Abstract
The Toll-like receptor (TLR) family consists of phylogenetically conserved transmembrane proteins, which function as mediators of innate immunity for recognition of pathogen-derived ligands and subsequent cell activation via the Toll/IL-1R signal pathway. Here, we show that human TLR9 (hTLR9) expression in human immune cells correlates with responsiveness to bacterial deoxycytidylate-phosphate-deoxyguanylate (CpG)-DNA. Notably “gain of function” to immunostimulatory CpG-DNA is achieved by expressing TLR9 in human nonresponder cells. Transfection of either human or murine TLR9 conferred responsiveness in a CD14- and MD2-independent manner, yet required species-specific CpG-DNA motifs for initiation of the Toll/IL-1R signal pathway via MyD88. The optimal CpG motif for hTLR9 was GTCGTT, whereas the optimal murine sequence was GACGTT. Overall, these data suggest that hTLR9 conveys CpG-DNA responsiveness to human cells by directly engaging immunostimulating CpG-DNA.
The immune system has developed germ-line encoded pattern recognition receptors that promote rapid responses to microbial pathogens. Cueing on conserved pathogen-associated molecular patterns not present in the host, cells of the innate immune system activate and direct the emanating anti-pathogen response (1). Toll-like receptors (TLRs) are transmembranal proteins and represent a newly recognized family of vertebrate pattern recognition receptors (2, 3). Subsequent to pathogen-associated molecular pattern engagement, TLRs initiate signaling via sequential recruitment of MyD88, IRAK and TRAF6, which in turn activate downstream mediators such as NF-κB and mitogen-activated protein kinases (4, 5). TLR2 and TLR4 transduce signal via their Toll/interleukin-1 receptor domain (TIR) and are the receptors for responses to Gram-positive and Gram-negative bacterial cell wall components (3, 6–10).
Similar to lipopolysaccharide (LPS), bacterial DNA induces acute inflammatory responses. Bacterial DNA acts as a pathogen-associated molecular pattern by virtue of a 20-fold greater frequency of unmethylated CG dinucleotides found in microbial DNA versus vertebrate DNA (11). Synthetic oligodeoxynucleotides (ODN) containing the proper CpG-DNA motif mimic the immunostimulatory effects of bacterial DNA (12). Cellular activation by CpG-DNA occurs via the IL-1R/TLR signal transduction pathway because it depends on MyD88 and TRAF-6, implying that the CpG-DNA signaling receptor utilizes a TIR domain (13, 14). Indeed TLR9-deficient mice express a nonresponsive phenotype to CpG-DNA, which suggests murine TLR9 (mTLR9) is a CpG-DNA receptor (15). On the other hand, there is also evidence that DNA-PKcs-deficient mice are nonresponsive to CpG-DNA and that the DNA-PKcs sequence specifically recognizes CpG motifs (16).
In mouse models, the potential of CpG-DNA to serve as a Th1 biasing adjuvant has been well documented (17–19). CpG-DNA also activates human cells, like dendritic cells and B cells (20, 21). Interestingly, the optimal CpG motif for activating human cells appears different from the effective mouse sequence (GTCGTT versus GACGTT) (11, 20, 22). We show here that expression of human TLR9 (hTLR9) is correlated with CpG-DNA responsiveness in primary human cells and that transfection of either hTLR9 or mTLR9 into nonresponsive cells reconstitutes MyD88-dependent CpG-DNA responses. Transfected hTLR9 and mTLR9 required distinct CpG motifs for signal initiation, implying they directly engage immunostimulatory CpG-DNA in a species-specific manner.
Materials and Methods
Cells and Reagents.
Human embryonic kidney 293 cells were from ATCC. Peripheral blood mononuclear cells (PBMC), human B cells, CD123+ dendritic cells (DC), and monocyte-derived dendritic cells (MDDC) were obtained and purified as described elsewhere (20). Escherichia coli LPS (serotype 0127:B8) was from Sigma, Bafilomycin A1 was from Calbiochem–Novabiochem, and IL-1α and TNF-α were from PeproTech (Boston). ODN were commercially synthesized by TIB MOLBIOL (Berlin, Germany) in a phosphothioate protected form (see Table 1 for names and sequences). The sequence of Me-CpG-ODN is identical to ODN 2006, but the cytosines at positions 2, 5, 13, and 21 are methylated. The sequence of the blocking ODN used for Fig. 3D was 5′-HHHHHHHHHHHHHHWGGGGG (H = A, T, C; W = A, T).
Table 1.
ODN sequences and ODN concentrations for half-maximal activation (Kac) of human and murine TLR9
| CpG-DNA | Sequence | 293-hTLR9 Kac, nM | 293-mTLR9 Kac, nM |
|---|---|---|---|
| 1668 | TCCATGACGTTCCTGATGCT | >10,000 | 70 |
| 1668-GC | TCCATGAGCTTCCTGATGCT | >10,000 | >10,000 |
| 2006 | TCGTCGTTTTGTCGTTTTGTCGTT | 400 | >10,000 |
| 2006-GC | TGCTGCTTTTGTGCTTTTGTGCTT | >10,000 | >10,000 |
| 5000 | TCCATGACGTTCTTGACGCT | 10,000 | 82 |
| 5001 | TCCATGACGTTCTTGACGTT | 7,000 | 55 |
| 5002 | TCCATGACGTTCTTGATGTT | 7,000 | 30 |
| 5003 | TCCATGACGTTTTTGATGTT | 10,000 | 30 |
| 5004 | TCCATGTCGTTCTTGATGTT | 5,000 | 400 |
| 5005 | TCCATGTCGTTTTTGATGTT | 3,000 | 2,000 |
| 5006 | TCCATGTCGTTTTTGTTGTT | 3,000 | 650 |
| 5007 | TCCATGTCGTTTTTGTCGTT | 700 | 1,000 |
| 5002 | TCCATGACGTTCTTGATGTT | NP | 30 |
| 5008 | TCCATGACGTTATTGATGTT | NP | 40 |
| 5009 | TCCATGACGTCCTTGATGTT | NP | >10,000 |
| 5010 | TCCATGACGTCATTGATGTT | NP | >10,000 |
293 cells stably transfected with a 6-fold NF-κB luciferase reporter plasmid and hTLR9 (293-hTLR9-luc) or mTLR9 (293-mTLR9-luc) were stimulated with listed ODN, and NF-κB activation was monitored after 12 h (refer to Fig. 4) (n = 2, mean ± SD). The concentration for half-mamimal activation (Kac) of human and murine TLR9 was calculated for each ODN. Results are representative of three independent experiments. NP, not performed.
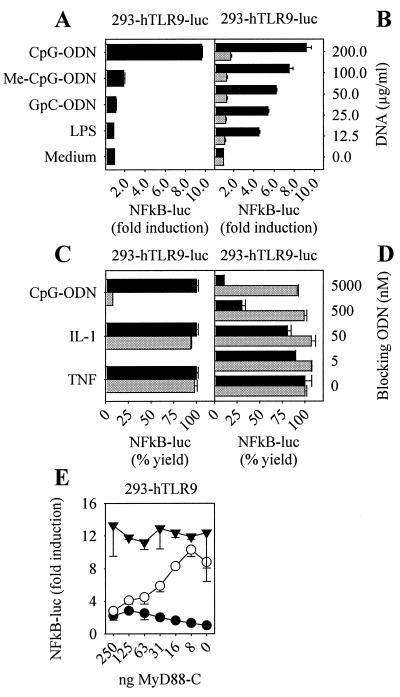
Figure 3.
Stable Reconstitution with human TLR9 recapitulates CpG-DNA mechanisms of action. The 293 cells stably transfected with hTLR9 and a 6-fold NF-κB luciferase reporter plasmid (293-hTLR9-luc) were stimulated with 2 μM CpG-ODN (2006), 2 μM Me-CpG-ODN (methylated 2006), 2 μM GpC-ODN (2006-GC), 100 ng/ml LPS, or medium (A) or with E. coli DNA (black bars) or E.coli DNA digested with DNase I (gray bars) at concentrations indicated (B). After 12 h, NF-κB activation was measured (n = 2, mean ± SD). (C) The 293-hTLR9-luc cells were preincubated with 10 nM Bafilomycin A (gray bars) or DMSO control (black bars) for 30 min and stimulated with 0.5 μM CpG-ODN (2006), 10 ng/ml IL-1α, or 10 ng/ml TNF-α. NF-κB activation was monitored after 12 h and is presented as % yield (fold NF-κB activation with Bafilomycin A treatment/fold NF-κB activation with DMSO control × 100) (n = 2, mean ± SD). (D) 293-hTLR9-luc cells were incubated with 0.5 μM CpG-ODN (2006) (black bars) or 10 ng/ml TNF-α (gray bars) and increasing concentrations of a blocking ODN (see sequence in Materials and Methods) as indicated. NF-κB activation was monitored after 12 h and is presented as % yield (fold NF-κB activation with blocking ODN/fold NF-κB activation without blocking ODN × 100) (n = 2, mean ± SD). (E) The 293 cells stably transfected with hTLR9 (293-hTLR9) were cotransfected with a 6-fold NF-κB luciferase reporter plasmid and increasing concentrations of dominant negative human MyD88 expression vector at concentrations indicated. Cells were not stimulated (●) or stimulated with 2 μM CpG-ODN (2006) (○) or 10 ng/ml TNF-α (▾), and NF-κB activation was monitored after 12 h (n = 2, mean ± SD). Results are representative of at least two independent experiments.
Reverse Transcriptase (RT)-PCR.
RNA from 106 cells was prepared by using RNAeasy kit from Qiagen (Chatsworth, CA). After DNase I treatment, 1 μg of RNA was reverse transcribed with M-MuLV reverse transcriptase from PeqLab, and fragments were amplified with Taq polymerase by using the following primer pairs: hTLR2, 5′-TGTGAACCTCCAGGCTCTG and 5′-GTCCATATTTCCCACTCTCAGG; hTLR4, 5′-ACAGAAGCTGGTGGCTGTG and 5′-TCTTTAAATGCACCTGGTTGG; hTLR9, 5′-GTGCCCCACTTCTCCATG and 5′ GGCACAGTCATGATGTTGTTG; mTLR9, 5′-CCGCAAGACTCTATTTGTGCTGG and 5′-TGTCCCTAGTCAGGGCTGTACTCAG; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′ACGGATTTGGTCGTATTGGGC and 5′-TTGACGGTGCCATGGAATTTG. cDNA amounts were normalized based on GAPDH amount determined by TaqMAN-PCR (TaqMan probe, 5′-FAM-CCTGGTCACCAGGGCTGCTTT-TAMRA; Applied Biosystems). RT-PCR was performed for 30 cycles on normalized cDNA diluted 1:5 for human TLR2, TLR4, TLR9 and murine TLR9 and diluted 1:125 for GAPDH.
Plasmids and cDNAs.
Expression plasmids for human CD14 (hCD14), human TLR4 (hTLR4), and dominant negative human MyD88 (hMyD88-C) were kindly provided from Tularik (South San Francisco, CA). Human MD2 (hMD2) expression plasmid and human TLR9 (hTLR9) cDNA were gifts from Kensuke Miyake (Saga Medical School, Japan) and B. Beutler (Scripps Research Institute, La Jolla, CA), respectively. The ORF of hTLR9 was cloned into pcDNA3.1 (Invitrogen). Murine TLR9 (mTLR9) was amplified from a RAW 264.5 macrophage cDNA using the primers 5′-GTCAGAGGGAGCCTCGGGAGAATCCTC and 5′-GCAGGCAGAGCAACTCGGGAACCAG and cloned into pcDNA3.1. Sequence information of the 5′ and 3′ untranslated regions for primer selection was obtained by race PCR on a spleen marathon cDNA from CLONTECH. The race PCR was performed utilizing sequence information deposited as est-sequence aa162495, which showed homology to hTLR9.
Transfection and Reporter Assays.
For monitoring transient NF-κB activation, 293 cells were seeded at 105 cells per well in a 12-well plate and transfected by the Ca2PO4 precipitation method (23) with 4 ng of hTLR4 and 4 ng of hMD2, or 50 ng of hTLR9 expression plasmids and 0.5 ng of 6-fold NF-κB luciferase reporter plasmid and 0.25 μg of β-galactosidase control plasmid for normalization. For testing the dominant negative effect of MyD88-C, 293 cells stably expressing hTLR9 were transiently transfected with increasing amounts of MyD88-C (8–250 ng), 0.5 ng of 6-fold NF-κB luciferase reporter plasmid, and 0.25 μg of β-galactosidase control plasmid. For measuring IL-8 production, 293 cells were seeded at 1.5 × 104 cells per well in a 96-well plate and transfected with 400 ng of DNA per well by Ca2PO4 transfection (24). The vectors expressing hTLR9, hTLR4, hMD2, and hCD14 were used at 4 ng per transfection plus noncoding vector DNA to give the final amount of 400 ng. After 18 h, the cells were treated with various stimuli for 48 h, and the supernatant was harvested. Stable clones expressing hTLR9, mTLR9, or TLR9 with a 6-fold NF-κB luciferase reporter plasmid were selected with 0.7 mg/ml G418. For monitoring of NF-κB activation or IL-8 production, cells were seeded at 104 cells per well and stimulated 16 h later; NF-κB activation or IL-8 production was monitored 12 or 48 h after stimulation, respectively. For monitoring NF-κB activation, 293 cells were lysed by using reporter lysis buffer (Promega), and lysate was assayed for luciferase activity by using a Berthold luminometer (Wildbad, Germany). Human IL-8 production was monitored by ELISA using matched-pair antibodies and standard IL-8 (BD Transduction Laboratories, San Diego) according to the manufacturer's protocol.
For monitoring the production of human TNF-α, human IL-8, and human or murine IL-12p40, CD123+ DC, MDCC, PBMC, or murine spleen cells were seeded in 24-well plates in 1 ml at 2 × 105 cells, 106 cells, 5 × 106 cells, and 2.5 × 105 cells per well, respectively. Supernatants were harvested after 12 h (for TNF-α) and 16 h (for IL-12 and IL-8), and cytokines were measured by ELISA using matched-pair antibodies and standard from BD Transduction Laboratories. Monitoring of B-cell proliferation has been described elsewhere (20).
Results
hTLR9 Expression Correlates with CpG-DNA Responsiveness.
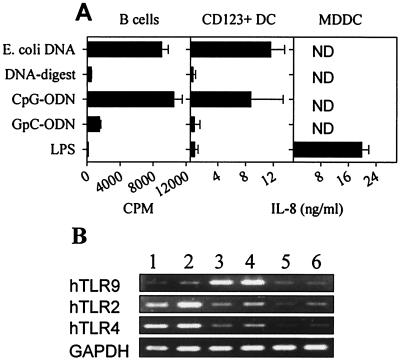
Bacterial DNA is mitogenic for human B cells; however, LPS is not. Fig. 1A demonstrates that human B cells proliferate after stimulation with E. coli DNA or a CpG-ODN but not in response to digested E. coli DNA, control GpC-ODN, or LPS. We also observed that plasmacytoid DC (CD123+DC) produced IL-8 and TNF (data not shown) in response to CpG-DNA in a CpG motif-restricted manner but not LPS (Fig. 1A). In contrast, MDDC demonstrated the converse response pattern (Fig. 1A), a response to LPS but not CpG-DNA. Thus, responses to CpG-DNA or LPS segregated between B cells and CD123+DC or MDDC, respectively. We therefore tested whether absence of responsiveness correlated to absence of TLR expression. By semiquantitative RT-PCR, both B cells and CD123+DC yielded positive signals for hTLR9, whereas MDDC, monocytes, and T cells appeared weak to negative (Fig. 1B). LPS-responsive MDDC and monocytes were positive for both TLR2 and TLR4, whereas B cells, T cells, and CD123+DC were weak to negative (Fig. 1B). These data suggest a clear correlation between hTLR9 mRNA expression and responsiveness to CpG-DNA or hTLR2/hTLR4 expression and LPS responsiveness.
Figure 1.
Human TLR9 expression correlates with CpG-DNA responsiveness. (A) Purified human B cells, CD123+ dendritic cells (DC), or monocyte-derived dendritic cells (MDDC) were stimulated with 50 μg/ml E. coli DNA, 50 μg/ml DNase I-digested E. coli DNA, 2 μM CpG-ODN (2006), 2 μM GpC-ODN (2006-GC), or 100 ng/ml LPS. B cell proliferation was monitored at day 2 by [H3]thymidine uptake (n = 4, mean ± SD). IL-8 concentration was determined by ELISA (n = 2, mean ± SD; ND = not detected). (B) cDNA was prepared from MDDC (lane 1), purified CD14+ monocytes (lane 2), B cells (lane 3), CD123+ DC (lane 4), CD4+ T cells (lane 5), and CD8+ T cells (lane 6). cDNA amounts were normalized based on the GAPDH amount determined by TaqMAN-PCR. RT-PCR was performed for 30 cycles on normalized cDNA diluted 1:5 for human TLR2, TLR4, and TLR9 and diluted 1:125 for GAPDH. Results are representative of three independent experiments.
hTLR9 Genetic Complementation Yields CpG-DNA Responsiveness.
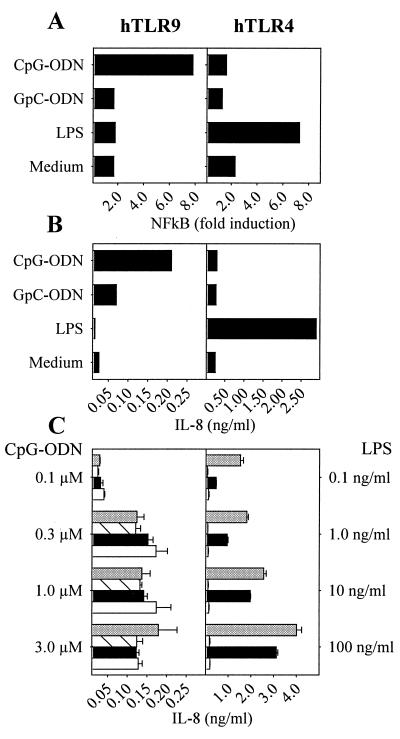
We next determined whether hTLR9 genetic complementation confers CpG-DNA responsiveness to nonresponder cells. hTLR9-transfected 293 cells were responsive to CpG-ODN but not the control GpC-ODN, whereas hTLR4-transfected cells gained responsiveness to LPS (Fig. 2 A and B). These data demonstrate the CpG-DNA specificity of the hTLR9-dependent response and noncrossreactivity between hTLR9 or hTLR4 and their respective ligands. Because CpG-DNA and LPS responsiveness segregated with TLR genetic complementation and because LPS responses are dependent on the coexpression of MD2 and CD14 (7, 25, 26), we asked whether hTLR9 shared CD14 or MD2 dependency. Cotransfection of TLR9 with either human MD2 or CD14, however, had little consequence on IL-8 production or NF-κB activation (Fig. 2C and data not shown). In contrast, hTLR4 cotransfection with MD2 alone dramatically improved LPS responsiveness, whereas the addition of CD14 was potentiating, especially at lower LPS concentrations. We concluded that unlike TLR4 responses to LPS, hTLR9 responses to CpG-DNA were independent of both MD2 and CD14.
Figure 2.
Reconstitution of human TLR9 yields CpG-DNA responsiveness, which is independent of MD2 and CD14. The 293 cells were transiently transfected with hTLR9 or hTLR4/hMD2 and a 6-fold NF-κB luciferase reporter plasmid (A) or with hTLR9 or hTLR4/hMD2 alone (B). After stimulation with 2 μM CpG-ODN (2006), 2 μM GpC-ODN (2006-GC), 100 ng/ml LPS or medium, NF-κB activation (A) or IL-8 production (B) was monitored. (C) The 293 cells were transiently transfected with hTLR9 or hTLR4 alone (white bars) or cotransfected with hMD2 (black bars), hCD14 (hatched bars), or hMD2 and hCD14 (gray bars). Transfected cells were stimulated with concentrations of CpG-ODN or LPS as indicated, and IL-8 production was measured by ELISA (n = 3, mean ± SD). Results are representative of at least two independent experiments.
Genetic Complementation of 293 Cells with hTLR9 Recapitulates CpG-DNA Mechanisms of Action.
We next established stable human TLR9 positive clones plus or minus an NF-κB luciferase reporter, 293-hTLR9 or 293-hTLR9-luc, respectively (see Materials and Methods). These cells showed reactivity to CpG-DNA and not LPS in a CpG motif-dependent manner as the methylation of CpG-DNA or inversion of the CpG ablated responsiveness (Fig. 3A). The 293-hTLR9-luc cells were also dose-dependently responsive to bacterial DNA and mute toward Dnase I-digested DNA (Fig. 3B). Similar data were obtained through monitoring IL-8 production and were reproducible with phosphodiester-linked ODN (data not shown). We also tested whether DNA uptake and endosomal maturation were required for signal initiation by CpG-DNA, as has been reported for wild-type cells (27). Interrupting endosomal maturation with Bafilomycin A (28) fully blocked CpG-ODN-mediated induction of NF-κB (Fig. 3C). The blockade was specific to CpG-DNA, as both IL-1 and TNF induction of NF-κB were unaffected. We have previously postulated that a receptor not dependent on a CpG motif translocates DNA into the endosomal/lysosomal compartment because nonactivating ODN block the cellular uptake of immunostimulatory CpG-ODN (27). Fig. 3D demonstrates that 293-hTLR9-luc cells were sensitive to blockade with a non-CpG-ODN. Based on these criteria, we concluded that 293 cells complemented with hTLR9 behave similar to CpG-DNA responsive wild-type cells.
In the mouse, CpG-DNA signaling has been shown to occur via the Toll/IL-1R signal pathway requiring sequential recruitment of MyD88, IRAK, and TRAF-6 (13, 14). We tested whether hTLR9 was MyD88 dependent. Fig. 3E shows that dominant negative MyD88 dose dependently blocked CpG-DNA driven NF-κB induction but not TNF-induced signal transduction in 293-hTLR9 cells. These data established a central role for MyD88 in CpG-DNA signaling and thus implied engagement of the Toll/IL-1R signal transduction pathway in human cells responsive to CpG-DNA.
TLR9 Confers Species-Specific CpG Motif Signaling.
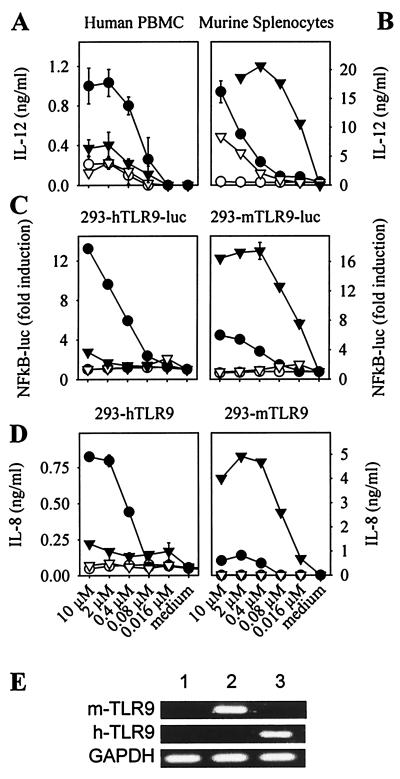
Human primary cells have been shown to respond to the CpG motif GTCGTT, whereas primary murine cells respond best to GACGTT (11, 20, 22). As shown in Fig. 4A, titration of the optimal human ODN-2006 on human PBMC dose dependently induced IL-12, whereas the optimal murine ODN-1668 was poor. In contrast, murine splenocytes responded best to ODN-1668 and to a lesser extent toward ODN-2006 (Fig. 4B). Assuming that the ODN concentration yielding half-maximal cytokine production reflects receptor affinity, the affinity of murine splenocytes for ODN-1668 was apparently greater than that of human PBMC for ODN-2006 (compare Fig. 4 A and B).
Figure 4.
TLR9 confers species-specific CpG motif signaling. PBMC (A) or murine splenocytes (B) were stimulated with ODN 2006 (●), ODN 2006-GC (○), ODN 1668 (▾), or ODN 1668-GC (▿) at the indicated concentrations, and IL-12 production was monitored by ELISA (n = 2, mean ± SD). The 293 cells stably transfected with a 6-fold NF-κB luciferase reporter plasmid and hTLR9 (293-hTLR9-luc) or mTLR9 (293-mTLR9-luc) (C) or with hTLR9 (293-hTLR9) or mTLR9 (293-mTLR9) alone (D) were stimulated with ODN 2006 (●), ODN 2006-GC (○), ODN 1668 (▾), or ODN 1668-GC (▿) at the indicated concentrations, and NF-κB activation or IL-8 production were measured after 12 and 48 h, respectively (n = 2, mean ± SD). Results are representative of at least two independent experiments. (E) cDNA was prepared from 293 cells (lane 1), 293-mTLR9 cells (lane 2), and 293-hTLR9 cells (lane 3), and RT-PCR for mTLR9, hTLR9, and GAPDH was performed.
To analyze whether TLR9 detected species-specific CpG-DNA motifs, we also established murine TLR9 clones (293-mTLR9), confirming TLR9 mRNA expression by RT-PCR (Fig. 4E). Fig. 4C shows the dose-dependent induction of NF-κB driven luciferase by the human ODN-2006 or the murine ODN-1668 in either 293-hTLR9-luc or 293-mTLR9-luc cells, whereas Fig. 4D depicts the respective IL-8 responses. Strikingly, CpG motif sequence specificity was granted in a species-specific manner by TLR9. In addition, the half-maximal concentration for either ODN-2006 or ODN-1668 appeared nearly identical to that determined by IL-12 release from primary cells (compare Fig. 4 A or B with C and D). These data suggested that TLR9 confers species-specific CpG motif responsiveness, implying that TLR9 acts as the CpG-DNA receptor.
TLR9 Determines CpG-ODN Affinity.
If indeed TLR9 acts as the CpG-DNA receptor, TLR9 ought to determine CpG-ODN affinity. Because all measurements of species specificity had been performed by using two dissimilar sequences, human ODN-2006 or murine ODN-1668 (22), we produced several sequences attempting a stepwise progression from the mouse sequence to the human sequence. The ODN-5002 is like ODN-1668 with the exception that Cs at positions 12 and 19 have been converted to Ts (Table 1). The last 16 nucleotides of ODN-5007 are the same as the last 15 nucleotides of ODN-2006 with the exception of an additional T (Table 1). Similar to ODN titrations in Fig. 4, we titrated these ODN versus either 293-hTLR9-luc or 293-mTLR9-luc cells to determine whether species specificity was preserved and to establish ODN concentration of half-maximal activation (Kac) (Table 1). The Kac displayed by the 293-hTLR9-luc clone strengthened with progressive nucleotide substitutions converting the mouse sequence toward the human sequence (Table 1, progression from 5000 to 5007). The reverse was true for the 293-mTLR9-luc clone, which showed a weakened Kac throughout the same progression. These results confirmed the notion that the preferred mouse motif contains ACG, whereas the human sequence is TCG. Of additional interest, we have observed that a CA substitution converting the mouse CpG motif from GACGTTC to GACGTCA was deleterious (data not shown). To extend our examination of the motif, we created three more ODN and measured Kac (5008–5010, Table 1). The change from CGTT to CGTC ablated 293-mTLR9-luc cell responsiveness. Overall, these data strongly suggest direct CpG motif engagement by TLR9.
Discussion
Activation of murine innate immune cells by CpG-DNA occurs via the Toll/IL-1R signal pathway, suggesting that the CpG-DNA receptor utilizes a TIR domain for the sequential recruitment of MyD88 and TRAF-6 (13, 14). Supporting this concept, TLR9-deficient mice express a nonresponsive phenotype toward CpG-DNA, implying TLR9 as a major component of the CpG-DNA receptor (15). This conclusion, however, has been subsequently challenged by the observation that DNA-PKcs deficient mice are also nonresponsive to CpG-DNA (16).
Based on the data presented here, it would seem clear that immunostimulatory CpG-DNA is engaged by TLR9. First, the distribution of hTLR9 in primary human cells correlated with CpG-DNA responsiveness (Fig. 1). Second, genetic complementation of CpG-DNA nonresponder cells with either hTLR9 or mTLR9 yielded a gain of function phenotype (Figs. 2–4). Last, both hTLR9 and mTLR9 appear to directly interact with CpG-DNA, as not only CpG restriction but also dose-dependent CpG motif species specificity was conferred through TLR9 (Fig. 4 and Table 1). Combined with the observations made in TLR9-deficient mice, these data clearly demonstrate TLR9 as necessary and sufficient for CpG-DNA driven responses. Because DNA-PKcs is ubiquitously expressed (29), one may assume TLR9−/− mice and 293 cells are positive for DNA-PKcs; yet, both were nonresponsive to CpG-DNA. Even though DNA-PKcs−/− mice showed a partial loss of CpG-DNA responder phenotype (16, 30), our genetic complementation data suggest that DNA-PKcs may be necessary for signaling but not sufficient. In support, mice deficient for CpG-DNA receptor signaling mediators, such as MyD88−/− mice, displayed a nonresponder phenotype (13, 31, 32). Loss of any downstream mediator would interrupt signaling, yet alone it could not restore function to a receptor-deficient system. The notion of DNA-PKcs as a candidate CpG-DNA receptor also conflicts with the finding that SCID mice, because of a mutation-induced truncation (33, 34), are deficient in DNA-PKcs yet respond in vivo to CpG-DNA with the release of serum cytokines and succumb to TNF-driven toxic shock (35, 36). Serum cytokine release and toxic shock after in vivo CpG-DNA challenge were absent in TLR9−/− mice (15). Whether DNA-PKcs is auxiliary to TLR9 in CpG-DNA signaling or part of an ill-defined signaling pathway is yet unanswered.
We observed in human immune cells a correlation between hTLR9 or hTLR2/hTLR4 expression and CpG-DNA or LPS sensitivity, respectively. For example, primary human plasmacytoid DCs and B cells expressed hTLR9 but minimal hTLR2/hTLR4 and were responsive to CpG-DNA but not to LPS. On the other hand, MDDCs showed converse TLR expression and pattern of responsiveness (Fig. 1). TLR2 and TLR4 mRNA expression in monocytes and MDDC but not B cells has been described (37–39). TLR1–5 were shown to be differentially expressed on various immune cells, whereas monocytes and immature MDDC differentially regulated TLR1–6 upon stimulation (37–39). It is thus tempting to speculate that the immune system can integrate infectious signals via TLR by virtue of receptor density and/or cellular distribution and thus tailor the emanating adaptive immune responses.
CpG-DNA nonresponsive 293 cells recapitulated upon TLR9 genetic complementation the bacterial DNA and CpG-DNA response phenotype previously established for primary DCs and macrophages (12). On stimulation with CpG-DNA, these cells activated NF-κB and produced the cytokine IL-8 (Figs. 2–4). Furthermore, CpG-DNA initiated signaling was ablated through blockade of cellular uptake by non-CpG-ODN or by Bafilomycin A, which blocks endosomal maturation (Fig. 3 C and D) (28). Finally, hTLR9 initiated signaling flows into the Toll/IL-1R signal pathway, as it depends on MyD88 (Fig. 3E). Similar data were obtained by hTLR9 complementation of the CpG-DNA nonresponsive human HL-60 cells (data not shown). Overall, these data suggested that hTLR9 grants “gain of function” to CpG-DNA with fidelity previously established for primary CpG-DNA responsive immune cells.
Signaling by hTLR9-reconstituted 293 cells was independent of the coreceptors MD2 and CD14 (Fig. 2), a characteristic that separates TLR9 from TLR4 (7, 25) and TLR2 (26). Expression of both MD2 and CD14 is regulated, which influences cellular responsiveness to bacterial substances. It is thus curious that TLR9 does not require either of these helper components. One explanation may be differences in the ligand character. For example, the well characterized ligands for TLR2 or TLR4 are hydrophobic in nature, forming micellular structures, whereas CpG-DNA is hydrophilic and fully water-soluble. Whether TLR9 is assisted by a modulated coreceptor is not clear; however, TLR9 alone was sufficient to reconstitute CpG-DNA responses in 293 cells.
The human optimal CpG motif GTCGTT differs from the optimal murine CpG motif GACGTT (11, 20, 22, 40). Because the genetic complementation system allowed the use of either human or murine TLR9, we were in the position to analyze whether TLR9 confers species-specific CpG motif signaling. Unambiguously, hTLR9 recognized the human CpG motif most efficiently, whereas mTLR9 had a high preference for the murine CpG motif (Fig. 4). This suggested direct interactions of the TLR9 receptor with its CpG-DNA ligand. In support, CpG-ODN point mutated in a manner that the individual ODN progressed from a murine to human CpG motif displayed inverse Kac toward hTLR9 and mTLR9, respectively (Table 1). Additionally, the extracellular region of TLR9 contains a DNA-binding motif described to occur in a family of methylated CpG-DNA binding proteins, MBD-1-4 (41, 42). Point mutations of this motif in TLR9 destroyed CpG-DNA driven responses (S.B. and G.B.L., unpublished data). Taken together, these data strongly implicate TLR9 as the receptor for CpG-DNA by directly engaging its ligand. Genetic complementation has also revealed direct interaction of TLR4 and LPS (43). In contrast to Drosophila where infection activates a proteolytic cascade producing an endogenous ligand (spaetzle) that engages Toll (44), in mammals the Toll receptors seem to directly interact with their respective microbial ligands.
The TLR9 complementation system described here may be useful to optimize species-specific immunostimulatory CpG motifs because we show that TLR9 prefers a mouse motif containing ACG, whereas hTLR9 prefers the sequence TCG. In addition, the conserved pyrimidine for pyrimidine change T to C in the mouse motif, ACGTT versus ACGTC (Table 1, ODN 5002 versus 5009), completely destroyed recognition by mTLR9. We therefore propose that the TLR9 genetic complementation system will allow definition of species-specific CpG motifs useful as vaccine adjuvants not only in man but also in diverse animal species.
Acknowledgments
We acknowledge B. Beutler for kindly providing human TLR9 cDNA, P. Baeuerle for providing 6-fold NF/κB luciferase reporter plasmid, and K. Miyake for providing human MD2 expression plasmid. We thank T. Gellert, F. Ampenberger, and S. Fichte for excellent technical assistance. We thank H. Hemmi for helpful discussion. This work was supported by Deutsche Forschungsgemeinschaft Grants BA1618/2-1 and KI591/1-2 Bundesministerium für Bildung und Forschung, Forschungsverbund Grundlagen Gentechnischer Verfahren (FORGEN II), Sonderforschungsbereich SFB-1738, and Coley Pharmaceutical Group GmbH.
Abbreviations
- TLR
Toll-like receptor
- LPS
lipopolysaccharide
- CpG
deoxycytidylate-phosphate-deoxyguanylate
- DC
dendritic cell
- ODN
oligodeoxynucleotides
- PBMC
peripheral blood mononuclear cells
- MDDC
monocyte-derived dendritic cells
- RT
reverse transcriptase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF348140).
References
- 1.Medzhitov R, Janeway C A., Jr Curr Opin Immunol. 1997;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- 2.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aderem A, Ulevitch R J. Nature (London) 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway C A., Jr Mol Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 5.Muzio M, Polentarutti N, Bosisio D, Manoj Kumar P P, Mantovani A. Biochem Soc Trans. 2000;28:563–566. doi: 10.1042/bst0280563. [DOI] [PubMed] [Google Scholar]
- 6.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 7.Lien E, Chow J C, Hawkins C D, McGuiness P D, Miyake K, Espevik T, Gusovsky F, Golenbock D T. J Biol Chem. 2001;276:1873–1880. doi: 10.1074/jbc.M009040200. [DOI] [PubMed] [Google Scholar]
- 8.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimura A, Lien E, Ingalls R R, Tuomanen E, Dziarski R, Golenbock D. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 10.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 11.Krieg A M, Yi A K, Matson S, Waldschmidt T J, Bishop G A, Teasdale R, Koretzky G A, Klinman D M. Nature (London) 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 12.Wagner H. Adv Immunol. 1999;73:329–368. doi: 10.1016/s0065-2776(08)60790-7. [DOI] [PubMed] [Google Scholar]
- 13.Hacker H, Vabulas R M, Takeuchi O, Hoshino K, Akira S, Wagner H. J Exp Med. 2000;192:595–600. doi: 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnare M, Holtdagger A C, Takeda K, Akira S, Medzhitov R. Curr Biol. 2000;10:1139–1142. doi: 10.1016/s0960-9822(00)00700-4. [DOI] [PubMed] [Google Scholar]
- 15.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al. Nature (London) 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 16.Chu W, Gong X, Li Z, Takabayashi K, Ouyang H, Chen Y, Lois A, Chen D J, Li G C, Karin M, et al. Cell. 2000;103:909–918. doi: 10.1016/s0092-8674(00)00194-x. [DOI] [PubMed] [Google Scholar]
- 17.Lipford G B, Bauer M, Blank C, Reiter R, Wagner H, Heeg K. Eur J Immunol. 1997;27:2340–2344. doi: 10.1002/eji.1830270931. [DOI] [PubMed] [Google Scholar]
- 18.Chu R S, Targoni O S, Krieg A M, Lehmann P V, Harding C V. J Exp Med. 1997;186:1623–1631. doi: 10.1084/jem.186.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carson D A, Raz E. J Exp Med. 1997;186:1621–1622. doi: 10.1084/jem.186.10.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer M, Heeg K, Wagner H, Lipford G B. Immunology. 1999;97:699–705. doi: 10.1046/j.1365-2567.1999.00811.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartmann G, Weiner G J, Krieg A M. Proc Natl Acad Sci USA. 1999;96:9305–9310. doi: 10.1073/pnas.96.16.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartmann G, Weeratna R D, Ballas Z K, Payette P, Blackwell S, Suparto I, Rasmussen W L, Waldschmidt M, Sajuthi D, Purcell R H, et al. J Immunol. 2000;164:1617–1624. doi: 10.4049/jimmunol.164.3.1617. [DOI] [PubMed] [Google Scholar]
- 23.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Okayama H. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dziarski R, Wang Q, Miyake K, Kirschning C J, Gupta D. J Immunol. 2001;166:1938–1944. doi: 10.4049/jimmunol.166.3.1938. [DOI] [PubMed] [Google Scholar]
- 27.Hacker H, Mischak H, Miethke T, Liptay S, Schmid R, Sparwasser T, Heeg K, Lipford G B, Wagner H. EMBO J. 1998;17:6230–6240. doi: 10.1093/emboj/17.21.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 29.Smith G C, Jackson S P. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 30.Aderem A, Hume D A. Cell. 2000;103:993–996. doi: 10.1016/s0092-8674(00)00201-4. [DOI] [PubMed] [Google Scholar]
- 31.Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 32.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Immunity. 1999;11:115–122. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 33.Blunt T, Gell D, Fox M, Taccioli G E, Lehmann A R, Jackson S P, Jeggo P A. Proc Natl Acad Sci USA. 1996;93:10285–10290. doi: 10.1073/pnas.93.19.10285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danska J S, Holland D P, Mariathasan S, Williams K M, Guidos C J. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipford G B, Sparwasser T, Bauer M, Zimmermann S, Koch E S, Heeg K, Wagner H. Eur J Immunol. 1997;27:3420–3426. doi: 10.1002/eji.1830271242. [DOI] [PubMed] [Google Scholar]
- 36.Sparwasser T, Miethke T, Lipford G, Erdmann A, Hacker H, Heeg K, Wagner H. Eur J Immunol. 1997;27:1671–1679. doi: 10.1002/eji.1830270712. [DOI] [PubMed] [Google Scholar]
- 37.Muzio M, Bosisio D, Polentarutti N, D'amico G, Stoppacciaro A, Mancinelli R, van't Veer C, Penton-Rol G, Ruco L P, Allavena P, et al. J Immunol. 2000;164:5998–6004. doi: 10.4049/jimmunol.164.11.5998. [DOI] [PubMed] [Google Scholar]
- 38.Muzio M, Polentarutti N, Bosisio D, Prahladan M K, Mantovani A. J Leukocyte Biol. 2000;67:450–456. doi: 10.1002/jlb.67.4.450. [DOI] [PubMed] [Google Scholar]
- 39.Visintin A, Mazzoni A, Spitzer J H, Wyllie D H, Dower S K, Segal D M. J Immunol. 2001;166:249–255. doi: 10.4049/jimmunol.166.1.249. [DOI] [PubMed] [Google Scholar]
- 40.Modlin R L. Nature (London) 2000;408:659–660. doi: 10.1038/35047207. [DOI] [PubMed] [Google Scholar]
- 41.Hendrich B, Bird A. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Mol Cell Biol. 2000;20:5107–5118. doi: 10.1128/mcb.20.14.5107-5118.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poltorak A, Ricciardi-Castagnoli P, Citterio S, Beutler B. Proc Natl Acad Sci USA. 2000;97:2163–2167. doi: 10.1073/pnas.040565397. . (First Published February 18, 2000; 10.1073/pnas.040565397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemaitre B, Nicolas E, Michaut L, Reichhart J M, Hoffmann J A. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]