Abstract
Early detection of breast cancer is a critical component in patient prognosis and establishing effective therapy regimens. Here, we developed an easily accessible yet potentially powerful sensor to detect cancer cell targets by utilizing seven dual-ligand cofunctionalized gold nanoclusters (AuNCs) as both effective cell recognition elements and signal transducers. On the basis of this AuNC multichannel sensor, we have successfully distinguished healthy, cancerous and metastatic human breast cells with excellent reproducibility and high sensitivity. Triple negative breast cancer cells (TNBCs), which exhibit low expression of the estrogen receptor, progesterone receptor, and human epidermal growth factor receptor-2, were identified. The high accuracy of the blind breast cell sample tests further validates the practical application of the sensor array. In addition, the versatility of the sensor array is further justified by identifying amongst distinct cell types, different cell concentrations and cell mixtures. Notably, the drug-resistant cancer cells can also be efficiently discriminated. Furthermore, the dual-ligand cofunctionalized AuNCs can efficiently differentiate different cells from the peripheral blood of tumor-free and tumor-bearing mice. Taken together, this fluorescent AuNCs based array provides a powerful cell analysis tool with potential applications in biomedical diagnostics.
1. Introduction
Breast cancer is the most common invasive malignancy diagnosed and the second leading cause of cancer fatality in women worldwide [1, 2]. Early breast cancer detection holds great promise for effective therapy [3–5]. Among them, triple negative breast cancers (TNBCs) are an aggressive breast cancer subtype defined by low expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor-2 (HER2) [6, 7]. Although TNBCs represent only 15 to 20% of all breast cancer cases [8, 9], they are responsible for a greater proportion of metastatic cases and deaths [9–11]. The high mortality rate appears to be due to the intrinsic aggressiveness of cancer cells, as well as the lack of effective diagnostic methods and targeted therapeutic strategies [12]. Therefore, the availability of rapid and sensitive methods to identify breast cancer cells, particularly TNBCs, may provide significant insight for predicting disease conditions and cancer treatment [13, 14].
Traditional techniques for cancer cell detection mainly apply molecular ligands (e.g., peptides, aptamers, and antibodies) that are highly specific to predetermined biomarkers of the target cell population [15, 16]. However, the identification of TNBCs by the representative approaches (e.g., ELISA-type tests [17], gel electrophoresis [18, 19], proteomics and related approaches coupled with mass spectrometry [20], RT-PCR [21], as well as immunotyping by flow cytometry [22, 23]) remains challenging due to the constrains in the availability of specific molecular biomarkers that can discriminate between TNBC cells and nonneoplastic cells. In addition, no biomarker is established as a cancer screening tool that has sufficient sensitivity to distinguish between normal, cancerous, and metastatic cell types [24]. Therefore, it is still highly appealing to develop facile and efficient methods for breast cell type analysis, especially for TNBCs.
Unbiased “chemical nose” array sensors may be considered as potential alternatives for cell discrimination, allowing identification through selective recognition [25, 26]. In the chemical nose strategy, a sensor array is developed to provide differential binding interactions with analytes via nonspecific receptors, generating fingerprint-like response patterns that can be statistically analyzed and utilized for discriminative identification [27, 28]. Analogous to our own noses, chemical nose sensors preclude the need of prior knowledge of the analytes and are instead “trained” to identify analytes [29, 30]. A wealth of applications of chemical nose sensors are demonstrated, including detection of metal ions [31], volatile organic compounds [32, 33], carbohydrates [34, 35], amino acids [36, 37], and proteins [38–45]. Recently, these strategies have been expanded to more complex systems, such as cell [46–55] and bacteria [56–61] sensing.
Various receptor systems have been employed for array-based sensing of cells, including fluorescent polymers [53], green fluorescent proteins [46, 50, 55], fluorescently labeled DNAs [52, 54], magnetic glyco-nanoparticles [51], and gold nanomaterials [48, 49]. Although these methods are capable of discerning cells, these systems generally require a large population of cells. For instance, Rotello and coworkers fabricated an array-based system for discrimination of normal, cancerous and metastatic cell types using conjugated polymer/gold nanoparticle constructs with a detection limit of higher than 20000 cells [53]. In addition, Fan and Hu applied adaptive “ensemble aptamers” that exploited the collective recognition abilities of a small set of rationally designed, nonspecific DNA sequences to identify a wide range of molecular or cellular targets discriminatively, including different cell lines with a limit of detection of 5000 cells [52]. Micrometastases remain undetectable by conventional means; detection methods for small numbers of cancer cells are a prerequisite to early intervention.
Generally, biosensors often possess two basic functional components: recognition units and transducers, which are usually divided into two parts and composed of two distinctive elements [30]. We here construct an array based cell identification system by using seven dual-ligand cofunctionalized gold nanoclusters (AuNCs) as both effective cell recognition elements and signal transducers (Scheme 1). Compared with organic dyes and quantum dots based fluorescent probes, AuNCs are more promising for bioanalysis due to their simple synthesis, favorable biocompatibility, strong photoluminescence and high stability for long-time observations [62–65]. This sensing system is designed utilizing one species with dual roles, which offers extra advantages in terms of function integration and low cost. As we know, cell membrane surfaces of different cell populations are comprised of distinctive types and amounts of integral membrane proteins, carbohydrates and amphipathic phospholipids [53, 66]. The as-prepared AuNCs showed similar fluorescence properties but distinctive surface properties, characterized by their unique surface charge states and chemistry. These differences of AuNCs may provide differential binding affinities and hence diverse interactions with individual cell lines [67], resulting in distinct fluorescence responses for each cell type. The additive responses offer an efficient means of identification. This sensing array is capable of efficiently distinguishing healthy, cancerous and metastatic human breast cells with excellent reproducibility and high sensitivity, including TNBC cells. In addition, the versatility of the sensor array is further demonstrated by identifying amongst distinct cell types, different cell concentrations, cell mixtures, and discrimination of unknown cell samples. Furthermore, the dual-ligand cofunctionalized AuNCs can identify nonresistant and drug-resistant cancer cells, and differentiate cells from the peripheral blood of tumor-free and tumor-bearing mice, providing new opportunities for cancer diagnosis.
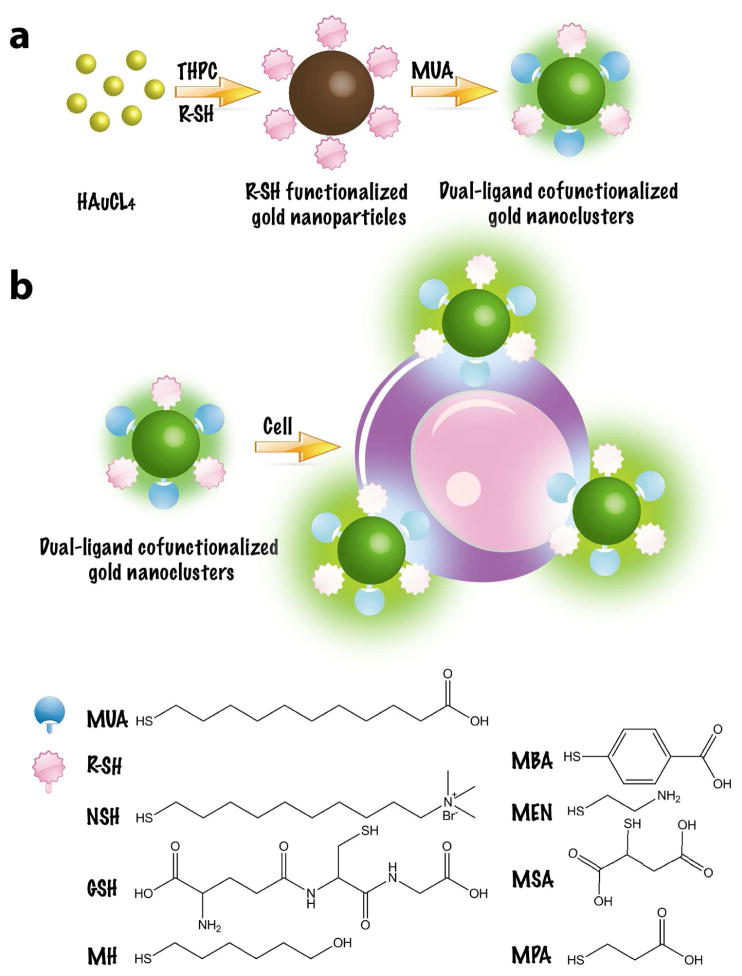
Scheme 1.
(a) Schematic illustration of the synthesis of the dual-ligand cofunctionalized gold nanoclusters. (b) Preparation of sensor array for cell identification based on dual-ligand cofunctionalized gold nanoclusters.
2. Materials and methods
2.1. Materials and instrumentation
11-mercaptoundecanoic acid (MUA), 6-mercapto-1-hexanol (MH), (11-mercaptoundecyl)-N,N,N-trimethylammonium bromide (NSH), 4-mercaptobenzoic acid (MBA), (2-mercaptoethyl)amine (MEN), glutathione (GSH), mercaptosuccinic acid (MSA), 3-mercaptopropionic acid (MPA), tetrakis (hydroxymethyl) phosphonium chloride (THPC), and gold(III) chloride hydrate (HAuCl4) were purchased from Sigma-Aldrich. Dulbecco’s phosphate buffered saline (PBS) was purchased from Invitrogen (Carlsbad, CA). All other reagents were all of analytical reagent grade and used as received. Nanopure water (18.2 MΩ; Millpore Co., USA) was used throughout the experiment. TEM images were recorded using a JEOL 2100 transmission electron microscope operating with an accelerating voltage of 200 kV. Zeta potential measurements were performed with a ZetaPALS zeta potential analyzer. Fluorescence measurements were carried out by using a microplate spectrofluorometer GeminiXPS. Mass spectrometry (MS) experiments were carried out using the Waters ESI-Q-TOF MS (m/z 70–1000), which can quantify the surface thiolate ligands on the dual-ligand cofunctionalized gold nanoclusters. NaCN (20 mM) was used to etch the purified dual-ligand cofunctionalized gold nanoclusters (100 nM, 1.0 mL) to liberate the thiolate ligands from the gold nanocluster surface. Each type of thiolates with the concentration range from 0 to 100 uM was prepared for the quantification curve. DTT (20 mM) was conducted to avoid the formation of disulfide bonds of thiolates. The etching sample (5.0 μL) was dispersed in 1:1 water/ethanol solution (5.0 μL). Each mixture was then injected into a capillary trap column. After that, the MS was operated with a spray potential of 3.0 kV. NanoLockSpray source was utilized for accurate mass measurement, with the lock mass channel sampled every 30 s. Data acquisition was carried out in data direct analysis mode. The concentration of thiolate was calculated according to the MS signal of sample and thiolate quantification curve.
2.2. Cell culture
MDA-MB-231, MDA-MB-436, MDA-MB-157, MCF7, SKBR3, MCF10A, HCC1806, HCC1569, Hs578T, MDA-MB-468, DU4475, HUVEC, PC3, 3T3 and 4T1 cells were all obtained from American Type Culture Collection (ATCC, Manassas, VA). MDA-MB-231, MCF7, MDA-MB-436, HUVEC, PC3 and 3T3 cells were cultured in DMEM (Corning, Corning, NY), supplemented with 10% FBS (Life Technologies, Carlsbad, CA) + 1% Pen/Strep (Life Technologies, Carlsbad, CA). SKBR3 cells were cultured in McCoy5a (Life Technologies, Carlsbad, CA) + 10% FBS. MCF10A cells were cultured in DMEM/F12 (Life Technologies, Carlsbad, CA) + 5% Horse Serum (Life Technologies, Carlsbad, CA) + 20 ng/ml of Epidermal Growth Factor (EGF) (Peprotech, NJ) + 0.5 mg/ml of Hydrocortisone (Sigma-Aldrich, St. Louis, MO) + 100 ng/ml Cholera Toxin (Sigma-Aldrich, St. Louis, MO) + 10 mg/ml of Insulin (Sigma-Aldrich, St. Louis, MO) + 1% Pen/Strep (Life Technologies, Carlsbad, CA). MDA-MB-157 cells were cultured in L-15 (ATCC) + 10% FBS. HCC1806, HCC1569, MDA-MB-468 and DU4475 cells were cultured in RPMI-1640 (Corning) + 10% FBS. Hs578T cells were cultured in DMEM, supplemented with 10% FBS + 10 mg/ml of Insulin. 4T1 cells were cultured in RPMI- 1640 + 10% FBS + 1% Pen/Strep. All cell lines were grown in a humidified atmosphere (5% CO2) at 37 °C.
2.3. Synthesis of dual-ligand cofunctionalized AuNCs
Dual-ligand cofunctionalized AuNCs were synthesized according to previous reports with slight modification [40, 68]. Firstly, 100 μL of NaOH (1 M) solution and 24 μL of THPC (8%, wt) solution were mixed with 8 mL of ultrapure water under violent stirring for 5 min, and then, 400 μL of HAuCl4 (Au(III), 24 mM) solution was added rapidly. The color of the solution turns from light-yellow to brown in 1 min, indicating the formation of small gold nanoparticles. At this point, 100 μL of thiolate (R-SH, 10 mM) solutions was added to obtain different thiolate-protected gold nanoparticles. After stirring for another 15 min at room temperature, the solution was cooled to 4 °C for further use. After aging for 1 day, 1 mL of thiolate-protected gold nanoparticles stock solution was mixed with 200 μL of carbonate buffer (0.1 M, pH 9.0) and 75 μL of MUA (0.1 M) ethanol solution in a thermomixer. The solution turns to light yellow (from brown) in color within 1 h and emits green light under UV (365 nm) light illumination, indicating the formation of fluorescent gold nanoclusters. At the 2 h point, the reaction was stopped. The resulted gold nanoclusters were purified by centrifugation at 13 000 rpm for 20 min to eliminate large aggregates and, then, was filtered with 10 kDa cutoff ultrafiltration centrifuge tubes to remove excess reactants (MUA, thiolates, salts, etc.). The purified gold nanoclusters were stored under a dark condition for further use.
2.4. Quantum Yield (QY) Measurements
The quantum yields (QYs) of the fluorescent AuNCs were detected [69]. Quinine sulfate in 0.1 M H2SO4 (QY = 0.577) was chosen as a standard with the dual-ligand cofunctionalized AuNCs. The QYs of the fluorescent AuNCs were measured by comparing the integrated photoluminescence intensities and the absorbency values with the quinine sulfate. The QYs of AuNCs are calculated using following equation: Φ = Φs(I/Is)(As/A)(ns/n)2. Where Φ is the quantum yield, I is the measured integrated emission intensity, n is the refractive index of the solvent (1.33 for water), and A is the optical density. The subscript “s” refers to the reference standard with known quantum yield. To minimize reabsorption effects, the optical density is kept below 0.1.
2.5. Endotoxin testing
The endotoxin levels in dual-ligand cofunctioanlized gold nanoclusters (25 nM) were determined using a Limulus Amebocyte Lysate (LAL) assay kit (Thermo scientific, USA) following the manufacturer's instructions.
2.6. Cytotoxicity assays
MTT assays were used to probe cellular viability. Cells were seeded at a density of 5000 cells well−1 (100 μL total volume well−1) in 96-well assay plates. After 24 h incubation, the as-prepared AuNCs, at the indicated concentrations, were added for further incubation of 24 h. To determine toxicity, 10 μL of MTT solution was added to each well of the microtiter plate and the plate was incubated in the CO2 incubator for an additional 4 h. Then the cells were lysed by the addition of 100 μL of DMSO. Absorbance values of formazan were determined with microplate reader at 490 nm (corrected for background absorbance at 630 nm). Three replicates were done for each treatment group.
2.7. Membrane protein, membrane lipid and carbohydrate extraction from the breast cells
Membrane proteins were extracted from the breast cells by using the membrane protein extraction kit (Thermo Scientific) according to the manufacturer's instructions. Membrane lipids and carbohydrates were extracted from the breast cell membranes according to previously reported method [70–72]. For membrane proteins extraction, the breast cell suspension (5 × 106 cells) was centrifuged at 1000 rpm for 5 min. After that, the cell pellet was washed with 3 mL of Cell Wash Solution and centrifuged at 1000 rpm for 5 min. Then the supernatant was carefully removed and discarded. The cells were then resuspended in 1.5 mL of Cell Wash Solution and transferred to a 2 mL centrifuge tube. Then the cell suspension was centrifuged and the supernatant was discarded. And 0.75mL of Permeabilization Buffer was added to the cell pellet. Then the homogeneous cell suspension was incubated 10 min at 4 °C. After that, the permeabilized cells were centrifuged for 15 min at 14000 rpm. The supernatant containing cytosolic proteins was carefully removed and transferred to a new tube. And 0.5 mL of Solubilization Buffer was added to the pellet. The pellet was resuspended by pipetting up and down and incubated at 4 °C for 30 min. Afterwards, the solution was centrifuged at 14000 rpm for 15 min at 4 °C. And the supernatant containing membrane-associated proteins was transferred to a new tube and proceeded to downstream application. The membrane protein solution was diluted with PBS with a protein concentration equivalent to 1000 cell/10 μL. The pellet was used for membrane lipids and carbohydrates extraction. 0.5 mL of methanol and 0.25 mL of chloroform were added to the pellet in the tube. Then the content of the tube was mixed for 2 min. An additional 0.25 mL of chloroform was added to the sample and the tube was mixed for 30 s. Finally, 0.25 mL of distilled water was added to the sample and then mixed for 30 s. Then mixture was centrifuged at 7600 rpm for 20 min. This provided complete separation with the chloroform/lipid layer at the bottom and the methanol/water/carbohydrate layer on the top. The bottom chloroform layer was removed using a glass pipette and placed into a glass bottle (10 mL). The methanol/water/carbohydrate layer was collected. These samples were further dried in a vacuum drying oven. Then the dried membrane lipids and carbohydrates were dissolved in the PBS with a concentration equivalent to 1000 cell/10 μL.
2.8. Ex vivo breast cancer discrimination
Female Balb/c mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). Female nude mice were provided by the Charles River Laboratories. 4T1 tumors were inoculated by subcutaneous injection of 2 × 106 4T1 cells in the flank of female Balb/c mice. And for MDA-MB-231 tumors, MDA-MB-231 cells (2 × 106 cells) were subcutaneously implanted into the flank of female nude mice. When the tumor volumes reached to about 200 mm3, whole blood was collected from tumor-free and tumor-bearing mice with 20 μL of heparin solution per milliliter of blood for anticoagulation. After the whole blood was centrifuged at 3000 rpm for 10 min at 4 oC, the plasma was carefully removed. The resulting cells were washed twice in cold PBS and then dissolved in PBS with a concentration of 10000 cell/10 μL. And the cells were used for the following detection [73–75]. All animal procedures were in accord with the guidelines of the Institutional Animal Care and Use Committee.
2.9. Analyte cell response
In the cell sensing study, seven fluorescent gold nanoclusters were first diluted with 1X PBS to get solutions with AuNC concentrations of 25 nM. After that, 190 μL of the resultant AuNCs were pipetted into the wells of the 96-well plate. And 10 μL of analyte cells at indicated concentrations were then added to produce a total volume of 200 μL/well. For control wells, 10 μL PBS was added instead of cells. Then the samples were mixed gently with the pipette tips and equilibrated for 30 min before a final reading. In all cases, fluorescence changes reported were in reference to the control samples. The cell targets were tested against seven fluorescent nanoprobes six times. The raw data matrix was processed using classical LDA. Similar procedures were also performed to identify analytical samples (membrane proteins, membrane lipids and carbohydrates extracted from the breast cells, various concentrations of cells and mixtures of cells).
2.10. Unknown identification
To test unknown cell samples, 50 samples were tested with the same procedures as for the training samples, and the resulting fluorescence response patterns were subjected to LDA. Identifications were made by evaluating Mahalanobis distance-square proximities to known group centers from the training matrix.
3. Results and discussion
3.1. Synthesis of the dual-ligand cofunctionalized AuNCs
Seven dual-ligand cofunctionalized AuNCs were synthesized, where one ligand (thiolate, RSH) served as the recognition probe and the other ligand (11-mercaptoundecanoic acid, MUA) acted as the fluorescence turn-on switch by using the one-pot, two-step strategy [68]. Seven fluorescent nanodots, including 6-mercapto-1-hexanol-MUA-AuNCs (MH-MUA-AuNCs), (11-mercaptoundecyl)-N,N,N-trimethylammonium bromide-MUA-AuNCs (NSH-MUA-AuNCs), 4-mercaptobenzoic acid-MUA-AuNCs (MBA-MUA-AuNCs), (2-mercaptoethyl)amine-MUA-AuNCs, (MEN-MUA-AuNCs), glutathione-MUA-AuNCs (GSH-MUA-AuNCs), mercaptosuccinic acid-MUA-AuNCs (MSA-MUA-AuNCs), and 3-mercaptopropionic acid-MUA-AuNCs (MPA-MUA-AuNCs), were prepared. The fluorescence properties and optical absorptions of the as-prepared dual-ligand cofunctionalized AuNCs were studied. As shown in Fig. S1, these AuNCs exhibited remarkably strong emissions, and were found to be stable for several months. The fluorescence spectra revealed that seven AuNCs showed similar fluorescence characteristics with excitation and emission maxima at approximately 380 and 510 nm, respectively. These AuNCs with similar fluorescent properties could minimize the interferences of the background signals and thus improve the detection sensitivity and accuracy [76]. The absorption spectra (Fig. S2) indicated that these AuNCs displayed strong absorption bands centered at around 375 nm, which were assigned to the AuNCs [77, 78]. Meanwhile, there were no absorption peaks around 520 nm, indicating the absence of large gold nanoparticles [79]. Quantum yields (QYs) of these AuNCs were calculated to range from 2.01% to 2.51% by using quinine sulfate as the reference (Table S1).
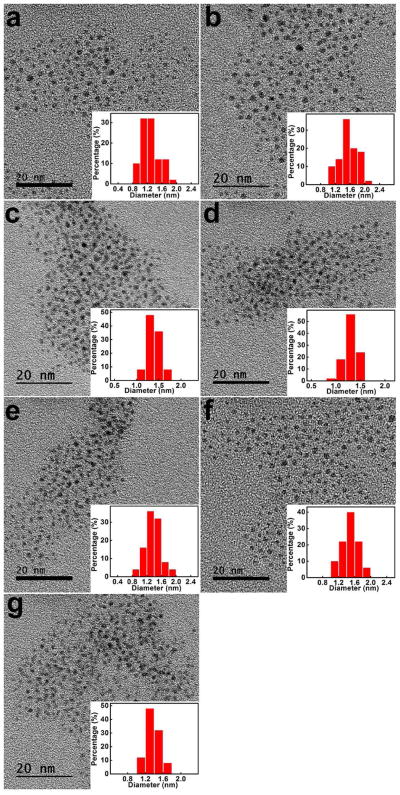
Fig. 1 displayed the high-resolution transmission electron microscopy (HRTEM) images of the seven AuNCs. All AuNCs exhibited spherical shapes and excellent size uniformity, with average sizes less than 2 nm. To further investigate the surface properties of these fluorescent AuNCs, Fourier transform infrared spectroscopy (FTIR) and zeta potential measurements were performed. FTIR spectra (Fig. S3) suggested that the C-S stretch bands (610–630 cm−1) were detected in all AuNCs, indicating the presence of thiolate ligands on the surface of AuNCs [80]. In addition, the emerging absorption bands at around 1650, 1560 and 1046 cm−1 in all AuNCs samples were able to be assigned to two C=O stretch bands and one C-O stretching of the carboxyl groups, further implicating the presence of MUA on the AuNC surfaces [81–83]. As for MEN-MUA-AuNCs and GSH-MUA-AuNCs, the successful grafting of MEN and GSH on the AuNCs could be validated by the appearance of bands at around 1150 cm−1, which were attributed to the amine C-N stretching vibration [84]. NSH-MUA-AuNCs exhibited the typical sharp peak of -CH3 (1340 cm−1), ascribing to the covalent NSH conjugation to NSH-MUA-AuNCs [85, 86]. The spectrum of MBA-MUA-AuNCs exhibited C=C stretch signal at around 1600 cm−1 that was characteristic of the benzene rings of MBA [87, 88]. Because MH, MPA and MSA showed similar FTIR spectra as compared to MUA, no more characteristic IR peaks were observed in these three AuNCs. In addition, MS measurements confirmed the successful dual-ligand cofunctionalization of AuNCs (Table 1). MS results indicated the low percentage of other thiolate ligands (≤12%) as compared to MUA ligand. The large amount of MUA ligand suggested the multilayer adsorption of MUA ligand on the AuNC surface [89]. In addition, the number of MUA ligand on the surface of dual-ligand cofunctionalized AuNCs was only half of number of MUA on MUA-AuNCs. These results indicated that the other thiolate ligands dramatically affect the surface properties of the dual-ligand cofunctionalized AuNCs. Zeta potential measurements (Fig. S4) indicated that all AuNCs were negatively charged owing to the anion form of MUA on the AuNC surfaces. Notably, the surface charges of the AuNCs were different from one another, further demonstrating the existence of other thiolate ligands on AuNCs other than MUA. After that, the stability of each AuNC was measured (Fig. S5–S11). The fluorescence and absorption spectra of all the AuNCs in PBS and cell culture media did not show any significant changes from the spectra of AuNCs in water (Fig. S5–S9). In addition, it was noted that no obvious fluorescent change in AuNCs occurred in aqueous solution even after 6 months (Fig. S10). Furthermore, no obvious photobleaching was observed over a broad pH range of 3–9 (Fig. S11). All these results revealed that the AuNCs had good stability. We also assessed endotoxin levels of the dual-ligand cofunctioanlized AuNCs by the LAL assay method. All the AuNCs exhibited endotoxin levels that are below the FDA standard for sterile water (Fig. S12). These measurements indicated that the dual-ligand cofunctionalized AuNCs exhibited similar fluorescence properties, excellent stabilities but differential surface characteristics, which might serve as powerful candidates for cell discrimination.
Fig. 1.
The high-resolution transmission electron microscopy (HRTEM) images and corresponding size distribution histograms of (a) MH-MUA-AuNCs, (b) NSH-MUA-AuNCs, (c) MBA-MUA-AuNCs, (d) MEN-MUA-AuNCs, (e) GSH-MUA-AuNCs, (f) MSA-MUA-AuNCs, (g) MPA-MUA-AuNCs.
Table 1.
Mass spectrometry characterization of surface ligands of AuNCs.
| AuNCs | thiolate number (per AuNC) | MUA number (per AuNC) | thiolate percentage (%) |
|---|---|---|---|
| MUA-AuNCs | 0 | 565 | 0 |
| MH-MUA-AuNCs | 25 | 356 | 6.6 |
| NSH-MUA-AuNCs | 15 | 262 | 5.4 |
| MBA-MUA-AuNCs | 18 | 285 | 5.9 |
| MEN-MUA-AuNCs | 38 | 278 | 12.0 |
| GSH-MUA-AuNCs | 22 | 254 | 8.0 |
| MSA-MUA-AuNCs | 19 | 292 | 6.1 |
| MPA-MUA-AuNCs | 28 | 295 | 8.7 |
3.2. Discrimination of different breast cancer cell lines
Prior to the application of the AuNCs in cell discrimination, we examined the cytotoxicity of AuNCs by a standard methyl thiazolyl tetrazolium (MTT) assay (Fig. S13), which evaluated the mitochondrial activity of viable cells [90, 91]. As expected, all breast cells maintained above 90% cell viability after incubation with AuNCs at tested concentrations for 24 h, demonstrating low cytotoxicity.
In order to demonstrate the performance of the sensing array, the fluorescence responses of this platform were investigated. The dual-ligand cofunctionalized AuNCs were incubated with ten different breast cell types (Table 2), followed by measuring changes in fluorescence intensities. These ten different human breast cell lines include five highly metastatic breast cancer cell lines (MDA-MB-231, MDA-MB-157, HCC1569, HCC1806 and Hs578T), four non- or low metastatic cancer cell lines (SKBR3, MDA-MB-436, MCF7 and MDA-MB-468), as well as one nonneoplastic, fibrocystic breast cell line (MCF10A). The final concentration of AuNCs for cell discrimination is 25 nM (Fig. S14). Cells interacting with AuNCs depended on not only the affinities of AuNCs but also the compositions and structures of the cells. The fluorescence changes for each cell type were unique due to the discrimination in interaction between AuNCs and cells. As shown in Fig. 2a, incubation of AuNCs with different breast cells resulted in a variety of fluorescence responses. And the fluorescence patterns of the dual-ligand cofunctionalized AuNCs from different synthetic batches were found to be reproducible (Fig. S15). Fluorescence enhancements of AuNCs could be observed for most of the cells while other cells attenuated the fluorescence of AuNCs. The increases of the fluorescence intensities may result from the better protection of the AuNCs by cells against the environment and inhibition of oxygen (O2)-mediated quenching [38, 92–97]. While the fluorescence quenching may be attributed to an energy or electron transfer process [98–101] or the aggregation of AuNCs [77, 102] induced by cells.
Table 2.
Ten different types of breast cell lines.
| Cell line | ER | PR | HER2 | Invasiveness |
|---|---|---|---|---|
| MCF10A | − | − | − | Low |
| MCF7 | + | − | − | Low |
| SKBR3 | − | − | + | Low |
| MDA-MB-436 | − | − | − | Medium |
| MDA-MB-468 | − | − | − | Low |
| MDA-MB-231 | − | − | − | High |
| MDA-MB-157 | − | − | − | Medium |
| HCC1806 | − | − | − | Low |
| HCC1569 | − | − | − | Low |
| Hs578T | − | − | − | High |
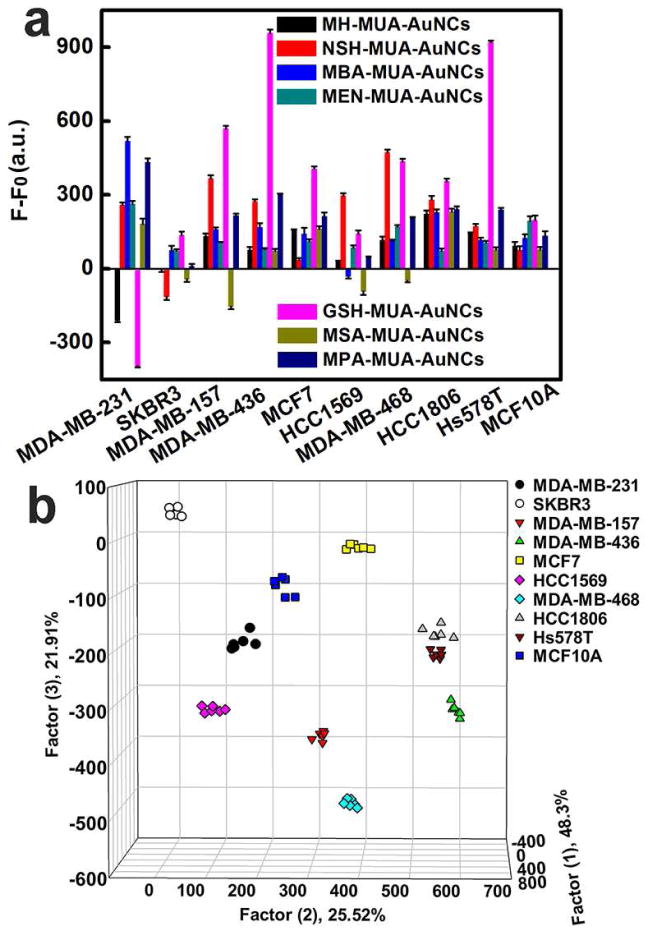
Fig. 2.
Breast cell identification with the fluorescent AuNCs based sensing platform. (a) Fluorescence response patterns of AuNCs against various cells (1000 cells): MDA-MB-231, SKBR3, MDA-MB-157, MDA-MB-436, MCF7, HCC1569, MDA-MB-468, HCC1806, Hs578T and MCF10A cells. Error bars represent standard deviations of six parallel measurements. F−F0 represents the fluorescence intensity change of AuNCs before and after addition of cells, where F0 represents original fluorescent intensity of AuNCs while F represents fluorescent intensity of AuNCs with different breast cells. (b) Canonical score plot for the fluorescent AuNCs sensor array. All ten cells were well separated and properly identified.
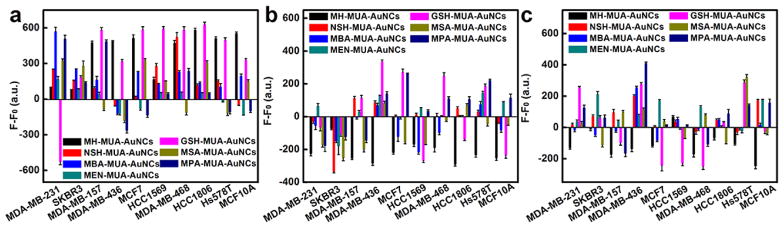
To further investigate the mechanism of breast cell identification based on the dual-ligand cofunctionalized AuNCs, we extracted cell membrane proteins, membrane lipids and carbohydrates from the breast cell membranes. The AuNCs were then incubated with the membrane proteins, membrane lipids and carbohydrates of different breast cell types, respectively, to determine changes in fluorescence intensities (Fig. 3). It can be seen that membrane proteins, membrane lipids and carbohydrates all contributed to the changes in fluorescence intensities. The exposure of different cell membrane proteins, membrane lipids or carbohydrates to AuNCs resulted in different levels of fluorescence changes. In addition, due to their distinctive surface charge states and chemistry, each fluorescent AuNC possesses a unique affinity to each cell type (each cell membrane proteins, lipids, and carbohydrates). All these results indicated that the different fluorescence changes of the AuNCs result from the unique affinities and hence diverse interactions with the membrane proteins, membrane lipids and carbohydrates on the cell surfaces. The cumulative response provides an efficient means of identification.
Fig. 3.
Fluorescence response patterns of AuNCs against (a) membrane proteins, (b) membrane lipids and (c) carbohydrates of different breast cell types (1000 cells): MDA-MB-231, SKBR3, MDA-MB-157, MDA-MB-436, MCF7, HCC1569, MDA-MB-468, HCC1806, Hs578T and MCF10A cells. Error bars represent standard deviations of six parallel measurements. F-F0 represents the fluorescence intensity change of AuNCs before and after addition of (a) membrane proteins, (b) membrane lipids and (c) carbohydrates, where F0 represents original fluorescent intensity of AuNCs while F represents fluorescent intensity of AuNCs with (a) membrane proteins, (b) membrane lipids and (c) carbohydrates of different breast cells.
The fluorescence responses for the dual-ligand cofunctionalized AuNCs depended on the class of cells and were reproducible. Confocal microscopy images of the breast cells treated with AuNCs showed that most AuNCs still remained extracellular during the cell detection process (Fig. S16). We tested the fluorescence responses of a variety of breast cells against seven kinds of AuNCs with six replicates, generating a 7 × 10 × 6 matrix [44]. Subsequently, the original response data obtained were statistically characterized by utilizing linear discriminant analysis (LDA), which maximized the ratio of between-class variance to the within-class variance, enabling maximal separability [44, 103]. This analysis reduced the size of the training matrix (7 fluorescent probes × 10 breast cell types × 6 replicates) and generated six canonical factors (48.3%, 25.52%, 21.91%, 2.74%, 1.09%, and 0.44% of the variation) that were linear combinations of the fluorescence response matrix. The first three most significant discrimination factors were employed to produce a 3D plot (Fig. 2b), in which each point represented the response pattern for an individual cell type against the sensor array. As illustrated in Fig. 2b, the ten different types of breast cells were clearly classified into ten nonoverlapping clusters, explicitly demonstrating the ability of these AuNCs to completely differentiate distinct breast cells. It can be seen that TNBC cells (MDA-MB-157, MDA-MB-436, MDA-MB-468, HCC1806, Hs578T), HER2-positive breast cancer cell lines (SKBR3, HCC1569) and ER-positive breast cancer cells (MCF7) could be clustered into three separate regions, revealing the potential correlations between cell surface properties and types and states of cells. Notably, this method could effectively identify the triple-negative breast cancer (TNBC) cells, which might facilitate its further applications in breast cancer theranostics. In addition, the discriminant ability of the dual-ligand co-functionalized AuNCs for TNBC cells was further proved by identifying TNBC cells from cell mixtures of cancer cells and normal cells (Fig. S17).
3.3. Unknown cell sample discrimination
To validate the recognition efficiency of our sensor array, we performed tests to identify unknown cell samples that were randomly chosen from the ten types of breast cells mentioned above (Table S2). The unknown samples were ranked in terms of Mahalonobis distance to the groups generated through the training matrix and the nearest samples were returned to the respective groups [58]. Notably, of the 50 unknown breast cell samples, all of them were correctly identified, which proved the diagnostic potential of our sensor platform.
3.4. Cell discrimination in different concentrations and in mixtures
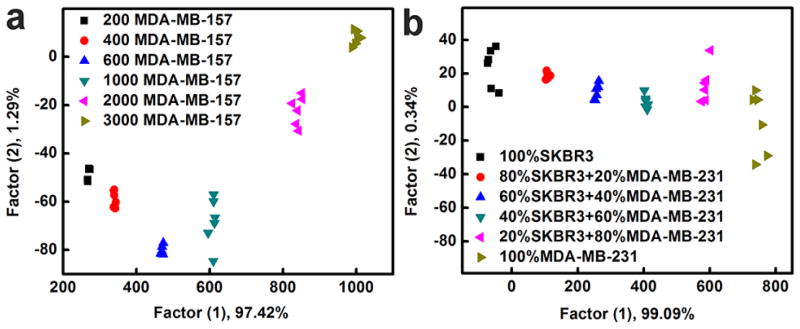
After the successful identification of breast cell types, the discriminant ability of our sensing platform was further tested by sensing cell analytes at different concentrations (200, 400, 600, 1000, 2000 and 3000 MDA-MB-157). As exhibited in Fig. 4a, six groups of samples were located explicitly in six isolated clusters, providing complete discrimination among different cell concentrations. This result indicated that AuNCs could not only be effectively utilized for cell concentration analysis, but were also able to detect MDA-MB-157 breast cells with excellent sensitivity as low as 200 cells. In addition, the cell recognition ability of these fluorescent AuNCs was maintained in mixtures. We tested mixtures of MDA-MB-231 and SKBR3 cells at different ratios (MDA-MB-231/SKBR3 = 80/20, 60/40, 40/60, and 80/20 with 1000 total cells). As illustrated in Fig. 4b, these mixtures, as well as individual MDA-MB-231 cells and SKBR3 cells, were distributed separately from each other in the LDA plot and properly arranged with the order of ratios in the dimension of the first factor, which exhibited a potential capability of this platform for analyzing cell samples with complex composition.
Fig. 4.
(a) Identification of cells at various concentrations using the fluorescent AuNCs based sensor array. Canonical score plot for fluorescence response patterns against different amounts of MDA-MB-157 cells (200, 400, 600, 1000, 2000 and 3000 cells). (b) Canonical score plot against cell mixtures, including pure SKBR3 cells; 80% SKBR3 + 20% MDA-MB-231; 60% SKBR3 + 40% MDA-MB-231; 40% SKBR3 + 60% MDA-MB-231; 20% SKBR3 + 80% MDA-MB-231; pure MDA-MB-231 cells. In each case, the total cell amount was 1000 cells.
3.5. Discrimination of drug resistant breast cancer cells
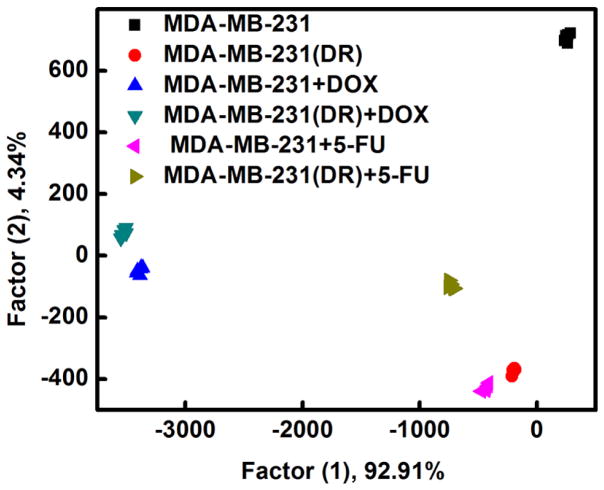
Multidrug resistance (MDR) is a major factor of cancer chemotherapy failure as more than 90% of malignant tumor patients die from complications arising due to multidrug resistance [104, 105]. Therefore, the identification of drug resistant cancer cells would be extremely helpful to provide an effective way to monitor progression of cancer diseases and facilitate the selection of efficient therapeutic strategies [106]. Previous studies indicated that the membrane properties and phospholipids of the cancer cells with MDR can be regulated by drugs [107–112]. In addition, as compared to the non-drug-resistant cancer cell lines, the MDR cancer cells also express different types and amounts of proteins in the membranes [113]. Traditional techniques for detection of drug resistant cancer cells are mainly based on upregulation of P-glycoprotein [106, 114–116]. Inspired by the exceptional performance of the AuNCs for cancer cell detection, we further tested if they could also be used to differentiate between nonresistant, drug-resistant, and chemotherapy-treated cancer cells. The concentrations of chemotherapeutic drugs doxorubicin and fluorouracil are 0.05 μg/mL and 0.5 μg/mL, respectively (Fig. S18). As shown in Fig. 5, MDA-MB-231 cells, doxorubicin-resistant MDA-MB-231 cells, and MDA-MB-231 cells treated with different anticancer drugs were well-separated from each other and properly arranged with the order of ratios in the dimension of the first factor, suggesting successful identification of nonresistant and drug-resistant cancer cells, as well as cells with or without exposure to chemotherapy. We proved for the first time the utility of the chemical nose strategy using dual-ligand cofunctionalized AuNCs for sensing MDR cells, presenting a promising method for MDR cancer diagnostics.
Fig. 5.
Identification of drug-resistant cells with the fluorescent AuNCs based sensing platform. Canonical score plot for fluorescence response patterns against cells (1000 cells): MDA-MB-231 cells, doxorubicin resistant MDA-MB-231 cells (MDA-MB-231(DR)), MDA-MB-231 cells treated with doxorubicin (MDA-MB-231+DOX), MDA-MB-231(DR) cells treated with doxorubicin (MDA-MB-231(DR)+DOX), MDA-MB-231 cells treated with fluorouracil (MDA-MB-231+5-FU), MDA-MB-231(DR) cells treated with fluorouracil (MDA-MB-231(DR)+5-FU).
3.6. Identification of different types of cells
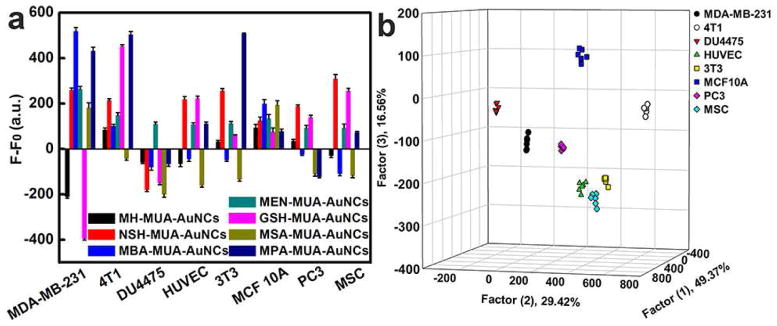
Furthermore, in order to improve the resolving power of this sensing platform in cell detection, we also tested the discriminatory ability of AuNCs against eight distinct types of cells. The eight different cells (Table S3), including both human and mouse cells, were employed as the target analytes for cell identification. As shown in Fig. 6a, the fluorescence responses demonstrated diversity. Through LDA transformation, the fluorescence pattern (7 fluorescent probes × 8 cell lines × 6 replicates) resulted in six canonical factors (49.37%, 29.42%, 16.56%, 3.12%, 1.1% and 0.43% of total variance), with the three most significant factors plotted in a well-clustered graph. As can be seen in Fig. 6b, the eight data groups each containing six points from each kind of cell were excellently classified and displayed 100% separation. This phenomenon proved that other types of cell lines, such as fibroblasts, epithelial and endothelial cells, as well as prostate cells and mesenchymal stem cells, could also be well discriminated from breast cells, confirming our array’s capability of cell discrimination. The AuNCs showed negligible cytotoxicity to these cells as shown by MTT results (Fig. S19) under our experimental conditions. All these results validated that our sensor array based on the dual-ligand cofunctionalized AuNCs could serve as an auxiliary strategy for the clinical standard method for cell discrimination, which would be beneficial for cancer diagnosis and treatment.
Fig. 6.
Cell identification with the fluorescent AuNCs based sensing platform. (a) Fluorescence response patterns of the probes against various cells (1000 cells): MDA-MB-231, MCF10A, DU4475, HUVEC, 3T3, 4T1, PC3 and MSC cells. Error bars represent standard deviations of six parallel measurements. F-F0 represents the fluorescence intensity change of AuNCs before and after addition of cells, where F0 represents original fluorescent intensity of AuNCs while F represents fluorescent intensity of AuNCs with different cells. (b) Canonical score plot for the fluorescent AuNCs sensor array. All eight cells were well separated and properly identified.
3.7. Ex vivo breast cancer discrimination
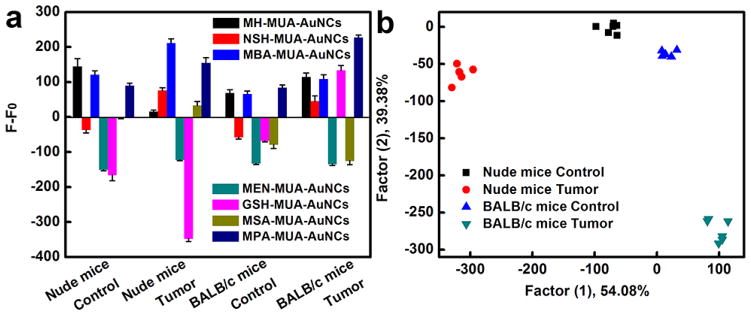
We also proved the practical application of the AuNC multichannel sensors with ex vivo experiments (Fig. 7). Recent researches have suggested that circulating tumor cells (CTCs), which are cancer cells that have shed from tumors, can spread into the bloodstream as the cellular origin of fatal metastasis [117–119]. In addition, in the presence of a growing tumor, changes in leukocyte compositions and levels in peripheral blood can also be observed [120–127]. Therefore, the types and populations of cells in peripheral blood of the tumor-bearing mice are supposed to be different from those of tumor-free mice. We isolated cells from the peripheral blood of tumor-free and tumor-bearing mice (4T1 breast tumor model in BALB/c mice and MDA-MB-231 tumor model in nude mice, respectively) and incubated the isolated cells with dual-ligand co-functionalized AuNCs. After that, we investigated the difference in fluorescence responses between the tumor-free and tumor-bearing groups. It can be seen that the exposure of cells from the peripheral blood of tumor-free and tumor-bearing mice to the dual-ligand co-functionalized AuNCs resulted in different levels of fluorescence changes due to their distinct interactions with AuNCs (Fig. 7a). LDA analysis indicated that the four different types of cells from the peripheral blood of tumor-free and tumor-bearing mice clustered into four nonoverlapping groups, explicitly proving the capability of these fluorescent AuNCs to differentiate cells from tumor-free and tumor-bearing mice (Fig. 7b). All these results indicate that the fluorescent AuNCs based array is a powerful cell analysis tool which may hold the promise of practical application in breast cancer diagnostics and therapy.
Fig. 7.
Identification of cells from the peripheral blood of tumor-free and tumor-bearing mice with the fluorescent AuNCs based sensing platform. (a) Fluorescence response patterns of the AuNCs against various cells (10000 cells): cells from the peripheral blood of tumor-free nude mice, cells from the peripheral blood of MDA-MB-231 tumor-bearing nude mice, cells from the peripheral blood of tumor-free BALB/c mice, cells from the peripheral blood of 4T1 tumor-bearing BALB/c mice. Error bars represent standard deviations of six parallel measurements. F-F0 represents the fluorescence intensity change of AuNCs before and after addition of cells, where F0 represents original fluorescent intensity of AuNCs while F represents fluorescent intensity of AuNCs with different cells. (b) Canonical score plot for the fluorescent AuNCs sensor array. All four different types of cells were well separated and properly identified.
3.8. Discussion
Chemical nose sensors have been successfully trained to recognize cancer cells [26, 46–50, 53–55]. For example, Rotello et al. were able to detect and differentiate isogenic normal and cancerogenic cell lines using conjugated polymer/gold nanoparticle constructs with a limit of detection of 20000 cells [53]. Green fluorescent protein was used instead of a fluorescent polymer as the negatively charged fluorophore to enhance cancer cell detection to levels as low as 5000 cells [50]. Recently, the utilization of an array of gold nanoparticle-green fluorescent protein was used to distinguish between normal and metastatic cells and tissues. Full differentiation between the analyte cell/tissue was achieved with as little as 200 ng of intracellular protein (~1000 cells) for each nanoparticle [46]. As compared to these previously published sensors, biosensing with dual-ligand cofunctionalized AuNCs provides an unparalleled detection limit of 200 cells for each AuNC without requiring specific biomarkers.
The dual-ligand cofunctionalized AuNCs, which serve as both effective cell recognition elements and signal transducers, offer extra merits in terms of function integration and low cost. Most of the current methods for TNBC detection are based on analysis of gene expression profiles [128, 129] or sensing of antibody array platforms [130], which are generally tedious and time consuming processes that require well-trained technical personnel in well-equipped laboratories. It is also clear that TNBCs are a diverse and heterogeneous group of tumors; a single molecule may not distinguish them. Our sensor array based on the dual-ligand cofunctionalized AuNCs offers both a rapid and sensitive measurement. Separation of different TNBC cells may be useful for enabling systematic and personalized therapies. Notably, this approach is the first successful application of the array-based systems for MDR cancer diagnostics. All these results reveal that the dual-ligand cofunctionalized AuNCs hold substantial promise for cell typing with high accuracy and sensitivity.
Despite the excellent performance of the dual-ligand cofunctionalized AuNCs based senor array in breast cell detection, some challenges still remain and need to be addressed before their true realistic applications can be fully realized. First, although a lot of chemical nose based sensors have been developed and have been widely utilized for cell discrimination, including our AuNCs based sensing array, they can only identify the cell types if they have been trained. Appropriate training of the system by the sensor array is vital [131]. If the analytes do not belong to the training set, it may be difficult to accurately identify the exact cell types. Hence, cell recognition can be coupled with other detection methods to expand the applications of chemical nose based sensors. Second, in contrast to other chemical nose sensors, the dual-ligand cofunctionalized AuNCs based senor array offers the advantages of easy synthesis, high biocompatibility and low cost. But as compared to fluorometric detection methods, colorimetry is easier to realize manufacturing portable and affordable diagnostic devices [132, 133]. Thus, efforts toward the construction of colorimetric chemical nose biosensors by using AuNCs with their intrinsic peroxidase-like activity will be made in our next works. Third, the seven dual-ligand cofunctionalized AuNCs based senor array here has been successfully utilized to distinguish ten different types of breast cells, including six TNBCs, explicitly proving the diagnostic potential of our sensor platform. But the sensor array should have better selectivity for analytes if we want to use it in more complicated training set (eg, more types of breast cells discrimination). Each analyte ought to have a distinct enough pattern of response to be recognized from other analytes that the system is trained to discriminate [131]. Therefore, it is necessary to involve more types of dual-ligand cofunctionalized AuNCs carrying additional electrostatic, aromatic, hydrophobic, or hydrogen-bonding interactions in order to identify more complex systems in future work. For example, we can synthesize distinctive dual-ligand cofunctionalized AuNCs with similar ligands as the recognition probe carrying different numbers of positive charges, or different numbers of hydroxyl groups, or different hydrophobic groups and then investigate the fluorescence responses of the cells against AuNCs, which may broaden the diagnostic applications of our sensor platform. In addition, AuNCs with bi- or multi-recognition probes may also be developed for additional important biomedical applications.
4. Conclusion
In conclusion, we have explored the usage of the seven dual-ligand cofunctionalized AuNCs as the easily accessible yet potentially powerful sensor to detect cell targets. As compared with conventional organic labels, these fluorescent AuNCs offer unique attributes such as simple synthesis, high brightness, favorable biocompatibility and high stability for long-time observations. On the basis of this AuNC multichannel sensor, we have successfully identified and differentiated normal, cancerous, and metastatic breast cells, including TNBC cells. This array may be used when specific cancer biomarkers have yet to be determined. The high accuracy of the blind breast cell sample tests further validates the practical application of our sensor array. In addition, the cell recognition ability of these fluorescent AuNCs is not sacrificed at low concentration (200 cells) and in mixtures. Furthermore, this biosensor can also be expanded to discriminate different types of cells, including both human and mouse cells, as well as drug-resistant cancer cells. Cells from the peripheral blood of tumor-free and tumor-bearing mice were efficiently identified. Taken together, the fluorescent AuNCs based array offers a potent cell analysis tool with promise applications in biomedical diagnostics.
Supplementary Material
Acknowledgments
The authors would like to acknowledge funding from the NIH (NCI DP2 CA174495-01).
Footnotes
Supporting Information Available: Supporting figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.He X, Bao X, Cao H, Zhang Z, Yin Q, Gu W, et al. Tumor-penetrating nanotherapeutics loading a near-infrared probe inhibit growth and metastasis of breast cancer. Adv Funct Mater. 2015;25:2831–9. [Google Scholar]
- 2.Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–44. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Srinivas PR, Kramer BS, Srivastava S. Trends in biomarker research for cancer detection. Lancet Oncol. 2001;2:698–704. doi: 10.1016/S1470-2045(01)00560-5. [DOI] [PubMed] [Google Scholar]
- 4.Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 5.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 7.Shu D, Li H, Shu Y, Xiong G, Carson WE, Haque F, et al. Systemic delivery of anti-miRNA for suppression of triple negative breast cancer utilizing RNA nanotechnology. ACS Nano. 2015;9:9731–40. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo P, Huang J, Wang L, Jia D, Yang J, Dillon DA, et al. ICAM–1 as a molecular target for triple negative breast cancer. Proc Natl Acad Sci USA. 2014;111:14710–5. doi: 10.1073/pnas.1408556111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan X, Peng J, Fu Y, An S, Rezaei K, Tabbara S, et al. MiR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014;16:435. doi: 10.1186/s13058-014-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toffoli S, Bar I, Abdel-Sater F, Delrée P, Hilbert P, Cavallin F, et al. Identification by array comparative genomic hybridization of a new amplicon on chromosome 17q highly recurrent in BRCA1 mutated triple negative breast cancer. Breast Cancer Res. 2014;16:466. doi: 10.1186/s13058-014-0466-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fadare O, Tavassoli FA. Clinical and pathologic aspects of basal-like breast cancers. Nat Clin Prac Oncol. 2008;5:149–59. doi: 10.1038/ncponc1038. [DOI] [PubMed] [Google Scholar]
- 12.McGowan PM, Mullooly M, Caiazza F, Sukor S, Madden SF, Maguire AA, et al. ADAM-17: a novel therapeutic target for triple negative breast cancer. Ann Oncol. 2013;24:362–9. doi: 10.1093/annonc/mds279. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, Yoon T-J, Figueiredo J-L, Swirski FK, Weissleder R. Rapid detection and profiling of cancer cells in fine-needle aspirates. Proc Natl Acad Sci USA. 2009;106:12459–64. doi: 10.1073/pnas.0902365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: A chemical biology approach. Acc Chem Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan W, Donovan MJ, Jiang J. Aptamers from cell-based selection for bioanalytical applications. Chem Rev. 2013;113:2842–62. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McPherson RA, Pincus MR. Henry's clinical diagnosis and management by laboratory methods. 21. Elsevier; Philadelphia: 2007. [Google Scholar]
- 18.O'Dwyer D, Ralton LD, O'Shea A, Murray GI. The proteomics of colorectal cancer: identification of a protein signature associated with prognosis. PLoS One. 2011;6:e27718. doi: 10.1371/journal.pone.0027718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J-W, Peng S-Y, Li J-T, Wang Y, Zhang Z-P, Cheng Y, et al. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281:71–81. doi: 10.1016/j.canlet.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198– 207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 21.Malek A, Catapano C, Czubayko F, Aigner A. A sensitive polymerase chain reaction-based method for detection and quantification of metastasis in human xenograft mouse models. Clin Exp Metastasis. 2010;27:261–71. doi: 10.1007/s10585-010-9324-1. [DOI] [PubMed] [Google Scholar]
- 22.Paredes-Aguilera R, Romero-Guzman L, Lopez-Santiago N, Burbano-Ceron L, Camacho-Del Monte O, Nieto-Martinez S. Flow cytometric analysis of cell-surface and intracellular antigens in the diagnosis of acute leukemia. Am J Hematol. 2001;68:69–74. doi: 10.1002/ajh.1155. [DOI] [PubMed] [Google Scholar]
- 23.Dunphy CH, Orton SO, Mantell J. Relative contributions of enzyme cytochemistry and flow cytometric immunophenotyping to the evaluation of acute myeloid leukemias with a monocytic component and of flow cytometric immunophenotyping to the evaluation of absolute monocytoses. Am J Clin Pathol. 2004;122:865–74. doi: 10.1309/BH58-8HVG-6UHN-2RF2. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez–Carbayo M. Antibody arrays: Technical considerations and clinical applications in cancer. Clin Chem. 2006;52:1651–9. doi: 10.1373/clinchem.2005.059592. [DOI] [PubMed] [Google Scholar]
- 25.Lavigne JJ, Anslyn EV. Sensing a paradigm shift in the field of molecular recognition: from selective to differential receptors. Angew Chem Int Ed. 2001;40:3118–30. doi: 10.1002/1521-3773(20010903)40:17<3118::AID-ANIE3118>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 26.Duncan B, Elci SG, Rotello VM. Beyond biomarkers: Identifying cell state using unbiased nanosensor arrays. Nano Today. 2012;7:228–30. doi: 10.1016/j.nantod.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, et al. Sensing of proteins in human serum using conjugates of nanoparticles and green fluorescent protein. Nat Chem. 2009;1:461–5. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda OR, Chen H-T, You C-C, Mortenson DE, Yang X-C, Bunz UHF, et al. Enzyme-amplified array sensing of proteins in solution and in biofluids. J Am Chem Soc. 2010;132:5285–9. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X, Cui Y, Levenson RM, Chung LWK, Nie S. In vivo cancer targeting and imaging with semiconductor quantum dots. Nat Biotech. 2004;22:969–76. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 30.Le NDB, Rana S, Rotello VM. Chemical nose sensors: an alternative strategy for cancer diagnosis. Expert Rev Mol Diagn. 2013;13:111–3. doi: 10.1586/erm.12.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JW, Lee J-S, Kang M, Su AI, Chang Y-T. Visual artificial tongue for quantitative metal-cation analysis by an off-the-shelf dye array. Chem-Eur J. 2006;12:5691–6. doi: 10.1002/chem.200600307. [DOI] [PubMed] [Google Scholar]
- 32.Rakow NA, Suslick KS. A colorimetric sensor array for odour visualization. Nature. 2000;406:710–3. doi: 10.1038/35021028. [DOI] [PubMed] [Google Scholar]
- 33.Xie Z, Cao K, Zhao Y, Bai L, Gu H, Xu H, et al. An optical nose chip based on mesoporous colloidal photonic crystal beads. Adv Mater. 2014;26:2413–8. doi: 10.1002/adma.201304775. [DOI] [PubMed] [Google Scholar]
- 34.Lee JW, Lee J-S, Chang Y-T. Colorimetric identification of carbohydrates by a pH indicator/pH change inducer ensemble. Angew Chem Int Ed. 2006;45:6485–7. doi: 10.1002/anie.200602055. [DOI] [PubMed] [Google Scholar]
- 35.Elci SG, Moyano DF, Rana S, Tonga GY, Phillips RL, Bunz UHF, et al. Recognition of glycosaminoglycan chemical patterns using an unbiased sensor array. Chem Sci. 2013;4:2076–80. [Google Scholar]
- 36.Folmer-Andersen JF, Kitamura M, Anslyn EV. Pattern-based discrimination of enantiomeric and structurally similar amino acids:3 An optical mimic of the mammalian taste response. J Am Chem Soc. 2006;128:5652–3. doi: 10.1021/ja061313i. [DOI] [PubMed] [Google Scholar]
- 37.Buryak A, Severin K. A chemosensor array for the colorimetric identification of 20 natural amino acids. J Am Chem Soc. 2005;127:3700–1. doi: 10.1021/ja042363v. [DOI] [PubMed] [Google Scholar]
- 38.Kong H, Lu Y, Wang H, Wen F, Zhang S, Zhang X. Protein discrimination using fluorescent gold nanoparticles on plasmonic substrates. Anal Chem. 2012;84:4258–61. doi: 10.1021/ac300718p. [DOI] [PubMed] [Google Scholar]
- 39.Xu S, Lu X, Yao C, Huang F, Jiang H, Hua W, et al. A visual sensor array for pattern recognition analysis of proteins using novel blue-emitting fluorescent gold nanoclusters. Anal Chem. 2014;86:11634–9. doi: 10.1021/ac502643s. [DOI] [PubMed] [Google Scholar]
- 40.Yuan Z, Du Y, Tseng Y-T, Peng M, Cai N, He Y, et al. Fluorescent gold nanodots based sensor array for proteins discrimination. Anal Chem. 2015;87:4253–9. doi: 10.1021/ac5045302. [DOI] [PubMed] [Google Scholar]
- 41.Lin M, Li W, Wang Y, Yang X, Wang K, Wang Q, et al. Discrimination of hemoglobins with subtle differences using an aptamer based sensing array. Chem Commun. 2015;51:8304–6. doi: 10.1039/c5cc00929d. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Kong H, Wen F, Zhang S, Zhang X. Lab-on-graphene: graphene oxide as a triple-channel sensing device for protein discrimination. Chem Commun. 2013;49:81–3. doi: 10.1039/c2cc37293b. [DOI] [PubMed] [Google Scholar]
- 43.Chou SS, De M, Luo J, Rotello VM, Huang J, Dravid VP. Nanoscale graphene oxide (nGO) as artificial receptors: Implications for biomolecular interactions and sensing. J Am Chem Soc. 2012;134:16725–33. doi: 10.1021/ja306767y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.You C-C, Miranda OR, Gider B, Ghosh PS, Kim I-B, Erdogan B, et al. Detection and identification of proteins using nanoparticle-fluorescent polymer 'chemical nose' sensors. Nat Nanotechnol. 2007;2:318–23. doi: 10.1038/nnano.2007.99. [DOI] [PubMed] [Google Scholar]
- 45.Yang X, Li J, Pei H, Li D, Zhao Y, Gao J, et al. Pattern recognition analysis of proteins using DNA-decorated catalytic gold nanoparticles. Small. 2013;9:2844–9. doi: 10.1002/smll.201202772. [DOI] [PubMed] [Google Scholar]
- 46.Rana S, Singla AK, Bajaj A, Elci SG, Miranda OR, Mout R, et al. Array-based sensing of metastatic cells and tissues using nanoparticle–fluorescent protein conjugates. Acs Nano. 2012;6:8233–40. doi: 10.1021/nn302917e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong H, Liu D, Zhang S, Zhang X. Protein sensing and cell discrimination using a sensor array based on nanomaterial-assisted chemiluminescence. Anal Chem. 2011;83:1867–70. doi: 10.1021/ac200076c. [DOI] [PubMed] [Google Scholar]
- 48.Lu Y, Liu Y, Zhang S, Wang S, Zhang S, Zhang X. Aptamer-based plasmonic sensor array for discrimination of proteins and cells with the naked eye. Anal Chem. 2013;85:6571–4. doi: 10.1021/ac4014594. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, Li J, Pei H, Zhao Y, Zuo X, Fan C, et al. DNA–gold nanoparticle conjugates-based nanoplasmonic probe for specific differentiation of cell types. Anal Chem. 2014;86:3227–31. doi: 10.1021/ac500381e. [DOI] [PubMed] [Google Scholar]
- 50.Bajaj A, Rana S, Miranda OR, Yawe JC, Jerry DJ, Bunz UHF, et al. Cell surface-based differentiation of cell types and cancer states using a gold nanoparticle-GFP based sensing array. Chem Sci. 2010;1:134–8. [Google Scholar]
- 51.El-Boubbou K, Zhu DC, Vasileiou C, Borhan B, Prosperi D, Li W, et al. Magnetic glyco-nanoparticles: A tool to detect, differentiate, and unlock the glyco-codes of cancer via magnetic resonance imaging. J Am Chem Soc. 2010;132:4490–9. doi: 10.1021/ja100455c. [DOI] [PubMed] [Google Scholar]
- 52.Pei H, Li J, Lv M, Wang J, Gao J, Lu J, et al. A graphene-based sensor array for high-precision and adaptive target identification with ensemble aptamers. J Am Chem Soc. 2012;134:13843–9. doi: 10.1021/ja305814u. [DOI] [PubMed] [Google Scholar]
- 53.Bajaj A, Miranda OR, Kim I-B, Phillips RL, Jerry DJ, Bunz UHF, et al. Detection and differentiation of normal, cancerous, and metastatic cells using nanoparticle-polymer sensor arrays. Proc Natl Acad Sci USA. 2009;106:10912–6. doi: 10.1073/pnas.0900975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Kong H, Gong X, Zhang S, Zhang X. Multicolor imaging of cancer cells with fluorophore-tagged aptamers for single cell typing. Anal Chem. 2014;86:8261–6. doi: 10.1021/ac501657g. [DOI] [PubMed] [Google Scholar]
- 55.Moyano D, Rotello V. Nanoparticle-GFP “chemical nose” sensor for cancer cell identification. Methods Mol Biol. 2013;991:1–8. doi: 10.1007/978-1-62703-336-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, Kong H, Mout R, Saha K, Moyano DF, Robinson SM, et al. Rapid identification of bacterial biofilms and biofilm wound models using a multichannel nanosensor. ACS Nano. 2014;8:12014–9. doi: 10.1021/nn505753s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen W, Li Q, Zheng W, Hu F, Zhang G, Wang Z, et al. Identification of bacteria in water by a fluorescent array. Angew Chem Int Ed. 2014;53:13734–9. doi: 10.1002/anie.201407606. [DOI] [PubMed] [Google Scholar]
- 58.Phillips RL, Miranda OR, You C-C, Rotello VM, Bunz UHF. Rapid and efficient identification of bacteria using gold-nanoparticle–poly(para-phenyleneethynylene) constructs. Angew Chem Int Ed. 2008;47:2590–4. doi: 10.1002/anie.200703369. [DOI] [PubMed] [Google Scholar]
- 59.Wan Y, Sun Y, Qi P, Wang P, Zhang D. Quaternized magnetic nanoparticles–fluorescent polymer system for detection and identification of bacteria. Biosens Bioelectron. 2014;55:289–93. doi: 10.1016/j.bios.2013.11.080. [DOI] [PubMed] [Google Scholar]
- 60.Ran X, Pu F, Ren J, Qu X. A CuS-based chemical tongue chip for pattern recognition of proteins and antibiotic-resistant bacteria. Chem Commun. 2015;51:2675–8. doi: 10.1039/c4cc08863h. [DOI] [PubMed] [Google Scholar]
- 61.Tao Y, Ran X, Ren J, Qu X. Array-based sensing of proteins and bacteria by using multiple luminescent nanodots as fluorescent probes. Small. 2014;10:3667–71. doi: 10.1002/smll.201400661. [DOI] [PubMed] [Google Scholar]
- 62.Tao Y, Li M, Ren J, Qu X. Metal nanoclusters: novel probes for diagnostic and therapeutic applications. Chem Soc Rev. 2015;44:8636–63. doi: 10.1039/c5cs00607d. [DOI] [PubMed] [Google Scholar]
- 63.Tao Y, Lin Y, Ren J, Qu X. A dual fluorometric and colorimetric sensor for dopamine based on BSA-stabilized Au nanoclusters. Biosens Bioelectron. 2013;42:41–6. doi: 10.1016/j.bios.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 64.Ju E, Liu Z, Du Y, Tao Y, Ren J, Qu X. Heterogeneous assembled nanocomplexes for ratiometric detection of highly reactive oxygen species in vitro and in vivo. Acs Nano. 2014;8:6014–23. doi: 10.1021/nn501135m. [DOI] [PubMed] [Google Scholar]
- 65.Liu C-L, Wu H-T, Hsiao Y-H, Lai C-W, Shih C-W, Peng Y-K, et al. Insulin-directed synthesis of fluorescent gold nanoclusters: preservation of insulin bioactivity and versatility in cell imaging. Angew Chem Int Ed. 2011;50:7056–60. doi: 10.1002/anie.201100299. [DOI] [PubMed] [Google Scholar]
- 66.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–31. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 67.Feng B, Xu Z, Zhou F, Yu H, Sun Q, Wang D, et al. Near infrared light-actuated gold nanorods with cisplatin–polypeptide wrapping for targeted therapy of triple negative breast cancer. Nanoscale. 2015;7:14854–64. doi: 10.1039/c5nr03693c. [DOI] [PubMed] [Google Scholar]
- 68.Yuan Z, Peng M, He Y, Yeung ES. Functionalized fluorescent gold nanodots: synthesis and application for Pb2+ sensing. Chem Commun. 2011;47:11981–3. doi: 10.1039/c1cc14872a. [DOI] [PubMed] [Google Scholar]
- 69.Sun H, Wu L, Gao N, Ren J, Qu X. Improvement of photoluminescence of graphene quantum dots with a biocompatible photochemical reduction pathway and its bioimaging application. Acs Appl Mater Inter. 2013;5:1174–9. doi: 10.1021/am3030849. [DOI] [PubMed] [Google Scholar]
- 70.Garoma T, Janda D. Investigation of the effects of microalgal cell concentration and electroporation, microwave and ultrasonication on lipid extraction efficiency. Renew Energy. 2016;86:117–23. [Google Scholar]
- 71.Zhang C, Zhou Y, Sun Z, Feng J, Wang Y. Polysaccharides extraction from Erythirna variegata, chemical characterization and its antioxidant activity. Int J Biol Macromol. 2014;68:267–73. doi: 10.1016/j.ijbiomac.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 72.Hua L, Guo L, Thakkar M, Wei D, Agbakpe M, Kuang L, et al. Effects of anodic oxidation of a substoichiometric titanium dioxide reactive electrochemical membrane on algal cell destabilization and lipid extraction. Bioresour Technol. 2016;203:112–7. doi: 10.1016/j.biortech.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 73.Hu C-MJ, Zhang L, Aryal S, Cheung C, Fang RH, Zhang L. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc Natl Acad Sci USA. 2011;108:10980–5. doi: 10.1073/pnas.1106634108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo Y, Wang D, Song Q, Wu T, Zhuang X, Bao Y, et al. Erythrocyte membrane-enveloped polymeric nanoparticles as nanovaccine for induction of antitumor immunity against melanoma. ACS Nano. 2015;9:6918–33. doi: 10.1021/acsnano.5b01042. [DOI] [PubMed] [Google Scholar]
- 75.Sacchetti C, Motamedchaboki K, Magrini A, Palmieri G, Mattei M, Bernardini S, et al. Surface polyethylene glycol conformation influences the protein corona of polyethylene glycol-modified single-walled carbon nanotubes: Potential implications on biological performance. ACS Nano. 2013;7:1974–89. doi: 10.1021/nn400409h. [DOI] [PubMed] [Google Scholar]
- 76.Pu K-Y, Liu B. Fluorescent conjugated polyelectrolytes for bioimaging. Adv Funct Mater. 2011;21:3408–23. [Google Scholar]
- 77.Huang CC, Yang Z, Lee KH, Chang HT. Synthesis of highly fluorescent gold nanoparticles for sensing Mercury(II) Angew Chem Int Ed. 2007;46:6824–8. doi: 10.1002/anie.200700803. [DOI] [PubMed] [Google Scholar]
- 78.Shibu ES, Pradeep T. Quantum clusters in cavities: Trapped Au15 in cyclodextrins. Chem Mater. 2011;23:989–99. [Google Scholar]
- 79.Yuan Z, Peng M, Shi L, Du Y, Cai N, He Y, et al. Disassembly mediated fluorescence recovery of gold nanodots for selective sulfide sensing. Nanoscale. 2013;5:4683–6. doi: 10.1039/c2nr33202g. [DOI] [PubMed] [Google Scholar]
- 80.Wijaya A, Hamad-Schifferli K. Ligand customization and DNA functionalization of gold nanorods via round-trip phase transfer ligand exchange. Langmuir. 2008;24:9966–9. doi: 10.1021/la8019205. [DOI] [PubMed] [Google Scholar]
- 81.Chen CE, Pu F, Huang ZZ, Liu Z, Ren JS, Qu XG. Stimuli-responsive controlled-release system using quadruplex DNA-capped silica nanocontainers. Nucleic Acids Res. 2011;39:1638–44. doi: 10.1093/nar/gkq893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu Q, Peng H, Meng L, Dong S, Dyson P, Fei Z. Fabrication of reduced graphene oxide hybrid materials that exhibit strong fluorescence. J Mater Chem. 2012;22:14868–73. [Google Scholar]
- 83.Niu W, Wu S, Zhang S. Utilizing the amidation reaction to address the "cooperative effect" of carboxylic acid/amine on the size, shape, and multicolor output of fluoride upconversion nanoparticles. J Mater Chem. 2011;21:10894–902. [Google Scholar]
- 84.Liu C, Wen J, Meng Y, Zhang K, Zhu J, Ren Y, et al. Efficient delivery of therapeutic miRNA nanocapsules for tumor suppression. Adv Mater. 2014;27:292–7. doi: 10.1002/adma.201403387. [DOI] [PubMed] [Google Scholar]
- 85.Huang Y, Ma W, Li J, Cheng M, Zhao J, Wan L, et al. A Novel β-CD-hemin complex photocatalyst for efficient degradation of organic pollutants at neutral pHs under visible irradiation. J Phys Chem B. 2003;107:9409–14. [Google Scholar]
- 86.Lan J, Zhang P, Wang TT, Chang Y, Lie SQ, Wu Zl, et al. One-pot hydrothermal synthesis of orange fluorescent silver nanoclusters as a general probe for sulfides. Analyst. 2014;139:3441–5. doi: 10.1039/c4an00505h. [DOI] [PubMed] [Google Scholar]
- 87.Rosendahl SM, Burgess IJ. Electrochemical and infrared spectroscopy studies of 4- mercaptobenzoic acid SAMs on gold surfaces. Electrochim Acta. 2008;53:6759–67. [Google Scholar]
- 88.Li H, Chen D-X, Sun Y-L, Zheng YB, Tan L-L, Weiss PS, et al. Viologen-mediated assembly of and sensing with carboxylatopilla [5]arene-modified gold nanoparticles. J Am Chem Soc. 2013;135:1570–6. doi: 10.1021/ja3115168. [DOI] [PubMed] [Google Scholar]
- 89.Chang H-Y, Chang H-T, Hung Y-L, Hsiung T-M, Lin Y-W, Huang C-C. Ligand effect on the luminescence of gold nanodots and its application for detection of total mercury ions in biological samples. RSC Adv. 2013;3:4588–97. [Google Scholar]
- 90.Moskovitz J, Maiti P, Lopes DHJ, Oien DB, Attar A, Liu T, et al. Induction of methionine-sulfoxide reductases protects neurons from amyloid β-protein insults in vitro and in vivo. Biochemistry. 2011;50:10687–97. doi: 10.1021/bi201426b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tao Y, Ju E, Li Z, Ren J, Qu X. Engineered CpG-antigen conjugates protected gold nanoclusters as smart self-vaccines for enhanced immune response and cell imaging. Adv Funct Mater. 2014;24:1004–10. [Google Scholar]
- 92.Le Guevel X, Trouillet V, Spies C, Auerbach D, Li K, Jung G, et al. High photostability and enhanced fluorescence of gold nanoclusters by silver doping. Nanoscale. 2012;4:7624–31. doi: 10.1039/c2nr30653k. [DOI] [PubMed] [Google Scholar]
- 93.Yu JH, Choi S, Dickson RM. Shuttle-based fluorogenic silver-cluster biolabels. Angew Chem Int Ed. 2009;48:318–20. doi: 10.1002/anie.200804137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Y, Wang Y, Zhou F, Kim P, Xia Y. Protein-protected Au clusters as a new class of nanoscale biosensor for label-free fluorescence detection of proteases. Small. 2012;8:3769–73. doi: 10.1002/smll.201201983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wen Q, Gu Y, Tang L-J, Yu R-Q, Jiang J-H. Peptide-templated gold nanocluster beacon as a sensitive, label-free sensor for protein post-translational modification enzymes. Anal Chem. 2013;85:11681–5. doi: 10.1021/ac403308b. [DOI] [PubMed] [Google Scholar]
- 96.Chen W-Y, Chen L-Y, Ou C-M, Huang C-C, Wei S-C, Chang H-T. Synthesis of fluorescent gold nanodot–liposome hybrids for detection of phospholipase C and its inhibitor. Anal Chem. 2013;85:8834–40. doi: 10.1021/ac402043t. [DOI] [PubMed] [Google Scholar]
- 97.Chen L-Y, Wang C-W, Yuan Z, Chang H-T. Fluorescent gold nanoclusters: recent advances in sensing and imaging. Anal Chem. 2015;87:216–29. doi: 10.1021/ac503636j. [DOI] [PubMed] [Google Scholar]
- 98.Lin S-Y, Chen N-T, Sun S-P, Chang JC, Wang Y-C, Yang C-S, et al. The protease-mediated nucleus shuttles of subnanometer gold quantum dots for real-time monitoring of apoptotic cell death. J Am Chem Soc. 2010;132:8309–15. doi: 10.1021/ja100561k. [DOI] [PubMed] [Google Scholar]
- 99.Liu J-M, Chen J-T, Yan X-P. Near infrared fluorescent trypsin stabilized gold nanoclusters as surface plasmon enhanced energy transfer biosensor and in vivo cancer imaging bioprobe. Anal Chem. 2013;85:3238–45. doi: 10.1021/ac303603f. [DOI] [PubMed] [Google Scholar]
- 100.Wang C, Li J, Amatore C, Chen Y, Jiang H, Wang X-M. Gold nanoclusters and graphene nanocomposites for drug delivery and imaging of cancer cells. Angew Chem Int Ed. 2011;50:11644–8. doi: 10.1002/anie.201105573. [DOI] [PubMed] [Google Scholar]
- 101.Zhang X, Wu F-G, Liu P, Gu N, Chen Z. Enhanced fluorescence of gold nanoclusters composed of HAuCl4 and histidine by glutathione: glutathione detection and selective cancer cell imaging. Small. 2014;10:5170–7. doi: 10.1002/smll.201401658. [DOI] [PubMed] [Google Scholar]
- 102.Nair LV, Philips DS, Jayasree RS, Ajayaghosh A. A near-infrared fluorescent nanosensor (AuC@Urease) for the selective detection of blood urea. Small. 2013;9:2673–7. doi: 10.1002/smll.201300213. [DOI] [PubMed] [Google Scholar]
- 103.Jurs PC, Bakken GA, McClelland HE. Computational methods for the analysis of chemical sensor array data from volatile analytes. Chem Rev. 2000;100:2649–78. doi: 10.1021/cr9800964. [DOI] [PubMed] [Google Scholar]
- 104.He Q, Shi J. MSN anti-cancer nanomedicines: Chemotherapy enhancement, overcoming of drug resistance, and metastasis inhibition. Adv Mater. 2014;26:391–411. doi: 10.1002/adma.201303123. [DOI] [PubMed] [Google Scholar]
- 105.He Q, Gao Y, Zhang L, Zhang Z, Gao F, Ji X, et al. A pH-responsive mesoporous silica nanoparticles-based multi-drug delivery system for overcoming multi-drug resistance. Biomaterials. 2011;32:7711–20. doi: 10.1016/j.biomaterials.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 106.Chandra P, Noh H-B, Pallela R, Shim Y-B. Ultrasensitive detection of drug resistant cancer cells in biological matrixes using an amperometric nanobiosensor. Biosens Bioelectron. 2015;70:418–25. doi: 10.1016/j.bios.2015.03.069. [DOI] [PubMed] [Google Scholar]
- 107.Wang CH, Zheng WB, Qiang O, Tang CW. Effects of non-cytotoxic drugs on the growth of multidrug-resistance human gastric carcinoma cell line. J Dig Dis. 2009;10:91–8. doi: 10.1111/j.1751-2980.2009.00370.x. [DOI] [PubMed] [Google Scholar]
- 108.Iida N, Takara K, Ohmoto N, Nakamura T, Kimura T, Wada A, et al. Reversal effects of antifungal drugs on multidrug resistance in MDR1-overexpressing HeLa cells. Biol Pharm Bull. 2001;24:1032–6. doi: 10.1248/bpb.24.1032. [DOI] [PubMed] [Google Scholar]
- 109.Maoret J-J, Anini Y, Rouyer-Fessard C, Gully D, Laburthe M. Neurotensin and a non-peptide neurotensin receptor antagonist control human colon cancer cell growth in cell culture and in cells xenografted into nude mice. Int J Cancer. 1999;80:448–54. doi: 10.1002/(sici)1097-0215(19990129)80:3<448::aid-ijc19>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 110.Pollak M, Schally A. Mechanisms of antineoplastic action of somatostatin analogs. Proc Soc Exp Biol Med. 1998;217:143–52. doi: 10.3181/00379727-217-44216. [DOI] [PubMed] [Google Scholar]
- 111.Pajeva I, Todorov DK, Seydel J. Membrane effects of the antitumor drugs doxorubicin and thaliblastine: comparison to multidrug resistance modulators verapamil and trans-flupentixol. Eur J Pharm Sci. 2004;21:243–50. doi: 10.1016/j.ejps.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 112.de Wolf FA, Staffhorst RWHM, Smits HP, Onwezen MF, de Kruijff B. Role of anionic phospholipids in the interaction of doxorubicin and plasma membrane vesicles: Drug binding and structural consequences in bacterial systems. Biochemistry. 1993;32:6688–95. doi: 10.1021/bi00077a023. [DOI] [PubMed] [Google Scholar]
- 113.Germann U, Ford P, Shlyakhter D, Mason V, Harding M. Chemosensitization and drug accumulation effects of VX-710, verapamil, cyclosporin A, MS-209 and GF120918 in multidrug resistant HL60/ADR cells expressing the multidrug resistance-associated protein MRP. Anticancer Drugs. 1997;8:141–55. doi: 10.1097/00001813-199702000-00005. [DOI] [PubMed] [Google Scholar]
- 114.Pétriz J, García-López J. Flow cytometric analysis of P-glycoprotein function using rhodamine 123. Leukemia. 1997;11:1124–30. doi: 10.1038/sj.leu.2400659. [DOI] [PubMed] [Google Scholar]
- 115.Emura I, Naito M, Kakihara T, Wakabayashi M, Hayashi N, Chou T. Identification of drug-resistant myeloid leukemic cells by measurement of DNA content, nuclear area, and detection of P-glycoprotein. Cancer. 1996;77:878–87. [PubMed] [Google Scholar]
- 116.Januchowski R, Zawierucha P, Ruciński M, Zabel M. Microarray-based detection and expression analysis of extracellular matrix proteins in drug-resistant ovarian cancer cell lines. Oncol Rep. 2014;32:1981–90. doi: 10.3892/or.2014.3468. [DOI] [PubMed] [Google Scholar]
- 117.Wen C-Y, Wu L-L, Zhang Z-L, Liu Y-L, Wei S-Z, Hu J, et al. Quick-response magnetic nanospheres for rapid, efficient capture and sensitive detection of circulating tumor cells. ACS Nano. 2014;8:941–9. doi: 10.1021/nn405744f. [DOI] [PubMed] [Google Scholar]
- 118.Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, et al. Specific capture and release of circulating tumor cells using aptamer-modified nanosubstrates. Adv Mater. 2013;25:2368–73. doi: 10.1002/adma.201300082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zheng F, Cheng Y, Wang J, Lu J, Zhang B, Zhao Y, et al. Aptamer-functionalized barcode particles for the capture and detection of multiple types of circulating tumor cells. Adv Mater. 2014;26:7333–8. doi: 10.1002/adma.201403530. [DOI] [PubMed] [Google Scholar]
- 120.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis- initiating breast cancer cells. Nature. 2015;528:413–7. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol. 2006;8:1369–75. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 122.Ishiwata I, Ishiwata C, Nozawa S, Iizuka R, Ishikawa H. Change in leukocyte count in peripheral blood of nude mouse bearing transplanted gynecological malignant tumors. Nihon Sanka Fujinka Gakkai Zasshi. 1988;40:1767–71. [PubMed] [Google Scholar]
- 123.Kinoshita J, Haga S, Shimizu T, Imamura H, Watanabe O, Nagumo H, et al. Effect of G-CSF (nartograstim) on neutropenia (leukopenia) induced by taxane in metastatic breast cancer--time-course changes in neutrophil and leukocyte counts. Gan To Kagaku Ryoho. 2001;28:815–9. [PubMed] [Google Scholar]
- 124.Garcia M, Chasseing N, Fernandez O, Bordenave H, Rumi L. Changes in leukocyte levels in patients with breast and lung-cancer. Medicina (B Aires) 1985;45:423. [Google Scholar]
- 125.Duggan C, Risques R, Alfano C, Prunkard D, Imayama I, Holte S, et al. Change in peripheral blood leukocyte telomere length and mortality in breast cancer survivors. J Natl Cancer Inst. 2014;106:dju035. doi: 10.1093/jnci/dju035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mathew P, Thall PF, Wen S, Bucana C, Jones D, Horne E, et al. Dynamic change in phosphorylated platelet-derived growth factor receptor in peripheral blood leukocytes following docetaxel therapy predicts progression-free and overall survival in prostate cancer. Br J Cancer. 2008;99:1426–32. doi: 10.1038/sj.bjc.6604706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Csontos Z, Nádasi E, Csejtey A, Illényi L, Kassai M, Lukács L, et al. Oncogene and tumor suppressor gene expression changes in the peripheral blood leukocytes of patients with colorectal cancer. Tumori. 2008;94:79–82. doi: 10.1177/030089160809400115. [DOI] [PubMed] [Google Scholar]
- 128.Miquel-Cases A, Steuten LMG, Retèl VP, van Harten WH. Early stage cost-effectiveness analysis of a BRCA1-like test to detect triple negative breast cancers responsive to high dose alkylating chemotherapy. The Breast. 2015;24:397–405. doi: 10.1016/j.breast.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 129.Komatsu M, Yoshimaru T, Matsuo T, Kiyotani K, Miyoshi Y, Tanahashi T, et al. Molecular features of triple negative breast cancer cells by genome-wide gene expression profiling analysis. Int J Oncol. 2012;42:478–506. doi: 10.3892/ijo.2012.1744. [DOI] [PubMed] [Google Scholar]
- 130.Li CI, Mirus JE, Zhang Y, Ramirez AB, Ladd JJ, Prentice RL, et al. Discovery and preliminary confirmation of novel early detection biomarkers for triple-negative breast cancer using preclinical plasma samples from the Women’s Health Initiative observational study. Breast Cancer Res Treat. 2012;135:611–8. doi: 10.1007/s10549-012-2204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Diehl KL, Anslyn EV. Array sensing using optical methods for detection of chemical and biological hazards. Chem Soc Rev. 2013;42:8596–611. doi: 10.1039/c3cs60136f. [DOI] [PubMed] [Google Scholar]
- 132.Tao Y, Lin Y, Ren J, Qu X. Self-assembled, functionalized graphene and DNA as a universal platform for colorimetric assays. Biomaterials. 2013;34:4810–7. doi: 10.1016/j.biomaterials.2013.03.039. [DOI] [PubMed] [Google Scholar]
- 133.Lin Y, Xu C, Ren J, Qu X. Using thermally regenerable cerium oxide nanoparticles in biocomputing to perform label-free, resettable, and colorimetric logic operations. Angew Chem Int Ed. 2012;51:12579–83. doi: 10.1002/anie.201207587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.