Abstract
Purpose
We evaluated the accuracy of magnetic resonance imaging in determining the size and shape of localized prostate cancer.
Materials and Methods
The subjects were 114 men who underwent multi-parametric magnetic resonance imaging before radical prostatectomy with patient specific mold processing of the specimen from 2013 to 2015. T2-weighted images were used to contour the prostate capsule and cancer suspicious regions of interest. The contours were used to design and 3-dimentional print custom molds, which permitted alignment of excised prostates with magnetic resonance imaging scans. Tumors were reconstructed in 3 dimensions from digitized whole mount sections. Tumors were then matched with regions of interest and the relative geometries were compared.
Results
Of the 222 tumors evident on whole mount sections 118 had been identified on magnetic resonance imaging. For the 118 regions of interest mean volume was 0.8 cc and the longest 3-dimensional diameter was 17 mm. However, for matched pathological tumors, of which most were Gleason score 3 + 4 or greater, mean volume was 2.5 cc and the longest 3-dimensional diameter was 28 mm. The median tumor had a 13.5 mm maximal extent beyond the magnetic resonance imaging contour and 80% of cancer volume from matched tumors was outside region of interest boundaries. Size estimation was most accurate in the axial plane and least accurate along the base-apex axis.
Conclusions
Magnetic resonance imaging consistently underestimates the size and extent of prostate tumors. Prostate cancer foci had an average diameter 11 mm longer and a volume 3 times greater than T2-weighted magnetic resonance imaging segmentations. These results may have important implications for the assessment and treatment of prostate cancer.
Keywords: prostatic neoplasms, magnetic resonance imaging, diagnostic errors, pathology, surgical, image processing, computer-assisted
Multiparametric MRI has become a valuable tool in the diagnosis of CaP.1 Although most large, high grade cancers are visible on mpMRI, intermediate grade and low volume cancers are often difficult to identify.2 Furthermore, the usefulness of MRI for determining the true size and shape of a tumor remains incompletely characterized.
Prior studies evaluating the accuracy of prostate MRI have largely relied on subjective methods to correlate whole mount histopathology slides with MRI. Since the gland is conventionally sliced posterior side down, the cutting plane may not reflect the imaging plane. Furthermore, prostate shape can be deformed by imaging coils, surgical resection and histopathology tissue processing.3–5 Even when image analysis software is used,6,7 the histological location relative to MRI can be confounded by variability in specimen sectioning.5,8 Thus, whole mount pathology slides may depict a different depth, angle and prostate shape from in vivo images.
In an attempt to improve registration accuracy guides or templates have been used to acquire uniform slices8–10 but this approach does not assure correct orientation of the specimen. Ex vivo MRI of specimens has also been done but only in a small number of cases.11,12 Turkbey et al first described the use of 3D printed, patient specific molds,13,14 which hold excised prostates in the same shape and orientation observed in vivo. Some prior work indicates that MRI provides an overestimate of tumor volume,15,16 although these studies were based on older technologies and did not use software registration. More recent publications indicate the opposite, that is that actual tumor volume may be up to 150% of the MRI estimate.14,17,18 Due to variations in imaging technology, contouring procedure and data analysis, available volume correlation studies have yielded conflicting results.
Nearly all prior attempts to define MRI/pathological relationships have relied on imprecise techniques such as manual registration, volume approximation and 2-dimensional measurements. In the current study we used patient specific molds to facilitate precise 3D co-registration of pathological tumors to MRI. This enabled a rigorous exploration of the relationship between MRI and pathological measurements of tumor size, shape and extent.
MATERIALS AND METHODS
Subjects were eligible for this institutional review board approved study if they underwent mpMRI within 6 months of surgery, were treated with radical prostatectomy at UCLA between August 2013 and November 2015, whole mount specimen processing was done with a custom printed mold and MRI predicted prostate volume was 110 cc (mold limit) or less. Data were prospectively acquired and retrospectively analyzed. During the study period 114 subjects meeting study criteria were consecutively accrued.
Magnetic Resonance Imaging
mpMRI studies were performed in a 3 Tesla Trio, Verio or Skyra scanner (Siemens, Erlangen, Germany). A transabdominal phased array was used in all cases and an endorectal coil was used in 47%. The scanning protocol was previously described.19 It included T1-weighted, high resolution 2-dimensional and 3D T2-weighted, diffusion-weighted with apparent diffusion coefficient and dynamic contrast enhanced sequences. In advance of prostatectomy a single fellowship trained genitourinary radiologist (DJM or SSR) with at least 10 years of experience reviewed the mpMRI series. While blinded to patient history and prior imaging, the radiologist identified each ROI, ie an image region suspicious for cancer. All ROIs were assigned an overall suspicion score between 2 and 5 based on a Likert-like scale predating and similar to PI-RADS™.19 Using ProFuse software (Eigen, Grass Valley, California) the prostate capsule and ROIs were manually contoured on T2-weighted images (fig. 1, A). Motion artifact was not found to influence ROI contouring in the current study.
Figure 1.
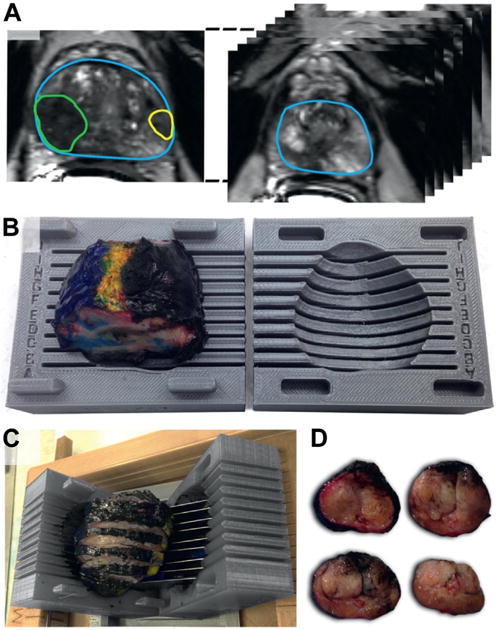
Design and use of patient specific mold. A, prostate capsule and ROIs on high resolution T2-weighted MRI. B, 3D printed mold with excised and inked prostate inside mold cavity. C, prostate sliced in 4.5 mm increments. D, prostate sections prior to fixation.
Histopathology Analysis
A mold was 3D printed prior to surgery based on the T2-weighted MRI prostate capsule (fig. 1, B). This mold was customized for each patient using SolidWorks® 2014 and Matlab® 2015. Molds were printed at 200 mm resolution using a consumer grade 3D printer (Makerbot Industries, Brooklyn, New York). The average mold required 15 minutes to design, 6 hours to print and an expense for materials of less than $4.00.
Each specimen was inked and trimmed before placement in the mold. Prostates were sliced at 4.5 mm intervals in the axial orientation from base to apex, corresponding to every third 1.5 mm T2-weighted MRI scan (fig. 1, C and D). After sectioning, the tissue slices were fixed in formalin, paraffin embedded, processed into slides and stained with hematoxylin and eosin. On each section regions of tumor, defined as disease foci seen on whole mount pathology, were contoured by a urological pathologist (JH) blinded to mpMRI findings (fig. 2, A). The use of elastic registration in this study obviated the need to adjust for formalin induced shrinkage, as detailed previously.20 Tumors were automatically reconstructed in 3D, matched to ROIs and overlaid with MRI (fig. 2, B and C).
Figure 2.
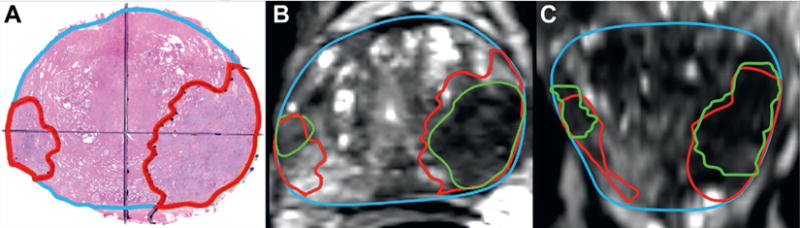
Example of registration of tumors (red outlines) to mpMRI ROI (green outlines). A, whole mount prostate section with 2 tumors delineated. B, tumor contours registered to corresponding axial MRI with 2 ROIs. C, tumor contours overlaid on coronal MRI. Note underestimation of actual tumor size by ROI.
Data Analysis
We recorded the volume, longest 3D diameter and position of all ROIs and tumors. For matched tumor/ROI pairs we measured the center offset, overlap volume and HDmax,18 the latter defined as the greatest distance between tumor and ROI surfaces. Two groups of tumors were considered potential drivers of clinical progression,21,22 including 1) index lesions, defined as the tumor with the highest GS or secondarily as the tumor with the greatest volume and 2) csCaP, defined as GS 3 + 4 or greater, or volume more than 0.5 cc.23,24
The Wilcoxon signed rank test was used for paired samples and the Mann-Whitney U test was used for unpaired samples. When comparing more than 2 groups, Kruskal-Wallis 1-way ANOVA and the post hoc Mann-Whitney U test were performed. The Pearson product-moment coefficient was used for linear correlations and the chi-square test was used to compare sensitivity. Statistical significance was considered at p <0.05.
RESULTS
In 114 men a total of 148 ROIs were present on mpMRI and 222 tumors were present on pathology evaluation. Table 1 lists patient characteristics. The excised whole prostates (mean 43 cc) were on average 7% larger than predicted on T2-weighted MRI (mean 40 cc, p <0.001). The longest prostate diameter was not significantly different than predicted by MRI (mean ± SD 50.2 ± 0.6 and 50.8 ± 0.6 mm, respectively).
Table 1.
Characteristics of 114 patients, 148 ROIs on MRI and 222 tumors seen pathologically
| Median age (IQR) | 62 | (56–67) |
| Median (ng/ml) PSA (IQR) | 6.5 | (4.6–8.7) |
| Median cc pathological prostate vol (IQR) | 39 | (32–51) |
| No. ethnicity (%): | ||
| White | 88 | (77) |
| African American | 8 | (7) |
| Asian, Hispanic or other | 18 | (16) |
| No. mpMRI ROI score (%):* | ||
| 3 or Less | 58 | (39) |
| 4 | 51 | (34) |
| 5 | 39 | (26) |
| No. final pathology GS (%): | ||
| 3 + 3 or Less | 99 | (46) |
| 3 + 4 | 83 | (37) |
| 4 + 3 or Greater | 38 | (17) |
UCLA score16 was concordant with PI-RADS in most cases (D.M. Margolis, unpublished data).
Table 2 shows the performance of mpMRI for detecting CaP. MRI detected 53% of tumors and 82% of index tumors. Missed tumors had a smaller volume (0.35 vs 2.5 cc, p <0.001) and a shorter diameter (16 vs 28 mm, p <0.001) than mpMRI visible tumors. Missed tumors were also lower grade than matched tumors (p <0.001). Of the missed tumors 73% were GS 3 + 3 whereas 79% of matched tumors were GS 3 + 4 or greater. Of 118 matched tumors 86 (73%) were primarily in the peripheral zone and 32 (27%) were primarily in the transition zone. The detection rate (sensitivity) of mpMRI was 54% for peripheral tumors and 49% for transition zone tumors (p not significant).
Table 2.
MRI predictive accuracy
| All CaP | csCaP | Index CaP | |
|---|---|---|---|
| No. tumors | 222 | 141 | 114 |
| No./total No. (%): | |||
| Sensitivity | 118/222 (53) | 107/141 (76) | 93/114 (82) |
| Specificity | 118/148 (80) | 107/148 (72) | 93/109 (85) |
| mpMRI invisible lesions | 63/114 (55) | 30/114 (26) | 20/114 (18) |
Tumor Size (Region of Interest vs Matched Pathology)
For the 118 matches mean tumor volume was 2.5 ± 0.3 cc, 3 times greater than the mean ROI volume of 0.8 ± 0.1 cc (table 3). Tumor volumes correlated with ROI volumes (r = 0.50, p <0.01, fig. 3, A). Bland-Altman analysis revealed that the magnitude of the disparity between MRI and tumor findings was directly related to tumor volume (p <0.01). Less than 0.5 cc (20%) of tumor volume was encompassed within the ROIs (table 4). These relationships held for csCaP and index tumors.
Table 3.
Size of tumors and matched ROIs
| Mean ± SD All Matches | Mean ± SD Gleason Score
|
p Value (3 + 4 vs 4 + 3 or greater) | |||
|---|---|---|---|---|---|
| 3 + 3 | 3 + 4 | 4 + 3 or Greater | |||
| No. tumors | 118 | 22 | 61 | 32 | – |
| Vol (cc): | |||||
| Tumor | 2.49 ± 0.26 | 1.1 ± 0.5 | 2.6 ± 0.3 | 3.2 ± 0.5 | 0.57 |
| ROI | 0.84 ± 0.11 | 0.31 ± 0.07 | 0.7 ± 0.1 | 1.2 ± 0.2 | 0.01 |
| Diameter (mm): | |||||
| Tumor | 28.4 ± 0.9 | 19.2 ± 2.3 | 30.0 ± 1.1 | 31.9 ± 1.3 | 0.38 |
| ROI | 17.0 ± 0.7 | 12.8 ± 1.0 | 16.5 ± 0.9 | 20.5 ± 1.3 | 0.01 |
| Max Hausdorff distance (mm) | 14.8 ± 0.7 | 10.9 ± 1.5 | 16.3 ± 0.9 | 15.2 ± 1.0 | 0.54 |
Tumor size exceeded ROI size in each Gleason score category and overall (p <0.05) andGleason scores significantly differed (p <0.05) but no difference was seen in tumor size when comparing Gleason scores 3 + 4 and 4 + 3 or greater.
Figure 3.
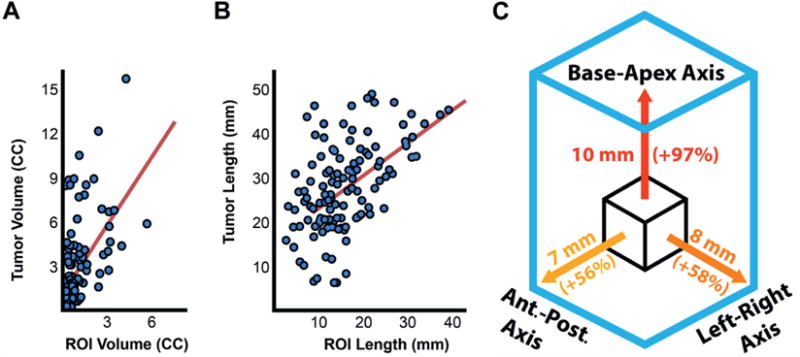
Size comparison between ROIs and tumors. A, MIR vs tumor volume. B, MRI vs pathology longest diameter. Red diagonal line indicates least squares trend line. C, mean difference in length and percent increase between ROIs and matched tumors along BA, AP (Ant.-Post.) and LR axes. For example, along BS diameter ROI would need to be expanded average of 10 mm or 97% to match actual tumor length.
Table 4.
Relationship between tumor location and matched ROIs
| All Tumors | Clinically Significant CaP | Index Tumor | |
|---|---|---|---|
| No. tumors | 118 | 107 | 94 |
| Mean ± SD overlap volume (cc)/% | 0.47 ± 0.7/19 | 0.52 ± 1.1/19 | 0.58 ± 0.8/20 |
| Mean ± SD center offset (mm) | 6.6 ± 0.3 | 6.5 ± 0.3 | 6.6 ± 0.3 |
| Mean ± SD max Hausdorff distance (mm): | 14.8 ± 0.7 | 15.6 ± 0.6 | 15.8 ± 0.6 |
| Base-apex | 9.4 ± 0.6 | 9.8 ± 0.6 | 9.9 ± 0.6 |
| Anteroposterior | 7.2 ± 0.6 | 7.7 ± 0.6 | 7.7 ± 0.6 |
| Lt-rt | 7.4 ± 0.6 | 7.8 ± 0.6 | 7.9 ± 0.6 |
No significant differences were observed between tumor groups.
Tumor diameter (eg the longest 3D axis) was underestimated by an average of 11 mm on T2-weighted MRI (p <0.001, fig 3, B). It was misaligned from ROI diameter by a median of 46 degrees. The median tumor extended 13.5 mm beyond the ROI contour (fig. 4). ROI diameter was directly related to the diameter of GS 4 + 3 or greater (r = 0.67) and GS 3 + 4 tumors (r = 0.46, each p <0.01). However, no significant correlation was found between GS 3 + 3 tumors and ROI diameter. Volume correlations followed a similar trend.
Figure 4.
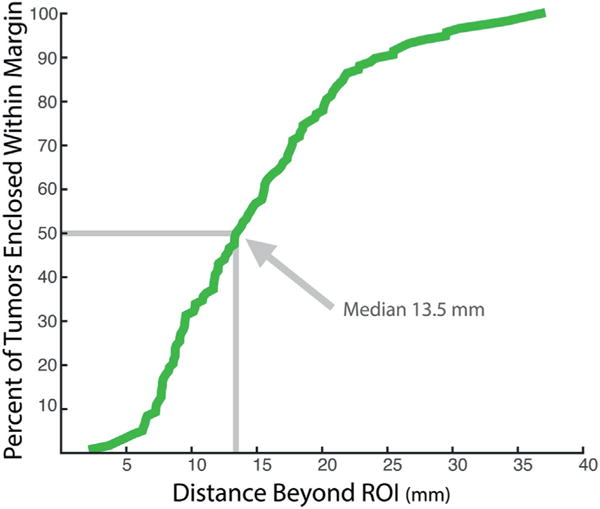
Frequency distribution shows MRI underestimation of tumor extent with percent of tumors extending beyond ROI surface when uniform margin was applied. With greater distance beyond ROI margin, percent of tumor enclosed within that distance was greater and 50% of tumors extended more than 13.5 mm beyond ROI.
Tumors were significantly larger than ROIs for all Gleason scores (p <0.001, table 3). GS 3 + 3 tumors were smaller than GS 3 + 4cHDmax values and less volume error (each p <0.01). The volume and diameter of ROIs were directly related to Gleason score (p = 0.01). However, no difference in tumor volume, diameter or HDmax was seen between GS 3 + 4 tumors and higher grade tumors.
The mean volume of transition zone tumors was greater than mean peripheral zone tumor volume (3.6 ± 0.7 vs 1.8 ± 0.2 cc, p = 0.01). Transition zone tumors were an average of 2.5 ± 0.7 cc greater than matched ROIs whereas peripheral zone tumors were only a mean of 1.0 ± 0.3 cc greater than matched ROIs (p <0.05). The longest diameter and HDmax values did not significantly differ between transition and peripheral zone tumors.
On separate analysis we found that using an endorectal vs an abdominal coil was not a significant factor in ROI size estimation.
Region of Interest Size Variation by Axis
Tumors were longer than ROIs on all anatomical axes, including BA, AP and LR (table 4). BA mean tumor length was almost double the mean ROI length (20.2 vs 10.1 mm), which was a greater error than that of the AP and the LR dimensions (p <0.001, fig. 3, C). ROI and tumor length better correlated along the AP (r = 0.43) and LR axes (r = 0.58, each p <0.01) than along the BA axis (r = 0.22, p <0.05). HDmax was also greatest along the BA axis at a mean of 9.4 mm (p <0.01).
DISCUSSION
The aim of our study was to determine the accuracy of MRI for predicting the location, size and shape of prostate tumors. We compared preoperative MRI to postoperative specimens using co-registration software and patient specific prostate molds in 114 men. The results indicate that T2-weighted MRI contours underestimated the volume, longest axis and extent of pathological tumors in nearly every case. Underestimation occurred for all Gleason scores and for all mpMRI suspicion levels.
More than half of the tumors were detected on mpMRI and detection rates were high for clinically significant and index disease. However, the average tumor diameter was underestimated by more than 1 cm. Due to differences in center, size and shape, tumors extended substantially beyond the ROI surface (median HDmax 13.5 cm). A 5 or 10 mm margin around the ROI is often cited as sufficient for focal treatment18,25 but our analysis indicates that tumor extent was underestimated by more than 10 mm in two-thirds of all cases. Thus, margins substantially wider than the ROI confines would be required for complete extirpation of a MRI visible tumor.
No significant difference in size was found between Gleason 3 + 4 and Gleason 4 + 3 or greater tumors. For example, average tumor diameter was 30 and 31.9 mm (p = not significant) but ROI diameter was 16.5 and 20.5 mm, respectively (p <0.01, table 3). Therefore, even if 2 tumors were the same size, the corresponding ROI would be larger and more accurate for the high grade tumor than for the lower grade tumor. This suggests that, in addition to suspicion score, ROI size may be a useful metric for evaluating disease severity.
MRI accuracy was highest in the axial plane and worst along the BA axis. Tumor measurements along the BA axis were truncated by the 4.5 mm spacing between tissue slices. Thus, tumors may be even larger and tumor underestimation may be more severe along this axis. ROI contouring was performed in the axial plane, which may explain the higher accuracy of axial measurements. Improving through-plane resolution or contouring on multiple image planes may help address this limitation.
Recently, groups have reported lesser degrees of tumor size underestimation. Turkbey et al noted a mean index volume underestimate of 0.16 cc (7%) and a correlation coefficient of 0.63.14 Although the strength of correlation was comparable to that in our series, the ROIs in the study by Turkbey et al were larger and they matched tumor volume more closely. However, the accuracy of tumor volume measurements was limited by inconsistent use of 3D molds as well as use of the ellipsoid formula, which does not account for actual tumor shape.
Le Nobin et al reported tumor volumes 1.5 times greater than ROIs and no HDmax greater than 9 mm.17,18 Their patients had a mean tumor volume of 1.38 cc compared to 2.5 cc in the current study. In addition, their work was restricted to a small number of men (33 and 37) who were retrospectively selected. Also, radiologists were informed of tumor location prior to ROI delineation. Perhaps most importantly, the investigators relied exclusively on in-plane measurements between MRI and pathology findings, whereas we observed that the largest errors were out of plane. Thus, the current work appears to more accurately reflect the accuracy of prostate MRI for predicting tumor size.
Some underestimation may be inevitable due to tumor heterogeneity and blending with healthy tissue.26 However, several steps can be taken to improve the accuracy of tumor contours. 1) Greater emphasis should be placed on segmentation with multiple image planes and mpMRI sequences used to aggressively contour MRI visible lesions. 2) Image processing software can be used to inform radiologists which regions are most likely to harbor cancer and contours can be modified accordingly.27,28 3) Targeted biopsy systems are able to confirm whether cancer is present along multiple vectors in the prostate gland.19 Registration of biopsy cores with MRI and adjustment of tumor contours in response have been shown to improve the characterization of cancer geometry.29
Interventions such as targeted biopsy and focal therapy can compensate for tumor size underestimation. During MRI-fusion biopsy additional cores can be taken around the outside of each ROI, allowing for more accurate estimation of tumor extent. During focal prostate therapy generous margins should be treated around the ROI, especially along the BA axis, where size underestimation appears to be greatest. Treatment margins can be made patient specific, accounting for ROI size, Gleason score, zonal anatomy and tracked biopsy results.
The current study has several limitations. 1) It is difficult to assess the error associated with transforming pathology tumor contours into MRI coordinates. Differences between the in vivo and the ex vivo prostate could have resulted in registration errors but prostate size was accurately predicted on T2-weighted MRI and the fit in patient specific molds was excellent. 2) Our study comprised patients treated with radical prostatectomy. Thus, generalizability to patients who do not undergo the operation is unknown. 3) ROI contours were made on the T2-weighted image only but segmenting on multiple co-registered sequences may improve accuracy.13,15,17 Incorporating additional sequences is complicated by geometric distortion and patient motion but advances in series registration may enable improvements in ROI delineation. 4) Interpretation of mpMRI and reporting guidelines have been tailored for suspicion grading and the guidance of targeted biopsy rather than for guiding targeted treatment.30 If mpMRI interpretation were amended to include greater emphasis on segmentation, ROI accuracy would likely improve. 5) Whole mount pathology contours did not account for blending between cancerous and benign tissue or distinguish between regions of high and low Gleason patterns in a tumor. T2-weighted images may reveal regions with high grade disease more accurately than regions with low grade disease or intermixed benign tissue.
Addressing these limitations and improving the accuracy of mpMRI tumor size characterization will be a future focus of our research.
CONCLUSIONS
Among 118 MRI visible tumors ROIs underestimated tumor length by an average of 11 mm and underestimated volume approximately threefold. The median tumor extended 13.5 mm beyond the ROI contour. Of index tumors 82% were identified on mpMRI but the size and shape of almost every tumor was underestimated on T2-weighted imaging.
Acknowledgments
Nima Nassiri, Shellee Ogawa, Jesse Le, James Garritano, Haerin Lee, Bryan Radosavcev, William Hsu and Brenda Brown assisted with study design, and data management and analysis.
Supported by National Cancer Institute Award R01CA158627, Prostate Cancer SPORE at University of California-Los Angeles P50CA092131, the Beckman Coulter Foundation, Jean Perkins Foundation and Steven C. Gordon Family Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Abbreviations and Acronyms
- 3D
3-dimensional
- AP
anteroposterior
- BA
base-apex
- CaP
prostate cancer
- csCaP
clinically significant CaP
- GS
Gleason score
- HDmax
maximum Hausdorff distance
- LR
left-right
- mpMRI
multiparametric MRI
- MRI
magnetic resonance imaging
- ROI
region of interest
Footnotes
The corresponding author certifies that, when applicable, a statement(s) has been included in the manuscript documenting institutional review board, ethics committee or ethical review board study approval; principles of Helsinki Declaration were followed in lieu of formal ethics committee approval; institutional animal care and use committee approval; all human subjects provided written informed consent with guarantees of confidentiality; IRB approved protocol number; animal approved project number.
References
- 1.Hegde JV, Mulkern RV, Panych LP, et al. Multi-parametric MRI of prostate cancer: an update on state-of-the-art techniques and their performance in detecting and localizing prostate cancer. J Magn Reson Imaging. 2013;37:1035. doi: 10.1002/jmri.23860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan N, Margolis DJ, Lu DY, et al. Characteristics of detected and missed prostate cancer foci on 3-T multiparametric MRI using an endorectal coil correlated with whole-mount thin-section histopathology. Am J Roentgenol. 2015;205:W87. doi: 10.2214/AJR.14.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim Y, Hsu ICJ, Pouliot J, et al. Expandable and rigid endorectal coils for prostate MRI: impact on prostate distortion and rigid image registration. Med Phys. 2005;32:3569. doi: 10.1118/1.2122467. [DOI] [PubMed] [Google Scholar]
- 4.Meyer C, Ma B, Kunju LP, et al. Challenges in accurate registration of 3-D medical imaging and histopathology in primary prostate cancer. Eur J Nucl Med Mol Imaging. 2013;40:72. doi: 10.1007/s00259-013-2382-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenster A, Ward A, Crukley C, et al. 3D prostate histology image reconstruction: quantifying the impact of tissue deformation and histology section location. J Pathol Inform. 2013;4:31. doi: 10.4103/2153-3539.120874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappelow J, Bloch BN, Rofsky N, et al. Elastic registration of multimodal prostate MRI and histology via multiattribute combined mutual information. Med Phys. 2011;38:2005. doi: 10.1118/1.3560879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park H, Piert MR, Khan A, et al. Registration methodology for histological sections and in vivo imaging of human prostate. Acad Radiol. 2008;15:1027. doi: 10.1016/j.acra.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto H, Nir D, Vyas L, et al. A workflow to improve the alignment of prostate imaging with whole-mount histopathology. Acad Radiol. 2014;21:1009. doi: 10.1016/j.acra.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 9.Drew B, Jones EC, Reinsberg S, et al. Device for sectioning prostatectomy specimens to facilitate comparison between histology and in vivo MRI. J Magn Reson Imaging. 2010;32:992. doi: 10.1002/jmri.22303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LH, Ho H, Lazaro R, et al. Optimum slicing of radical prostatectomy specimens for correlation between histopathology and medical images. Int J Comput Assist Radiol Surg. 2010;5:471. doi: 10.1007/s11548-010-0405-z. [DOI] [PubMed] [Google Scholar]
- 11.Reynolds HM, Williams S, Zhang A, et al. Development of a registration framework to validate MRI with histology for prostate focal therapy. Med Phys. 2015;42:7078. doi: 10.1118/1.4935343. [DOI] [PubMed] [Google Scholar]
- 12.Gibson E, Crukley C, Gaed M, et al. Registration of prostate histology images to ex vivo MR images via strand-shaped fiducials. J Magn Reson Imaging. 2012;36:1402. doi: 10.1002/jmri.23767. [DOI] [PubMed] [Google Scholar]
- 13.Turkbey B, Mani H, Shah V, et al. Multiparametric 3T prostate magnetic resonance imaging to detect cancer: Histopathological correlation using prostatectomy specimens processed in customized magnetic resonance imaging based molds. J Urol. 2011;186:1818. doi: 10.1016/j.juro.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turkbey B, Mani H, Aras O, et al. Correlation of magnetic resonance imaging tumor volume with histopathology. J Urol. 2012;188:1157. doi: 10.1016/j.juro.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazaheri Y, Hricak H, Fine S, et al. Prostate tumor volume measurement with combined T2-weighted imaging and diffusion-weighted MR: correlation with pathologic tumor volume. Radiology. 2009;252:449. doi: 10.1148/radiol.2523081423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaa A, Van De Thornbury JR, Barentsz J. Local staging of prostate cancer with endorectal MR imaging: correlation with histopathology. AJR Am J Roentgenol. 1996;166:845. doi: 10.2214/ajr.166.4.8610561. [DOI] [PubMed] [Google Scholar]
- 17.Le Nobin J, Orczyk C, Deng FM, et al. Prostate tumour volumes: evaluation of the agreement between magnetic resonance imaging and histology using novel co-registration software. BJU Int. 2014;114:E105. doi: 10.1111/bju.12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Nobin J, Rosenkrantz AB, Villers A, et al. Image guided focal therapy for magnetic resonance imaging visible prostate cancer: defining a 3-dimensional treatment margin based on magnetic resonance imaging histology co-registration analysis. J Urol. 2015;194:364. doi: 10.1016/j.juro.2015.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natarajan S, Marks LS, Margolis DJA, et al. Clinical application of a 3D ultrasound-guided prostate biopsy system. Urol Oncol Semin Orig Investig. 2011;29:334. doi: 10.1016/j.urolonc.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priester A, Natarajan S, Le JD, et al. A system for evaluating magnetic resonance imaging of prostate cancer using patient-specific 3D printed molds. Am J Clin Exp Urol. 2014;2:127. [PMC free article] [PubMed] [Google Scholar]
- 21.Wise AM, Stamey TA, McNeal JE, et al. Morphologic and clinical significance of multi-focal prostate cancers in radical prostatectomy specimens. Urology. 2002;60:264. doi: 10.1016/s0090-4295(02)01728-4. [DOI] [PubMed] [Google Scholar]
- 22.Epstein JI, Walsh PC, Carmichael M, et al. Pathologic and clinical findings to predict tumor extent of nonpalpable (stage T1c) prostate cancer. JAMA. 1994;271:368. [PubMed] [Google Scholar]
- 23.Stamey TA, Freiha FS, McNeal JE, et al. Localized prostate cancer. Relationship of tumor volume to clinical significance for treatment of prostate cancer. Cancer. 1993;71:933. doi: 10.1002/1097-0142(19930201)71:3+<933::aid-cncr2820711408>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed HU, Hu Y, Carter T, et al. Characterizing clinically significant prostate cancer using template prostate mapping biopsy. J Urol. 2011;186:458. doi: 10.1016/j.juro.2011.03.147. [DOI] [PubMed] [Google Scholar]
- 25.Groenendaal G, Moman MR, Korporaal JG, et al. Validation of functional imaging with pathology for tumor delineation in the prostate. Radiother Oncol. 2010;94:145. doi: 10.1016/j.radonc.2009.12.034. [DOI] [PubMed] [Google Scholar]
- 26.Rosenkrantz AB, Mendrinos S, Babb JS, et al. Prostate cancer foci detected on multiparametric magnetic resonance imaging are histologically distinct from those not detected. J Urol. 2012;187:2032. doi: 10.1016/j.juro.2012.01.074. [DOI] [PubMed] [Google Scholar]
- 27.Madabhushi A, Feldman MD, Metaxas DN, et al. Automated detection of prostatic adenocarcinoma from high-resolution ex vivo MRI. IEEE Trans Med Imaging. 2005;24:1611. doi: 10.1109/TMI.2005.859208. [DOI] [PubMed] [Google Scholar]
- 28.Langer DL, Van Der Kwast TH, Evans AJ, et al. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2009;30:327. doi: 10.1002/jmri.21824. [DOI] [PubMed] [Google Scholar]
- 29.Matsugasumi T, Baco E, Palmer S, et al. Prostate cancer volume estimation by combining magnetic resonance imaging and targeted biopsy proven cancer core length: Correlation with cancer volume. J Urol. 2015;194:957. doi: 10.1016/j.juro.2015.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinreb JC, Barentsz JO, Choyke PL, et al. PI-RADS Prostate Imaging-Reporting and Data System: 2015, version 2. Eur Urol. 2016;69:16. doi: 10.1016/j.eururo.2015.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]


