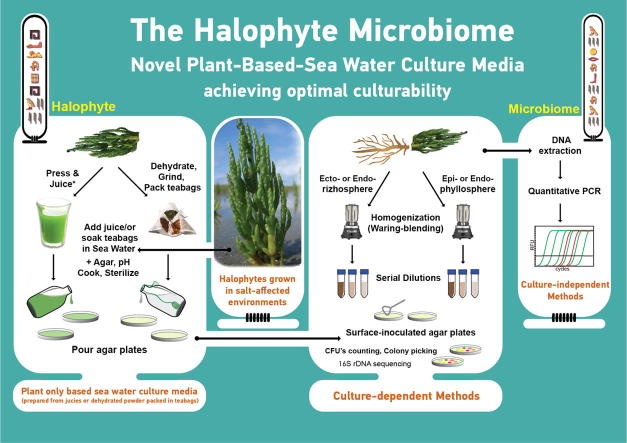
Graphical abstract
Keywords: Halophyte microbiome; Plant-based-sea water culture medium;; Lake Mariout, Alexandria- Egypt; 16S rRNA gene and qPCR; Bacillus spp., Halomonas spp. and Kocuria spp; Arthrocnemum macrostachyum, Halocnemum strobilaceum, Mesembryanthemum crystallinum, Mesembryanthemum forsskaolii and Suaeda pruinosa
Abstract
The plant-based-sea water culture medium is introduced to in vitro cultivation and in situ recovery of the microbiome of halophytes. The ice plant (Mesembryanthemum crystallinum) was used, in the form of juice and/or dehydrated plant powder packed in teabags, to supplement the natural sea water. The resulting culture medium enjoys the combinations of plant materials as rich source of nutrients and sea water exercising the required salt stress. As such without any supplements, the culture medium was sufficient and efficient to support very good in vitro growth of halotolerant bacteria. It was also capable to recover their in situ culturable populations in the phyllosphere, ecto-rhizosphere and endo-rhizosphere of halophytes prevailing in Lake Mariout, Egypt. When related to the total bacterial numbers measured for Suaeda pruinosa roots by quantitative-PCR, the proposed culture medium increased culturability (15.3–19.5%) compared to the conventional chemically-synthetic culture medium supplemented with (11.2%) or without (3.8%) NaCl. Based on 16S rRNA gene sequencing, representative isolates of halotolerant bacteria prevailed on such culture medium were closely related to Bacillus spp., Halomonas spp., and Kocuria spp. Seed germination tests on 25–50% sea water agar indicated positive interaction of such bacterial isolates with the germination and seedlings’ growth of barley seeds.
Introduction
Over 800 million hectares of land throughout the world are affected by salt, and according to global climate change scenarios, rising of the sea level will threaten agricultural production in large areas by increasing the salinity of the soil [1]. To tackle this problem, the use of traditional breeding, genetic engineering of halotolerant transgenic plants and application of halotolerant plant growth promoting (PGP) bacteria are among the major strategies proposed to improve cultivation of saline soil/water environments [2]. So far, members of the salt-tolerant plant microbiome, e.g. Arthrobacter spp., Azospirillum spp., Bacillus spp., Flavobacterium spp., Pseudomonas spp., and Rhizobium spp., have shown a great adaptation and beneficial interactions with plants in salt stressed environments [3]. Mechanisms involved are most similar among different taxa, and the main strategies include avoiding high salt concentration vis specific membrane or cell wall constructions, pumping ions out of the cell ‘salting out’ process or adjusting their intracellular environment by accumulating non-toxic organic osmolytes and the adaptation of proteins and enzymes to high concentrations of solute ions [4], [5], [6], [7]. Such adaptation mechanisms are partly related to their ability of expanding and regulating those genes required to survive and respond appropriately to the physical and chemical composition of these stressed habitats [6]. Microorganisms nesting roots and leaves of halophytes may contribute to their well-being and salinity tolerance. Directly, they promote plant growth by increasing the availability and efficient uptake of nutrients, e.g. fixing N2, solubilizing inorganic phosphate and producing siderophores [7]. They contribute, as well, to the modulation of plant hormone balance through the synthesis of hormone-like molecules; mainly auxins, cytokinins and gibberellins [8]. Indirect mechanisms include the prevention of attack of plant pathogens through the synthesis of antibiotics or antifungal compounds and through competition for nutrients [7]. On their side, plants noticeably contribute to the selection of the associated bacteria by releasing root exudates, which generate a positive selection pressure and increase competitiveness among bacteria in root colonization [9]. In addition, plants may protect themselves from drought and salt stresses by accumulating compatible solutes such as sugars and amino acids to osmotically adjust their environment [10]. Indeed, information is still limited on survival, physiological, and molecular responses of halotolerant microbiome to sea water intrusion, and consequently possible contribution to the salt-affected environments.
Increasing culturability of the plant microbiome under laboratory conditions represents a challenge to specialists, where cultivation on laboratory media has selective effects, and thus yields results that are not representative of the whole microbial community. Having in mind that the communities of rhizobacteria develop in concert with plant roots and, as well, are framed by the background and bulk soil community [11]. This has steered efforts towards tailoring culture media for increasing culturability of the plant microbiome. Including plant materials in the composition of used culture media was sporadic, and originally experimented through the use of plant infusion and extracts as additional supplements for cultivation of plant/soil microorganisms. Pathogenic and endophytic fungi as well as human pathogens were successfully grown on the extracts/juices of variety of plants and legume seed-proteins [12], [13], [14]. Furthermore, microbial metabolites were productively recovered from culture media based on plant substrates especially the by-products of agro-industries [15].
Our previous publications [16], [17] provided original results and evidences on the ability of crude plant slurry homogenates, juices and saps, as such without any supplements, to support culturability of rhizobacteria and to retrieve their in situ populations. For ease of application, plant dehydrated powders packed in teabags were used to prepare liquid infusions rich enough to cultivate rhizobacteria [18]. In fact, such plant teabags culture media do challenge standard chemically-synthetic culture media as they were adequate and capable to recover and mirror the complex and diverse communities of rhizobacteria. Based on Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (PCR-DGGE) of 16S rRNA gene fingerprints and sequencing, the plant teabags culture media proved to support higher diversity and significant increases in richness of endo-rhizobacteria, namely Gammaproteobacteria and dominantly Alphaproteobacteria. This culminated in more retrieval of the rhizobacteria taxa associated to the plant roots.
In this work, a number of the halophytes of the sea water-stressed environment of the western North Coast of Egypt was tested for the diversity and richness of associated halotolerant bacteria. In addition to plant phyllosphere, the two root compartments of ecto-rhizosphere (representing the root surface together with adhering soil particles) and endo-rhizosphere (representing endophytes in the outer and inner tissues of surface-sterilized roots) were included. Further, we present the original idea of the sole use of plant-based-sea water culture medium to in vitro cultivation and in situ recovery of the plant associated halotolerant microbiome. Culture-dependent (CFUs) and–independent (qPCR) analyses were performed on tested halophytes to expound how far such plant-based substrates would support halotolerant bacterial growth, and possibility to challenge the chemically-synthetic standard culture media supplemented with various types and amounts of salts. 16S rRNA gene analysis was used for identification and phylogenetic characterization of the halotolerant isolates secured from the tested salt-affected environments. For possible contribution to the nutritional status and establishment of tested halophytes, secured isolates were evaluated in relation to their potential to promote plant growth via N2-fixation, indole-acetic acid (IAA) production, and phosphate solubilization. Interaction of these isolates with germination indices of a salt tolerant cultivar of barley, nominated for cultivation in salt-affected Egyptian North Coast, was also monitored.
Material and methods
Sampling sites
Naturally-grown salt-affected plant environments along the northern coasts of Egypt were investigated. The site is located around Lake Mariout, 22 km southwest of Alexandria, Egypt (30°56′39.6″N 29°29′77.1″E).
Tested plants
Six representative salt-affected perennial shrubs were collected from the tested sea water-affected environments (Table 1 and Fig. 1A and B). Samples were obtained by first insertion and separation of the aerial parts of full-grown plants (phyllosphere) into sterilized plastic bags. Then, the root-soil system (intact roots with closely adherent soil) was carefully removed and transferred to sterilized plastic bags for microbiological analyses. Free soil samples nearby the roots were taken as well and subjected to physico-chemical analyses (Table 2) within 48 h of sampling. Plants were identified at “Cairo University Herbarium” based on the authentic herbarium specimens, and were found to belong to the families: Chenopdiaceae, Plumbaginaceae and Aizoaceae.
Table 1.
Tested plant species of the salt-affected environment of Lake Mariout, Alexandria, Egypt: Description, distribution and ecology.
| Tested plants | Species description | World distribution | Distribution in Egypt | Ecological habitat |
|---|---|---|---|---|
| 1- Arthrocnemum macrostachyum (Moric.) K. Koch (Family: Chenopodiaceae) | Halophytic perennial small shrub | North Africa, South Portugal, East Mediterranean region, Sinai to eastward to Iran and Indus River delta | Nile valley, Oases, Mediterranean region, desert, Red Sea and Sinai | Halophytic species grows in coastal salt marshes. The plant accumulates salts in its succulent young stems |
| 2- Halocnemum strobilaceum (Pall.) M. Bieb.(Family: Chenopodiaceae) | Halophytic glabrous shrub | Southern Europe, North Africa and Sinai to central Asia. | North Nile Delta, Mediterranean strip, Red Sea, Sinai and deserts | Grows as halophyte in coastal and desert salt marshes and saline plains |
| 3- Limoniastrum monopetalum (L.) Boiss(Family: Plumbaginaceae) | Halophytic low shrub | West Mediterranean region, Egypt, Crete, naturalized in Balearic islands. | Mediterranean strip and Sinai | Halophyte in coastal salt marshes. Dominate the salt marshes with high calcium concentration, this appears as calcareous scales on leaves |
| 4- Mesembryanthemum forsskaolii Hochst. ex (Family: Aizoaceae) | Annual succulent papillose herb | Egypt, Libya, Palestineand Saudi Arabia | Mediterranean strip, deserts, Sinai and Wadi Natrun | Grows in saline - sandy soil and salt affected deserts. Generally can grow in soil with lower salt concentrations than M. crystallinum. The plant is salt tolerant |
| 5- Mesembryanthemum crystallinum L.(Family: Aizoaceae) | Annual succulent recumbent herb | Mediterranean region, Macaronesia, Europe, South Africa, Naturalized in North and South America and Australia | Mediterranean strip, Nile valley, Eastern desert and Sinai | Maritime sand, coastal salt affected soil, edges of salt marches The plant is salt tolerant, accumulate salt in its root and stem, highest salt concentration stored in Epidermal cells (bladder cells giving the plant the crystalline shape). |
| 6- Suaeda pruinosa Lange (Family: Chenopodiaceae) | Halophytic shrub | Spain, Sicily and North Africa. | Mediterranean strip and Sinai coast | Grows in the edges of the salt marshes. |
Fig. 1.
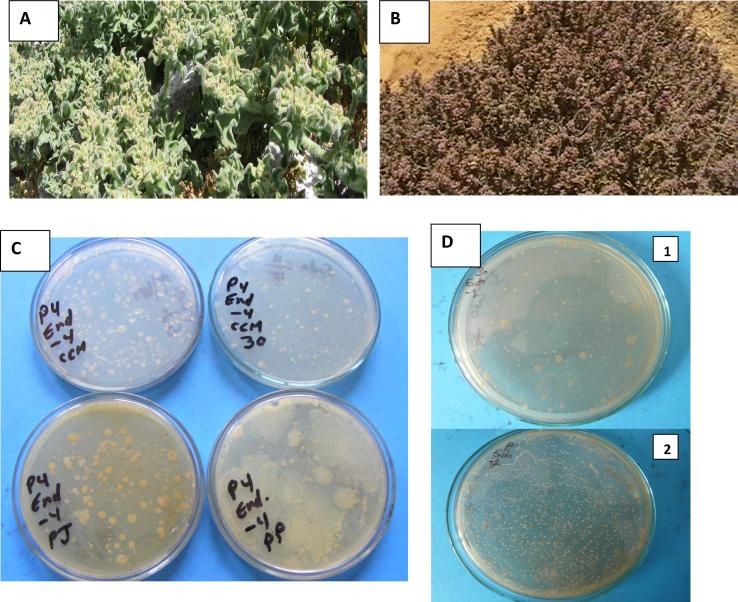
Very well-established vegetation of the salt-affected environment of Lake Mariout, Egypt; and CFUs development and morphologies of the endo-rhizosphere bacteria (endophytes) associated to the tested plants: A: Ice plant (Mesembryanthemum crystallinum), B: Suaeda pruinosa having very thick succulent leaves covered with salt crystals, C: CFUs (dilution 10−4) of endophytes of Mesembrynthemum crystallinum as developed on agar plates of: CCM standard culture medium without (CCM) or with NaCl (30 g L−1, CCM30), plant-based-seawater culture media prepared from juices (PJ) or teabags packed with dehydrated plant powder (PP) of ice plant; D: CFUs (dilution 10−1) of endophytes of Suaeda pruinosa as developed on agar plates of: 1, the chemically synthetic combined carbon sources medium supplemented with NaCl (30 g L−1, CCM30); 2, the plant-based seawater culture media prepared from the teabags of the dehydrated powder of ice plant.
Table 2.
Physico-chemical properties of collected samples representing free soils around tested plants of the salt-affected environment of Lake Mariout, Alexandria, Egypt; and physico-chemical properties of the nearby Mediterranean Sea water.
| Parameters | Salt-stressed free soils around the tested plants |
Mediterranean sea water | |||||
|---|---|---|---|---|---|---|---|
| L. monopetalum | S. pruinosa | H. strobilaceum | A. macrostachyum | M. crystallinuma | M. forsskaoliia | ||
| pH | 8.6 | 9.2 | 9.2 | 8.4 | 9.8 | 9.6 | 8.85 |
| EC (dS m−1) | 33.6 | 89.0 | 43.5 | 48.7 | 11.8 | 11.5 | 51.5 |
| Saturation perecentage (SP%) | 27.0 | 38.0 | 28.7 | 36.7 | 26.3 | 27.1 | ND b |
| Cations (meq L−1) | |||||||
| Ca++ | 59.0 | 24.6 | 22.1 | 21.0 | 7.4 | 7.4 | 19.6 |
| Mg++ | 151.0 | 36.1 | 44.5 | 57.0 | 2.4 | 2.6 | 107.0 |
| Na+ | 260.0 | 975.0 | 534.0 | 635.0 | 117.0 | 119.0 | 635.0 |
| K+ | 35.0 | 60.0 | 52.5 | 17.5 | 43.0 | 49.0 | 12.5 |
| Anions (meq L−1) | |||||||
| SO4−− | 117.0 | 457.0 | 60.0 | 148.0 | 12.9 | 12.5 | 147.0 |
| CO3−− | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 |
| HCO3− | 3.5 | 4.1 | 7.1 | 2.9 | 25.9 | 26.4 | 2.9 |
| Cl− | 383.0 | 635.0 | 586.0 | 580.0 | 131.0 | 126.0 | 623.0 |
Adjacent sand dunes.
ND, not determined.
Culture media
Chemically-synthetic standard culture media
We used the N-deficient combined carbon-sources medium (CCM) that was introduced by Hegazi et al. [19]. This particular culture medium was found to satisfy the nutritional requirements of a wide range of rhizobacteria because of its contents of limited N and diverse carbon sources that mimic the root milieu. It comprises of (g L−1): glucose, 2.0; malic acid, 2.0; mannitol, 2.0; sucrose, 1.0; K2HPO4, 0.4; KH2PO4, 0.6; MgSO4, 0.2; NaCl, 0.1; MnSO4, 0.01; yeast extract, 0.2; fermentol (a local product of corn-steep liquor), 0.2; KOH, 1.5; CaCl2, 0.02; FeCl3, 0.015; Na2MoO4, 0.002. In addition, CuSO4, 0.08 mg; ZnSO4, 0.25 mg; sodium lactate, 0.6 mL (50% v/v) were added per litter. The medium was used as such (CCM), or amended with NaCl: 30 g L−1 (513 mM; identified as CCM30).
Plant-based-sea water culture media
Plant juice culture media
The mature juicy shoots (leaves and stems) of H. strobilaceum, M. crystallinum, M. forsskaolii or S. pruinosa, were sliced and blended for 5 min in a Waring blender with the least possible amounts of sea water, except for M. crystallinum where no water was added because of its very juice nature. The resulting juices were thoroughly filtered through cheese cloth and stored at −20 °C for further use [17]. The crude plant juices, as such or diluted with sea water (juice diluted 1:10, 1:20 and 1:40 with sea water, v/v) were tested as liquid culture media. The used Mediterranean Sea water was of EC 51.5 dS m−1 (corresponding to 3.7% salts and 627 mM; Table 2). Agar culture medium was prepared by adding agar (2%, w/v), pH adjusted to 7.0, then autoclaved for 20 min at 121 °C.
Plant teabags powder culture media
The ice plant (M. crystallinum) was further used for media preparation because of its succulent and juicy nature, rich nutritional contents (Table 3) and abundance in the salt-affected sand dune environments of the northern coast of Egypt. According to Sarhan et al. [18], the vegetative parts of the ice plant were sun dried for 24 h, then oven-dried at 70 °C for 1–2 days. The dehydrated plant materials were mechanically ground to pass through a 2.0 mm sieve to obtain a fine dehydrated powder. Teabags were prepared by packing two grams of the dehydrated powder into empty teabags then sealed by stapling. Two teabags (each containing 2 g) were added to 1 liter of sea water to obtain liquid plant infusions. Agar culture medium was prepared by adding agar (2%, w/v), pH adjusted to 7.0, then autoclaved for 20 min at 121 °C. The teabags were left in the culture media during autoclaving for further plant extraction. Media were tested to ensure sterility before use.
Table 3.
Nutritional profilea of the dehydrated powder of the ice plant (M. crystallinum) used for the preparation of the plant-based-sea water culture media.
| Parameters | M. crystallinum (Sun dried) | Parameters | M. crystallinum (Sun dried) |
|---|---|---|---|
| Macronutrients (ppm) | Micronutrients (ppm) | ||
| Ca++ | 36.7 | Cu | 6.0 |
| Mg++ | 4.6 | Zn | 125.0 |
| K+ | 5.6 | Fe | 315.0 |
| Na+ | 246.9 | Mn | 31.0 |
| Se (ppb) | 2.3 | ||
| Pb (ppb) | 1070.0 | ||
| Total phosphate (%) | 2.20 | Total crude protein (%) | 12.30 |
| Total ash (%) | 44.7 | Moisture (%) | 8.0 |
| Total crude fiber (%) | 7.1 | ||
| Amino acids (mg/g) | Amino acids (mg/g) | ||
| Aspartic acid | 0.69 | Isoleucine | 0.45 |
| Threonine | 0.42 | Leucine | 0.78 |
| Serine | 0.56 | Tyrosine | 0.39 |
| Glutamic acid | 1.25 | Phenylalanine | 0.47 |
| Proline | 0.61 | Histidine | 0.38 |
| Glycine | 0.52 | Lysine | 0.71 |
| Alanine | 0.29 | Arginine | 0.69 |
| Valine | 0.61 | Cysteine | 0.58 |
| Methionine | 0.20 | ||
Methods used for analyses are those described in details by Youssef et al. [17].
In vitro growth of isolates of halotolerant rhizobacteria on plant-based-sea water culture media
The list of tested isolates included three halotolerant pure isolates, Bacillus megaterium, Bacillus pumilus, and Enterobacter spp. obtained from the culture collection of the Department of Microbiology, Faculty of Agriculture, Cairo University, Giza, Egypt. These particular isolates were selected because of their predominance in a number of tested Egyptian salt-affected environments. They were initially inoculated into semi-solid CCM30 test tubes, and microscopically examined for growth and purity. Aliquots of 100 µL were spread on surfaces of agar plates of various tested culture media. This included CCM amended with NaCl (CCM30) and plant-based-sea water culture media of various concentrations of plant juices (juice diluted 1:10, 1:20 and 1:40 with sea water v/v), and plant powder (2 g L−1 and 4 g L−1). After incubation at 30 °C for 4 days, the visual growth index recorded was: 1, scant (discontinued bacterial lawn, with scattered colonies); 2–3, good (continued bacterial lawn); and 4–5, very good (continued and denser bacterial lawn).
Culturability and recovery of plant halotolerant bacteria associated to tested plants
The efficiency of all tested culture media to recover the in situ halotolerant culturable populations associated to naturally grown halophytes was investigated. Three plant compartments were tested: the phyllosphere (representing all vegetative parts including leaves and stems), ecto-rhizosphere (representing the root surface together with closely-adhering soil particles), and endo-rhizosphere (representing endophytes in the outer and inner tissues of surface-sterilized roots). Samples of all tested spheres were prepared for microbiological analysis according to the methods described by Youssef et al. [17] and Sarhan et al. [18]. For endo-rhizosphere samples, roots were surface sterilized with 95% ethanol for 1 min followed by 3% sodium hypochlorite for 30 min, then washed 5 times with sterilized distilled water, 5 min for each wash, before crushing in Waring blender with adequate amount of sea water. Sea water was used as diluent for the preparation of additional serial dilutions of the phyllosphere, ecto- and endo-rhizosphere. Aliquots (200 µL) of suitable dilutions were surface inoculated on agar plates, with 3 replicates, representing the different plant-based culture media prepared from the ice plant juice/powder (juice diluted 1:10, 1:20 and 1:40 with sea water (v/v), and plant powder 2 g L−1 and 4 g L−1) as well as CCM with (3%, w/v) or without NaCl. Incubation took place at 30 °C for >2–7 days, and developed CFUs were counted (Fig. 1C and D). Suspended materials of shoots/roots were dried at 70 °C and weighed for calculations on dry basis of plant materials.
Pure isolates of halotolerant bacteria and determination of their plant growth promoting (PGP) functions
Throughout the microbiological analyses of tested halophytes, one hundred forty-six isolates were selected. Based on their cultural and morpho-physiological characteristics, forty-four representatives of various plants, spheres and culture media were selected for further characterisation. They were tested for PGP functions: nitrogen fixation, phosphate solubilization, indole acetic acid production, and salt tolerance. Based on results obtained, they were clustered (PAST3 software; https://folk.uio.no/ohammer/past), using Unweighted Pair Group Method with Arithmetic Mean (UPGMA). The resulting distance matrix was visualized in dendrogram, and reformatted using FigTree software (http://tree.bio.ed.ac.uk/software/figtree), and annotated using the online tool of Interactive Tree of Life (iTOL) (http://itol.embl.de).
Acetylene reduction assay (ARA)
Nitrogen fixation ability in the form of acetylene reducing activity was measured [20] for pure halotolerant isolates grown in semi solid CCM culture medium, supplemented with 3% NaCl (CCM30). Isolates produced more than 5 nmoles C2H4 culture−1 h−1 were considered positive and further maintained on CCM30 agar slants.
Indole-acetic acid (IAA) production
Tubes containing liquid CCM30 supplemented with L-tryptophan (0.5 g L−1) were inoculated with the selected isolates and incubated for 24–48 h at 30 °C. The resulting liquid cultures were centrifuged and 0.5 mL of Salkovisky’s reagent was added to the supernatant. Positive result was indicated with the change in colour to pink to deep purple and measured colorimetrically at 535 nm [20].
Phosphate solubilization
Isolates were grown on Pikovskaya's agar plates [21] that contained (g L−1): glucose, 10; Ca3(PO4)2, 5; (NH4)2SO4, 0.5; NaCl, 0.2; MgSO4·7H2O, 0.1; KCl, 0.2; yeast extract, 0.5; MnSO4·H2O, 0.002; and FeSO4·7H2O, 0.002; and agar, 20. The culture medium was additionally supplemented with NaCl (30 g L−1). The formation of clearance zone is considered positive result.
Salt tolerance
A number of tubes with liquid CCM amended with different NaCl concentrations (30, 50, 70, 100, 120, 150, 200, and 220 g L−1) was inoculated with the selected isolates. During incubation period of 2–7 days at 30 °C, growth turbidity confirmed by microscopic examination was considered an indication of positive growth and tolerance to the tested salt concentration.
Quantification of total bacterial counts using quantitative real-time PCR
Copy number quantification of 16S rRNA gene was performed by quantitative real-time PCR using the CFX96 Touch™ Detection System (Bio-Rad, CA, USA) in optical grade 96 well plates. Portions of the original root suspensions, prepared for CFUs plate counting were centrifuged at 9500g for 15 min., and then DNA was extracted from root pellets using the QIAGEN DNeasy plant mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted DNA was 1:10 (v/v) diluted and analyzed in duplicates [18]. The PCR reaction was performed in a total volume of 25 µL using SYBR® green master mix (Bio-Rad, CA, USA) containing 2 µL DNA (ca. 3–15 ng), 2.5 µL of 3.3 pmol of both primers of each of the universal forward 519f (CAGCMGCCGCGGTAANWC) and reverse 907r (CCGTCAATTCMTTTRAGTT) primers [18], and 5.5 µL PCR water. The standard curve was constructed using 407 bp length fragment of purified PCR product of the Escherichia coli 16S rRNA gene in tenfold dilutions with the range of 2.5E+2–2.5E+7. The amplification of DNA was done according to the thermal amplification cycling program: 3 min of initial denaturation at 95 °C, 40 thermal cycles of denaturation at 95 °C for 15 sec, annealing at 53 °C for 30 sec, and extension at 72 °C for 42 sec; followed by melting curve construction by increasing the temperature from 53 °C to 95 °C with fluorescence detection every 0.5 °C to verify the PCR quality. The bacterial cell numbers were obtained indirectly assuming 3.6 is the average number of rRNA operon [18], [22], [23].
16S rRNA gene sequencing and phylogenetic affiliation
Selected isolates were grown in liquid cultures of the corresponding culture media, then bacterial broth cultures were centrifuged at 9500g for 15 min., and DNA was extracted from bacterial pellets using the QIAGEN DNeasy plant mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The extracted DNA was used as a template to amplify the whole 16S rRNA gene using the primers 9bfm (GAGTTTGATYHTGGCTCAG) and 1512r (ACGGHTACCTTGTTACGACTT) [18]. The reaction was performed in a total volume of 25 µL with 2 µL template DNA (ca. 2–18 ng µL−1), 12.5 µL of QIAGEN TopTaq master mix (Qiagen, Hilden, Germany), 5.5 µL PCR water, and 2.5 µL of 3.3 pmol of both primers, using the Bio-Rad C1000 Thermal Cycler (Bio-Rad, CA, USA). The thermal cycling program was adjusted as follows: 4 min of initial denaturation at 95 °C, 30 thermal cycles of 1 min denaturation at 95 °C, 1 min annealing at 56 °C, and 1 min of extension at 74 °C; PCR was finished by a final extension step at 74 °C for 10 min. QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) was used to purify the PCR product according to the manufacturers’ instructions.
16S rRNA gene sequencing was performed according to Sanger enzymatic sequencing (Eurofins MWG Operon, Ebersberg, Germany). 16S rRNA gene sequences were compared with their closest matches in GenBank (www.ncbi.nlm.nih.gov/BLAST/) and GreenGenes (http://greengenes.lbl.gov/cgi-bin/nph-index.cgi) databases to determine the taxonomy of the bacterial strains. Together with 429 sequences representing all species of Bacillus spp. (280), Halomonas spp. (134), and Kocuria spp. (15), we constructed the phylogenetic tree using MUSCLE and the Neighbours-Joining methods based on the maximum composite likelihood model implemented in MEGA 6.0 [24]. The bootstrap values were calculated after 1000 replicates and indicated at each node. The 16S rRNA gene sequences identified in this study have been deposited in the GenBank database under the accession numbers: KU836856–KU836865
Interaction of halotolerant bacterial isolates with germination of barley seeds
This introductory experiment was carried out to report on the possible interaction of five tested PGP isolates, Bacillus spp. (PhS1), Bacillus subtilis (EcL2), Bacillus pumilus (EnS4), Bacillus spp. (EnM9), and Halomonas spp. (EnM10), with seed germination of barely. The salt tolerant cultivar Giza 126 was nominated and obtained from the Barley Department, Agricultural Research Centre (ARC), Giza, Egypt. Seeds were surface sterilized with 70% ethanol for 1 min, followed by soaking in 5% sodium hypochlorite for 10 min, then washed 5 times with sterilized distilled water, 5 min for each wash. Tested isolates were grown in liquid salt amended culture medium (CCM30) for 24 h at 25 °C. Seeds were submerged for 30 min in the resulting liquid cultures of the tested isolates (containing >107–108 cells/mL), and a set of seeds was submerged in sterilized liquid medium as a control [25]. The entire process was maintained under axenic conditions. Seed germination was carried out using agar plates (0.8% agar). Preliminary experiments indicated no germination on either undiluted sea water or 3% NaCl-amended tap water. Therefore, further germination experiments used tap water mixed with 25% or 50% sea water. For each salt concentration, three sets of plates were prepared; the set consists of three plates for each isolate with five seeds per plate. Plates were kept in dark at 25 °C, and number of germinated seeds was recorded daily up to 10 d. The following germination attributes were calculated [26]: germination percentage, coefficient of velocity of germination (CVG), germination rate index (GRI) and mean germination time (MGT) as follows:
where N is the number of seeds germinated on day i, and Ti is the number of days from sowing.
Shoot and root lengths as well as dry weights (oven dried at 70 °C overnight) were measured at the tenth day. Vigor index (VI) was calculated, VI = (mean root length + mean shoot length) × germination (%). Specific root length (SRL) was assessed as well, SRL = Root length (cm)/Root weight (g).
Statistical analysis
Analysis of Variance (ANOVA) and Fisher’s Least Significance Difference (LSD) were carried out using STATISTICA v10 (Statsoft, OK, USA).
Results
In vitro growth of pure isolates of halotolerant rhizobacteria on the plant-based-sea water culture media
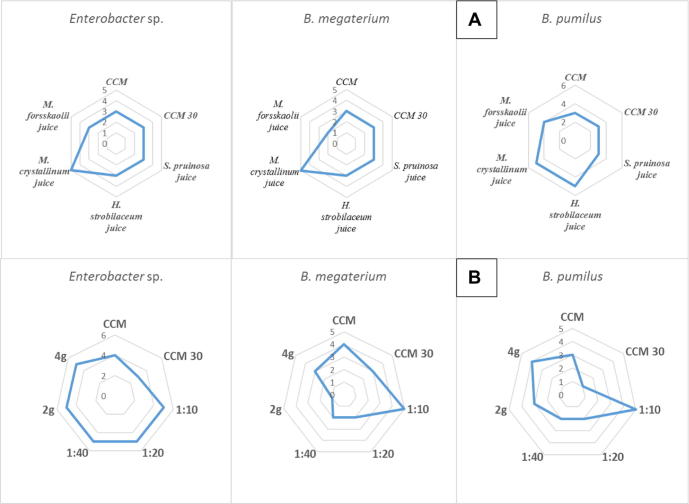
Preliminary experiments examined the possible preparation of culture media exclusively based on the crude juices and/or powders of tested plants, H. strobilaceum, S. pruinosa, M. forsskaolii, and M. crystallinum. Respectively, they were having juice contents of 5%, 17%, 47%, and 67%. Growth indices indicated that all plant juices were nutritionally rich to support good growth of the tested halotolerant bacterial isolates of Enterobacter spp., Bacillus pumilus, and Bacillus megaterium (Fig. 2A). Because of its widespread in salt-affected coastal environments of Egypt, its succulent nature and high content of juice (67%) that supports sufficient culture media preparation as well as better bacterial growth, the ice plant (M. crystallinum) was selected for further experiments (Fig. 1A). The plant was used in the form of juices, in different concentrations, and for ease of application as dehydrated plant powder packed in teabags. In general, the growth index of bacterial isolates measured on the plant-based-sea water culture media was good enough and very much comparable to the standard culture medium (CCM with or without salt amendment). The diluted plant juice (1:10, v/v) supported better growth compared to further diluted plant juices. Interestingly enough, the teabags of ice plant powder, in particular those of 4 g L−1, proved to be appropriate and rather practical (Fig. 2B).
Fig. 2.
Growth of halotolerant bacterial isolates on plant-based-sea water culture media compared to the chemically synthetic combined carbon sources medium (CCM). A, growth indices on various crude juices of tested plants; B, growth indices on various dilutions of the juice, and teabags of ice plant powder (Mesembryanthemum crystallinum); (0, no growth; 1, scant growth; 2–3, good growth; 4–5, very good growth.
The use of the plant-based-sea water culture media for in situ recovery of the halotolerant microbiome of tested halophytes
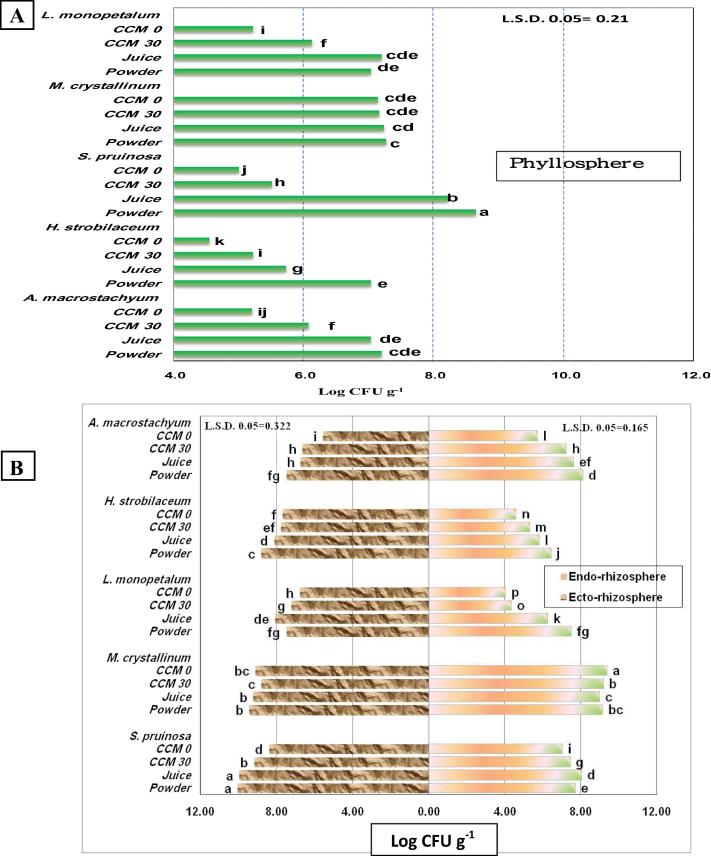
Compared to the chemically-synthetic CCM culture medium supplemented with 3% NaCl, the plant-based-sea water culture media supported well-developed CFUs of halotolerant bacteria (Fig. 1C and D). Irrespective of growth substrate, the culturable population in the ecto-rhizosphere (Fig. 3B) speaks well on the particular richness of the plants S. pruinosa and M. crystallinum (>108–1010 CFU g−1), while the poorest densities were reported for A. macrostachyum (<107 CFU g−1). As to culture media, the plant-based-sea water culture medium enriched with either juice or plant powder-teabags of ice plant, were as good as the chemically-synthetic CCM culture media, and in most cases recovered the highest culturable bacterial population.
Fig. 3.
Culturable bacterial loads (CFUs) of phyllosphere (A), and ecto-rhizosphere and endo-rhizosphere (B) of salt affected plants of Lake Mariout, developed on ice plant-seawater culture medium based on plant juice or dehydrated powder, compared to the chemically-synthetic combined carbon sources medium amended with salt (3%, CCM 30) or not (CCM). Different letters indicate significant differences among treatments (P ≤ 0.05).
Microbiological examination of surface sterilized roots (Fig. 3B), i.e. endo-rhizosphere, indicated the copious presence of endophytic halotolerant bacteria in the roots of tested halophytes; being in the wide range of >104–109 CFU g−1. The highest endophytic colonization was scored for the plant M. crystallinum (>109 CFU g−1) followed by S. pruinosa and A. macrostachyum (>105–108 CFU g−1); the lowest pattern of colonization was reported for L. monopetalum and H. strobilaceum (>104–107 CFU g−1). Again, the tested plant-based-sea water culture media recovered culturable endophytes with densities very much comparable, if not exceeding, to those developed on the salt-amended chemically-synthetic culture medium (CCM).
The bacterial load of the aerial parts, i.e. phyllosphere, of the tested plants was in the range of >104–109 CFU g−1 (Fig. 3 A). The phyllosphere load of the plants M. crystallinum, S. pruinosa and L. monopetalum was relatively higher to that of H. strobilaceum and A. macrostachyum. The plant-based-sea water culture media supported the highest recovery of both epiphytic and endophytic bacterial populations of the phyllosphere.
Using qPCR, the bacterial 16S rRNA gene copy numbers were determined per grams of dry weight of roots of S. pruinosa; the mean log number of bacterial cell calculated for 4 replicates was log 8.40 ± 0.007. The culture-dependent CFUs developed on agar plates represented 3.83–19.45% of qPCR bacterial cell numbers. The highest culturability was reported for the plant-sea water culture medium based on the ice plant juice (15.27%) or powder teabags (19.45%) compared to the chemically synthetic CCM either salted (11.22%) or not (3.83%) (Table 4). This is a strong indication on the capacity, together with practicability, of the introduced plant-based-sea water culture media to significantly increase culturability and recoverability of the in situ microbiome of tested halophytes.
Table 4.
The culturability of rhizobacteria in the endo-rhizosphere of S. pruinosa on various culture media, calculated as numbers of CFUs1 developed on agar plates, and related to the total bacterial numbers measured by qPCR.2
| Culture media | log CFU count g−1 root | % of culturability |
|---|---|---|
| CCM | 6.99 ± 0.009d,3 | 3.83% |
| CCM30 | 7.45 ± 0.008c | 11.22% |
| Ice plant juice | 7.59 ± 0.073b | 15.27% |
| Ice plant teabags | 7.69 ± 0.027a | 19.45% |
CFUs experiment of 3 replicates: Data are log means ± standard error (SE), n = 3.
qPCR experiment of 4 replicates of surface-sterilized roots: The mean value of qPCR cell numbers is log 8.40 ± 0.007 g−1 root dry weight, indirectly obtained by assuming that the average 16S rRNA gene copy number per bacterial cell is 3.6.
Statistical significant differences (LSD) are indicated by different letters (P value ≤ 0.05).
Characterization and identification of representative halotolerant bacterial isolates secured from various spheres of tested halophytes
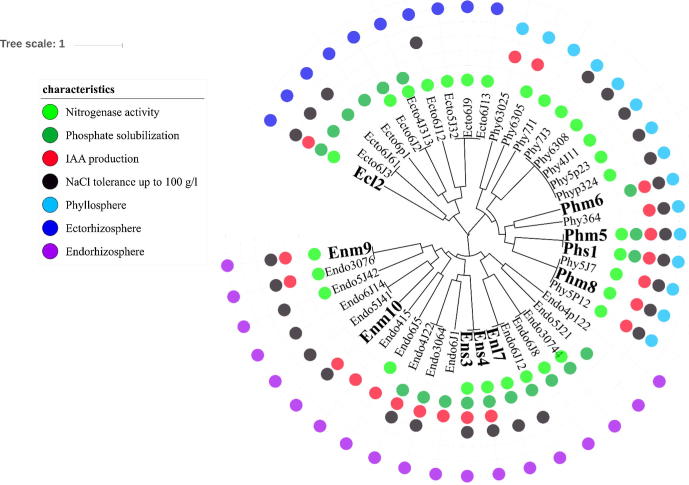
One hundred forty-six isolates representing phyllosphere (43 isolates), ecto-rhizosphere (47 isolates) and endo-rhizosphere (56 isolates) of the tested halophilic xerophytes were single-colony isolated from CFUs developed on various tested culture media. Based on their general cultural and morpho-physiological characteristics, forty-four representative isolates were further selected, tested and clustered according to their plant growth promoting potentials (acetylene reduction, IAA production, P-solubilization and salt tolerance; Fig. 4). In general, 40–80% of the isolates showed tolerance to higher concentrations of NaCl, particularly those found in the close proximity of the plant, i.e. phyllosphere (80.0%) and endo-rhizosphere (63.2%) compared to the ecto-rhizosphere (40.0%). Similarly, indole acetic acid production was a common function in the phyllosphere (60.0%) and endo-rhizosphere (52.6%) compared to the ecto-rhizosphere (10.0%). To the contrary, P-solubilization was reported higher in the root environment (50.0–60.0%) compared to the plant phyllosphere (20.0%). Nitrogen fixation, in terms of acetylene reduction activity, was the most predominant function representing 50.0–80.0%, being highest in the phyllosphere (80.0%) followed by ecto-rhizosphere (70.0%) and endo-rhizosphere (52.6%).
Fig. 4.
UPGMA cluster analysis of tested halotolerant bacterial isolates based on their plant growth promoting potential. Each circle represents a positive result of the tested traits: nitrogen fixation measured as acetylene reduction, phosphate solubilization, indole acetic acid production, salt tolerance; in addition to the plant sphere of origin (ecto-rhizosphere, endo-rhizosphere and phyllo-sphere). Isolates in bold are those selected for furthers tests of 16S rRNA gene sequencing and interaction with the germination of barley seeds.
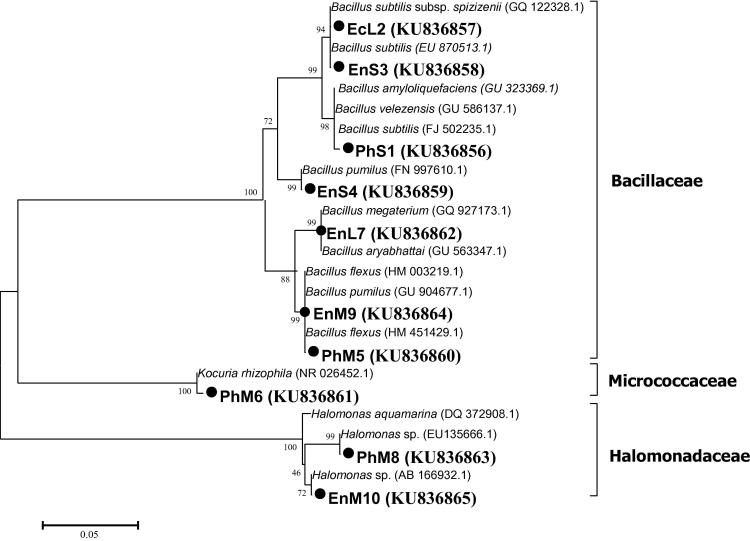
The ten most potential isolates of PGP multifunction (Table 5) were selected for further 16S rRNA gene sequencing. The constructed phylogenetic tree (Fig. 5) showed that they belonged to three families; Bacillaceae, Halomonadaceae and Micrococcaceae. The majority of isolates belonged to the genera Bacillus spp., followed by Halomonas spp. and Kocuria spp. All isolates shared more than 99% identity with their closest phylogenetic relatives.
Table 5.
Detailed information and plant growth promoting functions (PGP) of the selected halotolerant isolates associated to halophytes of Lake Mariout, Alexandria, Egypt.
| Isolate code | Host plant | Plant sphere | Culture media of isolation | ARA c | IAA e | Phosphate solubilization f | Salt tolerance g | Taxonomic position based on 16S rRNA gene sequence (best matched identity >99%) |
|---|---|---|---|---|---|---|---|---|
| EnS3 | S. pruinosa | Endorhizosphere | Juice-baseda | 159 | 8.2 | + | 150 | Bacillus subtilis |
| EnS4 | 19.7 | 16 | + | 100 | Bacillus pumilus | |||
| PhS1 | Phyllosphere | CCM30b | 35 | 13 | + | 100 | Bacillus spp. | |
| EcL2 | L. monopetalum | Ectorhizosphere | Juice-baseda | 38.1 | 7.5 | + | 150 | Bacillus subtilis |
| EnL7 | Endorhizosphere | CCM30b | 15.2 | NDd | + | 150 | Bacillus spp. | |
| EnM9 | M. crystallinum | Endorhizosphere | Juice-baseda | 17 | 21 | NDd | 100 | Bacillus spp. |
| EnM10 | Teabags of plant powdera | NDd | 88 | NDd | 100 | Halomonas spp. | ||
| PhM5 | Phyllosphere | Teabags of plant powdera | 13.9 | 24 | + | 150 | Bacillus flexus | |
| PhM6 | CCM30b | NDd | 7.3 | + | 100 | Kocuria rhizophila | ||
| PhM8 | Juice-baseda | 49.8 | 15 | NDd | 100 | Halomonas spp. |
Plant-based-sea water culture media of ice plant, using either juice or plant powder teabags.
N-deficient combined carbon sources medium (CCM) amended with 30 g L−1 NaCl.
nmoles C2H4 h−1 culture−1.
ND, not detected.
µg/mL culture.
Clear zone of solubilization.
Positive growth in CCM salted with NaCl (up to 100–150 g L−1).
Fig. 5.
Neighbour-joining tree based on 16S rRNA gene sequence. The tree shows the relationship of our isolates to closely related bacteria recovered from GenBank. Black circles indicate our PGP isolates, and values above each node are bootstrap percentages obtained from 1000 replicates. For more information on the bacterial isolates please refer to Table 5.
Interaction of multifunction PGP halotolerant isolates with germination of barley seeds
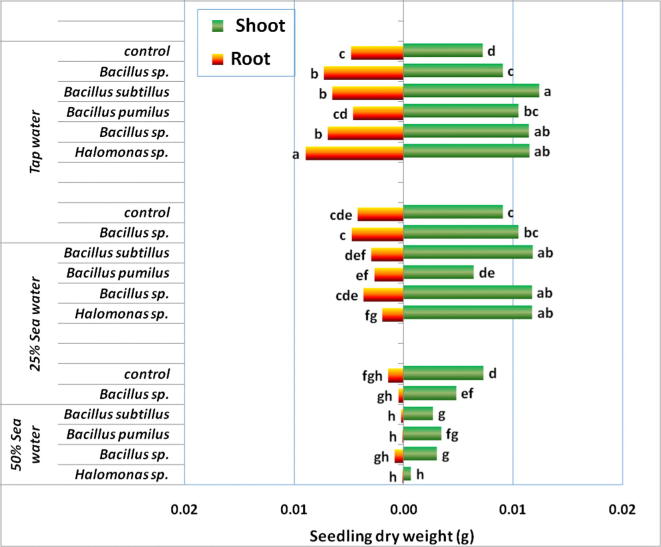
In absence of salt stress, majority of the tested bacterial isolates supported better germination and growth of barley seedlings; lengths of roots and shoots increased with corresponding percentages of 10–58% and 3–9% (data not shown). Increases in dry weights of shoots and roots of seedlings were 26–72% and 35–87%, respectively. Such positive interaction did persist in the environment of 25% sea water (corresponding to 157 mM), especially for shoot with increases ranging from 16% to 83% over control (Fig. 6). To the contrary, in the presence of 50% sea water (corresponding to 314 mM), growth of seedlings was very much retarded, with no positive interactions to any of the isolates tested.
Fig. 6.
Interaction of tested halotolerant bacterial isolates with growth (dry weights of shoots and roots) of barley seedlings developed on different concentrations of sea water (25% and 50% in water agar). Different letters indicate significant differences among treatments (P ≤ 0.05).
Discussion
Microorganisms represent the richest repository of molecular and chemical diversity in nature. They perform multiple functions vital to the sustainability of the biosphere, being abound in all kinds of habitat, viz, with extremes of pH, temperature, water stress and salinity. More recently, this largely unexplored reservoir of resources has become the focus of investigation for innovative application useful to mankind. In this respect, the widespread of halophilic microorganisms and shifts in their community composition with increasing salinity have been in focus, and research in functional interactions between plants and microorganisms contributing to salt stress is gaining interest [27], [28], [29], [30]. Bearing in mind that prokaryotic community composition of halophytes, compared to glucophytes, has only rarely been investigated and the phyllosphere even more sparsely than the rhizosphere [7].
The present study dealt with the plant cover of a well-known salt stressed environment in Egypt; namely Lake Mariout, western North Coast of Alexandria. This particular environment is under the salt stress of the Mediterranean Sea water. The prevailing halophytes are, certainly, possessing various physiological and biochemical mechanisms that allow optimal growth and persistence in such marginal conditions, and perhaps part of their adaptive success would depend at least on their ability to establish and maintain effective associations with endophytic and/or rhizospheric bacteria [7]. In this respect, the diversity of culturable halophilic bacteria, possibly of multiple plant growth promoting (PGP) functions, were documented in different environments all over the world [27], [28], [29], [30], [31].This was confirmed during the present study, as PGP multifunctions were reported among the tested isolates representing the culturable halotolerant population, being plant sphere-dependent. The majority of isolates (>75.0%) possessed more than two PGP functions, and 45.0% of more than three PGP functions. Of interest is that >40.0% of isolates in close proximity of the plant, both of endo-rhizosphere and phyllosphere origin, were of more than three PGP multifunctions, compared to only 10.0% for the ecto-rhizophere isolates. It appeared that those mechanisms of PGP functions account for the alleviating effects of microorganisms when host plants face unfavourable environmental conditions; encouraging further research on the development and future use of such halotolerant bacteria as biofertilizers. Supporting this conclusion is the findings of Nabti et al. [32] who reported that the salt-tolerant Azospirillum brasilense produced IAA under salt-stress conditions that may substantially contribute to the increased salt tolerance of inoculated wheat plants. Besides, the majority of bacterial isolates from saline habitats had phosphate-solubilizing abilities which are considered as a possible mechanism of PGPR to promote plant growth in salt-affected soils [7].
Irrespective of types of plants tested, higher populations of total culturable bacterial counts (>104–109 CFU g−1), were developed on N-deficient combined carbon-sources medium (CCM; [19]) supplemented with NaCl at a concentration of 513 mM. The overall richness of the ecto-rhizosphere of the tested plants supports the concept of the “rhizosphere effect”; it is the net result of the plant interweaving with the autochthonous soil community. The interacting factors include, beside plant exudates, flux of soluble salts into the rhizosphere under the effect of sea water salts, transpiration-driven movement of water, nutrient ion uptake and diffusional movement by root creating zones of nutrient depletion, diurnal water potential fluctuations in the soil adjacent to roots, creation of low O2 concentration zones and change of the pH of the rhizosphere [33]. Such fluctuations are considered as critical environmental characteristics for selecting rhizosphere microbial communities in quantity (compositional) and quality (functional). In addition, it is well established that the community structure of the plant microbiome is greatly determined by plant species, plant genotype and plant nutritional status [34], [35], [36]. The orchestral effect of plant through root exudates is very well documented [37], and >20% of photosynthetically assimilated carbon is released in the form of carbohydrates, organic acids, amino acids and amides as well as vitamins and other compounds [38].
At the phylogenetic level, Firmicutes, Actinobacteria, and Gammaproteobacteria were the dominant phyla reported among the culture-dependent and culture-independent populations nesting various root compartments of halophytes [27], [30], [39], [40], [41], [42], [43], [44], [45]. Among the strains classified to Firmicutes, Bacillus spp. was the most dominant genus of both endophytes and rhizosphere bacteria. Very common in marine environments were B. subtilis, B. licheniformis, B. pumilus and B. cereus [30], [39]. Similarly, many other bacilli have been isolated from a wide variety of salt-affected environments: B. vallismortis, Ammoniphilus sp, Halobacillus dabanensis, Oceanobacillus manasiensis sp. nov., and Pantibacillus spp. [40], [41], [42], [43], [45]. The genus Halomonas was often detected in the root environments of halophytes, and Marasco et al. [44] found that such salt-loving bacteria replaced the genera Marinobacter and Alcanivorax found in the supratidal rhizosphere of Salicornia strobilacea. A great part of these halophilic/halotolerant bacteria plays a prominent role in decomposition, biodegradation, carbon and nitrogen cycles, and promotion of plant growth due to various physiological and biochemical mechanisms [6], [27], [28], [29], [30], [31].
To improve culturability of rhizobacteria, we introduced the plant-based culture media. Crude plant juices, slurry homogenates and saps of plants were found to be rich enough, as such without any supplements, to support in vitro culturability and in situ recovery of rhizobacteria [16], [17]. For ease of application and practicability, we further recommended the use of plant dehydrated powders packed in teabags to prepare liquid infusions rich enough to cultivate endo-rhizobacteria [18]. Such plant-based culture media were able to resolve unique DGGE bands that were not detected by standard culture media. Results of 16S rRNA gene DGGE fingerprints and diversity indices concluded that plant teabags culture media supported higher diversity and significant increases in richness of endo-rhizobacteria. Furthermore, such plant media successfully recovered a number of not-yet-cultured bacteria, which most closely matched uncultured bacteria grouped to Novosphingobium spp., Lysobacter spp. and Pedobacter spp. Adopting this particular approach, the present study proposed the use of plant-based-sea water culture media for culturing and recovering of halotolerant bacteria associated to plants of salt-stressed environments. The tested combinations of plant materials, providing the diverse store of nutrients, and the sea water, exercising the necessary natural salt stress, supported excellent recovery of the halotolerant bacterial community compared to the chemically-synthetic CCM, salted with NaCl. The crude plant juices of almost all the tested plants, as such without any supplements, are rather rich in nutrients and sufficiently and efficiently supported in vitro growth of tested halotolerant bacterial isolates. Amongst tested halophytes, M. crystallinum favoured for further culture media preparations because of its juicy nature (67% juice extraction) and distinguished richness in protein, macro- and micro-nutrients, and amino acids as growth factors. In vitro growth of tested bacterial isolates was better reported with further dilutions of plant juice (juice: sea water; 1:10 v/v). Such positive dilution effect very possibly attributed to decreasing the osmotic impact of concentrated nutrients/salts as well as minimizing the inhibitory effect of antimicrobial compounds and/or antibiotics that might be present in the plant juice [17]. Further, the plant-based sea water culture media supported very good in situ recovery of the culturable haloterant microbiome of tested plants. The nice development of CFUs on such culture media indicated that the plant materials and sea water combinations are of promiscuous nature, and possessed the ability to support the general culturability of the salt-tolerant microbiome associated to the various spheres tested. This supports the idea that, on a coarse taxonomic scale, there is some degrees of commonality in the bacterial composition of plant sphere communities of many plants, ignoring a certain degree of specificity in the selection of these communities [46].
The constructed phylogenetic tree of our PGP isolates (Fig. 5) revealed that they belonged to three families: Bacillaceae (Bacillus subtilis, Bacillus pumilus, and Bacillus flexus), Halomoadaceae (Halomonas spp.), and Micrococcaceae (Kocuria rhizophila). Based on the available information in the literature (Table 6; [7], [44], [45]), the bacterial members isolated in the present investigation possess PGP multi-functions that mostly related to plant nutrition and survival of tested halophytes.
Table 6.
Multi-functions of the halotolerant bacterial isolates obtained during this study, related to those reported in literature.
| Our isolates | Plant growth promoting functions |
Similar isolates with corresponding functions previously reported in literature | |||
|---|---|---|---|---|---|
| ARAa | IAAb | Phosphate solubilization | Salt tolerance (% NaCl)c | Source of isolation (in literature) | |
| B. flexus (PhM5) | + | + | + | + | Industrial maize processing wastewater (Nejayote), Mexico |
| Plant roots, Mexico | |||||
| Banana tree root, Brazil | |||||
| B. pumilus (EnS4) | + | + | + | + | Wheat rhizosphere, India |
| Banana tree roots, Brazil | |||||
| Soil of barren fields and the rhizosphere of halophytes, Korea | |||||
| B. subtilis (EnS3 and Ecl2) | + | + | + | + | Banana tree roots, Brazil |
| Culture collection, India | |||||
| Halomonas sp. (EnM10 and Phm8) | + | + | NR | + | Salt lake, India |
| Salicornia brachiata rhizosphere, India | |||||
| K. rhizophila (PhM6) | NRd | + | NR | + | Marine sediment, East Siberian Sea |
| Ryegrass endophytes, Poland | |||||
ARA, acetylene reduction assay.
IAA, indole-acetic acid production.
Growth in CCM culture media in presence of NaCl ≥ 30 g L−1.
NR, not reported in literature.
Crop establishment comprises principally three processes; germination, emergence and early seedling growth. These growth stages are rather sensitive to salt stress and could be used as criteria to screen for salt tolerance. In this study, seed germination of the tested salt-tolerant variety of barley was partially injured in presence of 25% (v/v; 157 mM) sea water, further dramatic decreases were encountered with raising sea water level to 50% (v/v; 314 mM). In general, inoculation with halotolerant/halophilic bacteria alleviated the toxicity of salts. Common positive interactions of majority of the Bacillus spp. and Halomonas spp. are recognized in the absence of salts, and even extended to the environment of 25% sea water. Of interest is that the tested bacteria showed plant promotion at the shoot compared to the root level. An effect that was reported and explained as due to shoot length and biomass increases favouring accumulation of water in the tissue [44], [45]. Additionally, the PGP bacteria had transitional inhibitory effects over primary root growth followed by prominent simulation of lateral root formation. Such harmful effects might be caused by the high osmotic pressure of the solution slowing down the intake of necessary water for germination and by the toxic influence of high salt concentration on the embryo [47], [48]. Sayar et al. [49] reported that high salt levels inhibit the mobilization of the seed reserves and the growth of embryonic axis. Furthermore, Albacete et al. [50] reported that salt stress affects the hormonal equilibrium (cytokinins/auxins), which causes trouble in shoot growth, impairment and changes in the biomass partitioning. The decrease in dry biomass may be caused by the increase of Cl- concentration in the tissue [51].
Conclusions
For environmental and biotechnological necessities, the need arises to exploring the diversity of bacteria associated to the halophytes of sea water-stressed environments. The introduced plant-based-sea water culture media is a unique approach to increase in vitro culturability and in situ recovery of the halotolerant/halophilic microbiome. We recommend the use of sea water as a base for culture medium supplemented with the plant material of the predominant halophytic plants in a given salt-stressed environment. This will increase culturability, explore diversity and open the window for culturing the unculturable microbiome that failed to develop on conventional culture media artificially salted with NaCl. This will significantly contribute to our future utmost goal of employing the halotolerant/halophilic microbiome as a source of gene(s) that can increase salt tolerance in various crops possibly introduced to salt-stressed environments.
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
The present work was supported by the Research Grant NS-07-15 of the Egyptian Ministry of Agriculture and Land Reclamation. Hegazi acknowledges the support of both Alexander von Humboldt Stiftung for his research stay at IGZ, and of The German Academic Exchange Service (DAAD) for 2014 students- workshop/training on “Molecular Biological Techniques for Studying Microbial Ecology” at IGZ, Germany. We are grateful to all kinds of support provided by Prof. Eckhard George in his capacity as the research director of IGZ. Thanks are also extended to Birgit Wernitz for the excellent technical and lab support. Very much appreciated is the lab support of Hager G. El-Zayat, Aya M. Attia, Doaa M. Ali, Reham N. Mahmoud, Asmaa S. Eltahlawy and Ammar Abdalrahem.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.FAO. Global network on integrated soil management for sustainable use of salt-affected soils. Rome (Italy): Land and Plant Nutrition Management Services; 2005.
- 2.Mayak S., Tirosh T., Glick B.R. Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 2004;166(2):525–530. [Google Scholar]
- 3.Egamberdieva D. Survival of Pseudomonas extremorientalis TSAU20 and P. chlororaphis TSAU13 in the rhizosphere of common bean (Phaseolus vulgaris) under saline conditions. Plant, Soil Environ. 2011;57(3):122–127. [Google Scholar]
- 4.Kandowangko N.Y., Suryatmana G., Nurlaeny N., Simanungkalit R., Djonggi M. Proline and abscisic acid content in droughted corn plant inoculated with Azospirillum sp. and arbuscular mycorrhizae fungi. HAYATI. J Biosci. 2009;16(1):15–20. [Google Scholar]
- 5.Sandhya V., Ali S.Z., Grover M., Reddy G., Venkateswarlu B. Effect of plant growth promoting Pseudomonas spp. on compatible solutes, antioxidant status and plant growth of maize under drought stress. Plant Growth Regul. 2010;62(1):21–30. [Google Scholar]
- 6.Boor K.J. Bacterial stress responses: what doesn’t kill them can make them stronger. PLoS Biol. 2006;4(1):18–20. doi: 10.1371/journal.pbio.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruppel S., Franken P., Witzel K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct Plant Biol. 2013;40(9):940–951. doi: 10.1071/FP12355. [DOI] [PubMed] [Google Scholar]
- 8.Spaepen S., Vanderleyden J., Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev. 2007;31(4):425–448. doi: 10.1111/j.1574-6976.2007.00072.x. [DOI] [PubMed] [Google Scholar]
- 9.Couillerot O., Prigent-Combaret C., Caballero-Mellado J., Moënne-Loccoz Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol. 2009;48(5):505–512. doi: 10.1111/j.1472-765X.2009.02566.x. [DOI] [PubMed] [Google Scholar]
- 10.Sziderics A.H., Rasche F., Trognitz F., Sessitsch A., Wilhelm E. Bacterial endophytes contribute to abiotic stress adaptation in pepper plants (Capsicum annuum L.) Can J Microbiol. 2007;53(11):1195–1202. doi: 10.1139/W07-082. [DOI] [PubMed] [Google Scholar]
- 11.Hawkes C V., Deangelis K.M., Firestone M.K. The rhizosphere: an ecological perspective. 2007. Root interactions with soil microbial communities and processes; pp. 1–29. [Google Scholar]
- 12.Arulanantham R., Pathmanathan S., Ravimannan N., Niranjan K. Alternative culture media for bacterial growth using different formulation of protein sources. Nat Prod Plant Resour. 2012;2(6):697–700. [Google Scholar]
- 13.Osman Z.A., Elsanousi S.M., Elsheikh E.A.E. Plant materials as probable growth promoters for certain fungi. Asian J Plant Sci Res. 2013;3(1):87–93. [Google Scholar]
- 14.Murphy B.R., Batke S.P., Doohan F.M., Hodkinson T.R. Media manipulations and the culture of beneficial fungal root endophytes. Int J Biol. 2015;7(3):94–102. [Google Scholar]
- 15.Kosugi A., Tanaka R., Magara K., Murata Y., Arai T., Sulaiman O. Ethanol and lactic acid production using sap squeezed from old oil palm trunks felled for replanting. J Biosci Bioeng. 2010;110(3):322–325. doi: 10.1016/j.jbiosc.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Nour E.H., Hamza M.A., Fayez M., Monib M., Ruppel S., Hegazi N.A. The crude plant juices of desert plants as appropriate culture media for the cultivation of rhizospheric microorganisms. J Adv Res. 2012;3(1):35–43. [Google Scholar]
- 17.Youssef H.H., Hamza M.A., Fayez M., Mourad E.F., Saleh M.Y., Sarhan M.S. Plant-based culture media: efficiently support culturing rhizobacteria and correctly mirror their in-situ diversity. J Adv Res. 2016;7(2):305–316. doi: 10.1016/j.jare.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarhan M.S., Mourad E.F., Hamza M.A., Youssef H.H., Scherwinski A.C., El-Tahan M. Plant powder teabags: a novel and practical approach to resolve culturability and diversity of rhizobacteria. Physiol Plant. 2016;157(4):403–413. doi: 10.1111/ppl.12469. [DOI] [PubMed] [Google Scholar]
- 19.Hegazi N.A., Hamza M.A., Osman A., Ali S., Sedik M.Z., Fayez M. Modified combined carbon N-deficient medium for isolation, enumeration and biomass production of diazotrophs. In: Malik K.A., Mirza M.S., Ladha J.K., editors. Nitrogen fixation with non-legumes. Springer; Dordrecht (Netherlands): 1998. pp. 247–253. [Google Scholar]
- 20.Hegazi N.A., Fayez M., Amin G., Hamza M.A., Abbas M., Youssef H. Diazotrophs associated with non-legumes grown in sandy soils. In: Malik K.A., Mirza M.S., Ladha J.K., editors. Nitrogen fixation with non-legumes. Springer; Dordrecht (Netherlands): 1998. pp. 209–222. [Google Scholar]
- 21.Pikovskaya R.I. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologya. 1948;17:362–370. [Google Scholar]
- 22.Klappenbach J.A., Dunbar J.M., Thomas M., Schmidt T.M. RRNA operon copy number reflects ecological strategies of bacteria. Appl Envir Microbiol. 2000;66(4):1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schippers A., Neretin L.N., Kallmeyer J., Ferdelman T.G., Cragg B.A., Parkes R.J. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433(7028):861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- 24.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El Goumi Y., Fakiri M., Lamsaouri O., Benchekroun M. Salt stress effect on seed germination and some physiological traits in three Moroccan barley (Hordeum vulgare L.) cultivars. J Mater Environ Sci. 2014;5(2):625–632. [Google Scholar]
- 26.Kader M. A Comparison of seed germination calculation formulae and the associated interpretation of resulting data. J Proc Res Soc New South Wales. 2005;138:65–75. [Google Scholar]
- 27.Szymańska S., Płociniczak T., Piotrowska-Seget Z., Złoch M., Ruppel S., Hrynkiewicz K. Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiol Res. 2016;182:68–79. doi: 10.1016/j.micres.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Sass A.M., McKew B.A., Sass H., Fichtel J., Timmis K.N., McGenity T.J. Diversity of Bacillus-like organisms isolated from deep-sea hypersaline anoxic sediments. Saline Syst. 2008;4(1):8. doi: 10.1186/1746-1448-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith S.A., Benardini J.N., Strap J.L., Crawford R.L. Diversity of aerobic and facultative alkalitolerant and halotolerant endospore formers in soil from the Alvord Basin. Oregon Syst Appl Microbiol. 2009;32(4):233–244. doi: 10.1016/j.syapm.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Ettoumi B., Raddadispi N., Borin S., Daffonchio D., Boudabous A., Cherif A. Diversity and phylogeny of culturable spore-forming Bacilli isolated from marine sediments. J Basic Microbiol. 2009;49(1):13–23. doi: 10.1002/jobm.200800306. [DOI] [PubMed] [Google Scholar]
- 31.Sgroy V., Cassán F., Masciarelli O., Del Papa M.F., Lagares A., Luna V. Isolation and characterization of endophytic plant growth-promoting (PGPB) or stress homeostasis-regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera. Appl Microbiol Biotechnol. 2009;85(2):371–381. doi: 10.1007/s00253-009-2116-3. [DOI] [PubMed] [Google Scholar]
- 32.Nabti E., Sahnoune M., Ghoul M., Fischer D., Hofmann A., Rothballer M. Restoration of growth of durum wheat (Triticum durum var. waha) under saline conditions due to inoculation with the rhizosphere bacterium Azospirillum brasilense NH and extracts of the marine alga Ulva lactuca. J Plant Growth Regul. 2010;29(1):6–22. [Google Scholar]
- 33.Hinsinger P., Plassard C., Tang C., Jaillard B. Origins of root-mediated pH changes in the rhizosphere and their responses to environmental constraints: a review. Plant Soil. 2003;248(1–2):43–59. [Google Scholar]
- 34.Stephan A., Meyer A.H., Schmid B. Plant diversity affects culturable soil bacteria in experimental grassland communities. J Ecol. 2000;88(6):988–998. [Google Scholar]
- 35.Smith K.P., Goodman R.M. Host variation for interactions with beneficial plant-associated microbes. Annu Rev Phytopathol. 1999;37(1):473–491. doi: 10.1146/annurev.phyto.37.1.473. [DOI] [PubMed] [Google Scholar]
- 36.Yang C, Crowley DE. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status 2000;66(1):345–51. [DOI] [PMC free article] [PubMed]
- 37.Bürgmann H., Meier S., Bunge M., Widmer F., Zeyer J. Effects of model root exudates on structure and activity of a soil diazotroph community. Environ Microbiol. 2005;7(11):1711–1724. doi: 10.1111/j.1462-2920.2005.00818.x. [DOI] [PubMed] [Google Scholar]
- 38.Hütsch B.W., Augustin J. Plant rhizodeposition—an important source for carbon turnover in soils. J Plant Nutr Soil Sci. 2002;165(4):397–407. [Google Scholar]
- 39.Miranda C.A.C., Martins O.B., Clementino M.M. Species-level identification of Bacillus strains isolates from marine sediments by conventional biochemical, 16S rRNA gene sequencing and inter-tRNA gene sequence lengths analysis. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol. 2008;93(3):297–304. doi: 10.1007/s10482-007-9204-0. [DOI] [PubMed] [Google Scholar]
- 40.Spring S., Ludwig W., Marquez M.C., Ventosa A., Schleifer K.-H.H. Halobacillus gen. nov., with descriptions of Halobacillus litoralis sp. nov. and Halobacillus trueperi sp. nov., and transfer of Sporosarcina halophila to Halobacillus halophilus comb. nov. Int J Syst Bacteriol. 1996;46(2):492–496. [Google Scholar]
- 41.Zaitsev G.M., Tsitko I.V., Rainey F.A., Trotsenko Y.A., Uotila J.S., Stackebrandt E. New aerobic ammonium-dependent obligately oxalotrophic bacteria: description of Ammoniphilus oxalaticus gen. nov., sp. nov. and Ammoniphilus oxalivorans gen. nov., sp. nov. Int J Syst Bacteriol. 1998;48(1):151–163. doi: 10.1099/00207713-48-1-151. [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Liu W.Y., Gu Z.J., Chen S.F., Yang S.S. Oceanobacillus manasiensis sp. nov., a moderately halophilic bacterium isolated from the salt lakes of Xinjiang, China. J Microbiol. 2010;48(3):312–317. doi: 10.1007/s12275-010-0135-5. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y.-G., Zhang Y.-Q., Xiao H.-D., Liu Z.-X., Yi L.-B., Shi J.-X. Pontibacillus halophilus sp. nov., a moderately halophilic bacterium isolated from a sea urchin. Int J Syst Evol Microbiol. 2009;59(7):1635–1639. doi: 10.1099/ijs.0.002469-0. [DOI] [PubMed] [Google Scholar]
- 44.Marasco R., Mapelli F., Rolli E., Mosqueira M.J., Fusi M., Bariselli P. Salicornia strobilacea (synonym of Halocnemum strobilaceum) grown under different tidal regimes selects rhizosphere bacteria capable of promoting plant growth. Front Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.01286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Palacio-Rodríguez R., Coria-Arellano J.L., López-Bucio J., Sánchez-Salas J., Muro-Pérez G., Castañeda-Gaytán G. Halophilic rhizobacteria from Distichlis spicata promote growth and improve salt tolerance in heterologous plant hosts. Symbiosis. 2017;17:1–11. [Google Scholar]
- 46.Konstantinidis K.T., Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci. 2005;102(7):2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishac Y.Z., El-Haddad M.E., Daft M.J., Ramadan E.M., El-Demerdash M.E. Effect of seed inoculation, mycorrhizal infection and organic amendment on wheat growth. Plant Soil. 1986;90(1–3):373–382. [Google Scholar]
- 48.Munns R., Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59(1):651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 49.Sayar R., Bchini H., Mosbahi M., Ezzine M. Effects of salt and drought stresses on germination, emergence and seedling growth of Durum wheat (Triticum durum Desf.) J Agric Res. 2010;5(15):2008–2016. [Google Scholar]
- 50.Albacete A., Ghanem M.E., Martinez-Andujar C., Acosta M., Sanchez-Bravo J., Martinez V. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J Exp Bot. 2008;59(15):4119–4131. doi: 10.1093/jxb/ern251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tavakkoli E., Fatehi F., Coventry S., Rengasamy P., McDonald G.K. Additive effects of Na+ and Cl− ions on barley growth under salinity stress. J Exp Bot. 2011;62(6):2189–2203. doi: 10.1093/jxb/erq422. [DOI] [PMC free article] [PubMed] [Google Scholar]