Abstract
Type 2 diabetes (T2D) and insulin resistance are associated with reduced glucose utilization in the muscle and poor exercise performance. Here we find that depletion of an epigenome modifier, histone deacetylase 3 (HDAC3), specifically in skeletal muscle causes severe systemic insulin resistance in mice, but markedly enhances exercise endurance and muscle fatigue resistance, despite reducing muscle force. This seemingly paradoxical phenotype is due to lower glucose utilization and greater lipid oxidation in HDAC3-depleted muscles, a fuel switch caused by the activation of anaplerotic reactions driven by AMP deaminase 3 (Ampd3) and branched-chain amino acid catabolism. These findings highlight the pivotal role of amino acid catabolism in muscle fatigue and T2D pathogenesis. Further, as genome occupancy of HDAC3 in skeletal muscle is controlled by the circadian clock, these results delineate an epigenomic regulatory mechanism through which the circadian clock governs skeletal muscle bioenergetics. These findings suggest that physical exercise at certain times of the day or pharmacological targeting of HDAC3 could potentially be harnessed to alter systemic fuel metabolism and exercise performance.
Keywords: circadian clock, insulin sensitivity, amino acid metabolism, exercise endurance, HDAC
INTRODUCTION
Skeletal muscle is the major tissue of glucose consumption and clearance, and as such insulin resistance in this organ is believed to be a key mediator of T2D pathogenesis1. Ectopic lipid accumulation in muscles can cause insulin resistance by activating cytosolic kinase cascades that disrupt molecular insulin signaling, a process referred to as lipotoxicity2. However, endurance athletes have higher intramuscular lipid contents associated with higher insulin sensitivity, a phenomenon known as the “athlete’s paradox”3, suggesting that how the muscle handles lipid storage is more important than the lipid content itself. In addition, muscle-specific knockout of the insulin receptor does not increase blood glucose levels despite severe muscle insulin resistance4, 5, suggesting that disruption of the cytosolic insulin signaling in muscle may not be sufficient to cause systemic glucose intolerance.
Mitochondrial dysfunction as a cause of muscle insulin resistance is debated. Reduced skeletal muscle mitochondrial content or impaired mitochondrial oxidative function correlates with T2D, leading to speculation that mitochondrial dysfunction underlies insulin resistance. However, depletion of mitochondrial oxidative phosphorylation (OXPHOS) genes in the muscle actually improves glucose tolerance and insulin sensitivity, presumably through adaptive enhancement of glucose utilization6–9. These results argue against muscle mitochondrial deficiency as the root cause of T2D, although such severe genetic mitochondrial dysfunction in animal models is different from the relatively mild acquired impairments in mitochondrial function that occur in humans. The reciprocal competition between glucose and lipid for muscle fuel sources is known as the ‘Randle cycle’, and likely involves mechanisms beyond the allosteric enzymatic regulation as originally defined by Randle10, 11. An emerging paradigm is that metabolic inflexibility, caused by nutrition overload and heightened fuel competition, prevents efficient utilization of any fuel, which leads to accumulation of toxic intermediates and insulin resistance12, 13. Reduced mitochondrial activity in this model is acquired and contributes to T2D although it is not the root cause.
Metabolism is intrinsically rhythmic. Circadian behaviors of feeding/fasting and activity/sleep are controlled by the central circadian clock in the brain, which is entrained by light. Meanwhile, transcription of many metabolic genes display robust oscillation in peripheral tissues such as liver and skeletal muscle14. This circadian rhythm is mainly dictated by the molecular clock within peripheral tissues15. Misalignment of the peripheral circadian clock and feeding behavior underlies the pathogenesis of diabetes in animal models16. The core molecular clock is composed of several transcription factors and co-regulators17. The nuclear receptors Rev-erbα and Rev-erbβ, key components of the molecular clock, repress gene transcription by recruiting nuclear receptor co-repressors (NCORs) and HDAC318. We have demonstrated that HDAC3 and Rev-erb orchestrates epigenomic circadian rhythm in liver, which coordinates reciprocal competition of lipogenesis and gluconeogenesis19–21. How the circadian clock in skeletal muscle regulates fuel catabolism remains largely unexplored22, 23.
As the primary site of the gene-environment interaction, epigenomic regulation of gene expression is increasingly recognized as a key component in T2D pathogenesis24. Histone acetylation and deacetylation is a major epigenomic modification that is dictated by acetyltransferases and deacetylases. Distinct partnership with different corepressor complexes determines the functional non-redundancy between HDAC3 and other zinc-dependent class I, II, and IV HDACs. Class IIa HDACs (HDAC4, 5, 7, and 9) also associate with the NCOR complex but have no intrinsic enzyme activity25. As a result, the enzymatic activity of class IIa HDACs is dependent on HDAC326. Class IIa HDACs play important roles in development and muscle remodeling27. However, how zinc-dependent HDACs regulate muscle fuel metabolism has not been addressed in vivo. In this study, we examined the metabolic role of HDAC3 in skeletal muscle.
RESULTS
HDAC3 depletion decreases muscle insulin sensitivity and glucose utilization
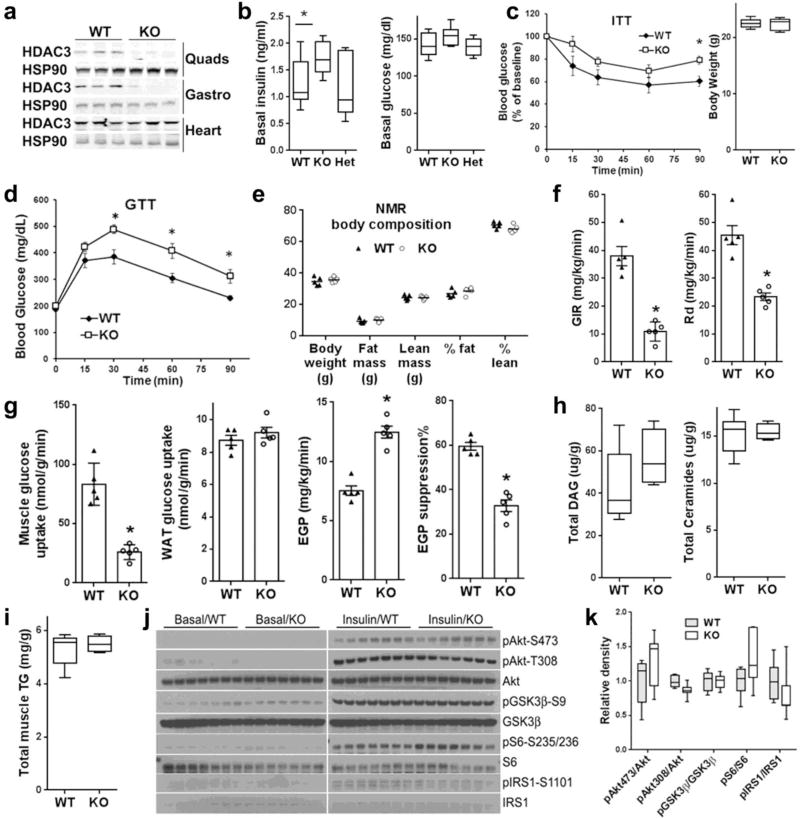
Skeletal muscle-specific HDAC3 knockout mice (HDAC3-SkMKO) were generated by crossbreeding HDAC3loxP/loxP mice with myosin light chain 1f-Cre (MLC-Cre) mice28. HDAC3loxP/loxP (WT) or HDAC3loxP/+, MLC-Cre (Het) littermates were used as controls. HDAC3-SkMKO mice were born at a normal Mendelian ratio and do not exhibit obvious abnormal phenotypes. Western blot and RT-qPCR analysis confirmed efficient depletion of HDAC3 in multiple skeletal muscles, but not in heart or liver (Fig. 1a and Supplementary Fig. 1a). Compared to WT mice, HDAC3-SkMKO mice showed mildly elevated basal insulin levels without changes in basal blood glucose levels (Fig. 1b), which led to a higher index of insulin resistance (HOMA-IR) (Supplementary Fig. 1b). This was confirmed by a blunted insulin response in insulin tolerance tests (ITTs) (Fig. 1c), which led to significant glucose intolerance without a change in body weight or body composition compared to WT (Fig. 1d,e and Supplementary Fig. 1c). A hyperinsulinemic euglycemic clamp analysis revealed a notably lower glucose infusion rate (GIR) and glucose rate of disposition (Rd) in HDAC3-SkMKO mice compared to WT mice, demonstrating pronounced systemic insulin resistance (Fig. 1f). Metabolic tracing with 14C-deoxyglucose identified that the skeletal muscle, but not white adipose tissue (WAT), had impaired glucose uptake, which accounted for the defective glucose clearance in HDAC3-SkMKO mice compared to WT (Fig. 1g). 3H-glucose tracing showed that endogenous glucose production was higher in HDAC3-SkMKO mice compared to WT mice during insulin clamping, demonstrating hepatic insulin resistance that was secondary to altered muscle metabolism (Fig. 1g). The insulin resistance and glucose intolerance was observed in young adult mice in both normal chow diet and high fat diet regardless of the sex (Supplementary Fig. 1d). Total diacylglycerols, ceramides, and triglycerides remain unaltered (Fig. 1h,i), although some species of free fatty acids were slightly higher in HDAC3-depleted muscles compared to WT (Supplementary Fig. 1e). Expression of inflammation index genes remained unaltered (Supplementary Fig. 1f). Molecular insulin signaling was not impaired in HDAC3-depleted muscles compared to WT upon either persistent hyperinsulinemia (Fig S1g) or bolus insulin injection (Fig. 1k,j). These results demonstrated that the lower glucose uptake in HDAC3-depleted muscle compared to WT is not due to lipotoxicity or impaired cytosolic insulin signaling.
Figure 1. Skeletal muscle HDAC3 deletion reduces glucose uptake and insulin sensitivity independently of the upstream insulin signalling.
(a) Western blot analysis (n = 2) of quadriceps femoris (Quads), gastrocnemius (Gastro), and cardiac muscles. (b) Basal insulin and glucose levels, n = 8. (c) Insulin tolerance test (ITT) and body weight on chow at the age of 4 months, n = 8. (d) Glucose tolerance test (GTT) at the age of 5 months after feeding high fat diet for two weeks, n = 8. (e) Body composition measured by nuclear magnetic resonance (NMR), n = 5. (f,g) Hyperinsulinemic euglycemic clamp analysis, GIR, glucose infusion rate; Rd, rate of disposition; EGP, endogenous glucose production; WAT, white adipose tissue. n = 5. (h) Mass spectrometry-based lipidomics analysis of diacylglycerol (DAG) and ceramides in quadriceps muscles, n = 5. (i) Total muscle triglycerides (TG) measurement, n = 5. (j) Western blot analysis (n = 2) of molecular insulin signaling in quadriceps muscles harvested at 20 min after a bolus insulin injection. Phosphorylation on Akt (pAkt), glycogen synthase kinase (GSK3β), S6 ribosomal protein (S6), and insulin receptor substrate (IRS1) at indicated sites were shown. (k) Quantification of the western blot densities in the insulin treatment condition, n = 7. Box-plots center line, limits, and whiskers represent median, quartile, and minimum/maximum values respectively. All other graphs were presented as the mean ± S.E.M. *P < 0.05 between groups by 2-sided t-test.
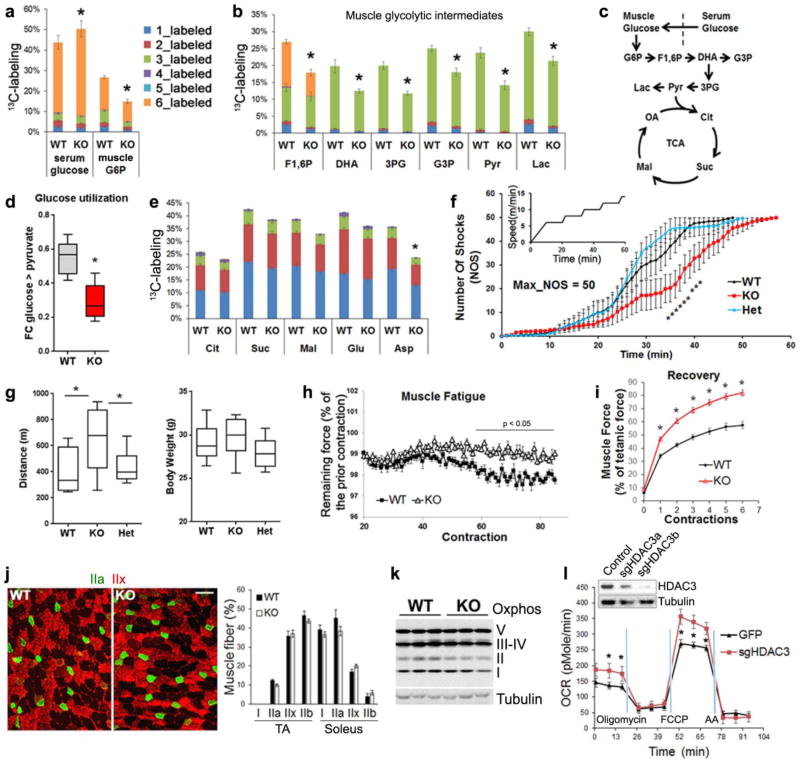
Exercise induces muscle glucose uptake through a different mechanism from that mediated by insulin signaling. To address whether HDAC3 depletion alters exercise-induced glucose utilization, we intravenously infused mice with uniform-13C-labeled glucose at all 6 carbons (U-13C-glucose) at a constant rate for 2.5 h while the mice were running on treadmill (Supplementary Fig. 2a). Upon tissue harvest, isotope labeling of glycolysis and tricarboxylic acid (TCA) cycle intermediates in quadriceps muscles were analyzed by liquid chromatography and tandem mass spectrometry (LC-MS/MS) (Supplementary Table 1). Compared to WT mice, HDAC3-SkMKO mice showed lower labeling of glucose-6-phosphate in muscle, despite greater labeling of its precursor, blood glucose (Fig. 2a), demonstrating a defect in exercise-induced glucose uptake. HDAC3-depleted muscles also showed lower labeling than WT in glycolytic intermediates, including fructose-1,6-phosphate, dihydroxyacetone phosphate, 3-phosphoglycerate, glycerol-3-phosphate, lactate, and pyruvate (Fig. 2b,c). This demonstrated reduced utilization of circulating glucose in HDAC3-SkMKO mice compared to WT mice during exercise (Fig. 2d). HDAC3-depleted muscles showed more similar labeling pattern with WT in TCA intermediates citrate, succinate-, and malate (Fig. 2e). The carbon 2 (C2)- versus C4-labeled TCA intermediates are not significantly altered (Supplementary Table 1). Considering lower pyruvate labeling, these data suggested simultaneous increase in anaplerosis and TCA turning in HDAC3-depleted muscles compared to WT. In contrast to the TCA intermediates, labeling of aspartate is notably lower in KO than WT (Fig 2e). This suggested a less net cataplerotic flux from TCA intermediates to aspartate synthesis, and therefore a higher predominant reverse flux of amino acid catabolism as anaplerotic reactions in HDAC3-depleted muscles. Total amounts of all detected metabolites remained unaltered except higher glutamate and lower aspartate in HDAC3-depleted muscle compared to WT, which further supports higher aspartate catabolism (Supplementary Table 1). Taken together, HDAC3 deletion reduces muscle glucose uptake and utilization during muscle contraction.
Figure 2. HDAC3 deletion reduces glucose utilization during muscle contractions, but enhances exercise endurance and oxidative metabolism.
(a,b) 13C-enrichment in metabolites from serum or muscles after infusion of U-13C-glucose through jugular vein catheter while mice are running on treadmill, n = 6 for WT, n = 5 for KO. Total number of labelled carbon atoms in a given metabolite is indicated by color. G6P, glucose-6-phosphate, F1,6,P, fructose-1, 6-phosphate, DHA, dihydroxyacetone phosphate, 3PG, 3-phosphoglycerate, G3P, glycerol-3-phosphate, Lac, lactate, and Pyr, pyruvate. (c) Diagram of glycolysis and TCA cycle pathways with detected intermediates highlighted. (d) Relative utilization rate of circulating glucose. Fractional contribution (FC) is derived from muscle pyruvate labeling after correction with blood precursor labeling. (e) 13C-enrichment in muscle TCA intermediates. Cit, citrate, Suc, succinate, Mal, malate, Glu, glutamate, and Asp, aspartate. (f) Treadmill speed profile and real-time shock detection during running, n = 10. (g) Distance run when mice received 50 shocks and body weight, n = 10. (h,i) Muscle fatigue and recovery in ex vivo contraction study with extensor digitorum longus (EDL) muscles, n = 8. Scale bar, 50 µm. (j) Representative immunofluorescence staining (n = 8 images) of tibialis anterior (TA) with indicated MHC isoform-specific antibodies, and quantification of staining in TA and soleus, n = 8 animals. (k) Western blot analysis (n = 3) of mitochondrial OXPHOS complexes in quadriceps complexes. (l) Respirometry analysis in fully-differentiated C2C12 myotubes after treatment with adenoviral vectors for HDAC3 knockdown, n = 6 wells of cells. Oligomycin, inhibitor of ATP synthase. FCCP, uncoupling agent. Antimycin A (AA) and rotenone, inhibitors of electron transport. Box-plots center line, limits, and whiskers represent median, quartile, and minimum/maximum values respectively. All other graphs were presented as the mean ± S.E.M. *P < 0.05 between genotypes under the same condition by 2-sided t-test.
HDAC3 depletion enhances exercise endurance and oxidative metabolism
Glucose is a major fuel for muscle, and reduced glucose utilization during contraction would predict impaired exercise endurance. To assess this, mice were put on a treadmill equipped with electronic shock grids. The speed of treadmill gradually ramped up from 0 to 14 m/min (Fig. 2f). Compared to WT mice, HDAC3-SkMKO mice receive fewer shocks during running (Fig. 2f) and ran longer distance at the time of exhaustion that is defined as when a mouse receives 50 accumulating shocks (Fig. 2g). The enhanced exercise endurance was also observed in younger mice or female mice (Supplementary Fig. 2b). Consistent with this in vivo finding, extensor digitorum longus (EDL) muscles isolated from HDAC3-SkMKO mice displayed a slower rate of force dropping compared to WT, when subjected to repeated field electric stimulations in an ex vivo test29 (Fig. 2h), demonstrating enhanced fatigue resistance. In addition, HDAC3-depleted muscles recovered more quickly from fatigue than WT (Fig. 2i). These data suggest that intrinsic changes in muscles, other than endocrine or neural factors, underlie the exercise endurance. Despite enhanced fatigue resistance and recovery, HDAC3-depleted muscles generated less force than WT muscles (Supplementary Fig. 2c), suggesting a trade-off between muscle strength and fatigue resistance.
Concurrence of defective glucose uptake and superior endurance is intriguing. One possible explanation is altered muscle fiber-type composition. Skeletal muscles are composed of slow-contracting, fatigue-resistant, oxidative fibers (type I) and fast-contracting, fatigue-prone, glycolytic fibers (type IIb) as well as intermediate fibers (type IIa and IIx)30, 31. The molecular determinant of the fiber type is the expression of myosin heavy chains (MHCs) isoforms. Immunostaining and western blot analysis of HDAC3-depleted skeletal muscles with isoform-specific MHC antibodies did not reveal obvious changes in fiber type composition (Fig 2j and Supplementary Fig. 2d,e). RT-qPCR and proteomics analysis did not identify obvious changes in expression levels of different MHC isoforms except moderate decrease of MHC-IIb in HDAC3-depleted gastrocnemius compared to WT (Supplementary Fig. 2f,g). Succinate dehydrogenase staining also failed to identify obvious changes (Supplementary Fig. 2h). These results suggest that fiber type switch is very limited and unlikely account for the endurance phenotype. Another possible explanation for fatigue resistance is enhanced mitochondrial oxidative bioenergetics. Mitochondrial DNA copy number is indeed higher in HDAC3-depleted muscles compared to WT (Supplementary Fig. 3a), but mitochondrial protein content remained unaltered for oxidative phosphorylation (OXPHOS) complexes (Fig. 2k) and translocase of the inner membrane 23 complexes (Supplementary Fig. 3b). Electron microscopy did not reveal obvious morphological changes of mitochondria (Supplementary Fig. 3c). Despite the lack of obvious structural changes, mitochondrial respiration rate was increased in differentiated C2C12 myotubes cultured in media containing amino acids after HDAC3 was depleted with adenovirus-mediated CRISPR constructs (Fig. 2l), indicating enhanced bioenergetics and higher oxidative capacity in a cell-autonomous manner. These data suggest that HDAC3 depletion enhances mitochondrial oxidative capacity without altering overall mitochondrial structures.
HDAC3 controls muscle fuel preference and amino acid catabolism
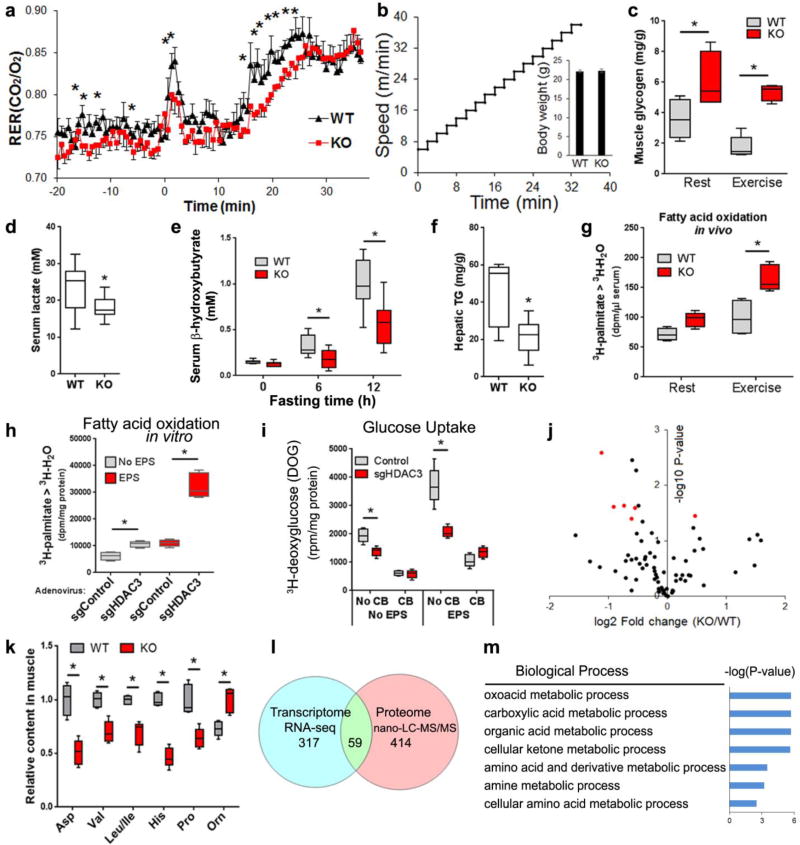
To address the mechanism for how HDAC3 depletion enhances mitochondrial oxidative capacity, we asked what the fuel source is for HDAC3-depleted muscles to sustain the superior endurance. Given the defective glucose uptake, the major fuel(s) for the increased muscle endurance could include glycogen or lipid. Indirect calorimetry analysis of mice running on a treadmill revealed lower respiratory exchange ratio (RER) in HDAC3-SkMKO mice compared to WT mice, suggesting a preference towards lipid oxidation (Fig. 3a,b). Although muscle triglycerides remained unaltered during rest or exercise (Supplementary Fig. 4b), higher muscle glycogen and lower blood lactate levels in HDAC3-SkMKO mice compared to WT mice suggest that endogenous carbohydrate is unlikely to be the preferred fuel source for improved muscle endurance (Fig. 3c,d). Lower blood ketone levels and lower liver triglyceride (Fig. 3e,f) further support the notion that HDAC3-KO muscles develop a fuel preference towards lipids compared to WT, and therefore consume more lipid or ketone bodies under fasting conditions.
Figure 3. HDAC3 regulates muscle fuel preference and controls amino acid metabolism.
(a) Respiratory exchange ratio (RER) in mice running on treadmill with increasing speed to exhaustion, n = 6. (b) Speed profile of the treadmill and body weight. (c) Muscle glycogen at rest or immediately after exercise, n = 5. (d) Blood lactate after exercise, n = 8. (e) Blood ketone body after fasting, n = 8. (f) Liver triglyceride (TG) after fasting, n = 8. (g) Fatty acid oxidation rate measured by 3H-H2O generation in blood at 20 min after i.p. injection of 3H-palmitate, n = 5. (h) Fatty acid oxidation in differentiated C2C12 myotubes treated with adenovirus to knock down HDAC3, n = 6 wells of cells. EPS, electric pulse stimulation. (i) Glucose uptake assay in differentiated C2C12 myotubes treated with adenovirus. Cytochalasin B (CB), an inhibitor of glucose transport, n = 6 wells of cells. (j) Volcano plot of metabolomics profiling in quadriceps muscles harvested at rest. Altered amino acids highlighted in red. (k) Amino acid contents from metabolomics profiling in quadriceps muscles, n = 5. (l) Venn diagram of differentially expressed genes and proteins (KO versus WT) as identified by RNA-seq (q<0.1, fold change>1.5) and proteomics profiling using nano-liquid chromatography mass spectrometry (nano-LC-MS) with label-free and tandem mass tag (TMT) quantitation (p<0.05 in label-free quantification and p<0.1 in TMT quantification). (m) Enriched biological pathway of overlapping genes/proteins. Box-plots center line, limits, and whiskers represent median, quartile, and minimum/maximum values respectively. All other graphs were presented as the mean ± S.E.M. *P < 0.05 between genotypes under the same condition by 2-sided t-test.
To directly measure lipid oxidation, we i.p. injected a bolus of 3H-palmitate in resting or exercising mice, obtained blood after 20 min, and measured the end product 3H-H2O. HDAC3-SkMKO mice showed a higher degree of fatty acid oxidation compared to WT mice (Fig. 3g). To further address whether this is cell autonomous, we adapted an in vitro exercise model32. Electric pulse stimulation (EPS) and adenovirus was applied in fully-differentiated C2C12 myotubes to induce contraction and gene deletion, respectively. The EPS contraction protocol and adenoviral treatment did not cause cell toxicity or de-differentiation (Supplementary Fig. 4b,c). HDAC3 deletion dramatically increased basal and contraction-induced fatty acid oxidation compared to the mock control (Fig. 3h). On the other hand, glucose uptake was inhibited by HDAC3 deletion, as measured by 3H-deoxyglucose (Fig. 3i). This demonstrates that HDAC3 regulates fuel preference in a myocyte-autonomous manner. There were minimal changes in expression of several well-regarded genes in glucose transport, fatty acid oxidation, the TCA cycle, and oxidative phosphorylation, including PGC1 (Supplementary Fig. 4d,e). GLUT4 protein levels, AMP-activated protein kinase (AMPK) activities, and phosphorylation on its substrate HSL also remained unaltered between HDAC3-depleted and WT muscles (Fig. S4f). These data suggest that the enhanced oxidative bioenergetics in HDAC3-depleted muscles is unlikely due to direct regulation of fatty acid oxidation genes.
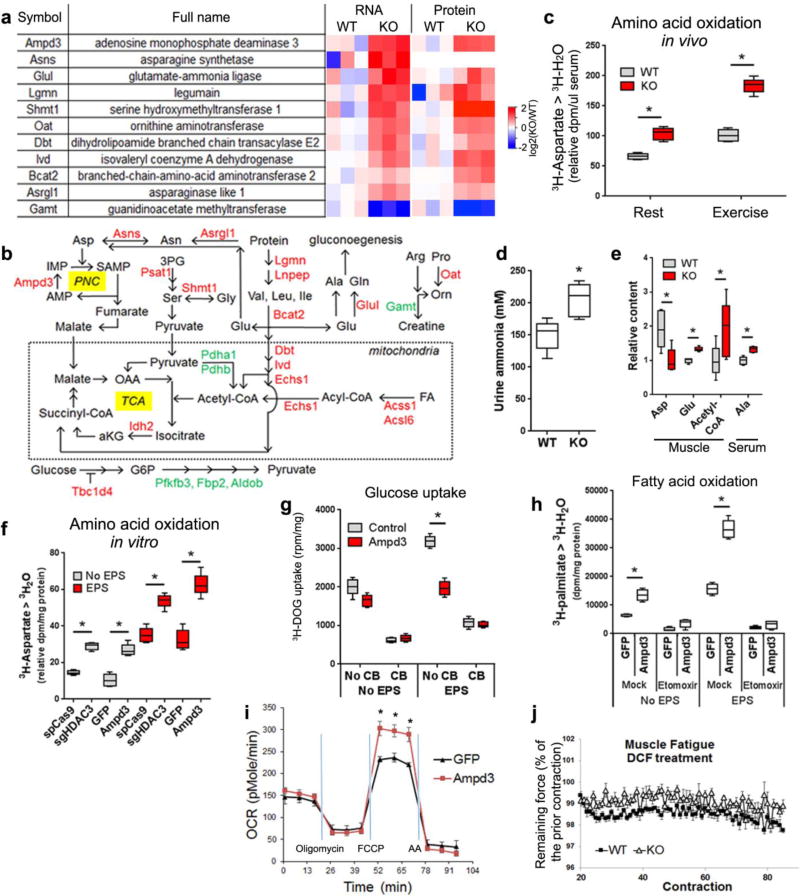
To further elucidate the underlying mechanism of enhanced oxidative bioenergetics, we integrated metabolomics, transcriptomics, and proteomics approaches. A metabolomics study identified that the most prominently altered metabolites in HDAC3-depleted muscles versus WT muscles were amino acids (Fig. 3j). Aspartate, proline, histidine, and branched-chain amino acids (BCAA) were reduced in HDAC3-depleted muscles compared to WT (Fig. 3k). RNA-seq and nano-LC-MS/MS proteomics identified many differentially expressed genes or proteins (Fig. 3l and Supplementary Table 2,3). The overlapping targets were once again dominated by enzymes involved in amino acid catabolism (Fig. 3m). Key amino acid catabolism enzymes were upregulated at both mRNA levels and protein levels upon HDAC3 deletion (Fig. 4a). The most prominently upregulated gene was AMP deaminase 3 (Ampd3) that catalyzes the first and rate-limiting step of the purine nucleotide cycle (PNC), an anaplerotic reaction in skeletal muscles.
Figure 4. Enhanced amino acid catabolism in HDAC3-depleted muscles underlies the fuel switch.
(a) Heat map of altered genes or proteins from RNA-seq and proteomics analysis. Data for Asns protein is missing. (b) Metabolic pathway with genes upregulated (red) or downregulated (green) in HDAC3-depleted muscles (KO) versus WT control. (c) Aspartate catabolism measured by 3H-H2O accumulation in blood at 20 min after i.p. injection of 3H-aspartate, n = 6. (d) Urine ammonia measurement, n = 8. (e) In vivo metabolites measurement by LC-MS/MS after exercise, n = 5. (f) Aspartate catabolism assay in C2C12 myotubes treated with indicated adenovirus, n = 6 wells of cells. (g) Glucose uptake assay in C2C12 myotubes after overexpression of Ampd3, n = 6 wells of cells. (h) Fatty acid oxidation assay in C2C12 myotubes, n = 6 wells of cells. (i) Respirometry analysis of mitochondrial oxidative capacity in C2C12 myotubes. OCR, oxygen consumption rate; n = 6 wells of cells. (j) Ex vivo contraction analysis of EDL muscles fatigue resistance in the presence of AMP deaminase inhibitor deoxycoformycin (DCF), n = 8. Box-plots center line, limits, and whiskers represent median, quartile, and minimum/maximum values respectively. All other graphs were presented as the mean ± S.E.M. *P < 0.05 between genotypes under the same condition by 2-sided t-test.
Anaplerotic amino acid catabolism promotes lipid oxidation
During exercise when AMP level is high in muscles, the PNC consumes aspartate and connects BCAA catabolism to the TCA cycle, which creates an anaplerotic reaction that increases TCA flux and oxidative capacity in handling acetyl-CoA, the end product of fatty acid oxidation (Fig. 4b). Another anaplerotic reaction is driven by catabolism of BCAA through BCAA aminotransferase 2, dihydrolipoamide branched-chain transacylase (the E2 component of the branched-chain alpha-keto acid dehydrogenase complex), isovaleryl-CoA dehydrogenase, and enoyl Coenzyme A hydratase, short chain, 1, mitochondrial, which leads to production of succinyl-CoA. These anaplerotic reactions, in alliance with upregulated isocitrate dehydrogenase and several genes in acyl-CoA metabolism, could explain the enhanced mitochondrial oxidative metabolism in HDAC3-depleted muscle compared to WT. Lower expression of glycolytic genes in HDAC3-depleted muscle compared to WT (Fig. 4b and Supplementary Fig. 5a), together with the enhanced oxidative metabolism, could thus account for the fuel preference switch. Glutamate generated from BCAA catabolism can be released into circulation as alanine or glutamine that serve as gluconeogenic precursors in liver or kidney, which could explain the higher endogenous glucose production in HDAC3-SkMKO mice compared to WT mice as observed in the previous insulin clamp study (Fig. 1g).
In support of this hypothesis derived from –omics studies, the kinetic flux of amino acid catabolism was found markedly upregulated in HDAC3-SkMKO mice compared to WT mice as measured using 3H-aspartate (Fig. 4c). Elevated ammonia in urine in HDAC3-SkMKO mice compared to WT mice is also consistent with enhanced amino acid catabolism in vivo (Fig. 4d). In light of the altered enzymes in the metabolic pathway (Fig. 4b), we predict enhanced catabolism of fatty acids, proteins, and amino acids as well as associated amino acid release from HDAC3-depleted muscles. This is supported by the higher levels of muscle glutamate (an intermediate between amino acid catabolism and excretion), serum alanine, and muscle acetyl-CoA during exercise in HDAC3-SkMKO mice compared to WT mice (Fig. 4e). Regulation of amino acid catabolism by HDAC3 was cell autonomous since it is increased in cultured myotubes upon acute depletion of HDAC3 (Fig. 4f). Overexpression of Ampd3 to a similar level as in HDAC3-depleted myotubes was sufficient to cause such flux change, demonstrating that PNC is the major driver for aspartate catabolism (Fig 4f and S5b). We also predict that persistent amino acid catabolism could drive up muscle protein breakdown and lead to muscle mass loss over time, even considering the enormous protein synthesis capacity. Indeed, muscle mass from HDAC3-SkMKO mice became lower than WT as mice age (Supplementary Fig. 5c,d). However, the insulin resistance and exercise endurance phenotypes were fully developed before there was a significant change in muscle mass, suggesting that the metabolic change causes, rather than results from, a change in muscle mass. Taken together, in vivo –omics studies, together with in vivo and in vitro tracing studies, identified amino acid catabolism as a major process that is directly suppressed by HDAC3.
Can enhanced amino acid catabolism explain the fuel switch in HDAC3-depleted muscles? Overexpression of Ampd3 to a similar level as in HDAC3 depletion reduces glucose uptake in differentiated myotubes compared to GFP control (Fig. 4g and Supplementary Fig. 5b), suggesting that enhanced PNC and amino acid catabolism has glucose-sparing effect. More importantly, overexpression of Ampd3 strikingly increases fatty acid oxidation (Fig. 4h) and mitochondrial oxygen consumption rate compared to GFP control (Fig. 4i), demonstrating that the anaplerotic PNC and associated BCAA catabolism is sufficient to induce the enhanced mitochondrial oxidative capacity and lipid preference. To address the requirement of PNC in fatigue resistance, we utilized adenine deaminase inhibitor deoxycoformycin (DCF) in an ex vivo contraction analysis. As previously demonstrated, HDAC3-depleted EDL muscles retain higher percentage of force during repetitive fatigue-inducing contractions compared to WT muscles (Fig. 2h). Such fatigue difference was diminished in the presence of DCF (Fig. 4j) when all other factors remain the same, including mouse age, sex, and experimental procedures. DCF does not disrupt the difference in muscle force (Supplementary Fig. 5e). These data demonstrate the requirement of Ampd3 and the anaplerotic PNC reaction for the fatigue resistance of HDAC3-depleted muscles. Taken together, increased BCAA catabolism in alliance with anaplerotic reactions drives the fuel preference toward lipid over glucose.
HDAC3 couples circadian clock with muscle fuel preference
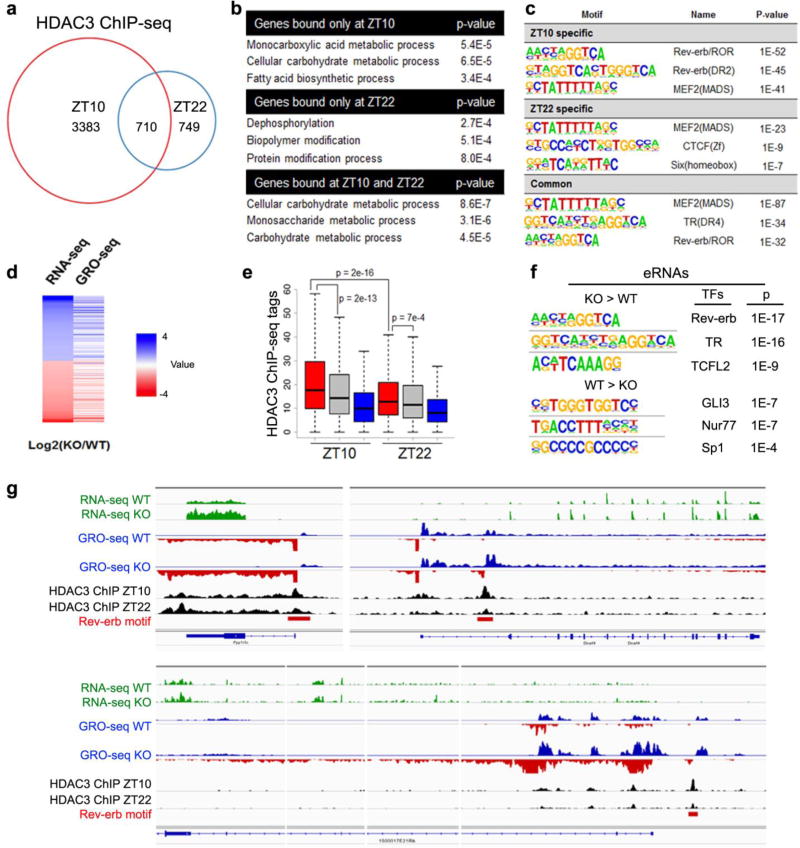
How is HDAC3 itself regulated? Our previous studies showed that the majority of the DNA-binding events of HDAC3 in liver occur at late afternoon19, 21. To address where HDAC3 binds to the muscle genome, we performed HDAC3 ChIP-seq in muscles at Zeitgeber time 10 (ZT10, late afternoon) and ZT22 (early morning). Interestingly, like in liver, HDAC3 displayed a clear time-dependent binding pattern with the majority of the binding occurred at ZT10 (Fig. 5a). ZT10-specific binding sites or overlapping sites highly enriched in metabolic genes, while ZT22-specific binding sites were not (Fig. 5b). The top enriched motif at ZT10 sites was the binding site for nuclear receptor Rev-erb or RAR-related orphan receptor (ROR), key components of the circadian clock (Fig. 5c). This suggests that HDAC3 is regulated by the muscle circadian clock.
Figure 5. Nonbiased identification of the circadian clock as an upstream regulator of muscle HDAC3.
(a) Venn diagram of HDAC3 ChIP-seq binding sites in mouse quadriceps muscles at Zeitgeber time 10 (ZT10) versus ZT22 (>1rpm, filtered by HDAC3-KO, >1bp overlap for common sites). (b) Top enriched biological processes of genes near HDAC3 binding sites. (c) Top enriched motif in HDAC3 binding sites at ZT10 versus ZT22. (d) Heat map correlation of RNA-seq and GRO-seq performed at ZT10 in HDAC3-depleted quadriceps muscle versus WT control. Red indicates upregulation (KO versus WT), and blue indicate downregulation. (e) HDAC3 binding at upregulated (red), unchanged (gray), or downregulated (blue) bidirectional eRNAs in KO versus WT. p value by t-test. (f) Top enriched motif in bidirectional eRNAs that are up- or down- regulated in HDAC3-depleted muscles. (g) Browser tracks showing HDAC3 ChIP-seq at ZT10 and ZT22 in WT muscles as well as RNA-seq and GRO-seq in WT and KO muscles at ZT10. Same scales were used for each type of experiment.
One caveat of crosslinking-based assays such as ChIP, is that they do not distinguish functional versus non-functional binding because of the ‘trial and error’ mode of protein-DNA interaction. Gene transcription is initiated by enhancers looping to transcription start sites (TSS), followed by recruitment of transcription initiation complexes containing RNA polymerase that generates RNAs from both gene bodies and enhancers. While transcripts at gene bodies are followed with productive elongation, bidirectional transcripts from the enhancers (eRNA) are short and transient, but are detectable by global nuclear run-on assay (GRO-seq). eRNAs are superior markers of active enhancers because of their small size, high dynamic range, and tight correlation with the expression of their target genes33. Using GRO-seq and RNA-seq, we found that nascent transcription changes at gene bodies correlated well with mature mRNA changes upon HDAC3 depletion (Fig. 5d), suggesting that altered transcription initiation underlies gene expression changes. At eRNA sites that were upregulated in HDAC3-depleted muscles compared to WT, HDAC3 binding was stronger than that at unchanged or downregulated eRNA sites at ZT10, and this difference was less obvious at ZT22 (Fig. 5e). This suggests that the ZT10 binding contributes to gene repression, consistent with the primary role of Rev-erb as transcription repressor and its high expression level at ZT10. The top motif enriched in the upregulated enhancers was again the binding site of Rev-erb (Fig. 5f). In comparison, the downregulated eRNAs were enriched with other motifs with much lower p-values (Fig. 5f). Many upregulated genes were bound in a circadian fashion by HDAC3 at TSS or upregulated eRNA sites that contain Rev-erb motifs (Fig. 5g). These nonbiased analyses of the transcriptome and cistrome suggest that Rev-erb recruits HDAC3 at ZT10 to enhancers where HDAC3 represses nearby metabolic genes in skeletal muscle.
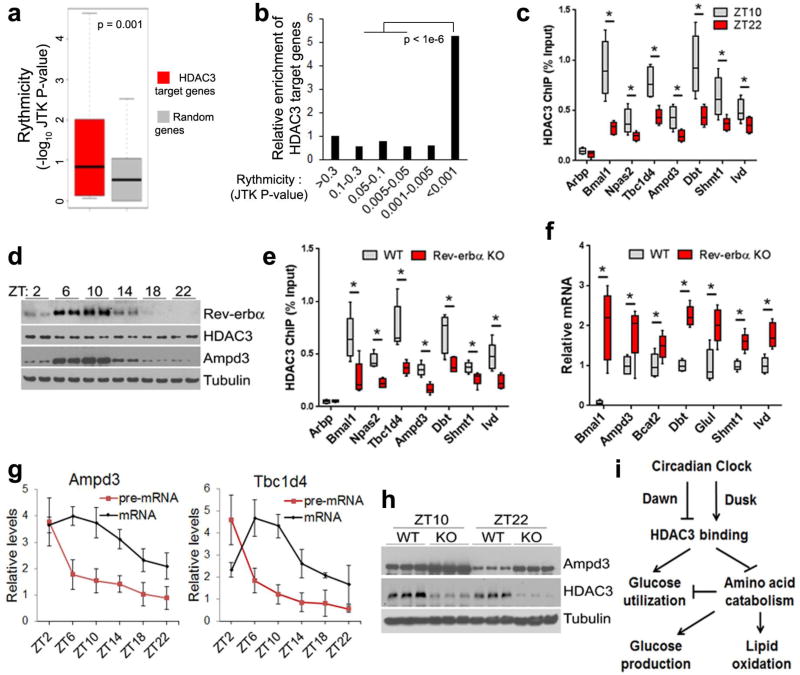
Compared to random genes, HDAC3 target genes identified in our RNA-seq and ChIP-seq analysis were more likely to display circadian expression as determined by others14 (Fig. 6a). Conversely, genes with more robust rhythmic expression were more enriched in HDAC3 target genes (Fig. 6b). HDAC3 ChIP-qPCR further confirmed the circadian binding pattern of HDAC3 on key genes involved in amino acid catabolism and glucose uptake (Fig 6c). Regardless of its constant protein level, HDAC3 binding on the genome peaked at ZT10, in phase with the maximum protein level of Rev-erbα (Fig. 6d), and was significantly reduced in muscles from Rev-erbα null mice19 compared to WT mice (Fig. 6e), demonstrating that Rev-erbα is required for maximum genomic recruitment of HDAC3. The residual HDAC3 binding in the absence of Rev-erbα could be attributed to Rev-erbβ and other transcription factors. Ampd3 and several BCAA catabolism genes were upregulated in Rev-erbα-depleted muscles, further supporting a pivotal role of Rev-erb in HDAC3-mediated gene repression (Fig. 6f). In addition, expression of these key metabolic genes regulated by HDAC3 showed clear circadian expression pattern in WT muscles, with pre-mRNA elevated shortly after HDAC3 was released from the chromatin at ZT22 (Fig. 6g). Protein and mRNA levels peaked at later time points (Fig. 6g,h). Such a circadian pattern was replaced by constant high level expression when HDAC3 was depleted (Fig. 6h), although circadian locomotor activities were not affected (Supplementary Fig. 6). Collectively, these data establish HDAC3 as a master transcriptional regulator that couples circadian cues with muscle fuel metabolism.
Figure 6. HDAC3 couples circadian cues with the regulation of genes in anaplerotic reactions and amino acid catabolism.
(a) Enrichment of circadian expressed genes in HDAC3 targets. HDAC3 target genes are defined by their upregulation in KO versus WT in RNA-seq and the presence of HDAC3 binding sites within 50 kb of their transcription start sites. (b) Enrichment of HDAC3 target genes in circadian expressed genes in skeletal muscle. (c) ChIP-qPCR analysis at ZT10 and ZT22 in WT quadriceps muscles, n = 6. (d) Western blot analysis in WT quadriceps harvested at the indicated time points. (e) HDAC3 ChIP-qPCR analysis in Rev-erbα-KO quadriceps muscles at ZT10, n = 6. (f) RT-qPCR analysis in Rev-erbα-KO quadriceps muscles at ZT14, n = 4. (g) Circadian expression of Ampd3 and Tbc1d4 pre-mRNA and mRNA in WT quadriceps muscles, n = 6. (h) Western blot analysis of Ampd3 in HDAC3-depleted quadriceps muscles at ZT10 and ZT22. (i) Summary model of how HDAC3 connects circadian cues to muscle metabolism. Box-plots center line, limits, and whiskers represent median, quartile, and minimum/maximum values respectively. All other graphs were presented as the mean ± S.E.M. *P < 0.05 between genotypes by 2-sided t-test.
DISCUSSION
Our results delineate an intricate regulatory network through which circadian clock governs skeletal muscle intermediary metabolism and bioenergetics (Fig. 6i). Before dawn, the muscle circadian clock in nocturnal animals anticipates the upcoming fasting period and clears HDAC3 from the genome at several metabolic genes through the change of Rev-erb protein levels. On one hand, this activates amino acid catabolism and anaplerotic reactions, which increases mitochondrial oxidative capacity, enhances lipid oxidation, and inhibits glucose utilization. On the other hand, the absence of HDAC3 suppresses genes in glycolysis. Together, these mechanisms in muscle spare glucose for its utilization by brain or red blood cells. Breakdown of muscle amino acids and proteins provides precursors for gluconeogenesis in liver or kidney, which helps maintain blood glucose levels during the fasting cycle. Before dusk, elevated Rev-erb protein level leads to recruitment of HDAC3 to the genome, which prepares the muscle to utilize glucose in alliance of systemic assimilation of carbohydrates into protein or lipid during the feeding cycle.
Insulin resistance and reduced glucose utilization in HDAC3-SkMKO mice is a combined outcome of at least three factors: (1) carbohydrate-sparing effects of enhanced amino acid catabolism and lipid oxidation at the level of substrate competition; (2) increased gluconeogenesis in liver or kidney due to increased gluconeogenic precursors from muscle amino acid catabolism; (3) inhibition of muscle glucose uptake and glycolysis through regulated expression of several carbohydrate metabolism genes such as Tbc1d4, a negative regulator of GLUT4 translocation. HDAC3-depleted muscles do not show dramatic changes in lipid contents and maintain normal molecular insulin signaling, suggesting that the physiological insulin resistance is not due to lipotoxicity or disruption of insulin signaling. Enhanced oxidative metabolism in HDAC3-depleted muscles also argues against mitochondrial deficiency as a contributor of its insulin resistance. The increase in oxidative metabolism and reciprocal decrease in glucose utilization is in keeping with the idea that mitochondrial fuel inflexibility causes physiological insulin resistance12, 13. In addition, our study highlights several underappreciated concepts regarding the molecular mechanisms that coordinate fuel competition in muscle. (1) Mitochondrial pathway usage, i.e. relative flux rates across different catabolic pathways, rather than mitochondrial content or muscle fiber composition, can contribute to metabolic remodeling. (2) Metabolism of different fuels can be oppositely and simultaneously controlled at the gene expression level by one factor. (3) Mitochondrial fuel preference not only passively responds to feeding behavior, but also anticipates fuel availability from cues provided by the circadian clock.
Under normal physiological conditions, the anticipatory regulation conferred by HDAC3 can go hand in hand with responsive mechanisms, such as those mediated by PPARα and its coactivator PGC1. In response to fasting, PPARα senses lipid influx from adipose lipolysis and activates fatty acid oxidation in muscles. This is coincident with inhibition of glucose uptake and activation of amino acid catabolism due to clearance of HDAC3 from the genome. Overexpression PPARα or PGC1α increases fatty acid oxidation, but decreases glucose utilization and cause insulin resistance34–39. There is a clear segregation of downstream target genes between PPARα/PGC1 and Rev-erb/HDAC3 signaling. The former upregulates fatty acid oxidation and OXPHOS genes, while the latter regulates amino acid catabolism and carbohydrate metabolism. Overexpression of PPARα or PGC1α disrupts insulin signaling due to incomplete oxidation and lipotoxicity because lipid uptake exceeds lipid oxidation, while HDAC3 depletion does not disrupt upstream insulin signaling. The clearance of HDAC3 from the genome occurs immediately before fasting, while PPARα activation occurs during late fasting when lipolysis dominates over glycogenolysis. Thus, the anticipatory and responsive regulations are mechanistically distinct, but are temporally connected. Together, they coordinate catabolism of different fuels by skeletal muscles in normal circadian physiology.
The Rev-erb motif was the top hit in both HDAC3 binding sites in the ChIP-seq analysis and activated enhancer regions in the GRO-seq analysis. This pinpoints Rev-erb as a major contributor to HDAC3 binding in muscles, which is further supported by decreased HDAC3 binding on key metabolic genes in Rev-erbα-depleted muscles. We do not suggest Rev-erb as the only transcription factor (TF) that regulates HDAC3 in muscles. On the contrary, our epigenomic profiling identified quite a few other TFs that also likely play a role. Circadian feeding cycles could contribute to the rhythmic binding pattern of HDAC3 in muscle, since peripheral circadian clocks are entrained by feeding. HDAC3 is dispensable for the circadian clock itself, and serves as an output arm of the clock in regulation of metabolic genes40. This is further supported by the current RNA-seq result showing that expression of most core clock genes remain unaltered in HDAC3-depleted muscles. We found here that voluntary wheel-running activity is not changed by muscle-specific manipulation of HDAC3. Interestingly, Rev-erbα null mice show reduced mitochondrial content and oxidative capacity in muscle41. It is possible that other TFs interact with HDAC3 and account for the different phenotypes. It is also possible that the nearly 90% reduction in voluntary locomotor activity in the whole-body Rev-erbα null mice could lead to disuse-induced deterioration of muscle function. The physiological changes in HDAC3-depleted muscles are overall consistent with those from muscle-specific knockout of NCOR142, although that mouse model does not show glucose intolerance but instead shows altered expression of mitochondrial OXPHOS genes. It is possible that NCOR2 could partially compensate for loss of NCOR1 or oppose NCOR1 function. RORα-KO mice display an opposite metabolic phenotype to the HDAC3-SkMKO mice, with improved systemic glucose tolerance43. This is consistent with RORs being anti-phase with Rev-erb/HDAC3 in the molecular clock machinery. However, RORα-KO muscles show altered GLUT4 expression while HDAC3-KO muscles do not, suggesting that RORα regulates a separate molecular output signaling that is independent of HDAC3. It is likely that different components of the core clock dictate distinct output signals that regulate different aspects of metabolism.
Amino acid catabolism in HDAC3-SkMKO mice increases lipid oxidation, which underlies exercise endurance. This is supported by several observations: (1) Activation of amino acid catabolism by overexpression of a PNC rate-limiting enzyme Ampd3 is sufficient to increase fatty acid oxidation (FAO) flux and mitochondrial respiratory capacity. (2) Inhibition of AMP deaminase masked the muscle fatigue resistance phenotype in ex vivo muscle contraction studies. (3) Transcriptomic analysis did not identify changes in major FAO genes, so there is no alternative or direct explanation for increased FAO flux. The endurance phenotype in HDAC3-SkMKO mice was characterized using a low-intensity exercise protocol. Higher intensity exercise requires greater carbohydrate utilization, and could lead to decreased exercise tolerance in HDAC3-SkMKO mice. More vigorous exercise is also more demanding on muscle strength that is compromised in HDAC3-depleted muscles. The perturbed force production in HDAC3-depleted muscles could be due to disrupted acetylation of non-histone proteins involved in contraction44.
The concept of anaplerosis was raised in the 1960s, and amino acid catabolism is a major source for anaplerotic reactions for the TCA cycle during muscle contractions45, 46. PNC is an important component in connecting protein and BCAA catabolism to the TCA47. Although anaplerosis originally referred to increases in the pool size of TCA intermediates, it is now accepted that the anaplerotic flux rate, rather than the absolute pool size, is important in sustaining TCA cycle flux and oxidative metabolism48. Genetic alterations of BCAA catabolism and PNC affect exercise endurance in both mouse and human49–52. Selectively bred rodents with high running capacity burn more lipid than control animals during low-intensity exercise, which is associated with enhanced BCAA catabolism53, 54. These studies and our current work suggest that amino acid catabolism could be a general mechanism that facilitates FAO. Our study further showed that this process is under delicate circadian regulation at the gene expression level. Given the temporal insight from our study, low-intensity exercise at night time coupled with nighttime fasting could be an efficient way to burn lipid for the purpose of losing body fat.
While amino acid catabolism can facilitate lipid oxidation, its over-activation impairs glucose utilization. Elevated BCAA levels are strongly associated with T2D. It is unclear whether defective BCAA catabolism per se causes insulin resistance. The body consumes BCAA during fasting and burns glucose when feeding. Such temporal segregation argues against the dependence of glucose utilization on BCAA catabolism. In addition, genetic disruption of BCAA catabolism ameliorates, rather than exacerbates, insulin resistance in animal models55, 56. The elevated BCAA in T2D patients is most likely to be an outcome of nutrient overload, and could contribute to repression of glucose utilization through fuel competition, either directly or via facilitating fat entry or burning in muscle57.
The paradoxical concurrence of superior exercise endurance and muscle insulin resistance in HDAC3-SkMKO mice challenges the common belief that muscle’s ability to utilize carbohydrates underlies its exercise endurance. Carbohydrate loading is widely practiced and recommended to improve endurance exercise performance58. This carbohydrate supremacy view does not make evolutionary sense before invention of agriculture that made sustained supply of carbohydrates possible59. Intuit people in the polar area get most of their energy from lipid and protein without compromised exercise endurance. In recognition that lipid is a better fuel for endurance exercise due to its high calorie-to-mass ratio, ketogenic diets have been used with some success to increase exercise endurance by nudging muscles to burn more lipid60. Identification of HDAC3 as a pivotal regulator of exercise endurance and the availability of small-molecule HDAC inhibitors makes it possible to increase endurance through a combination of pharmacological and dietary interventions. In summary, enhancing mitochondrial oxidative capacity could lead to physiological insulin resistance, and blocking muscle metabolic flexibility could benefit exercise endurance. These two seemingly counterintuitive phenomena can be readily unified by considering the rhythmic nature of muscle fuel metabolism.
ONLINE METHODS
Animals
C57BL/6 HDAC3fl/fl mice20, MLC-Cre mice28, and Rev-erbα null mice19 were previously described. Male mice at the age of 2 months to 8 months were used for all experiments unless indicated otherwise in figure legends or the main text. GTT and ITT assays were performed after fasting for 6 h since 9 a.m. Glucose was i.p. injected at 1.5 g/kg or 2 g/kg. Insulin (Novolin R) was i.p. injected at 0.6U/kg for ITT and at 1U/kg for western blot analysis of insulin signaling. Blood glucose was measured by glucometer (Onetouch) via sampling from the tail. HFD containing 60 kcal % fat was purchased from Research Diets Inc. (D12492i). Treadmill running were performed with Columbus instrument connected with a shock detection module. All the animal care and use procedures followed the guidelines of the Institutional Animal Care and Use Committee in UPenn and Baylor.
DNA construct, western blot, and RT-qPCR
Plasmid backbone for cloning sgRNA was previously described 61. Adenovirus was produced using Invitrogen’s system. For Western blot, tissues or cells were lysed in RIPA buffer supplemented with phosphatase and protease inhibitors. Lysates were resolved by Tris-glycine SDS-PAGE, transferred to PVDF membrane, and blotted with antibodies against HDAC3 (Abcam 7030), Ampd3 (Abcam 194361), MHC isoform I (BA-F8), IIa (2F7), IIx (6H1), IIb (BF-F3) (Developmental Studies Hybridoma Bank at the University of Iowa), phospho-Akt at S473 (pAkt-S473) (Cell Signaling Tech CST9271), pAkt-T308 (CST4056), total Akt (CST9272), pIRS1-S1101 (CST2385), IRS1 (CST2390), pGSK3β-S9 (CST5558), GSK3β (CST9315), pS6-S235/236 (CST4858), S6 (CST2217), Hsp90 (CST4874), pAMPKα-T172 (CST4188), pAMPKβ1-S108 (CST4181), AMPKβ1/2 (CST4150), pHSL-S660 (CST4126), HSL (CST4107), GLUT4 (Millipore 07-1404), and GAPDH (CST2118). For quantification of western blot results, images were acquired and analyzed using Quantity One (Bio-Rad). For RT-qPCR or RNA-seq, total RNA was extracted using TRIzol (Invitrogen) and High Pure RNA tissue kit (Roche). Reverse transcription and quantitative PCR was performed with High Capacity RT kit, SYBR Green PCR Master Mix, and the Quant Studio 6 instrument (Life Science) using absolute quantification method with standard curves. 36B4 (Arbp) or 18S RNA was used as the housekeeping control.
Histology and tissue metabolites measurement
Muscles were freshly embedded in OTC and dipped into isopentane cooled by liquid nitrogen. 10-µm cross sections were prepared and stained with indicated antibodies. For measuring triglyceride, tissues were lysed in lysis buffer (140 mM NaCl, 50 mM Tris and 1% Triton X-100, pH 8.0) followed by triglyceride assay using LiquiColor kit (Stanbio). For measuring glycogen, tissues were homogenized in 0.5 N KOH. Glycogen was precipitated by ethanol and digested with 0.25 mg/ml amyloglycosidase (Sigma), followed by glucose assay using HK kit (Sigma).
Energy expenditure, body composition, and hyperinsulinemic clamp
Oxygen consumption, NMR body composition, and the insulin clamp study was performed at the Mouse Phenotyping Core at UPenn. For insulin clamp, mice were allowed to recover a week from single catheterization surgery. Human insulin was infused at 2.5 mU/kg/min into unrestrained mice, and blood glucose levels were maintained between 120–140 mg/dl by infusing 20% glucose at various rates. 3H-glucose and 14C-deoxyglucose was used to trace endogenous glucose production and tissue-specific glucose uptake respectively.
Cellular assays and pulse stimulation
C2C12 myocytes (from ATCC, tested negative for mycoplasma contamination) were plated in 12-well plate in complete DMEM medium. Differentiation was induced in 2% horse serum. C2C12 cells were not listed in the database of commonly misidentified cell lines maintained by ICLAC. Adenovirus was applied after cells were fully differentiated. Electric pulse stimulation (EPS) was applied using the IonOptix C-pace system with 10–15 volts at 1 Hz for 6 to 12 hours. For fatty acids oxidation, cells were incubated in PBS supplemented with [9,10-3H(N)]-palmitate, conjugated on BSA and carnitine for 15–120 min. The resultant 3H2O in the incubation solution was separated from precursors using ion-exchange columns (DOWEX 1X4-400), and was measured by scintillation counter. Amino acid oxidation was done with D-[2, 3-3H]-aspartate tracer in a similar procedure. For cellular glucose uptake, 2-[3H(N)]-deoxy-D-glucose was used in the presence or absence of 20 µM cytochalasin B, an inhibitor of glucose transport. The cells were washed two times with PBS and harvested. Intracellular glucose was counted for 3H. Cellular respirometry was performed using Seahorse XF24 analyzer with the mitochondrial stress test kit. The culture media in Seahorse analysis is based on Dulbecco’s Modified Eagle’s Medium (DMEM). It contains multiple vitamins, inorganic salts, and (in mg/L) pyruvate 110, glucose 200, together with the following amino acids: glycine 30, arginine hydrochloride 84, cystine-2HCl 63, valine 94, histidine hydrochloride-H2O 42, isoleucine 105, leucine 105, threonine 95, lysine hydrochloride 146, serine 42, methionine 30, phenylalanine 66, tryptophan 16, and tyrosine disodium salt dihydrate 104.
Isotope tracing in vivo and metabolomics
A bolus of 0.4mg/g 13C6-glucose (Sigma 389374) was injected into mice via jugular vein catheter, followed by continuous infusion of 13C6-glucose at 12 mg/kg/min and 3.3 ul/min for 150 min. Mice ran on treadmill at 10 m/m for the last 45 min before tissue harvest. About 30 mg of frozen tissue samples were weighed and then pulverized in a CryoMill machine (Retsch, Germany) with stainless ball at liquid nitrogen temperature. For water-soluble metabolites, the pulverized tissue powder was mixed by vortexing with 1 mL of -80°C 80:20 methanol:water, and sit on dry ice for 10 min. Samples were centrifuged at 13,000 rpm for 10 min and the supernatants collected as the first extract. The insoluble material was extracted one more time with 1 mL of -80°C 80:20 methanol:water. The supernatants from two rounds of extraction were combined, dried under nitrogen flow, and re-dissolved in LC-MS grade water using a ratio of 1 mL of water per 25 mg initial tissue weight and analyzed using Liquid chromatography mass spectrometry. Negative charged metabolites were analyzed via reverse-phase ion-pairing chromatography coupled to an Exactive orbitrap mass spectrometer (Thermo Fisher). The mass spectrometer was operated in negative ion mode with resolving power of 100,000 at m/z 200, scanning range being m/z 75-1000. The LC method has been described before 62, using a Synergy Hydro-RP column (100 mm × 2 mm, 2.5 µm particle size, Phenomenex, Torrance, CA) with a flow rate of 200 µL/min. The LC gradient was 0 min, 0% B; 2.5 min, 0% B; 5 min, 20% B; 7.5 min, 20% B; 13 min, 55% B; 15.5 min, 95% B; 18.5 min, 95% B; 19 min, 0% B; 25 min, 0% B. Solvent A is 97:3 water:methanol with 10 mM tributylamine and 15 mM acetic acid; solvent B is methanol. Data analyses were performed using MAVEN software for peak alignment and extracted ion chromatogram visualization 63. Ion signal of each metabolites, both unlabeled and 13C-labeled forms, were further processed to correct for the natural isotope abundance. Fractional contribution of pyruvate from circulating glucose was calculated by dividing muscle 13C-Pyr % with blood 13C-glucose %. For measuring in vivo fatty acid oxidation or aspartate oxidation using radioactive isotopes, mice were i.p. injected with 1mCi [9,10-3H(N)]-palmitate or [2, 3-3H]-aspartate respectively. 30ul serum was collected at 20 min after injection from tail. 3H-H2O in the serum was separated from precursors using ion-exchange columns followed by scintillation counting. Mice in the exercise group ran on treadmill at 10m/min for 30 mins before injection and continued to run treadmill for 20 min after injection before tissue harvest.
Ex vivo muscle physiology
Muscle physiological analysis was performed on isolated EDL muscles using an Aurora Mouse 1200A System equipped with Dynamic Muscle Control v.5.415 software. EDL muscles were dissected and analyzed in constantly oxygenated Ringer's solution (100 mM NaCl, 4.7 mM KCl, 3.4 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 25 mM HEPES, 5.5 mM D-glucose and 2% BSA) without or with 200 µM deoxycoformycin (DCF) at 24°C. Maximal isometric twitch and tetanic contractions were obtained using a stimulation frequency of 2500 Hz for 0.2 msec or 120 Hz for 500 msec respectively. Five minutes were allowed between two tetanic contractions to ensure muscle recovery. For induction of fatigue, five minutes after the last maximal tetanic contraction, muscles were stimulated every second for 8 minutes using 40 Hz pulses lasting 330 ms (Loro et al. 2015). The fatigue index, was expressed as EDL % force drop (the percentage difference in force between the first contraction and every subsequent contraction) or as % remaining force (the percentage of force left after the preceding contraction). Following the fatigue protocol, a burst of 50 maximal tetanic contractions (120 Hz for 500 ms) was applied to maximize the fatigue to comparable levels between the samples. The recovery protocol started one second after the last burst contraction. A maximal tetanic stimulation (120 Hz for 500 ms) was given every 5 minutes for 30 minutes and the force recovery was expressed as % of the maximal isometric tetanic force.
Proteomics
Protein was extracted from muscle tissue of triplicates for each wildtype and HDAC3 knock-out mice with frozen grinding in 8M urea. 100 µg of protein from each sample were precipitated with acetone, and protein pellets were re-suspended and denatured in urea and reduced with DTT prior to alkylation of cysteines with iodoacetamide. Proteins were digested with LysC (Wako Chemicals) followed by trypsin (Promega) and desalted with Empore C18 STaGETips (3M). Peptides were resuspended in 100 mM triethylammonium bicarbonate, and peptides were labeled with 6-plex TMT (ThermoScientific) following the manufacturer’s instructions. After checking for labeling efficiency (>97%), samples were mixed together, and peptides were fractionated into 7 fractions by strong anion exchange (SAX). 60 ug of peptides were loaded at pH11 onto 8 layers of SAX StaGE-Tips, and pH steps of 11 (flow-through), 8, 6, 5, 4, and 3 were collected and desalted. One-tenth of each fraction was injected for nano-LC-MS/MS analysis (QExactive, ThermoScientific). Peptides were separated using a 12 cm×75µm C18 column (Nikkyo Technos Co., Ltd. Japan) at a flow rate of 200 nL/min, with a 5–40% gradient over 160 minutes (buffer A 0.1% formic acid, buffer B 0.1% formic acid in acetonitrile). The QExactive was operated in data-dependent mode, with a top 20 method. Nano-LC-MS/MS data were analyzed using MaxQuant and Perseus software, searching against a Uniprot Mus musculus database, allowing oxidation of methionine and protein N-terminal acetylation, and filtering at a 1% false discovery rate at the peptide and protein level.
RNA-seq, ChIP-seq, and GRO-seq
RNA-seq was performed using total RNA extracted from tibialis anterior muscles at ZT10 in 4-month old mice when they are resting (n = 3). ChIP and ChIP-seq were described previously 64. Briefly, tissues were grounded in liquid N2 and cross-linked in 1% Formaldehyde for 20 min. Whole cell extracts were sonicated followed by immunoprecipitation with antibodies for HDAC3 (Abcam). For ChIP-seq, ChIP was performed independently on muscles from 5 WT mice harvested at ZT10 and ZT22. The precipitated DNA were then pooled and amplified according to the guide of Illumina, followed by deep sequencing on Illumina Genome Analyzer IIx. HDAC3-KO muscles were included in ChIP-seq as negative input for peak calling. GRO-seq was performed using quadriceps muscles from the same cohort of mice as RNA-seq, following the protocol described previously 33. Briefly, nuclei were extracted from muscles using hypotonic buffer. Nuclear run-on was performed in the presence of Br-UTP followed by enrichment with anti-BrUTP antibodies, reverse transcription, and library preparation. Run-on reactions from 5 mice were pooled to make one sequencing library.
GRO-seq data processing
GRO-seq sequencing reads were aligned to the mm9 genome using Bowtie v0.12.7. Uniquely mapped reads were extended to 150bp in the 5′to 3′direction and used for downstream analysis. Genome browser tracks were generated using Homer v4.8 and visualized in Integrative Genome Browser (IGV). eRNA identification and quantification were performed according to previously established protocols (Fang et al. 2014).
ChIP-seq data processing
Sequenced reads were aligned to the mm9 genome using Bowtie v0.12.7. Non-redundant and uniquely mapped reads were extended to 150bp in the 5′ to 3′ direction and used for downstream analysis. Genome browser tracks were generated using Homer v4.8and visualized in Integrative Genome Browser (IGV). Peak calling was performed with Homer using HDAC3 knockout sample as input control. Peaks higher than 1 read per million (rpm) were used for comparison between ZT10 and ZT22. Common and differential peaks at ZT10 and ZT22 were found using the intersect function of Bedtools v2.21.0 and a Venn diagram was generated using the R package Vennerable. Motif mining was performed using Homer in 200bp regions surrounding peak centers. Peaks were annotated with Homer using the nearest mapping within 50kb of gene TSS and gene ontology was performed using DAVID Bioinformatics Resources 6.7.
RNA-seq data processing
Sequenced reads from biological replicates were aligned to the mm9 genome using Tophat v2.0.11 using default parameters. Genome browser tracks were generated using Homer v4.8 and visualized in Integrative Genome Browser (IGV). Normalized expression values and differential gene expression were obtained using cuffdiff from Cufflinks-v2.2.1.
Statistics
n ≥ 5 in each mouse group were used for all metabolic studies considering individual variations based on previous studies. Littermates were used as control mice for all experiments without randomization. No mice were excluded. Metabolomics, proteomics, CLAMP, body composition, fiber quantification, indirect calorimetry, muscle fatigue, and RNA-seq were performed by experimenters who were blinded to the mouse genotype. Student's two-tail t-test was performed for all experiments to determine significance of differences between two groups except indicated otherwise. Data met the assumption of the statistic tests. Variances were similar between the groups being statistically compared. All experiments were replicated at least twice in the laboratory except the CLAMP, RER, and –omics studies that were performed once. All statistical analysis and S.E.M. calculations were based on individual mice or wells of cells.
Data availability
ChIP-seq, RNA-seq, and GRO-seq data are available in GEO (GSE79696).
Supplementary Material
Acknowledgments
We thank Dr. Steve Burden for MLC-Cre mouse, Dr. John Hogenesch for processed data from circaDB, Drs. Morris Birnbaum and Joseph Baur for helpful discussion, Dr. Shaodong Guo for help with the EPS procedure, Dr. Pumin Zhang for adenoviral plasmids, Drs. Sheng Hui and Junyoung Park for analysis of metabolomics data, Dr. Vihang Narkar for immunostaining protocol, Dr. Marcus Goncalves for help with muscle histology, and Cristina Lanzillotta for technical assistance. We thank the Penn Diabetes Center (DK19525) Functional Genomics Core for nucleotide sequencing, Mouse Metabolic Phenotyping Core for clamp experiments, Penn Muscle Institute Muscle Physiology Assessment Core for muscle contraction study, and Princeton/Penn Regional Metabolomics Core for flux and lipid analysis. We thank Baylor Diabetes Center Metabolism Core for Seahorse analysis, Vanderbilt MMPC (DK59637) for lipidomics analysis, Leona M. and Harry B. Helmsley Charitable Trust for supporting Rockefeller Proteomics Center, and NIH (CA211437) for supporting W.L. The work was supported by NIH grant DK043806 (M.A.L.) and DK099443 (Z.S.).
Footnotes
ACCESSIOIN CODES
ChIP-seq, RNA-seq, and GRO-seq data are available in GEO (GSE79696).
AUTHOR CONTRIBUTION
ZS and MAL conceived the study and designed experiments. SH, WZ, WL, EL, GD, JJ, SZ, YZ, DF, QC, BDD, and ZS conducted experiments. SH, WZ, BF, WL MD, GD, SZ, BDD, HM, and ZS analyzed the data. EL, TSK, JDR, MAL, and ZS interpreted the data. MAL and ZS acquired funding. ZS wrote the manuscript with the input from other authors.
COMPETING FINANCIAL INTERESTS STATEMENT
The authors disclose no competing financial conflict of interest.
References
- 1.DeFronzo RA, Tripathy D. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 2009;32(Suppl 2):S157–163. doi: 10.2337/dc09-S302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubé JJ, et al. Exercise-induced alterations in intramyocellular lipids and insulin resistance: the athlete’s paradox revisited. Am. J. Physiol. Endocrinol. Metab. 2008;294:E882–888. doi: 10.1152/ajpendo.00769.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brüning JC, et al. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell. 1998;2:559–569. doi: 10.1016/s1097-2765(00)80155-0. [DOI] [PubMed] [Google Scholar]
- 5.Kim JK, et al. Redistribution of substrates to adipose tissue promotes obesity in mice with selective insulin resistance in muscle. J. Clin. Invest. 2000;105:1791–1797. doi: 10.1172/JCI8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holloszy JO. ‘Deficiency’ of mitochondria in muscle does not cause insulin resistance. Diabetes. 2013;62:1036–1040. doi: 10.2337/db12-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307:384–387. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 8.Szendroedi J, Phielix E, Roden M. The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2012;8:92–103. doi: 10.1038/nrendo.2011.138. [DOI] [PubMed] [Google Scholar]
- 9.Muoio DM, Neufer PD. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 2012;15:595–605. doi: 10.1016/j.cmet.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randle PJ, Garland PB, Hales CN, Newsholme EA. The glucose fatty-acid cycle. Its role in insulin sensitivity and the metabolic disturbances of diabetes mellitus. Lancet. 1963;1:785–789. doi: 10.1016/s0140-6736(63)91500-9. [DOI] [PubMed] [Google Scholar]
- 11.Hue L, Taegtmeyer H. The Randle cycle revisited: a new head for an old hat. Am. J. Physiol. Endocrinol. Metab. 2009;297:E578–591. doi: 10.1152/ajpendo.00093.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelley DE, Mandarino LJ. Fuel selection in human skeletal muscle in insulin resistance: a reexamination. Diabetes. 2000;49:677–683. doi: 10.2337/diabetes.49.5.677. [DOI] [PubMed] [Google Scholar]
- 13.Muoio DM. Metabolic inflexibility: when mitochondrial indecision leads to metabolic gridlock. Cell. 2014;159:1253–1262. doi: 10.1016/j.cell.2014.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc. Natl. Acad. Sci. U.S.A. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatori M, et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol. Endocrinol. Baltim. Md. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 19.Feng D, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Z, et al. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nat. Med. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Feng D, Everett LJ, Bugge A, Lazar MA. Circadian epigenomic remodeling and hepatic lipogenesis: lessons from HDAC3. Cold Spring Harb. Symp. Quant. Biol. 2011;76:49–55. doi: 10.1101/sqb.2011.76.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harfmann BD, Schroder EA, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. J. Biol. Rhythms. 2015;30:84–94. doi: 10.1177/0748730414561638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayeuf-Louchart A, Staels B, Duez H. Skeletal muscle functions around the clock. Diabetes Obes. Metab. 2015;17(Suppl 1):39–46. doi: 10.1111/dom.12517. [DOI] [PubMed] [Google Scholar]
- 24.Kang S, Tsai LT-Y, Rosen ED. Nuclear Mechanisms of Insulin Resistance. Trends Cell Biol. 2016 doi: 10.1016/j.tcb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bottomley MJ, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 27.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Selective expression of Cre recombinase in skeletal muscle fibers. Genes. N. Y. N 2000. 2000;26:165–166. [PubMed] [Google Scholar]
- 29.Loro E, et al. IL-15Rα is a determinant of muscle fuel utilization, and its loss protects against obesity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;309:R835–844. doi: 10.1152/ajpregu.00505.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zierath JR, Hawley JA. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2004;2:e348. doi: 10.1371/journal.pbio.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 32.Nedachi T, Fujita H, Kanzaki M. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1191–1204. doi: 10.1152/ajpendo.90280.2008. [DOI] [PubMed] [Google Scholar]
- 33.Fang B, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159:1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 35.Calvo JA, et al. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J. Appl. Physiol. Bethesda Md 1985. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- 36.Choi CS, et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19926–19931. doi: 10.1073/pnas.0810339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong KE, et al. Muscle-Specific Overexpression of PGC-1α Does Not Augment Metabolic Improvements in Response to Exercise and Caloric Restriction. Diabetes. 2015;64:1532–1543. doi: 10.2337/db14-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finck BN, et al. A potential link between muscle peroxisome proliferator- activated receptor-alpha signaling and obesity-related diabetes. Cell Metab. 2005;1:133–144. doi: 10.1016/j.cmet.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Pagel-Langenickel I, Bao J, Pang L, Sack MN. The role of mitochondria in the pathophysiology of skeletal muscle insulin resistance. Endocr. Rev. 2010;31:25–51. doi: 10.1210/er.2009-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348:1488–1492. doi: 10.1126/science.aab3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woldt E, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 2013;19:1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto H, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau P, Fitzsimmons RL, Pearen MA, Watt MJ, Muscat GEO. Homozygous staggerer (sg/sg) mice display improved insulin sensitivity and enhanced glucose uptake in skeletal muscle. Diabetologia. 2011;54:1169–1180. doi: 10.1007/s00125-011-2046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samant SA, Pillai VB, Sundaresan NR, Shroff SG, Gupta MP. Histone Deacetylase 3 (HDAC3)-dependent Reversible Lysine Acetylation of Cardiac Myosin Heavy Chain Isoforms Modulates Their Enzymatic and Motor Activity. J. Biol. Chem. 2015;290:15559–15569. doi: 10.1074/jbc.M115.653048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagenmakers AJ. Muscle amino acid metabolism at rest and during exercise: role in human physiology and metabolism. Exerc. Sport Sci. Rev. 1998;26:287–314. [PubMed] [Google Scholar]
- 46.Gibala MJ, Young ME, Taegtmeyer H. Anaplerosis of the citric acid cycle: role in energy metabolism of heart and skeletal muscle. Acta Physiol. Scand. 2000;168:657–665. doi: 10.1046/j.1365-201x.2000.00717.x. [DOI] [PubMed] [Google Scholar]
- 47.Arinze IJ. Facilitating understanding of the purine nucleotide cycle and the one-carbon pool: Part I: The purine nucleotide cycle. Biochem. Mol. Biol. Educ. Bimon. Publ. Int. Union Biochem. Mol. Biol. 2005;33:165–168. doi: 10.1002/bmb.2005.494033032469. [DOI] [PubMed] [Google Scholar]
- 48.Bowtell JL, Marwood S, Bruce M, Constantin-Teodosiu D, Greenhaff PL. Tricarboxylic acid cycle intermediate pool size: functional importance for oxidative metabolism in exercising human skeletal muscle. Sports Med. Auckl. NZ. 2007;37:1071–1088. doi: 10.2165/00007256-200737120-00005. [DOI] [PubMed] [Google Scholar]
- 49.She P, et al. Disruption of BCAA metabolism in mice impairs exercise metabolism and endurance. J. Appl. Physiol. Bethesda Md 1985. 2010;108:941–949. doi: 10.1152/japplphysiol.01248.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ginevičienė V, et al. AMPD1 rs17602729 is associated with physical performance of sprint and power in elite Lithuanian athletes. BMC Genet. 2014;15:58. doi: 10.1186/1471-2156-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van den Berghe G, Bontemps F, Vincent MF, Van den Bergh F. The purine nucleotide cycle and its molecular defects. Prog. Neurobiol. 1992;39:547–561. doi: 10.1016/0301-0082(92)90006-z. [DOI] [PubMed] [Google Scholar]
- 52.Operti MG, Vincent MF, Brucher JM, van den Berghe G. Enzymes of the purine nucleotide cycle in muscle of patients with exercise intolerance. Muscle Nerve. 1998;21:401–403. doi: 10.1002/(sici)1097-4598(199803)21:3<401::aid-mus16>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 53.Templeman NM, Schutz H, Garland T, McClelland GB. Do mice bred selectively for high locomotor activity have a greater reliance on lipids to power submaximal aerobic exercise? Am. J. Physiol. Regul. Integr. Comp. Physiol. 2012;303:R101–111. doi: 10.1152/ajpregu.00511.2011. [DOI] [PubMed] [Google Scholar]
- 54.Overmyer KA, et al. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 2015;21:468–478. doi: 10.1016/j.cmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.She P, et al. Disruption of BCATm in mice leads to increased energy expenditure associated with the activation of a futile protein turnover cycle. Cell Metab. 2007;6:181–194. doi: 10.1016/j.cmet.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng J, et al. AMPD1: a novel therapeutic target for reversing insulin resistance. BMC Endocr. Disord. 2014;14:96. doi: 10.1186/1472-6823-14-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang C, et al. A branched-chain amino acid metabolite drives vascular fatty acid transport and causes insulin resistance. Nat. Med. 2016;22:421–426. doi: 10.1038/nm.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hawley JA, Schabort EJ, Noakes TD, Dennis SC. Carbohydrate-loading and exercise performance. An update. Sports Med. Auckl. NZ. 1997;24:73–81. doi: 10.2165/00007256-199724020-00001. [DOI] [PubMed] [Google Scholar]
- 59.Phinney SD. Ketogenic diets and physical performance. Nutr. Metab. 2004;1:2. doi: 10.1186/1743-7075-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paoli A, Bianco A, Grimaldi KA. The Ketogenic Diet and Sport: A Possible Marriage? Exerc. Sport Sci. Rev. 2015;43:153–162. doi: 10.1249/JES.0000000000000050. [DOI] [PubMed] [Google Scholar]
- 61.Cheng R, et al. Efficient gene editing in adult mouse livers via adenoviral delivery of CRISPR/Cas9. FEBS Lett. 2014;588:3954–3958. doi: 10.1016/j.febslet.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 62.Lu W, et al. Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 2010;82:3212–3221. doi: 10.1021/ac902837x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 2010;82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun Z, et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell. 2013;52:769–782. doi: 10.1016/j.molcel.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq, RNA-seq, and GRO-seq data are available in GEO (GSE79696).