. Shox2 hypomorphic mice exhibit bradycardia and die a few days after birth [59]. Expression of Tbx3, Isl1 and Hcn4 is downregulated, whereas Nkx2–5, Nppa and Cx40 are ectopically expressed in Shox2 mutant SAN, demonstrating a failure to repress an atrial myocardial program [57, 58]. Tbx18 expression in Shox2 mutant SANs is not changed. Overexpression of Shox2 in Xenopus embryos results in loss of cardiac progenitors and Nkx2–5 expression, and mutant embryos exhibit a smaller heart tube, and slow and irregular heartbeat, similar to that of Nkx2–5 null mice [58].
Expression of Shox2 and Nkx2–5 in the IFT is largely complementary, except within the SAN tail/periphery and in PV myocardium, where they are strongly coexpressed. Recent studies have revealed a Shox2–Nkx2–5 antagonistic mechanism for regulating cell fate between pacemaker and working myocardium. Disturbing the balance of gene expression in favor of Shox2 or Nkx2–5 shifts cell fate between pacemaker and working myocardium, respectively [14, 15]. Deletion of Shox2 in the SAN tail/periphery with Nkx2–5-Cre results in loss of Tbx3, Hcn4 and Isl1 expression, but ectopic Cx40 activation, and adult mutant mice exhibit sick sinus syndrome characterized by severe bradycardia, irregular heartbeat and sinoatrial (SA) block. Concomitant Nkx2–5 hypomorphism in Shox2 mutants re-establishes expression of Hcn4, Tbx3, Isl1 and Cx40 in the SAN tail/periphery. Similarly, Nkx2–5 hypomorphism results in a pacemaker-like phenotype (Hcn4+/Cx40−) in PV myocardium, which is reverted to a working myocardial fate (Cx40+/Hcn4−) when Shox2 is simultaneously deleted. These studies suggest that Shox2 plays a permissive role for activation of the pacemaker gene program by antagonizing inhibitory effects of Nkx2–5. When Nkx2–5 is absent, Shox2 is dispensable for expression of Isl1, Tbx3 and Hcn4, suggesting that these genes are not direct targets of Shox2, although Isl1 has been shown to be a direct target of Shox2 [60]. These studies suggest a complex cross-regulatory network during SAN formation, where expression of Isl1 and Shox2 is mutually dependent, both acting upstream of Tbx3, and together regulating an SAN gene program. Expression of Isl1, Shox2 and Tbx3 (SAN genes) is repressed by Nkx2–5, whereas Shox2 and Tbx3 can repress Nkx2–5 expression (Fig. 2). In addition, deletion of Shox2 in the SAN tail/periphery leads to loss of the SAN tail and severe SAN dysfunction, arguing again that the SAN tail plays an essential role in pacemaking.
Shox2 directly interacts with Nkx2–5 and Tbx5. ChIP-Seq analyses have revealed substantial genomewide co-occupancy of binding peaks for Shox2, Nkx2–5 and Tbx5, suggesting co-regulation of target genes by Shox2, Nkx2–5 and Tbx5 directly, and a mechanism for the antagonistic action between Shox2 and Nkx2–5 in SAN and PV development. Tbx5 is an upstream activator of Shox2 in IFT development. Tbx5 heterozygous mutant mice and Tbx5 deficient zebrafish exhibit marked reduction in expression of Shox2 and Bmp4 in the IFT, and Bmp4 is completely absent from IFT of Shox2 mutant mice and zebrafish, although its expression in the OFT remains unchanged [57, 61]. Tbx5 acts cooperatively with Nkx2–5 to activate Shox2 expression, which in turn activates Bmp4 in the inflow tract of the embryonic heart [61]. ChIP and reporter gene assays have revealed that Shox2 binds to a promoter/enhancer of Bmp4 and promotes its expression [61]. Pitx2c is an upstream suppressor of the pacemaker gene program in the left SHF, left atria and pulmonary vein [30, 62–65]. Pitx2 directly binds to the Shox2 promoter in vivo and regulates its expression [64]. Pitx2 can also regulate Shox2 expression indirectly via microRNAs (miR-17-92 and miR-106b-25) that directly repress Shox2 and Tbx3 expression [65].
Overexpression of human Shox2 in mouse ES cells results in increased expression of Cx45, Hcn4 and endogenous Shox2, but decreased Nkx2–5 and Cx43 expression. These Shox2-EBs exhibit an increased automaticity and can function as a biological pacemakers when injected into rat heart in vivo [66]. Shox2 promoter-driven neomycin gene or GFP have been used as selection strategies to enrich and isolate pacemaker cells from ESCs [67, 68]. In contrast to results with adenoviral transduction with TBX18 in neonatal rat ventricular cardiomyocytes and pig heart [53], lentiviral Shox2 transduction of neonatal rat ventricular myocytes failed to induce pacemaker activity [53].
Tbx3
Expression of Tbx3 is first observed in the IFT of the heart, and in the pharyngeal region at E8.5. From E9.5 onward, Tbx3 expression is largely confined to atrioventricular (AV) canal and a subregion of the sinus horn, the forming SAN, complementary to the expression of working myocardial markers Cx40 and Cx43 [69]. Like Shox2, Tbx3 functions predominantly as a transcriptional repressor in the SAN, repressing the atrial working myocardial gene program, and indirectly promoting the pacemaker gene program. Ablation of Tbx3 leads to embryonic lethality at E11.5–E15.5, with ectopic Cx43 expression in the SAN. However, the size of the SAN, Hcn4 expression, and cardiac rhythm in Tbx3 mutant embryos appear to be normal. No changes in cell death and proliferation were observed in Tbx3 mutant SANs. Atrial working myocardial genes Cx40, Nppa and Scn5a are not ectopically expressed within the SAN before E12.5 [70]. However, from E13.5 onward, these working myocardial genes (Cx40, Nppa) start to be ectopically expressed in SAN of Tbx3 mutants, correlated with upregulation of Nkx2–5 expression in the SAN/tail periphery region [15, 45, 70]. Tbx3 expression persists in adult SAN, and adult mice that are hypomorphic for Tbx3 exhibit various lethal arrhythmias, including AV block, bradycardia, sinus pauses and sudden death. The SAN of Tbx3 hypomorphs is hypoplastic with ectopic Cx43 expression, suggesting that continued expression of Tbx3 during later development and adulthood is required for SAN morphogenesis and homeostasis [71]. In Tbx3 hypomorph SANs, expression of Tbx5 and Tbx18 is normal, suggesting Tbx3 acts downstream or independently of Tbx5 and Tbx18 [71]. Consistent with this, compound mutants for Tbx3 and Tbx18 exhibit additive phenotypes of individual Tbx3 and Tbx18 mutants.
Overexpression of Tbx3 in embryonic mouse hearts using Nppa-Cre induces a switch to an SAN gene program and ectopic pacemaker sites within atria [70]. Expression of working myocardial genes Cx40, Cx43, Nppa and ion channels (Scn5a and Kir2.1, kir2.1 and Kir3.1) are downregulated, whereas pacemaker genes are ectopically induced in atria of Tbx3 mutants, including Hcn1, Hcn2, Hcn4 and Cx30.2. However, Nkx2–5 expression is not repressed, suggesting that ectopic Tbx3 can induce a pacemaker-like phenotype in immature atrial myocytes.
Overexpression of Tbx3 in adult hearts using a tamoxifen inducible Cre line (Myh6-MerCreMer) similarly downregulates working myocardial genes responsible for intercellular coupling and inward-rectifying current in both atria and ventricles [72]. However, induction of SAN enriched genes, including a number of pacemaker enriched ion channels (Hcn4, Ca2+ channel Cacna1d and Cacna1g), is less efficient compared to the upregulation observed consequent to Tbx3 overexpression in immature atrial myocytes. Accordingly, no ectopic pacemaker activity is induced in an atrial preparation of adult Tbx3 overexpressing mutants. Similarly, lentiviral transduction of Tbx3 overexpression in neonatal rat ventricular myocytes failed to induce pacemaker activity [53, 72]. However, Tbx3 overexpression in ES cells plus an additional Myh6 promoter-based antibiotic selection results in a large fraction (>80 %) of selected cells characteristic of pacemaker-like cells, and aggregates of these cells are able to pace myocardium ex vivo [73].
Tbx3 ChIP-Seq analyses on heart extracts of Tbx3 overexpressing adult mice revealed that Tbx3 competes with Tbx5 for binding with Nkx2–5, thus repressing activation of Nkx2–5 and Tbx5 target genes, including Cx40, Cx43 and Scn5α [69, 70, 74, 75]. Many enhancers of ion channel genes are directly repressed by Tbx3, including Scn5a and Scn10a. Tbx3 may complex with Baf250a and HDAC3 to bind to the promoter/enhancers of Nkx2–5 and repress Nkx2–5 expression directly [76]. Tbx3 binding to the Nkx2–5 promoter is dependent on physical association with Baf250a, since ablation of Baf250a abolishes Tbx3 binding to the Nkx2–5 promoter, results in increased expression of Nkx2–5, and subsequently Gata4 and Tbx5, suggesting conversion from a pacemaker to working myocardial gene program [76]. However, ablation of Tbx3 at either embryonic or adult stages does not lead to ectopic Nkx2.5 expression, suggesting other factor(s) may exist that are sufficient to repress Nkx2–5 expression. Interestingly, a recent study has revealed that Tbx3 may cooperate with Shox2 to repress Nkx2–5 expression, and mice compound hypomorphic for Shox2 and Tbx3 display significant higher levels of Nkx2–5 in SAN compared to littermate controls [14]. Tbx3 acts downstream of Shox2 and Isl1, and deletion of Shox2 or Isl1 leads to absence of Tbx3 expression [58, 59, 77]. Isl1 ChIP-seq analysis together with a recently published Hi-C seq data suggested that Isl1 may bind two putative long-range enhancers that looped to the Tbx3 promoter to regulate Tbx3 expression [77–79].
Isl1
Isl1 is a LIM homeodomain transcriptional factor that marks undifferentiated cardiac progenitors of the SHF and is required for normal heart development [24]. Isl1 mRNA expression is first observed at around E7.0 in lateral plate mesoderm and endoderm [24, 31, 80]. As discussed above, definitive SV or sinus horn progenitors/myocardium of the caudal-lateral most IFT coexpresses Tbx18 and Isl1, but not Nkx2–5 [31, 44]. Initial expression of Isl1 in the IFT is bilateral, however, after E8.5 Isl1 expression in the left IFT is downregulated, thus coexpression of Isl1 and Tbx18 is confined to the right sinus horn that will form the SAN and sinus horn myocardium [30, 31, 44, 52]. Isl1 expression persists in SAN pacemaker cells during embryonic development, but is gradually downregulated postnatally, and persists only in a small subpopulation of pacemaker cells in adult SAN [31, 52, 77, 81]. However, a potential role for Isl1 in adult SAN remains to be addressed. Given its role at embryonic stages in regulation of expression of genes essential for pacemaker cell proliferation and function, it is tempting to speculate that Isl1 may be required in a subset of pacemaker cells in adult SAN, and may confer distinct properties on those cells that fulfill critical functions in pacemaking activity. Lineage studies have revealed that the majority of SAN pacemaker cells are labeled by Isl1-Cre.
Isl1 progenitors are multipotent and can differentiate into multiple cell types within the heart, including cardiomyocytes, smooth muscle cells, endothelial cells and pacemaker cells [24, 52, 82]. Isl1 null mice die at E10.5, with cardiac structures derived from the SHF being severely affected, exhibiting loss of OFT and RV, and severely hypoplastic atria [24]. Isl1 is required for pacemaker cell proliferation, survival and pacemaker function. In zebrafish, Isl1 mutation results in bradycardia and irregular heart beat with frequent pauses [83, 84]. ln mice, reduced Isl1 expression, or ablation of Isl1 in pacemaker cells using HCN4-CreERT2 induced at several distinct embryonic stages, results in bradycardia, increased heart-rate variability, and prolonged sinus pauses. Isl1 mutant mice exhibit hypoplastic SANs due to increased cell death and reduced proliferation, and markedly reduced expression of Shox2, Tbx3 and Hcn4 [77]. RNA-seq analyses revealed that Isl1 is a key upstream regulator of ion channels (Cacna1a, Cacna1d, Cacnb1, Hcn4, Kcnn1, and Ank2) and transcription factors (Shox2, Tbx3, Ehmt2, Hdac7, Smyd, and Arid1b) required for SAN function [77, 85]. A number of genes that play important roles in heart or SAN development are downregulated in Isl1 mutant pacemaker cells, including genes associated with cell cycle (Arid1b, Wdr62, Kras, and Myc) and signaling pathways (Bmp4, Rgs4, Calcitonin, Calcrl, Klotho, Sema3c, and Sema3d). Despite marked downregulation of Shox2 and Tbx3, there is no ectopic Nkx2–5 expression in Isl1 mutant pacemaker cells, although a number of atrial myocardial specific genes (Nppa, Nppb, Cx43, and Cx40) are upregulated. Overexpression of Isl1 in mESCs results in upregulation of cardiac progenitor markers and increased cardiomyocyte generation, suggesting a role of Isl1 in cardiac progenitor specification and early myocardial lineage differentiation [86]. Overexpression of Isl1 appears to favor a pacemaker phenotype, as evidenced by increased beating rate, upregulation of a number of pacemaker cell enriched ion channels including Hcn4 and calcium channels, and downregulation of working myocardial genes (Cx40, Cx43, Nppa, Myl2, Scn5a).
RNA-seq and ChIP-seq analyses have revealed that Isl1 acts predominantly as a transcriptional activator within pacemaker cells. Among direct targets of Isl1 are genes important for pacemaker function or implicated in human SND including Ank2, Kl, Tbx3, Calcrl and Flrt2 [13, 71, 77, 87–89], suggesting Isl1 mutations may underlie SND. Analyses of DNA binding motifs enriched within Isl1 binding peaks in pacemaker cells have revealed enrichment for other homeodomain and FOX transcription factor binding sites, suggesting that members of these transcription factor families may cooperate with Isl1 to regulate expression of SAN genes.
Together, these studies suggest that Isl1 plays an upstream role in the transcriptional hierarchy that coordinate the specification of cardiac progenitors and subsequent diversification of distinct myocardial lineages including pacemaker cells. However, similar to studies of Tbx3 and Shox2, overexpression of Isl1 results in only partial activation of the SAN program [86]. Thus, to generate a faithful biological pacemaker from ES cells or other adult somatic cells, it is necessary to explore combinations of cardiac transcription factors important in SAN identity and function, and appropriate selection strategies.
Nkx2–5
The Nk2 homeodomain transcription factor Nkx2–5 is among the earliest cardiogenic factors expressed in all myocardial progenitors of the FHF and SHF, with the exception of a subset of progenitors that give rise to the sinus horn and definitive pacemaker cells [13, 31, 44, 45]. Expression of Nkx2–5 increases with myocardial differentiation and is required for myocardial specification and maturation. In the anterior SHF, Nkx2–5 is activated in Isl1 progenitors and plays critical roles in promoting SHF proliferation and OFT morphogenesis. Nkx2–5 acts as a feedback repressor of Bmp2, a gene responsible for cardiac specification, and can directly bind to an Isl1 enhancer and suppress its expression [86, 90]. Ablation of Nkx2–5 leads to upregulation and ectopic expression of cardiac progenitor genes (Bmp2 and Isl1), resulting in over-specification of cardiac progenitors, but reduced SHF proliferation [86, 90]. Conversely, overexpression of Nkx2–5 in differentiating mESCs leads to delayed expression of key SHF factors, including Isl1, Tbx20, Gata4, Fgf10 and Kdr, partially mediated by direct inhibition of Isl1 by Nkx2–5 [90].
Lineage studies with Nkx2–5-Cre demonstrated that all myocardium is labeled by Nkx2–5-Cre lineages except the sinus horns and SAN head [44, 45]. At E10.5 and thereafter, Nkx2–5 is expressed in atrial and PV myocardium, and SAN tail/periphery. Nkx2–5 acts to repress Hcn4 and Tbx3, and is required to establish the SAN-atrial boundary [15, 45]. Ablation of Nkx2–5 leads to ectopic expression of Hcn4 and Tbx3 throughout the heart tube, and a failure to express atrial chamber genes Cx40 and Nppa, direct targets of Nkx2–5, Tbx5 and Tbx3 [45, 69, 91, 92]. Reduced Nkx2–5 expression in Nkx2–5 hypomorphic mice results in reduced expression of Cx40, and an expansion of SAN genes such Tbx3 and Hcn4 into surrounding atrial myocardium [45], demonstrating a critical role of Nkx2–5 in regulation and maintenance of the atrial chamber gene program at the SAN-atria boundary.
Nkx2–5 is expressed in the SAN tail/periphery and the PV, and its expression in these domains increases and expands with growth and maturation of the SAN [15, 45]. As discussed above in detail, a recent study uncovered an antagonism between Shox2 and Nkx2–5 in control of pacemaker versus working myocardial cell fate in SAN and PV [15]. Nkx2–5 upregulation is correlated with development of atrial strands that interdigitate and penetrate into the SAN and function as effective conduction paths [17, 19] (Fig. 1). Sarcolipin-Cre marks more mature atrial myocardium and IFT at E12.5 and thereafter [93]. Ablation of Nkx2–5 in IFT after its initial morphogenesis using Sarcolipin-Cre results in hyperplastic atrial myocardium, expansion of Hcn4 expression, and increased proliferation of both atria and SAN tail/periphery, suggesting a suppressive role for Nkx2–5 in myocardial proliferation of atria and SAN tail/periphery, partially attributed to upregulation of the Notch pathway [93].
Genomewide association studies have identified Nkx2–5 and Tbx5 as heart-rate associated loci, and mutation of Nkx2–5 and Tbx5 causes SND [15, 89, 94, 95]. Scn5a is the most common gene mutated in human SND and is also implicated in aging-related SND [4, 96–98]. Scn5a mutant mice exhibit SND with SAN exit block [99]. Scn5a is expressed in working myocardium and SAN tail/periphery, but not within the SAN head. Similar to the other atrial working myocardial genes such as Nppa and Cx40, Scn5a is directly activated by Nkx2–5 and Tbx5, but repressed by Tbx3 [69, 74, 75].
Pitx2
The homeobox transcription factor Pitx2 is a laterality gene responsible for establishment of the left–right body axis, asymmetrical gene expression, and organ morphogenesis. Pitx2 is expressed on the left side of multiple organs including the heart. Asymmetric expression of Pitx2 in mice is first observed at E8.0 (6 somites) in the left cardiac crescent, the left side of the SHF and heart tube [32, 100–102]. Subsequently, Pitx2 continues to be expressed on the left side of the developing heart from the venous pole to arterial pole, including the left SV, the PV, left atrium, and the ventral region of both ventricles, interventricular septum and ventral/left OFT [100–102]. At later developmental and postnatal stages, Pitx2 expression is gradually diminished, with a low level of expression being maintained within left atrium, the PV and RV [64].
Lineage studies revealed that Pitx2 lineages are confined largely to second heart lineages [62]. Consistent with this, ablation of Pitx2 in the second heart lineage with Isl1-Cre or Mef2c-Cre results in cardiac defects similar to those seen in Pitx2 null mice, including double outflow right ventricle, transposition of the great arteries (TGA), and atrial (ASD) and ventricular (VSD) septal defects. However, expression of Isl1 itself in the OFT of Pitx2 mutants is unchanged, suggesting that Isl1 is not regulated by Pitx2 [62].
Mutation of Pitx2 has been associated with atrial arrhythmias such as atrial fibrillation. At E8.5 Pitx2 is expressed in the entire left SV with strongest expression in caudal-most mesenchyme, where it is coexpressed with Tbx18 and Isl1 [30, 32, 101, 102]. Pitx2 acts to prevent proliferation and expansion of left SV myocardium [32], and is required for repressing the pacemaker gene program in left SV, left atria and PV. Global or myocardial specific loss of Pitx2 results in formation of bilateral SANs that are morphologically and molecularly indistinguishable [30, 45], suggesting a default program for SAN formation. Pitx2 loss or haplo-insufficiency causes atrial fibrillation, and microarray analysis of RNA from mutant hearts revealed upregulation of SAN genes (Hcn4, Tbx3 and Shox2) and a number of ion channels, among which Kcnq1 is implicated in familial atrial fibrillation [64]. ChIP analysis demonstrated that Pitx2 directly binds to and represses the Shox2 promoter in vivo [64]. Pitx2 can also regulate Shox2 expression indirectly via microRNAs (miR-17-92 and miR-106b-25) that directly repress Shox2 and Tbx3 expression [65].
Conclusion
The SAN is a complex and well-coupled biological system, composed of subdomains and multiple cell types with distinct functions. Formation of the SAN is under tight control of a network of transcription factors acting as both activators and repressors. Despite great progress achieved, important questions remain to be addressed, including the role of these pacemaker specific transcription factors in adult SAN structure and functional homeostasis, both under physiological and pathological conditions. What are the mechanisms underlying the role of Tbx18 and Pitx2 in pacemaker specification, given their profound roles in SAN induction and formation? What are target genes of Tbx18 and Pitx2 in pacemaker progenitors? What is the cell identity of the transitional cells within the SAN? Answers to these questions will further facilitate the development of regenerative therapies, including development of biological pacemakers.
Acknowledgments
YFS was supported by grants from the Ministry of Science and Technology China (2013CB967400) and National Natural Science Foundation of China (NSFC) (81570285); SME by grants from NIH (HL123747, HL119967). XQL by grants from NSFC (81370196, 81670448, 81221001).
Compliance with ethical standards
Conflict of interest
The authors confirm that there are no conflicts of interest.
References
- 1.Keith A, Flack M. The form and nature of the muscular connections between the primary divisions of the vertebrate heart. J Anat Physiol. 1907;41(Pt 3):172–189. [PMC free article] [PubMed] [Google Scholar]
- 2.Dobrzynski H, Boyett MR, Anderson RH. New insights into pacemaker activity: promoting understanding of sick sinus syndrome. Circulation. 2007;115(14):1921–1932. doi: 10.1161/CIRCULATIONAHA.106.616011. [DOI] [PubMed] [Google Scholar]
- 3.Monfredi O, Boyett MR (2015) Sick sinus syndrome and atrial fibrillation in older persons—a view from the sinoatrial nodal myocyte. J Mol Cell Cardiol 83:88–100 [DOI] [PubMed]
- 4.Choudhury M, Boyett MR, Morris GM. Biology of the sinus node and its disease. Arrhythm Electrophysiol Rev. 2015;4(1):28–34. doi: 10.15420/aer.2015.4.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Froese A, Breher SS, Waldeyer C, Schindler RF, Nikolaev VO, Rinne S, Wischmeyer E, Schlueter J, Becher J, Simrick S, et al. Popeye domain containing proteins are essential for stress-mediated modulation of cardiac pacemaking in mice. J Clin Invest. 2012;122(3):1119–1130. doi: 10.1172/JCI59410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schindler RF, Scotton C, French V, Ferlini A, Brand T. The Popeye domain containing genes and their function in striated muscle. J Cardiovasc Dev Dis. 2016;3(2):22. doi: 10.3390/jcdd3020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schindler RF, Scotton C, Zhang J, Passarelli C, Ortiz-Bonnin B, Simrick S, Schwerte T, Poon KL, Fang M, Rinne S, et al. POPDC1(S201F) causes muscular dystrophy and arrhythmia by affecting protein trafficking. J Clin Invest. 2016;126(1):239–253. doi: 10.1172/JCI79562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dobrzynski H, Anderson RH, Atkinson A, Borbas Z, D’Souza A, Fraser JF, Inada S, Logantha SJ, Monfredi O, Morris GM, et al. Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol Ther. 2013;139(2):260–288. doi: 10.1016/j.pharmthera.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Csepe TA, Kalyanasundaram A, Hansen BJ, Zhao J, Fedorov VV (2015) Fibrosis: a structural modulator of sinoatrial node physiology and dysfunction. Frontiers Physiol 6:37 [DOI] [PMC free article] [PubMed]
- 10.Fedorov VV, Glukhov AV, Chang R. Conduction barriers and pathways of the sinoatrial pacemaker complex: their role in normal rhythm and atrial arrhythmias. Am J Physiol Heart Circ Physiol. 2012;302(9):H1773–H1783. doi: 10.1152/ajpheart.00892.2011. [DOI] [PubMed] [Google Scholar]
- 11.Verheijck EE, van Kempen MJ, Veereschild M, Lurvink J, Jongsma HJ, Bouman LN. Electrophysiological features of the mouse sinoatrial node in relation to connexin distribution. Cardiovasc Res. 2001;52(1):40–50. doi: 10.1016/S0008-6363(01)00364-9. [DOI] [PubMed] [Google Scholar]
- 12.Monfredi O, Dobrzynski H, Mondal T, Boyett MR, Morris GM. The anatomy and physiology of the sinoatrial node—a contemporary review. Pacing Clin Electrophysiol. 2010;33(11):1392–1406. doi: 10.1111/j.1540-8159.2010.02838.x. [DOI] [PubMed] [Google Scholar]
- 13.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM (2009) Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res 104(3):388–397 [DOI] [PubMed]
- 14.Ye W, Song Y, Huang Z, Zhang Y, Chen Y. Genetic regulation of sinoatrial node development and pacemaker program in the venous pole. J Cardiovasc Dev Dis. 2015;2(4):282–298. doi: 10.3390/jcdd2040282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye W, Wang J, Song Y, Yu D, Sun C, Liu C, Chen F, Zhang Y, Wang F, Harvey RP, et al. A common Shox2–Nk2–5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 2015;142(14):2521–2532. doi: 10.1242/dev.120220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Quintana D, Cabrera JA, Farre J, Climent V, Anderson RH, Ho SY. Sinus node revisited in the era of electroanatomical mapping and catheter ablation. Heart. 2005;91(2):189–194. doi: 10.1136/hrt.2003.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang X, Cui X. The functions of atrial strands interdigitating with and penetrating into sinoatrial node: a theoretical study of the problem. PLoS One. 2015;10(3):e0118623. doi: 10.1371/journal.pone.0118623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Dobrzynski H, Yanni J, Boyett MR, Lei M. Organisation of the mouse sinoatrial node: structure and expression of HCN channels. Cardiovasc Res. 2007;73(4):729–738. doi: 10.1016/j.cardiores.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Boyett MR, Honjo H, Kodama I. The sinoatrial node, a heterogeneous pacemaker structure. Cardiovasc Res. 2000;47(4):658–687. doi: 10.1016/S0008-6363(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 20.Verheijck EE, Wessels A, van Ginneken AC, Bourier J, Markman MW, Vermeulen JL, de Bakker JM, Lamers WH, Opthof T, Bouman LN. Distribution of atrial and nodal cells within the rabbit sinoatrial node: models of sinoatrial transition. Circulation. 1998;97(16):1623–1631. doi: 10.1161/01.CIR.97.16.1623. [DOI] [PubMed] [Google Scholar]
- 21.Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107(12):1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336(2):137–144. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly RG, Buckingham ME. The anterior heart-forming field: voyage to the arterial pole of the heart. Trends Genet. 2002;18(4):210–216. doi: 10.1016/S0168-9525(02)02642-2. [DOI] [PubMed] [Google Scholar]
- 24.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bressan M, Liu G, Mikawa T. Early mesodermal cues assign avian cardiac pacemaker fate potential in a tertiary heart field. Science. 2013;340(6133):744–748. doi: 10.1126/science.1232877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang X, Evans SM, Sun Y. Insights into cardiac conduction system formation provided by HCN4 expression. Trends Cardiovasc Med. 2015;25(1):1–9. doi: 10.1016/j.tcm.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113(4):399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franco D, Christoffels VM, Campione M. Homeobox transcription factor Pitx2: the rise of an asymmetry gene in cardiogenesis and arrhythmogenesis. Trends Cardiovasc Med. 2014;24(1):23–31. doi: 10.1016/j.tcm.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, et al. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94(3):299–305. doi: 10.1016/S0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 30.Ammirabile G, Tessari A, Pignataro V, Szumska D, Sutera Sardo F, Benes Jr J, Balistreri M, Bhattacharya S, Sedmera D, Campione M. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc Res. 2012;93(2):291–301. doi: 10.1093/cvr/cvr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, Christoffels VM (2010) The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res 87(1):92–101 [DOI] [PubMed]
- 32.Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, Buckingham ME. Atrial myocardium derives from the posterior region of the second heart field, which acquires left–right identity as Pitx2c is expressed. Development. 2008;135(6):1157–1167. doi: 10.1242/dev.014563. [DOI] [PubMed] [Google Scholar]
- 33.Hirota A, Fujii S, Kamino K. Optical monitoring of spontaneous electrical activity of 8-somite embryonic chick heart. Jpn J Physiol. 1979;29(5):635–639. doi: 10.2170/jjphysiol.29.635. [DOI] [PubMed] [Google Scholar]
- 34.Kamino K, Hirota A, Fujii S. Localization of pacemaking activity in early embryonic heart monitored using voltage-sensitive dye. Nature. 1981;290(5807):595–597. doi: 10.1038/290595a0. [DOI] [PubMed] [Google Scholar]
- 35.Van Mierop LH. Location of pacemaker in chick embryo heart at the time of initiation of heartbeat. Am J Physiol. 1967;212(2):407–415. doi: 10.1152/ajplegacy.1967.212.2.407. [DOI] [PubMed] [Google Scholar]
- 36.Sakai T, Hirota A, Fujii S, Kamino K. Flexibility of regional pacemaking priority in early embryonic heart monitored by simultaneous optical recording of action potentials from multiple sites. Jpn J Physiol. 1983;33(3):337–350. doi: 10.2170/jjphysiol.33.337. [DOI] [PubMed] [Google Scholar]
- 37.Hirota A, Kamino K, Komuro H, Sakai T, Yada T (1985) Early events in development of electrical activity and contraction in embryonic rat heart assessed by optical recording. J Physiol 369:209–227 [DOI] [PMC free article] [PubMed]
- 38.Yi T, Wong J, Feller E, Sink S, Taghli-Lamallem O, Wen J, Kim C, Fink M, Giles W, Soussou W, et al. Electrophysiological mapping of embryonic mouse hearts: mechanisms for developmental pacemaker switch and internodal conduction pathway. J Cardiovasc Electrophysiol. 2012;23(3):309–318. doi: 10.1111/j.1540-8167.2011.02191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelder TP, Vicente-Steijn R, Harryvan TJ, Kosmidis G, Gittenberger-de Groot AC, Poelmann RE, Schalij MJ, DeRuiter MC, Jongbloed MR. The sinus venosus myocardium contributes to the atrioventricular canal: potential role during atrioventricular node development? J Cell Mol Med. 2015;19(6):1375–1389. doi: 10.1111/jcmm.12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viragh S, Challice CE. The development of the conduction system in the mouse embryo heart. Dev Biol. 1980;80(1):28–45. doi: 10.1016/0012-1606(80)90496-0. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Frigola C, Shi Y, Evans SM. Expression of the hyperpolarization-activated cyclic nucleotide-gated cation channel HCN4 during mouse heart development. Gene Expr Patterns. 2003;3(6):777–783. doi: 10.1016/S1567-133X(03)00125-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ludwig A, Zong X, Jeglitsch M, Hofmann F, Biel M. A family of hyperpolarization-activated mammalian cation channels. Nature. 1998;393(6685):587–591. doi: 10.1038/31255. [DOI] [PubMed] [Google Scholar]
- 43.Spater D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, Chien KR. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 2013;15(9):1098–1106. doi: 10.1038/ncb2824. [DOI] [PubMed] [Google Scholar]
- 44.Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, et al. Formation of the venous pole of the heart from an Nk2–5-negative precursor population requires Tbx18. Circ Res. 2006;98(12):1555–1563. doi: 10.1161/01.RES.0000227571.84189.65. [DOI] [PubMed] [Google Scholar]
- 45.Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, et al. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100(3):354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 46.Moorman AF, Anderson RH. Development of the pulmonary vein. Int J Cardiol. 2011;147(1):182. doi: 10.1016/j.ijcard.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 47.Lescroart F, Mohun T, Meilhac SM, Bennett M, Buckingham M. Lineage tree for the venous pole of the heart: clonal analysis clarifies controversial genealogy based on genetic tracing. Circ Res. 2012;111(10):1313–1322. doi: 10.1161/CIRCRESAHA.112.271064. [DOI] [PubMed] [Google Scholar]
- 48.Rentschler S, Vaidya DM, Tamaddon H, Degenhardt K, Sassoon D, Morley GE, Jalife J, Fishman GI. Visualization and functional characterization of the developing murine cardiac conduction system. Development. 2001;128(10):1785–1792. doi: 10.1242/dev.128.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson RH, Brown NA, Moorman AF. Development and structures of the venous pole of the heart. Dev Dyn. 2006;235(1):2–9. doi: 10.1002/dvdy.20578. [DOI] [PubMed] [Google Scholar]
- 50.Jongbloed MR, Schalij MJ, Poelmann RE, Blom NA, Fekkes ML, Wang Z, Fishman GI, Gittenberger-De Groot AC. Embryonic conduction tissue: a spatial correlation with adult arrhythmogenic areas. J Cardiovasc Electrophysiol. 2004;15(3):349–355. doi: 10.1046/j.1540-8167.2004.03487.x. [DOI] [PubMed] [Google Scholar]
- 51.Moorman AF, Christoffels VM, Anderson RH. Anatomic substrates for cardiac conduction. Heart Rhythm. 2005;2(8):875–886. doi: 10.1016/j.hrthm.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai CL, Chen J, Evans SM. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 2007;304(1):286–296. doi: 10.1016/j.ydbio.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31(1):54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu YF, Dawkins JF, Cho HC, Marban E, Cingolani E (2014) Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci Transl Med 6(245):245ra94 [DOI] [PMC free article] [PubMed]
- 55.Greulich F, Trowe MO, Leffler A, Stoetzer C, Farin HF, Kispert A (2016) Misexpression of Tbx18 in cardiac chambers of fetal mice interferes with chamber-specific developmental programs but does not induce a pacemaker-like gene signature. J Mol Cell Cardiol 97:140–149 [DOI] [PubMed]
- 56.Kapoor N, Galang G, Marban E, Cho HC. Transcriptional suppression of connexin43 by TBX18 undermines cell–cell electrical coupling in postnatal cardiomyocytes. J Biol Chem. 2011;286(16):14073–14079. doi: 10.1074/jbc.M110.185298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115(14):1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 58.Espinoza-Lewis RA, Yu L, He F, Liu H, Tang R, Shi J, Sun X, Martin JF, Wang D, Yang J, et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nk2–5. Dev Biol. 2009;327(2):376–385. doi: 10.1016/j.ydbio.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu H, Chen CH, Espinoza-Lewis RA, Jiao Z, Sheu I, Hu X, Lin M, Zhang Y, Chen Y. Functional redundancy between human SHOX and mouse Shox2 genes in the regulation of sinoatrial node formation and pacemaking function. J Biol Chem. 2011;286(19):17029–17038. doi: 10.1074/jbc.M111.234252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmann S, Berger IM, Glaser A, Bacon C, Li L, Gretz N, Steinbeisser H, Rottbauer W, Just S, Rappold G. Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res Cardiol. 2013;108(2):339. doi: 10.1007/s00395-013-0339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129(19):4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 62.Ai D, Liu W, Ma L, Dong F, Lu MF, Wang D, Verzi MP, Cai C, Gage PJ, Evans S, et al. Pitx2 regulates cardiac left–right asymmetry by patterning second cardiac lineage-derived myocardium. Dev Biol. 2006;296(2):437–449. doi: 10.1016/j.ydbio.2006.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nowotschin S, Liao J, Gage PJ, Epstein JA, Campione M, Morrow BE. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133(8):1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107(21):9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XH, Martin JF. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci USA. 2014;111(25):9181–9186. doi: 10.1073/pnas.1405411111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ionta V, Liang W, Kim EH, Rafie R, Giacomello A, Marban E, Cho HC. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Rep. 2015;4(1):129–142. doi: 10.1016/j.stemcr.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hashem SI, Claycomb WC. Genetic isolation of stem cell-derived pacemaker-nodal cardiac myocytes. Mol Cell Biochem. 2013;383(1–2):161–171. doi: 10.1007/s11010-013-1764-x. [DOI] [PubMed] [Google Scholar]
- 68.Hashem SI, Lam ML, Mihardja SS, White SM, Lee RJ, Claycomb WC. Shox2 regulates the pacemaker gene program in embryoid bodies. Stem Cells Dev. 2013;22(21):2915–2926. doi: 10.1089/scd.2013.0123. [DOI] [PubMed] [Google Scholar]
- 69.Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62(3):489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- 70.Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21(9):1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels VM, Moon AM. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci USA. 2012;109(3):E154–E163. doi: 10.1073/pnas.1115165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, et al. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94(3):439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 73.Jung JJ, Husse B, Rimmbach C, Krebs S, Stieber J, Steinhoff G, Dendorfer A, Franz WM, David R. Programming and isolation of highly pure physiologically and pharmacologically functional sinus-nodal bodies from pluripotent stem cells. Stem Cell Rep. 2014;2(5):592–605. doi: 10.1016/j.stemcr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, t Hoen PA, Bakkers J, Barnett P, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122(7):2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arnolds DE, Liu F, Fahrenbach JP, Kim GH, Schillinger KJ, Smemo S, McNally EM, Nobrega MA, Patel VV, Moskowitz IP. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122(7):2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu M, Peng S, Yang J, Tu Z, Cai X, Cai CL, Wang Z, Zhao Y. Baf250a orchestrates an epigenetic pathway to repress the Nkx2.5-directed contractile cardiomyocyte program in the sinoatrial node. Cell Res. 2014;24(10):1201–1213. doi: 10.1038/cr.2014.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liang X, Zhang Q, Cattaneo P, Zhuang S, Gong X, Spann NJ, Jiang C, Cao X, Zhao X, Zhang X, et al. Transcription factor ISL1 is essential for pacemaker development and function. J Clin Invest. 2015;125(8):3256–3268. doi: 10.1172/JCI68257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen CA, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Weerd JH, Badi I, van den Boogaard M, Stefanovic S, van de Werken HJ, Gomez-Velazquez M, Badia-Careaga C, Manzanares M, de Laat W, Barnett P, et al. A large permissive regulatory domain exclusively controls Tbx3 expression in the cardiac conduction system. Circ Res. 2014;115(4):432–441. doi: 10.1161/CIRCRESAHA.115.303591. [DOI] [PubMed] [Google Scholar]
- 80.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133(12):2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sizarov A, Devalla HD, Anderson RH, Passier R, Christoffels VM, Moorman AF. Molecular analysis of patterning of conduction tissues in the developing human heart. Circ Arrhythm Electrophysiol. 2011;4(4):532–542. doi: 10.1161/CIRCEP.111.963421. [DOI] [PubMed] [Google Scholar]
- 82.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127(6):1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 83.de Pater E, Clijsters L, Marques SR, Lin YF, Garavito-Aguilar ZV, Yelon D, Bakkers J. Distinct phases of cardiomyocyte differentiation regulate growth of the zebrafish heart. Development. 2009;136(10):1633–1641. doi: 10.1242/dev.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tessadori F, van Weerd JH, Burkhard SB, Verkerk AO, de Pater E, Boukens BJ, Vink A, Christoffels VM, Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS One. 2012;7(10):e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vedantham V, Galang G, Evangelista M, Deo RC, Srivastava D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for islet-1 in cardiac pacemaker cells. Circ Res. 2015;116(5):797–803. doi: 10.1161/CIRCRESAHA.116.305913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dorn T, Goedel A, Lam JT, Haas J, Tian Q, Herrmann F, Bundschu K, Dobreva G, Schiemann M, Dirschinger R, et al. Direct nk2–5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cells. 2015;33(4):1113–1129. doi: 10.1002/stem.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Le Scouarnec S, Bhasin N, Vieyres C, Hund TJ, Cunha SR, Koval O, Marionneau C, Chen B, Wu Y, Demolombe S, et al. Dysfunction in ankyrin-B-dependent ion channel and transporter targeting causes human sinus node disease. Proc Natl Acad Sci USA. 2008;105(40):15617–15622. doi: 10.1073/pnas.0805500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takeshita K, Fujimori T, Kurotaki Y, Honjo H, Tsujikawa H, Yasui K, Lee JK, Kamiya K, Kitaichi K, Yamamoto K, et al. Sinoatrial node dysfunction and early unexpected death of mice with a defect of klotho gene expression. Circulation. 2004;109(14):1776–1782. doi: 10.1161/01.CIR.0000124224.48962.32. [DOI] [PubMed] [Google Scholar]
- 89.den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, Nolte IM, Segre AV, Holm H, Handsaker RE, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45(6):621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. An Nk2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dupays L, Jarry-Guichard T, Mazurais D, Calmels T, Izumo S, Gros D, Theveniau-Ruissy M. Dysregulation of connexins and inactivation of NFATc1 in the cardiovascular system of Nk2–5 null mutants. J Mol Cell Cardiol. 2005;38(5):787–798. doi: 10.1016/j.yjmcc.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 92.Linhares VL, Almeida NA, Menezes DC, Elliott DA, Lai D, Beyer EC, Campos de Carvalho AC, Costa MW. Transcriptional regulation of the murine Connexin40 promoter by cardiac factors Nk2–5, GATA4 and Tbx5. Cardiovasc Res. 2004;64(3):402–411. doi: 10.1016/j.cardiores.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakashima Y, Yanez DA, Touma M, Nakano H, Jaroszewicz A, Jordan MC, Pellegrini M, Roos KP, Nakano A. Nk2–5 suppresses the proliferation of atrial myocytes and conduction system. Circ Res. 2014;114(7):1103–1113. doi: 10.1161/CIRCRESAHA.114.303219. [DOI] [PubMed] [Google Scholar]
- 94.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42(2):153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Basson CT, Cowley GS, Solomon SD, Weissman B, Poznanski AK, Traill TA, Seidman JG, Seidman CE. The clinical and genetic spectrum of the Holt–Oram syndrome (heart-hand syndrome) N Engl J Med. 1994;330(13):885–891. doi: 10.1056/NEJM199403313301302. [DOI] [PubMed] [Google Scholar]
- 96.Benson DW, Wang DW, Dyment M, Knilans TK, Fish FA, Strieper MJ, Rhodes TH, George AL., Jr Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112(7):1019–1028. doi: 10.1172/JCI200318062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Makiyama T, Akao M, Tsuji K, Doi T, Ohno S, Takenaka K, Kobori A, Ninomiya T, Yoshida H, Takano M et al (2005) High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol 46(11):2100–2106 [DOI] [PubMed]
- 98.Butters TD, Aslanidi OV, Inada S, Boyett MR, Hancox JC, Lei M, Zhang H. Mechanistic links between Na+ channel (SCN5A) mutations and impaired cardiac pacemaking in sick sinus syndrome. Circ Res. 2010;107(1):126–137. doi: 10.1161/CIRCRESAHA.110.219949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lei M, Goddard C, Liu J, Leoni AL, Royer A, Fung SS, Xiao G, Ma A, Zhang H, Charpentier F, et al. Sinus node dysfunction following targeted disruption of the murine cardiac sodium channel gene Scn5a. J Physiol. 2005;567(Pt 2):387–400. doi: 10.1113/jphysiol.2005.083188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tessari A, Pietrobon M, Notte A, Cifelli G, Gage PJ, Schneider MD, Lembo G, Campione M. Myocardial Pitx2 differentially regulates the left atrial identity and ventricular asymmetric remodeling programs. Circ Res. 2008;102(7):813–822. doi: 10.1161/CIRCRESAHA.107.163188. [DOI] [PubMed] [Google Scholar]
- 101.Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, et al. The homeobox gene Pitx2: mediator of asymmetric left–right signaling in vertebrate heart and gut looping. Development. 1999;126(6):1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- 102.Campione M, Ros MA, Icardo JM, Piedra E, Christoffels VM, Schweickert A, Blum M, Franco D, Moorman AF. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231(1):252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]