Abstract
Methionine-S-sulfoxide reductase (MsrA) protects against high-fat diet-induced insulin resistance due to its antioxidant effects. To determine whether its counterpart, methionine-R-sulfoxide reductase (MsrB) has similar effects, we compared MsrB1 knockout and wild-type mice using a hyperinsulinemic-euglycemic clamp technique. High-fat feeding for 8 weeks increased body weights, fat masses, and plasma levels of glucose, insulin, and triglycerides to similar extents in wild-type and MsrB1 knockout mice. Intraperitoneal glucose tolerance test showed no difference in blood glucose levels between the two genotypes after 8 weeks on the high-fat diet. The hyperglycemic-euglycemic clamp study showed that glucose infusion rates and whole body glucose uptakes were decreased to similar extents by the high-fat diet in both wild-type and MsrB1 knockout mice. Hepatic glucose production and glucose uptake of skeletal muscle were unaffected by MsrB1 deficiency. The high-fat diet-induced oxidative stress in skeletal muscle and liver was not aggravated in MsrB1-deficient mice. Interestingly, whereas MsrB1 deficiency reduced JNK protein levels to a great extent in skeletal muscle and liver, it markedly elevated phosphorylation of JNK, suggesting the involvement of MsrB1 in JNK protein activation. However, this JNK phosphorylation based on a p-JNK/JNK level did not positively correlate with insulin resistance in MsrB1-deficient mice. Taken together, our results show that, in contrast to MsrA deficiency, MsrB1 deficiency does not increase high-fat diet-induced insulin resistance in mice.
Keywords: Methionine sulfoxide B1, High-fat diet, Insulin resistance, Hyperinsulinemic-euglycemic clamp, Antioxidant
Introduction
The prevalence of type 2 diabetes has increased dramatically in parallel with the global obesity epidemic, and has placed health care systems under severe pressure. Insulin resistance links obesity and type 2 diabetes [1] and is also associated with a variety of metabolic diseases, such as hypertension, dyslipidemia, and atherosclerosis [2].
Although various mechanisms have been proposed to explain obesity-induced insulin resistance, oxidative stress and inflammation have recently received much attention. Obese and diabetic subjects exhibit increased oxidative stress markers in plasma and peripheral tissues [3-5]. Furthermore, in normal subjects, high-fat consumption induces insulin resistance and reduces antioxidant capacity [5]. In addition, it has been shown that increased expression of the antioxidant enzyme superoxide dismutase (SOD) 2 in skeletal muscle attenuates high-fat diet-induced insulin resistance in mice [6].
Methionine sulfoxide reductases (Msrs) are important protein repair enzymes that catalyze the reduction of oxidized methionines back to methionines [7,8]. Msrs also act as antioxidants and protect cells (from bacteria to mammals) from oxidative stress [9]. Two Msr classes, that is, MsrA and MsrB, stereospecifically reduce methionine sulfoxide residues in proteins. MsrA is specific for the reduction of the S-form of methionine sulfoxide, whereas MsrB is specific for the R-form. Mammals possess a single MsrA gene, and its products are present in mitochondria, cytosol, and nuclei [10-12]. On the other hand, mammals possess three MsrB genes, the products of which localize to different cellular compartments [13,14]: MsrB1 is present in cytosol and nuclei, MsrB2 is a mitochondrial protein, and MsrB3 localizes to endoplasmic reticulum.
A number of studies have shown that MsrA is associated with a variety of physiopathologic processes, which include oxidative stress and age-related neurodegenerative diseases. MsrA knockout mice show increased oxidative stress, behavioral/cognitive impairments, and lower locomotor activities [15,16], and are more susceptible to kidney ischemia/reperfusion (I/R) injury [17], whereas MsrA transgenic mice show resistance to myocardial I/R injury [18]. Recently, Styskal et al reported that modulation of the MsrA gene can affect insulin resistance [19]. In particular, it was found that MsrA deficiency aggravates insulin resistance induced by high-fat diet in mice by facilitating the oxidation of insulin receptor, thereby reducing its activity. Accordingly, the above findings suggest that MsrA plays a protective role against obesity-induced insulin resistance.
Since MsrA has been shown to be involved in insulin signaling, it would be interesting to know whether MsrB also influences insulin resistance. Of the three MsrBs, MsrB1 is a selenoprotein and the major MsrB enzyme in liver [20]. MsrB1-deficient mice have recently been developed and reported to exhibit increased markers of protein and lipid oxidation in peripheral tissues, suggesting that MsrB1 plays a role in the alleviation of oxidative stress [21]. In the present study, we examined the role of MsrB1 in high-fat diet-induced insulin resistance using MsrB1 knockout mice.
Materials and methods
MsrB1 knockout mice
The generation of MsrB1 knockout (MsrB1−/−) mice has been described elsewhere [21]. The breeding lines for MsrB1 knockout mice were obtained by backcrossing into the C57BL/6N genetic background for >8 generations. Eight-week-old male C57BL/6N wild-type and MsrB1 knockout mice were housed in a room under a 12:12-h light/dark cycle. Mice were fed a chow diet or a high-fat diet for 8 weeks and given ad libitum access to water. The laboratory chow diet (AIN-93G) provided 64% energy as carbohydrate, 20% as protein, and 16% as fat, while the high-fat diet 60% calorie from fat) provided 20% energy as carbohydrate, 20% as protein, and 60% as fat (Feedlab, Korea). Compositions of the chow and high-fat diets are summarized in supplementary Table S1. The mice were anesthetized via an intraperitoneal injection of avertin (a mixture of 2,2,2-tribromoethanol and tert-amyl alcohol) for vein cannulation in the hyperinsulinemic-euglycemic clamp study. Retroperitoneal fat was excised for fat mass measurement. After all experiments, the mice were sacrificed with an overdose of avertin (>1.0 g•kg, i.p.) and death was verified by cervical dislocation. The study was conducted in strict accordance with the guidelines and protocols approved by Institutional Animal Care and Use Committee of Yeungnam University College of Medicine (permit number: YUMC-AEC2012-004).
Intraperitoneal glucose tolerance test
After 8 weeks on the high-fat diet, mice were fasted overnight, and blood samples were collected for basal blood glucose levels using microcapillary tubes from tail vessels. Glucose (1.5 g/kg body weight) was injected intraperitoneally, and blood samples were collected at 15, 30, 60, 90, and 120 min. Blood glucose levels were measured using OneTouch blood glucose meters (LifeScan Europe, Switzerland).
Hyperinsulinemic-euglycemic clamp study
A hyperinsulinemic-euglycemic clamp study was performed 4 days after vein cannulation in conscious mice as described previously [22]. Briefly, after overnight fasting, a 2-h hyperinsulinemic-euglycemic clamp was performed with a 900 pmol•kg−1 priming dose and a continuous infusion of human regular insulin (Novolin, Denmark) at 15 pmol•kg−1•min−1, and 20% glucose was infused at variable rates to maintain glucose at constant concentrations of 5∼6 mM. Insulin-stimulated rates of whole body glucose uptake were estimated using a continuous infusion of [3-3H] glucose (PerkinElmer Life and Analytical Sciences, USA) throughout clamps (0.1 μCi/min). To estimate insulin-stimulated glucose uptake in individual tissues, 2-deoxy-D-[1-14C] glucose (2-[14C]DG; PerkinElmer Life and Analytical Sciences) was administered as a bolus (10 μCi) at 75 min after starting clamps. Plasma glucose concentrations were measured using a glucose analyzer (Analox, UK), and plasma insulin concentrations were measured using an enzyme-linked immunosorbent assay (ELISA; Merck Millipore, USA). To determine tissue 2-[14C]DG 6-phosphate (2-[14C]DG-6-P) contents, the supernatants of homogenized tissue samples were introduced into an ion exchange column to separate 2-DG-6-P from 2-DG. Rates of insulin-stimulated whole-body glucose uptake were calculated by dividing [3H] glucose infusion rates [disintegrations per minute (dpm/min)] by specific activities of plasma glucose (dpm/μmol) during the final 30 min of clamps. Hepatic glucose production (HGP) during clamps was determined by subtracting glucose infusion rate from whole-body glucose uptake rate. Glucose uptake in individual tissues was calculated from the plasma 2-[14C]DG profile, which was fitted with a double exponential or linear curve using MLAB (Civilized Software, USA) and tissue 2-[14C]DG-6-P content.
Quantitative real time polymerase chain reaction (qRT-PCR)
qRT-PCR was performed as previously described [23]. Briefly, 25 mg tissue samples from hyperinsulinemic-euglycemic clamp study were homogenized in TRI reagent (Sigma–Aldrich, USA) and RNAs were reverse transcribed to cDNA using a reverse transcription kit (Applied Biosystems, USA). qRT-PCR was performed using the Real-Time PCR 7500 System and Power SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer's instructions. Validation of β-actin as a reference gene for sample normalization was tested by its Ct levels and stability index. Each reaction mixture was incubated at 95°C for 10 min and amplified over 45 cycles of 95°C for 15 s, 55°C for 20 s, and 72°C for 35 s. The primer sequences were designed using the Primer Express Program (Applied Biosystems). The primers used were: β-actin (121 bp: forward, 5′-TGGACAGTGAGGCAAGGATAG-3′; reverse, 5′-TACTGCCCTGGCTCCTAGCA-3′), SOD2 (71 bp: forward, 5′-CTGCTCTAATCAGGACCCATT-3′; reverse, 5′-GTGCTCCCACACGTCAATC-3′), glutathione peroxidase 1 (GPX1; 71 bp: forward, 5′-GAAGTGCGAAGTGAATGGTG-3′; reverse, 5′-TGGGTGTTGGCAAGGC-3′), c-Jun N-terminal kinase (JNK) 1 (100 bp: forward, 5′-GCTCATGGATGCAAATCTTT-3′; reverse, 5′-AAGGTGCTTGATTCCACACA-3′); JNK2 (100 bp: forward, 5′-CACCACAAAAAACGCTAGAAG-3′; reverse, 5′-TCATGGTCCAGTTCCATATGA-3′).
Western blotting
Antibodies against JNK, extracellular signal-regulated kinase (ERK), p38, and their phosphorylated forms were obtained from Cell Signaling Technology, antibodies against 4-hydroxy-2-nonenal (4-HNE) from Abcam, and antibodies against SOD2 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) from Santa Cruz Biotechnology. Antibodies against MsrA, MsrB1, MsrB2, and MsrB3 have been described elsewhere [17,21]. Tissue samples (25 mg) from hyperinsulinemic-euglycemic clamp study were homogenized in lysis buffer containing 50 mM HEPES, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM bezamide, 1 mM Na3VO4, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 1% NP40, 10% glycerol, β-glycerophosphate, aprotinin, leupeptin, and pepstatin A. Proteins were then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) using a 10% gel or a NuPAGE 4–12% Bis-Tris gel (Invitrogen, USA). Resolved proteins were then transferred to 0.45 μm polyvinylidene fluoride membranes (PVDF; Millipore). After blocking with a solution containing 5% skim milk in TBST (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.1% Tween-20), membranes were incubated overnight at 4°C with primary antibodies (1:1000 dilution), except for GAPDH antibody (1:1000 dilution), in which case, membranes were incubated for 1 h at room temperature. Specific antibody binding was detected using sheep anti-rabbit IgG horseradish peroxidase (Bio-Rad, USA) for 1 h at room temperature. After addition of chemiluminescence detection regent (Millipore), signals were recorded and quantified using a LAS-3000 image analyzer and Multi Gauge 3.0 software (Fujifilm, Japan). GAPDH was used as a loading control since the protein levels were found to be not significantly different among the experimental groups.
Protein carbonylation
Protein-carbonyl contents were measured using an OxyBlot Protein Oxidation Detection kit (Chemicon, USA) as previously described [22]. Briefly, tissues were homogenized in lysis buffer (Invitrogen) containing 50 mM DTT. Protein (10 μg) in 3 μl of lysis buffer was added to 3 μl of 12% SDS. Samples were derivatized by adding 6 μl of 2,4-dinitrophenylhydrazine solution, incubated at room temperature for 15 min, and then 4.5 μl of neutralization solution was added. Protein samples were separated by 10% SDS-PAGE and transferred to PVDF membranes, which were blocked with 1% bovine serum albumin (BSA) in TBST, incubated for 1 h at room temperature with rabbit anti-2,4-dinitrophenyl antibody diluted 1:150 in 1% BSA/TBST, washed with TBST, and incubated for 1 h at room temperature with goat anti-rabbit IgG coupled to horseradish peroxidase. Carbonylated proteins were visualized using the enhanced chemiluminescence detection reagent.
MsrA and MsrB activities
Reaction mixtures (100 μl) contained 50 mM sodium phosphate (pH 7.5), 50 mM NaCl, 20 mM DTT, 200 μM dabsylated methionine-S-sulfoxide (for MsrA) or methionine-R-sulfoxide (for MsrB), and 200 μg of crude protein. Reactions were carried out at 37°C for 30 min, after which the reaction product, dabsyl-Met, was analyzed by HPLC as previously described [24]. The enzyme activity was expressed as pmole•min•mg of crude protein.
Hydrogen peroxide
Hydrogen peroxide levels in tissue samples were determined using the ferric-sensitive dye, xylenol orange (Sigma–Aldrich, USA) as previously described [24].
Glutathione to oxidized glutathione (GSH/GSSG) ratio
GSH/GSSG ratios were measured using a Glutathione Fluorescent Detection kit (Arbor Assays, USA) according to the instructions of the manufacturer. Briefly, skeletal muscle samples were homogenized in ice-cold 5% sulfosalicilic acid, incubated for 10 min at 4°C, and centrifuged at 14,000 rpm for 10 min at 4°C to remove the precipitated proteins. The supernatant was diluted 5 times with the assay buffer, and reacted with a ThioStar reagent to determine the free GSH levels (λ emission 510 nm and excitation 390 nm). The sample was added to the reaction mixture containing NADPH and GSH reductase to produce total GSH. The GSSG levels were calculated using the difference between the total and free GSH levels.
Statistics
The results are expressed as mean±SEM. Statistical analyses were performed with SPSS software (version 22; IBM, USA) using one-way ANOVA followed by Tukey's post hoc test unless stated otherwise. Statistical details are provided in figure legends. A p value <0.05 was considered significant.
Results
Body weight, fat mass, and plasma triglyceride levels
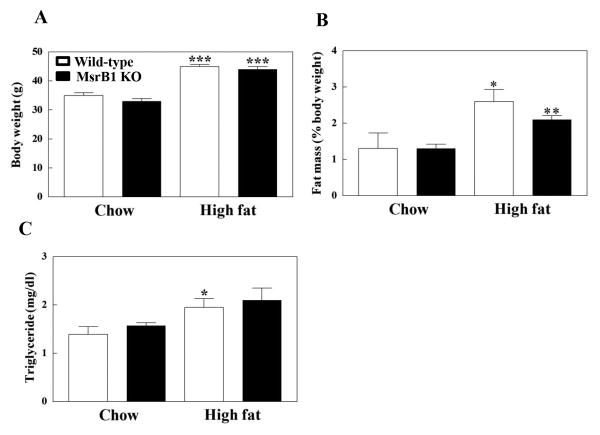
Body weights, fat masses, and plasma levels of triglyceride were not significantly different between chow diet-fed wild-type and MsrB1 knockout mice (p=0.12, p=0.85, and p=0.34 in body weights, fat masses, and triglyceride levels, respectively). High-fat diet significantly increased body weights and fat masses in wild-type and MsrB1 knockout mice; however, no significant differences in body weights (p=0.98) and fat masses (p=0.19) were observed between the two genotypes (Figs. 1A and B). Plasma triglyceride levels were significantly increased by the high-fat diet in wild-type mice and seemed to be increased in MsrB1 knockout mice (p=0.1) (Fig. 1C).
Fig. 1. Body weights (A), fat masses (B), and plasma triglyceride levels (C) in chow- and high-fat-fed wild-type and MsrB1 knockout mice.
Retroperitoneal fat was excised and the fat mass was standardized with body weight. Results are presented as mean±SEM (n=6, chow-fed wild-type; n=5, chow-fed MsrB1 knockout; n=6, high-fat-fed wild-type; n=8, high-fat-fed MsrB1 knockout). *p<0.05, **p<0.01, and ***p<0.001 vs. wild-type chow diet or knockout chow diet. KO, knockout.
Expression and activity levels of MsrA and MsrBs in skeletal muscle and liver
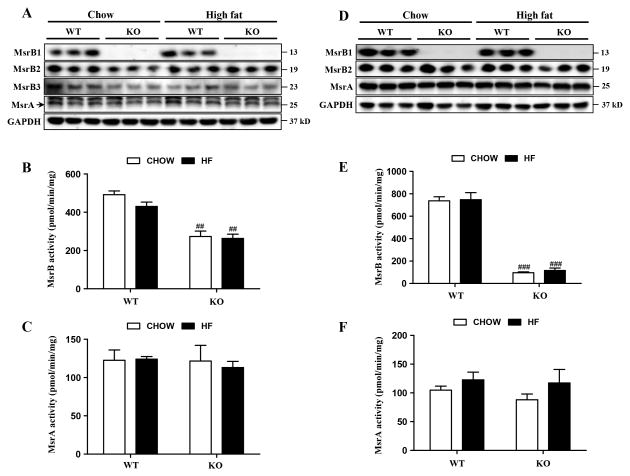
In skeletal muscle, MsrB1 and MsrB2 protein levels were similar in high-fat diet- and chow diet-fed wild-type mice (Fig. 2A); however, MsrB3 protein levels were significantly reduced by the high-fat diet in wild-type mice (p<0.05). MsrB activities in wild-type mice were insignificantly decreased by the high-fat diet (p=0.16) (Fig. 2B). In MsrB1 knockout mice, MsrB2 and MsrB3 protein levels were significantly reduced in a chow-fed group (p<0.05), compared to those in chow-fed wild-type mice. The high-fat diet significantly increased the MsrB2 levels in MsrB1 knockout mice (p<0.05), which were similar to those in high-fat-fed wild-type mice. MsrB3 expressions in MsrB1 knockout mice were unaffected by diet. MsrB activity was significantly lower (by 60%) in MsrB1 knockout mice than in wild-type mice, and no difference in MsrB activity was observed between high-fat and chow dietary animals. The residual MsrB activity was attributed to MsrB2 and MsrB3 isozymes. The high-fat diet did not change MsrA expressions in the skeletal muscles of wild-type or MsrB1 knockout mice (Fig. 2A). In accordance with the lack of diet-induced MsrA protein level changes in wild-type and MsrB1 knockout mice, MsrA activities were also unaffected by diet in both genotypes (Fig. 2C). In livers, MsrB1 protein and MsrB activity levels were similar in high-fat diet- and chow diet-fed wild-type mice (Figs. 2D and E). The expression levels of MsrB2 were not altered by diet in the livers of wild-type or MsrB1 knockout mice. Notably, MsrB3 protein was hardly detectable in the livers of both mice fed chow or high-fat diets. MsrB activity in the livers of MsrB1 knockout mice was ∼15% of activity observed in wild-type mice, consistent with previous findings that MsrB1 is the major MsrB enzyme in liver [20], and was similar in chow and high-fat dietary groups. The expression and activity levels of MsrA in livers were not altered by the high-fat diet in wild-type or MsrB1 knockout mice (p=0.31) (Figs. 2D and F).
Fig. 2. Expression and activities of MsrA and MsrBs in skeletal muscle (A–C) and liver (D–F) tissues.
Protein levels of MsrA and MsrBs were analyzed by Western blotting (A, D); GAPDH was used as a loading control. MsrB3 in livers was hardly detectable. A strong non-specific band against MsrA antibody with size of approximately 27 kD was detected on the blot of skeletal muscle. The activities of MsrB (B, E) and MsrA (C, F) were determined as described in Materials and Methods. Results are expressed as mean±SEM (n=3 for each group). ##p<0.01 and ###p<0.001 vs. wild-type chow diet or wild-type high-fat diet. WT, wild-type; KO, MsrB1 knockout; HF, high-fat.
Intraperitoneal glucose tolerance test
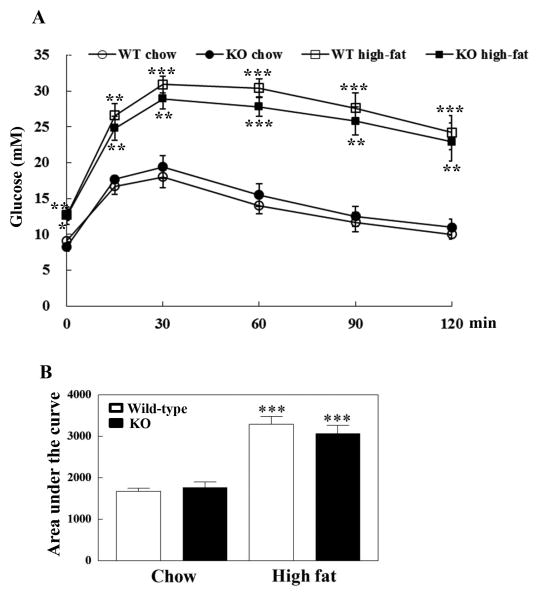
Basal blood glucose levels were similar in wild-type and MsrB1 knockout mice fed chow diets after overnight fasting, but the high-fat diets significantly increased basal blood glucose levels in both groups of mice (Fig. 3A). Blood glucose levels were significantly higher at 15, 30, 60, 90, and 120 min after an intraperitoneal injection of 1.5 g/kg glucose in high-fat-fed mice than in chow-fed wild-type and MsrB1 knockout mice, and no difference was observed between wild-type and MsrB1 knockout mice (p=0.93, p=0.49, p=0.32, p=0.22, p=0.57, and p=0.74 at 15, 30, 60, 90 and 120 min, respectively) (Fig. 3A). The high-fat diet significantly increased area under the curve, but no difference was found between wild-type and MsrB1 knockout mice (p=0.45) (Fig. 3B).
Fig. 3. Intraperitoneal glucose tolerance test in chow- and high-fat-fed wild-type and MsrB1 knockout mice.
Blood glucose levels (A). Area under the curve of blood glucose level (B). Results are presented as mean±SEM (n=5 per genotype in chow-fed groups; n=6 per genotype in high-fat diet groups). *p<0.05, **p<0.01, and ***p<0.001 vs. wild-type chow diet or knockout chow diet. WT, wild-type; KO, MsrB1 knockout.
Hyperinsulinemic-euglycemic clamp
We next performed a hyperinsulnemic-euglycemic clamp study, the gold-standard method to assess insulin sensitivity, to confirm the results from glucose tolerance test and to collect additional data as to the insulin resistance of peripheral tissues, liver and skeletal muscle. Since blood glucose levels may be affected by diverse factors including stress from experimental procedure, the hyperinsulinemic-euglycemic clamp study would strengthen the data of insulin sensitivity. Plasma glucose levels in the basal state were similar in wild-type and MsrB1 knockout mice fed a chow diet (p=0.40). The high-fat diet similarly increased plasma glucose levels in the basal state in both wild-type and MsrB1 knockout mice. Plasma insulin levels at basal state were not significantly different between wild-type and MsrB1 knockout mice fed a chow diet (p=0.22). Although the high-fat diet significantly increased plasma insulin levels in wild-type and in MsrB1 knockout mice, no difference was observed between these two genotypes (p=0.95) (Table 1). During the hyperinsulinemic-euglycemic clamp study, plasma glucose levels were maintained at 5∼6 mM in wild-type and in MsrB1 knockout mice on both diets. Plasma insulin levels were elevated to approximately 300 pM in both genotypes fed a chow diet. These plasma insulin levels were significantly increased by the high-fat diet, but no significant difference was observed between the wild-type and MsrB1 knockout mice groups (p=0.69) (Table 1).
Table 1. Plasma levels of glucose and insulin in MsrB1 knockout (MsrB1 KO) and wild-type mice during the basal and hyperinsulinemic-euglycemic clamp periods.
| Basal period | Clamp period | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Diet | Animal | n | Glucose (mM) | Insulin (pM) | Glucose (mM) | Insulin (pM) |
| Chow | Wild-type | 6 | 4.1±0.2 | 54±15 | 5.8±0.5 | 281±8 |
| MsrB1 KO | 5 | 3.9±0.3 | 96±27 | 5.7±0.6 | 318±30 | |
| High-fat | Wild-type | 6 | 5.6±0.4* | 218±60* | 5.3±0.3 | 558±44** |
| MsrB1 KO | 8 | 5.5±0.4* | 221±33** | 5.4±0.4 | 594±75* | |
p<0.05, and
p<0.01 vs. wild-type chow diet or knockout chow diet.
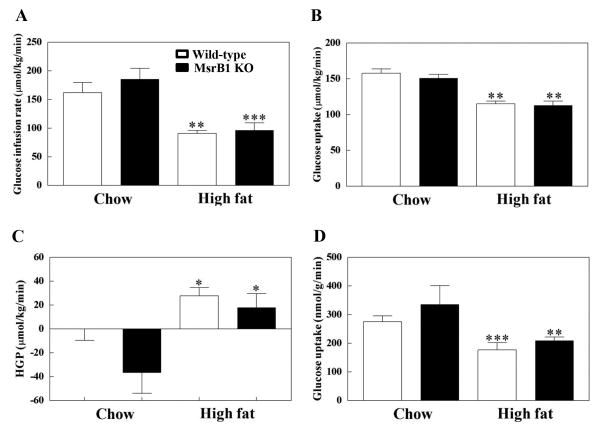
Glucose infusion rates required to maintain euglycemia were similar for wild-type and MsrB1 knockout mice fed a chow diet (p=0.40). The high-fat diet significantly reduced glucose infusion rates in both wild-type and MsrB1 knockout mice, but no difference was observed between these two groups (p=0.80) (Fig. 4A). Insulin-stimulated whole body glucose uptakes were also similar in wild-type and MsrB1 knockout mice on a chow diet (p=0.38). The high-fat diet decreased whole body glucose uptake similarly in wild-type and MsrB1 knockout mice (p=0.97) (Fig. 4B).
Fig. 4. Glucose metabolism as determined using the hyperinsulinemic-euglycemic technique in chow- and high-fat-fed wild-type and MsrB1 knockout mice.
Glucose infusion rates (A), whole body glucose uptakes (B), insulin-stimulated hepatic glucose production (HGP) (C), and skeletal muscle glucose uptakes (D) were measured as described in Materials and Methods. Results are presented as mean±SEM (n=6, chow-fed wild-type; n=5, chow-fed MsrB1 knockout; n=6, high-fat-fed wild-type; n=8, high-fat-fed MsrB1 knockout). *p<0.05, **p<0.01, and ***p<0.001 vs. wild-type chow diet or knockout chow diet. KO, knockout.
Insulin-stimulated HGP levels were not significantly different between the wild-type and MsrB1 knockout mice in the chow diet group (p=0.11). The high-fat diet significantly increased HGP levels in wild-type and MsrB1 knockout mice, but no significant difference was observed between the two groups (p=0.54) (Fig. 4C). Skeletal muscle glucose uptake was similar for wild-type and MsrB1 knockout mice fed a chow diet (p=0.28). The high-fat diet reduced glucose uptake in both wild-type and MsrB1 knockout mice, but no significant difference was observed between genotypes (p=0.97) (Fig. 4D). We analyzed if there were interactions of factors between diet and genotype using a two-way ANOVA, but found no statistically significant interactions from any data of the hyperinsulinemic-euglycemic clamp study. Collectively, these results show that high-fat diet-induced insulin resistance in liver and skeletal muscle is not increased by MsrB1 gene deletion.
Oxidative stress status in skeletal muscle and liver
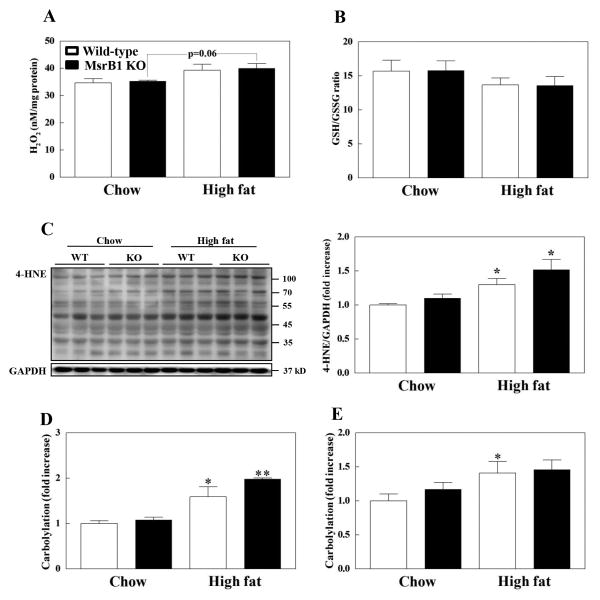
Since MsrB1 functions as an antioxidant enzyme and a high-fat diet is known to induce oxidative stress, we investigated oxidative stress status of skeletal muscle and liver by measuring hydrogen peroxide, GSH/GSSG ratio, 4-HNE, and protein-carbonyl levels. Basal hydrogen peroxide levels in skeletal muscle were similar in wild-type and MsrB1 knockout mice. The high-fat diet seemed to increase hydrogen peroxide levels in wild-type (p=0.15) and MsrB1 knockout mice (p=0.06) with no difference between the two genotypes (p=0.83) (Fig. 5A). GSH/GSSG ratio in skeletal muscle was also similar in wild-type and MsrB1 knockout mice in chow diet (p=0.95) and high-fat diet groups (p=0.96). Although high-fat diet did not significantly affect GSH/GSSG ratio in wild-type (p=0.34) and MsrB1 knockout mice (p=0.27), it decreased GSH/GSSG ratio by ∼15% in both genotype of mice (Fig. 5B). No significant increase in 4-HNE (p=0.19) and protein-carbonyl levels (p=0.34) was detected in the skeletal muscles of MsrB1 knockout mice versus the wild-type mice fed a chow diet (Fig. 5C and D). The high-fat diet significantly increased 4-HNE and protein-carbonyl levels, but no significant differences were observed between the two genotypes (p=0.24 for HNE and p=0.42 for protein-carbonyl levels) (Figs. 5C and D). Protein-carbonyl levels were similar in the livers of MsrB1 knockout and wild-type mice fed the chow diet. The high-fat diet significantly increased carbonyl contents in livers of wild-type mice but not in livers of MsrB1 knockout mice (p=0.19); however, no difference was observed between the two genotypes (p=0.83) (Fig. 5E). Collectively, these results suggest that high-fat diet-induced oxidative stress is not aggravated in MsrB1 knockout mice.
Fig. 5. Oxidative stress markers in the skeletal muscles (A-D) and livers (E) of chow- and high-fat-fed wild-type and MsrB1 knockout mice.
H2O2 levels (A). GSH/GSSG ratio (B). 4-hydroxy-2-nonenal (HNE) levels (C). Protein-carbonyl levels (D, E). Each oxidative stress marker was measured as described in Materials and Methods. Results are presented as mean±SEM. Experimental cases in each group are as follows: (A) n=3 for each group; (B-E) n=6, chow-fed wild-type; n=5, chow-fed MsrB1 knockout; n=6, high-fat-fed wild-type; n=6, high-fat-fed MsrB1 knockout. *p<0.05 and **p<0.01 vs. wild-type chow diet or knockout chow diet. KO, knockout.
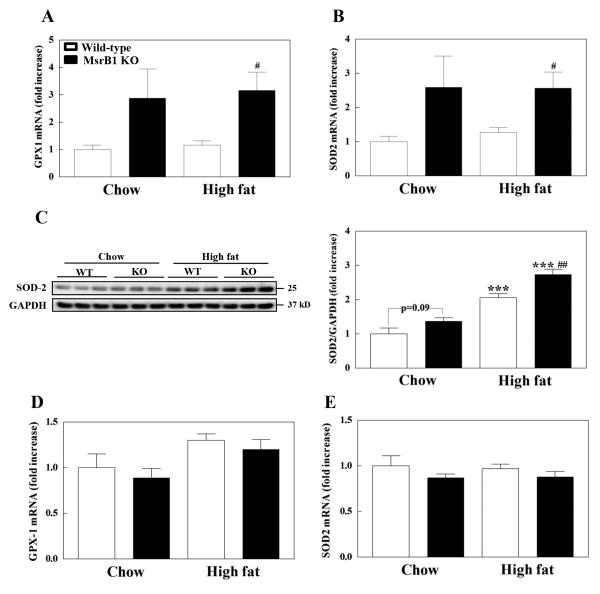
We also examined the gene expression levels of GPX1 and SOD2, which are representative antioxidant enzymes localized predominantly in the cytoplasm and mitochondria, respectively. Basal mRNA levels of both GPX1 and SOD2 in skeletal muscle seemed to increase (p=0.12) in MsrB1 knockout mice (Figs. 6A and B). The high-fat diet had no significant effect on GPX1 or SOD2 mRNA levels in the skeletal muscles of wild-type (p=0.49 for GPX1 and p=0.48 for SOD2) or MsrB1 knockout mice (p=0.82 for GPX1 and p=0.98 for SOD2). However, the mRNA levels of GPX1 or SOD2 in MsrB1 knockout mice were found to be significantly higher than those in wild-type mice fed the high-fat diet (Figs. 6A and B). Basal protein levels of SOD2 in skeletal muscle seemed to be higher (p=0.09) in MsrB1 knockout mice (Fig. 6C), consistent with the mRNA data. The high-fat diet significantly increased the protein levels of SOD2 in both wild-type and MsrB1 knockout mice (Fig. 6C). Significantly higher protein levels of SOD2 were observed in MsrB1 knockout mice, consistent with the transcript data. Notably, while the high-fat diet had no effect on SOD2 transcription, it positively regulated the protein levels of SOD2. These results suggest that SOD2 expression may be post-transcriptionally regulated in high-fat diet group. MsrB1 knockout and wild-type mice had similar mRNA levels of GPX1 (p=0.57) and SOD2 (p=0.35) in liver tissues (Figs. 6D and E). The high-fat diet did not affect the transcription levels of SOD2 in liver, while it seemed to increase GPX1 mRNA levels in both wild-type (p=0.1) and MsrB1 knockout mice (p=0.07). There were no statistically significant interactions of factors between diet and genotype in oxidative stress and antioxidant expression data (two-way ANOVA).
Fig. 6. Expression of GPX1 and SOD2 in skeletal muscles (A-C) and livers (D,E) of chow- and high-fat-fed wild-type and MsrB1 knockout mice.
mRNA levels of GPX1 (A, D) and SOD2 (B, E) and protein levels of SOD2 (C) were measured as described in Materials and Methods. Results are presented as mean±SEM (n=6, chow-fed wild-type; n=5, chow-fed MsrB1 knockout; n=6, high-fat-fed wild-type; n=6, high-fat-fed MsrB1 knockout). ***p<0.001 vs. wild-type chow diet or knockout chow diet; #p<0.05 and ##p<0.01 vs. wild-type high-fat diet. KO, knockout.
Phosphorylations of JNK and ERK in skeletal muscle and liver
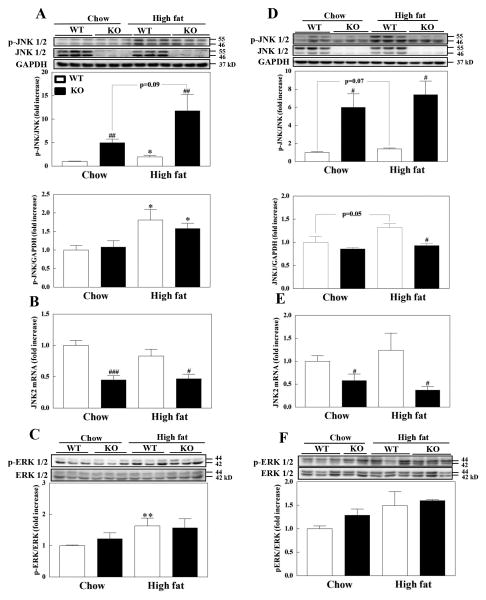
We examined whether MsrB1 status affects the activation of JNK and ERK, members of mitogen activated protein kinase (MAPK) family, in skeletal muscle and liver, as altered activities of JNK and ERK are associated with insulin resistance [25-27]. Interestingly, the protein levels of JNK were significantly lower in the skeletal muscles of MsrB1 knockout mice than in wild-type mice in both chow- and high-fat-fed animals (Fig. 7A). To explore this further, we measured the mRNA levels of JNK1 and JNK2. We found that the mRNA levels of JNK1 were not significantly different in wild-type and MsrB1 knockout mice (data not shown; p=0.27 and p=0.64 between wild-type and MsrB1 knockout mice in chow diet and high-fat diet groups, respectively), but the mRNA levels of JNK2 were significantly lower in MsrB1 knockout mice than in the wild-type mice (Fig. 7B). More interestingly, the levels of phosphorylated JNK forms in MsrB1 knockout mice fed a chow diet were remarkably comparable to those of wild-type mice. Therefore, the level of JNK phosphorylation calculated by p-JNK/JNK was significantly higher in the MsrB1 knockout mice (Fig. 7A, upper graph), suggesting activation of JNK protein by MsrB1 deficiency. The high-fat diet increased the phosphorylation of JNK in both wild-type (p=0.03) and MsrB1 knockout mice (p=0.09) with a significant difference between these mice groups (Fig. 7A, upper graph).
Fig. 7. Phosphorylation of JNK and ERK in skeletal muscles (A–C) and livers (D–F) of chow- and high-fat-fed wild-type and MsrB1 knockout mice.
JNK phosphorylation (A, D), JNK2 mRNA levels (B, E), and ERK phosphorylation (C, F) were measured as described in Materials and Methods. Results are presented as mean±SEM (n=6, chow-fed wild-type; n=5, chow-fed MsrB1 knockout; n=6, high-fat-fed wild-type; n=6, high-fat-fed MsrB1 knockout). *p<0.05 and **p<0.01 vs. wild-type chow diet or knockout chow diet; #p<0.05, ##p<0.01, ###p<0.001 vs. wild-type chow diet or wild-type high-fat diet. WT, wild-type; KO, MsrB1 knockout.
ERK phosphorylation was not different in wild-type and MsrB1 knockout mice fed a chow diet (p=0.36). The high-fat diet significantly increased ERK phosphorylation in wild-type mice but not in MsrB1 knockout mice (p=0.21). However, no difference was observed between wild-type and MsrB1 knockout mice (p=0.84) (Fig. 7C). Phosphorylation of p38 was unaffected by MsrB1 deficiency or a high-fat diet (data not shown).
As was found for skeletal muscle, JNK protein levels in livers were also significantly reduced in MsrB1 knockout mice (Fig. 7D). While the mRNA levels of JNK1 were similar in wild-type and MsrB1 knockout mice (data not shown; p=0.99 and p=0.37 between wild-type and MsrB1 knockout mice in chow diet and high-fat diet groups, respectively), the mRNA levels of JNK2 were significantly lower in MsrB1 knockout mice than in wild-type mice (Fig. 7E). Like skeletal muscle, the levels of JNK phosphorylation in liver were significantly elevated in MsrB1 knockout mice fed a chow diet (Fig. 7D, upper graph). The high-fat diet seemed to increase the phosphorylation of JNK in wild-type mice (p=0.07) but not in MsrB1 knockout mice (p=0.53). JNK phosphorylation was significantly higher in MsrB1 knockout mice than in wild-type mice following high-fat diet. ERK phosphorylation was not significantly affected by MsrB1 deficiency (p=0.10 and p=0.78 between wild-type and MsrB1 knockout mice in chow-fed and high-fat-fed groups, respectively) or high-fat diet (p=0.22 and p=0.10 between chow-fed and high-fat-fed mice in wild-type and MsrB1 knockout groups)(Fig. 7F).
Discussion
In a previous study, it was reported that MsrA deficiency aggravates insulin resistance induced by high-fat diet in mice [19], which suggests an important role of MsrA in insulin sensitivity. In contrast, in the present study we found that high-fat-fed MsrB1 knockout and wild-type mice exhibited similar whole body and skeletal muscle glucose uptakes and hepatic glucose production rates. These results suggest that MsrB1 does not play a role in high-fat diet-induced insulin resistance.
MsrA prevents high-fat diet-induced insulin resistance and this protective role is directly related to its antioxidant function [19]. MsrA deficiency results in increased oxidation of insulin receptor in skeletal muscle, and thus, reduces receptor function. However, although MsrB1 plays a protective role against oxidative stress in transgenic flies and mammalian cells [28,29], no significantly greater accumulation of oxidative stress was observed in the skeletal muscles or livers of chow-fed MsrB1 knockout mice than in chow-fed wild-type mice. In addition, MsrB1 deficiency had no effect on the high-fat diet-induced oxidative stress in these tissues. These results suggest that MsrB1 does not play an antioxidant role in high-fat diet-induced insulin resistance in mice. In a recent study, overexpression of MsrA targeted to mitochondria protected from high-fat diet-induced insulin resistance, whereas its overexpression in the cytoplasm had no effect, suggesting that mitochondrial oxidative stress plays a crucial role in the development of high-fat diet-induced insulin resistance [30]. In contrast to abundant mitochondrial distribution of MsrA [10-12], MsrB1 is not found in the mitochondria but is present in the cytosol [13,14]. These different subcellular localizations between MsrA and MsrB1 may be responsible for the different responses to high-fat diet-induced insulin resistance.
However, it is also possible that the effect of MsrB1 deficiency on oxidative stress is compromised by the compensatory overexpressions of other antioxidant enzymes. Indeed, this notion is supported by the observations that the expression levels of GPX1 and SOD2 were elevated in the skeletal muscles of MsrB1 knockout mice. Enhanced expression of antioxidants by a compensatory effect has been previously reported in GPX1 knockout mice [31,32]. However, this overexpression was not observed in livers. Susceptibility of skeletal muscle to oxidative injury that causes insulin resistance may be higher than that of liver [33].
In a previous study, it was reported that MsrB1 knockout mice show elevated levels of protein carbonyls in liver and kidney, but no change in levels in heart and testis [21]. In the present study, carbonyl content was not significantly elevated in livers of MsrB1 knockout mice. The different levels of oxidative stress observed in the livers of MsrB1 knockout mice could have been due to different genetic backgrounds of the mice used and/or different housing conditions.
MAPK activation is a mechanism by which oxidative stress induces insulin resistance. Reactive oxygen species activate MAPK members, such as JNK and ERK, and it has been established that activation of JNK by increased oxidative stress in skeletal muscle inhibits insulin signaling pathways by increasing serine phosphorylation of insulin receptor substrate, whereas suppression of JNK by gene modulation or inhibitors improves insulin sensitivity [26]. In addition, ERK activation has recently been demonstrated in the skeletal muscles of obese insulin resistant mice [34]. ERK-deficient mice are protected from insulin resistance without affecting obesity [35], whereas high basal ERK activity induces mature onset obesity and insulin resistance [36]. The increased oxidative stress and phosphorylation of JNK and ERK in skeletal muscle of wild-type mice-fed a high-fat diet, which were observed in the present study, are consistent with previous studies. This result supports the association between high-fat diet-induced oxidative stress and insulin resistance.
An interesting finding in the present study is that JNK protein levels are dramatically diminished in MsrB1 knockout mice, which has not been described previously. It seems that the reduced mRNA levels of JNK2 but not JNK1 attributes to the diminished JNK protein levels. Redox status of methionine is involved in the regulation of protein function in transcription factors. For example, methionine oxidation of HypT increases its transcriptional activity in Escherichia coli [37], whereas methionine oxidation of NirA makes this transcription factor inactive in fungus [38]. Thus, MsrB1 deficiency may alter methionine redox status of a transcription factor of JNK2, which leads to decreased transcriptional process of JNK2 gene.
In contrast to a great reduction of JNK protein, unexpectedly, phosphorylated JNK forms in MsrB1 knockout mice remain in a high level that is similar to wild-type mice, leading to significantly increased p-JNK/JNK levels versus wild-type mice. These results suggest that MsrB1 deficiency induces activation of JNK protein in skeletal muscles and livers to compensate for reduced JNK protein levels, and that this increased basal JNK protein activation may be necessary to maintain homeostasis of JNK signaling pathway. However, no direct or indirect interaction between JNK and MsrB1 has been demonstrated. As mentioned above, reversible oxidation and reduction of methionine is a protein modification to regulate protein activity [39]. In particular, MsrB1 regulates F-actin dynamics by reversing the Mical-mediated oxidation of actin protein [40,41]. It would be interesting to investigate how MsrB1 is involved in JNK protein activation.
Since MsrB1 depletion does not aggravate insulin resistance induced by a high-fat diet, increased JNK phosphorylation based on a p-JNK/JNK level in MsrB1-deficient mice seems to be independent of high-fat diet-induced insulin resistance. However, due to the very low protein levels of JNK found in MsrB1 knockout mice, JNK phosphorylation level was additionally normalized to an internal control protein GAPDH and would be considered as another surrogate of total JNK activity. On this basis, JNK phosphorylation in skeletal muscle was found to be similar in wild-type and MsrB1 knockout mice fed a chow diet (Fig. 7A, lower graph). The high-fat diet significantly increased JNK phosphorylation, but no significant difference was observed between wild-type and MsrB1 knockout mice (Fig. 7A, lower graph). Thus, this JNK phosphorylation based on a p-JNK/GAPDH level might explain the correlation of JNK activation with insulin resistance in MsrB1-deficient mice.
Unlike skeletal muscle, the association between JNK phosphorylation based on a p-JNK/GAPDH level and insulin resistance did not match well in liver. JNK phosphorylation level normalized to GAPDH was not different in wild-type and MsrB1 knockout mice in the chow-fed animals (Fig. 7D, lower graph). Whereas the high-fat diet increased JNK phosphorylation level in wild-type mice, it did not affect JNK phosphorylation in MsrB1 knockout mice (Fig. 7D, lower graph), although insulin sensitivity was similarly impaired after high-fat diet in MsrB1 knockout and wild-type mice. Inconsistent data on the correlation of JNK activity with hepatic insulin resistance have been reported previously [25-27].
In conclusion, the present study shows MsrB1 deficiency does not affect insulin resistance induced by a high-fat diet in mice, which contrasts with the protective effect of MsrA on insulin resistance. Based on the findings of the present and previous studies, it appears that methionine-S-sulfoxide reduction plays an important role in insulin resistance and that methionine-R-sulfoxide reduction does not. However, to clarify whether stereospecific methionine sulfoxide reduction is involved in insulin resistance, further studies are needed to investigate the roles of the other two MsrB isozymes, MsrB2 and MsrB3.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea (2014R1A1A2057525 and 2015R1A5A2009124), and the Korean Health Technology R&D Project, Korean Ministry of Health & Welfare (A111345).
This work was supported by National Institutes of Health (AG021518).
Footnotes
Declaration of interest: The authors report no declarations of interest.
References
- 1.Eckel RH, Kahn SE, Ferrannini E, Goldfine AB, Nathan DM, Schwartz MW, Smith RJ, Smith SR, Endocrine S, American Diabetes A, et al. Obesity and type 2 diabetes: what can be unified and what needs to be individualized? Diabetes Care. 2011;34(6):1424–30. doi: 10.2337/dc11-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106(4):473–81. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torres SH, De Sanctis JB, de LBM, Hernandez N, Finol HJ. Inflammation and nitric oxide production in skeletal muscle of type 2 diabetic patients. J Endocrinol. 2004;181(3):419–27. doi: 10.1677/joe.0.1810419. [DOI] [PubMed] [Google Scholar]
- 4.Furukawa S, Fujita T, Shimabukuro M, Iwaki M, Yamada Y, Nakajima Y, Nakayama O, Makishima M, Matsuda M, Shimomura I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J Clin Invest. 2004;114(12):1752–61. doi: 10.1172/JCI21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson EJ, Lustig ME, Boyle KE, Woodlief TL, Kane DA, Lin CT, Price JW, 3rd, Kang L, Rabinovitch PS, Szeto HH, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009;119(3):573–81. doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boden MJ, Brandon AE, Tid-Ang JD, Preston E, Wilks D, Stuart E, Cleasby ME, Turner N, Cooney GJ, Kraegen EW. Overexpression of manganese superoxide dismutase ameliorates high-fat diet-induced insulin resistance in rat skeletal muscle. Am J Physiol Endocrinol Metab. 2012;303(6):E798–805. doi: 10.1152/ajpendo.00577.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta. 2005;1703(2):203–12. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Hansel A, Heinemann SH, Hoshi T. Heterogeneity and function of mammalian MSRs: enzymes for repair, protection and regulation. Biochim Biophys Acta. 2005;1703(2):239–47. doi: 10.1016/j.bbapap.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Kim HY, Gladyshev VN. Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem J. 2007;407(3):321–9. doi: 10.1042/BJ20070929. [DOI] [PubMed] [Google Scholar]
- 10.Vougier S, Mary J, Friguet B. Subcellular localization of methionine sulphoxide reductase A (MsrA): evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem J. 2003;373(Pt 2):531–7. doi: 10.1042/BJ20030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim G, Cole NB, Lim JC, Zhao H, Levine RL. Dual sites of protein initiation control the localization and myristoylation of methionine sulfoxide reductase A. J Biol Chem. 2010;285(23):18085–94. doi: 10.1074/jbc.M110.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HY, Gladyshev VN. Role of structural and functional elements of mouse methionine-S-sulfoxide reductase in its subcellular distribution. Biochemistry. 2005;44(22):8059–67. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- 13.Kim HY. The methionine sulfoxide reduction system: selenium utilization and methionine sulfoxide reductase enzymes and their functions. Antioxid Redox Signal. 2013;19(9):958–69. doi: 10.1089/ars.2012.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15(3):1055–64. doi: 10.1091/mbc.E03-08-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oien DB, Osterhaus GL, Latif SA, Pinkston JW, Fulks J, Johnson M, Fowler SC, Moskovitz J. MsrA knockout mouse exhibits abnormal behavior and brain dopamine levels. Free Radic Biol Med. 2008;45(2):193–200. doi: 10.1016/j.freeradbiomed.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc Natl Acad Sci U S A. 2001;98(23):12920–5. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JI, Choi SH, Jung KJ, Lee E, Kim HY, Park KM. Protective role of methionine sulfoxide reductase A against ischemia/reperfusion injury in mouse kidney and its involvement in the regulation of trans-sulfuration pathway. Antioxid Redox Signal. 2013;18(17):2241–50. doi: 10.1089/ars.2012.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, Sun J, Deschamps AM, Kim G, Liu C, Murphy E, Levine RL. Myristoylated methionine sulfoxide reductase A protects the heart from ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2011;301(4):H1513–8. doi: 10.1152/ajpheart.00441.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Styskal J, Nwagwu FA, Watkins YN, Liang H, Richardson A, Musi N, Salmon AB. Methionine sulfoxide reductase A affects insulin resistance by protecting insulin receptor function. Free Radic Biol Med. 2013;56:123–32. doi: 10.1016/j.freeradbiomed.2012.10.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novoselov SV, Kim HY, Hua D, Lee BC, Astle CM, Harrison DE, Friguet B, Moustafa ME, Carlson BA, Hatfield DL, et al. Regulation of selenoproteins and methionine sulfoxide reductases A and B1 by age, calorie restriction, and dietary selenium in mice. Antioxid Redox Signal. 2010;12(7):829–38. doi: 10.1089/ars.2009.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fomenko DE, Novoselov SV, Natarajan SK, Lee BC, Koc A, Carlson BA, Lee TH, Kim HY, Hatfield DL, Gladyshev VN. MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J Biol Chem. 2009;284(9):5986–93. doi: 10.1074/jbc.M805770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon MJ, Ju TJ, Heo JY, Kim YW, Kim JY, Won KC, Kim JR, Bae YK, Park IS, Min BH, et al. Deficiency of clusterin exacerbates high-fat diet-induced insulin resistance in male mice. Endocrinology. 2014;155(6):2089–101. doi: 10.1210/en.2013-1870. [DOI] [PubMed] [Google Scholar]
- 23.Song SE, Kim YW, Kim JY, Lee DH, Kim JR, Park SY. IGFBP5 mediates high glucose-induced cardiac fibroblast activation. J Mol Endocrinol. 2013;50(3):291–303. doi: 10.1530/JME-12-0194. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Seok YM, Jung KJ, Park KM. Reactive oxygen species/oxidative stress contributes to progression of kidney fibrosis following transient ischemic injury in mice. Am J Physiol Renal Physiol. 2009;297(2):F461–70. doi: 10.1152/ajprenal.90735.2008. [DOI] [PubMed] [Google Scholar]
- 25.Hussey SE, Lum H, Alvarez A, Cipriani Y, Garduno-Garcia J, Anaya L, Dube J, Musi N. A sustained increase in plasma NEFA upregulates the Toll-like receptor network in human muscle. Diabetologia. 2014;57(3):582–91. doi: 10.1007/s00125-013-3111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanti JF, Jager J. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr Opin Pharmacol. 2009;9(6):753–62. doi: 10.1016/j.coph.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Sabio G, Cavanagh-Kyros J, Ko HJ, Jung DY, Gray S, Jun JY, Barrett T, Mora A, Kim JK, Davis RJ. Prevention of steatosis by hepatic JNK1. Cell Metab. 2009;10(6):491–8. doi: 10.1016/j.cmet.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia Y, Li Y, Du S, Huang K. Involvement of MsrB1 in the regulation of redox balance and inhibition of peroxynitrite-induced apoptosis in human lens epithelial cells. Exp Eye Res. 2012;100:7–16. doi: 10.1016/j.exer.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Shchedrina VA, Kabil H, Vorbruggen G, Lee BC, Turanov AA, Hirosawa-Takamori M, Kim HY, Harshman LG, Hatfield DL, Gladyshev VN. Analyses of fruit flies that do not express selenoproteins or express the mouse selenoprotein, methionine sulfoxide reductase B1, reveal a role of selenoproteins in stress resistance. J Biol Chem. 2011;286(34):29449–61. doi: 10.1074/jbc.M111.257600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunnicut J, Liu Y, Richardson A, Salmon AB. MsrA Overexpression Targeted to the Mitochondria, but Not Cytosol, Preserves Insulin Sensitivity in Diet-Induced Obese Mice. PLoS One. 2015;10(10):e0139844. doi: 10.1371/journal.pone.0139844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Haan JB, Witting PK, Stefanovic N, Pete J, Daskalakis M, Kola I, Stocker R, Smolich JJ. Lack of the antioxidant glutathione peroxidase-1 does not increase atherosclerosis in C57BL/J6 mice fed a high-fat diet. J Lipid Res. 2006;47(6):1157–67. doi: 10.1194/jlr.M500377-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Lewis P, Stefanovic N, Pete J, Calkin AC, Giunti S, Thallas-Bonke V, Jandeleit-Dahm KA, Allen TJ, Kola I, Cooper ME, et al. Lack of the antioxidant enzyme glutathione peroxidase-1 accelerates atherosclerosis in diabetic apolipoprotein E-deficient mice. Circulation. 2007;115(16):2178–87. doi: 10.1161/CIRCULATIONAHA.106.664250. [DOI] [PubMed] [Google Scholar]
- 33.Szczesny B, Tann AW, Mitra S. Age- and tissue-specific changes in mitochondrial and nuclear DNA base excision repair activity in mice: Susceptibility of skeletal muscles to oxidative injury. Mech Ageing Dev. 2010;131(5):330–7. doi: 10.1016/j.mad.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng ZX, Wang L, Xiao Y, Lin JD. The Baf60c/Deptor pathway links skeletal muscle inflammation to glucose homeostasis in obesity. Diabetes. 2014;63(5):1533–45. doi: 10.2337/db13-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jager J, Corcelle V, Gremeaux T, Laurent K, Waget A, Pages G, Binetruy B, Le Marchand-Brustel Y, Burcelin R, Bost F, et al. Deficiency in the extracellular signal-regulated kinase 1 (ERK1) protects leptin-deficient mice from insulin resistance without affecting obesity. Diabetologia. 2011;54(1):180–9. doi: 10.1007/s00125-010-1944-0. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3(3):211–22. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Drazic A, Miura H, Peschek J, Le Y, Bach NC, Kriehuber T, Winter J. Methionine oxidation activates a transcription factor in response to oxidative stress. Proc Natl Acad Sci U S A. 2013;110(23):9493–8. doi: 10.1073/pnas.1300578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallmetzer A, Silvestrini L, Schinko T, Gesslbauer B, Hortschansky P, Dattenbock C, Muro-Pastor MI, Kungl A, Brakhage AA, Scazzocchio C, et al. Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA. PLoS Genet. 2015;11(7):e1005297. doi: 10.1371/journal.pgen.1005297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drazic A, Winter J. The physiological role of reversible methionine oxidation. Biochim Biophys Acta. 2014;1844(8):1367–82. doi: 10.1016/j.bbapap.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Lee BC, Peterfi Z, Hoffmann FW, Moore RE, Kaya A, Avanesov A, Tarrago L, Zhou Y, Weerapana E, Fomenko DE, et al. MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation. Mol Cell. 2013;51(3):397–404. doi: 10.1016/j.molcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol. 2013;15(12):1445–54. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.