Abstract
Genetically encoded small molecule sensors are important tools for revealing the dynamics of metabolites and other small molecules in live cells over time. We recently developed RNA-based sensors that exhibit fluorescence in proportion to a small molecule ligand. One class of these RNA-based sensors are termed Spinach riboswitches. These are RNAs that are based on naturally occurring riboswitches, but have been fused to the Spinach aptamer. The resulting RNA is a fluorogenic riboswitch, producing fluorescence upon binding the cognate small molecule analyte. Here we describe how to design and optimize these sensors by adjusting critical sequence elements, guided by structural insights from the Spinach aptamer. We provide a step-wise procedure to characterize sensors in vitro and to express sensors in bacteria for live-cell imaging of metabolites. Spinach riboswitch sensors offer a simple method for fluorescence measurement of a wide range of metabolites for which riboswitches exist, including nucleotides and their derivatives, amino acids, cations, and anions.
1. INTRODUCTION
Measuring the flux of cellular metabolites is important because metabolites exert a direct influence on cell physiology (Patti, Yanes, & Siuzdak, 2012). Metabolic pathways are highly linked to both signaling pathways and nutrient availability, and various lines of evidence show that alterations in cellular metabolite dynamics is a mediator of diverse diseases. In addition to imaging endogenously generated metabolites, there is considerable interest in imaging exogenously derived molecules such as antibiotics, drugs, and other xenobiotics.
Traditional methods for quantifying metabolite levels rely on liquid chromatography, NMR spectroscopy or mass spectrometry. However, these techniques require harvesting cells, and thus are not “continuous” assays. They do not enable quantification of metabolite levels in the same cells over time. Additionally, they do not reveal cell-to-cell variation in metabolite levels, which is important for understanding population heterogeneity.
Fluorescence imaging approaches can provide a continuous assay of metabolite concentrations in cells. There are two popular approaches for designing metabolite sensors. One approach is fluorescent protein (FP)-based sensors, which typically depend on the FRET signal generated when two fused FPs become closer together. This is regulated by a ligand-binding domain, such as calmodulin (Tian et al., 2009), that changes structure in response to binding the target ligand. The advantages of this approach are that it can be easily genetically encoded and that the fluorescence signal is ratiometric (Palmer, Qin, Park, & McCombs, 2011); however, the changes in signal are often not very pronounced. Another noteworthy approach is to derivatize organic dyes such that the dye fluorescence is activated by a metabolite-recognizing group, as is the case for Fura-2 and many reactivity-based sensors (Chan, Dodani, & Chang, 2012; Malgaroli, Milani, Meldolesi, & Pozzan, 1987; Valeur, 2000). Despite producing a robust signal, this approach can suffer from difficulty in delivering sensors intracellularly, toxicity problems, and the labor-intensive task of synthesizing novel dye-based sensors.
Recently a new type of sensor technology has been developed based on fluorogenic aptamers (Paige, Nguyen-Duc, Song, & Jaffrey, 2012; You, Litke, & Jaffrey, 2015) (Figure 1). These fluorescent RNA aptamers have been synthetically evolved to activate the fluorescence of an otherwise non-fluorescent dye that easily crosses membranes, shows minimal nonspecific fluorescence in cells, and is nontoxic. As an example, Spinach (Paige, Wu, & Jaffrey, 2011) binds to 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI), an analogue of the chromophore formed in green fluorescent protein. Binding of the target metabolite is linked to the folding of Spinach, leading to fluorescence. This approach is genetically encodable since the RNA can be expressed within the cell.
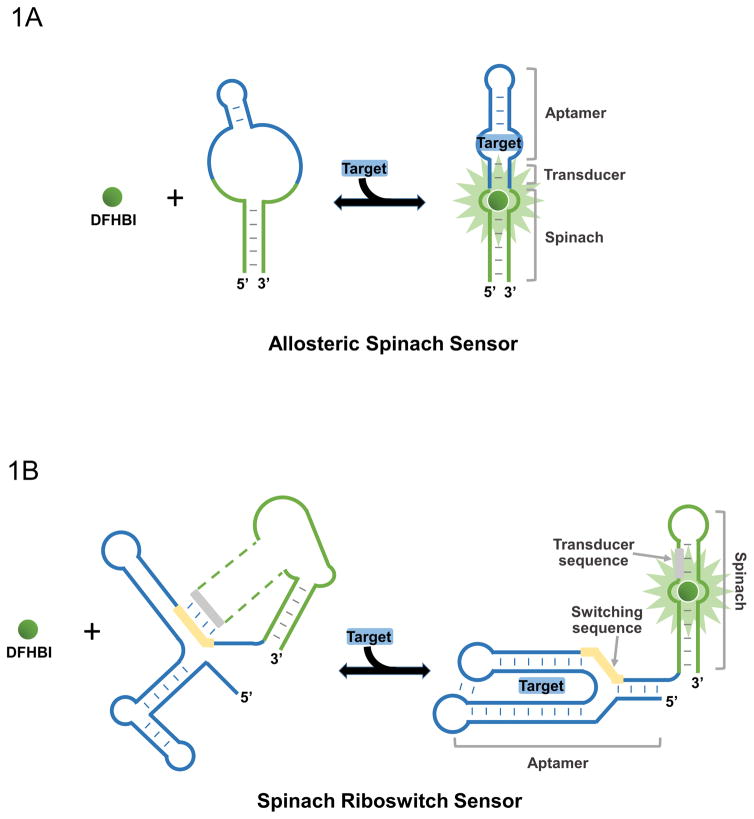
Figure 1.
Two types of metabolite sensors based on fluorogenic aptamers. (A) Spinach-RNA-based sensor using an allosteric mechanism of metabolite (target) detection. Spinach can only bind DFHBI when the target is available to correctly fold Spinach’s binding pocket. (B) Structure switching approach to sensor design using Spinach and riboswitch RNA. Transducer sequence of Spinach and switching sequence of riboswitch interact such that Spinach cannot bind DFHBI until target is bound, which precludes this interaction, such that Spinach can bind DFHBI and fluoresce.
Two types of approaches have been described: allosteric Spinach sensors (Paige et al., 2012), and Spinach riboswitches (You et al., 2015). The structure of allosteric Spinach sensors is shown in Figure 1A. The metabolite-binding aptamer is linked to a critical stem in Spinach via a short transducer region (Paige et al., 2012). In the absence of the metabolite target, the aptamer is unstructured and prevents Spinach from folding. However, metabolite binding results in hybridization of the transducer domain and folding of Spinach, leading to fluorescence. Optimal performance requires optimizing the transducer sequence. This approach has led to the development of sensors for adenosine diphosphate, cyclic-di-GMP, cyclic-di-AMP, etc. (Kellenberger, Wilson, Sales-Lee, & Hammond, 2013; Song, Strack, & Jaffrey, 2013; Strack, Song, & Jaffrey, 2013). These allosteric Spinach sensors allow rapid and dynamic monitoring of intracellular dynamics of metabolites. However, one key limitation is to identify a metabolite-binding aptamer that has suitable selectivity in a complex cellular environment.
The Spinach riboswitch approach takes advantage of the high specificity of naturally occurring riboswitches. Riboswitches are found in the 5′-UTR of certain mRNAs from many lower organisms, and undergo a conformational switch upon binding metabolites that affects mRNA translation or stability (Mandal & Breaker, 2004; Winkler, Nahvi, & Breaker, 2002). The key element of this regulation mechanism is the switching of a single strand, i.e., switching sequence, between binding to the riboswitch’s aptamer domain and to a semi-structured region adjacent that influences mRNA translation or stability, known as the expression platform.
Given the known structure of Spinach and several riboswitches, we modified several known riboswitches so that the switching triggered the formation of the DFHBI-binding pocket of Spinach (Figure 1B). This strategy takes advantage of the similarity between the structure of Spinach and the expression platform. The strategy also takes advantage of the similarity between sequences in the switching strand and the fluorophore-binding pocket in Spinach. The straightforward process of developing Spinach riboswitch sensors (Figure 2) allows the development of an array of sensors for many important cellular metabolites recognized by riboswitches. Here, we describe the procedure to design, optimize and apply Spinach riboswitch sensors for imaging of metabolite dynamics in bacteria cells.
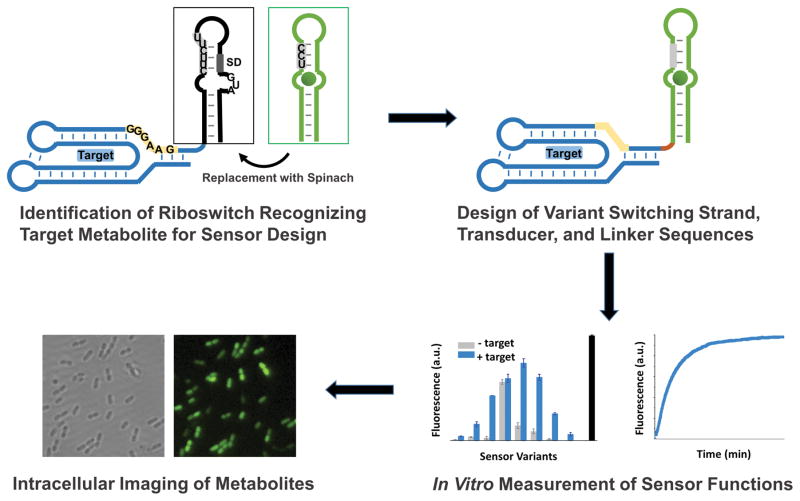
Figure 2.
Scheme for development of RNA-based structure switching sensor. A riboswitch or aptamer for the target metabolite is identified and the expression platform is replaced with the Spinach sequence. The switching sequence of the riboswitch is identified that can interact with Spinach’s transducer sequence or a slightly modified transducer sequence. Variants of the putative RNA sensors with changes in the switching strand transducer and linker regions are tested in vitro for properties such as signal response, affinity, and kinetics of binding. Cloning of best sensors variants into vector for bacterial T7-induced expression allows imaging of metabolite concentration changes in the presence of DFHBI.
2. STRUCTURE-GUIDED DESIGN OF SPINACH RIBOSWITCH SENSORS
For rational design of RNA Spinach riboswitch sensors, structural information is very useful in finding RNA sequences with sensor-like properties and optimizing their behavior, shown in Figure 3A. The 24 different classes of riboswitches are comprised of sequences that recognize nucleotides, amino acids, sugars, and cations/anions (Baker et al., 2012; Breaker, 2011). To date, the Nucleic Acid Database Project lists over 180 different crystal structures of known riboswitches (http://ndbserver.rutgers.edu/). Some riboswitches have structures in multiple conformations, which can help indicate which sequences might make suitable Spinach riboswitch sensor candidates.
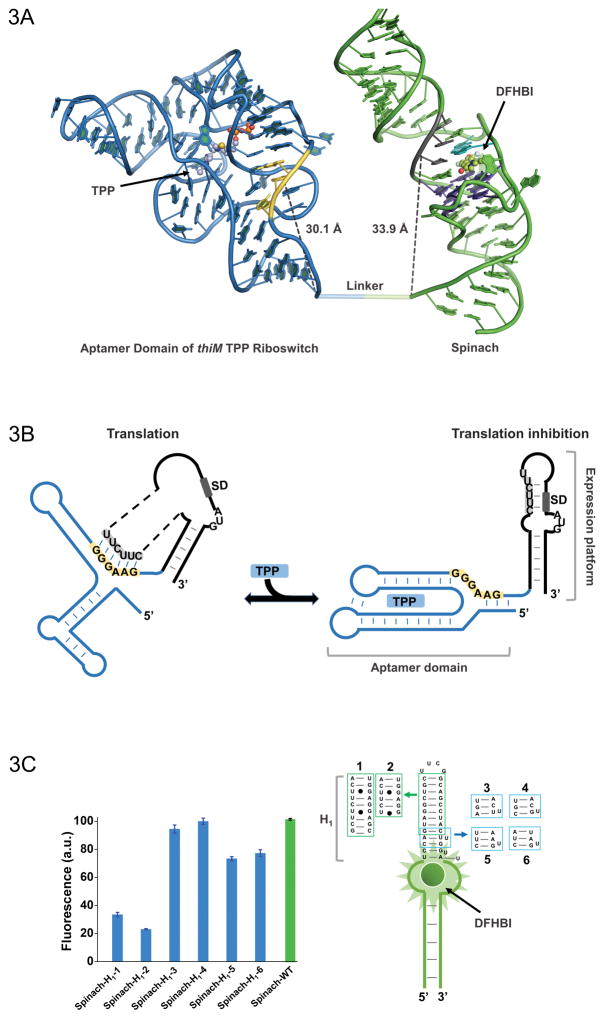
Figure 3.
(A) Model of the thiM-based Spinach riboswitch sensor for TPP. Riboswitch in blue with its switching sequence in yellow, while Spinach in green with transducer sequence in gray. Spinach U•A•U base triple is shown in cyan (overlap with most proximal residue of transducer sequence) and G-quadruplex shown in purple. TPP and DFHBI are shown in ball and stick representative. The distance between the switching sequence and transducer sequence to the base of thiM and Spinach, respectively, is quite similar, which would not be apparent in a two dimensional representation. (B) Mechanism of translational control by the thiM riboswitch in response to TPP. The transducer sequence shows complementarity to the 5′-AGGAGG-3′ Shine-Dalgarno sequence and switching sequence of the riboswitch’s aptamer domain. (C) H1 stem of Spinach tolerates certain modifications with only modest fluorescence changes. Such mutants expand the modularity of the approach and the range of transducer sequences that can be used for Spinach-based sensor design.
2.1 Structure-guided design of switching sequences and transducer sequences
Spinach forms a unique G-quadruplex structure (Warner et al., 2014), which along with a Hoogsteen U•A•U base triple, forms a binding pocket for DFHBI that activates the fluorophore’s fluorescence. In Spinach, the stem adjacent to the fluorophore-binding pocket (H1 stem) holds a 5′-UCC-3′ sequence (Figure 2 and Figure 3A). This sequence, called the transducer sequence, overlaps with the base triple that helps form the DFHBI-binding pocket and is critical for Spinach fluorescence (Paige et al., 2012; You et al., 2015). If this transducer sequence interacts with a complementary sequence from the riboswitch, then we would expect destabilization the U•A•U triplet and reduced DFHBI binding and fluorescence. If this complementary sequence also occupies different conformations in the riboswitch structure when the metabolite is bound or unbound, then we have the basis for a sensor.
Nature has already evolved the necessary complementarity in the switching sequence of many riboswitches. The key element of the riboswitch mechanism is the switching of this sequence between binding to the aptamer domain and to the expression platform, which, for example, contains a Shine-Dalgarno (SD) sequence. This switching is mediated by riboswitch conformation, such that metabolite binding to the riboswitch releases the SD sequence and enables ribosome binding, illustrated in Figure 3A. Furthermore, the switching sequence can be optimized to allow efficient switching between interaction with Spinach and the riboswitch in response to metabolite concentration.
Knowledge of riboswitch structure is immensely useful in identifying a switching sequence that will interact with Spinach’s transducer sequence. PyMOL is an invaluable open-source software tool for making structure-based hypotheses and is available for free from Schrodinger. A community-supported wiki (http://pymolwiki.org/) along with various online sources contain tutorials for the skills needed to observe and measure aspects of a riboswitch structure as in Figure 3B. An algorithmic prediction of RNA structure, such as mFold (http://unafold.rna.albany.edu/?q=mfold) can also help guide identification of a good switching sequence. Structural information has also been collected in the apo-states of certain riboswitches (Garst, Héroux, Rambo, & Batey, 2008; Huang, Serganov, & Patel, 2010) and many have been probed using 2′-hydroxyl probing (SHAPE) to identify bases that have altered conformations upon metabolite binding (Steen, Malhotra, & Weeks, 2010). In several riboswitch classes and putative classes, genomic alignment of homologous riboswitches (Barrick & Breaker, 2007; Weinberg et al., 2010) suggests which bases are more conserved and perhaps more essential for control of gene regulation. Without much prior knowledge of the aptamer domain, a stem that holds the 5′ and/or 3′ ends together in a riboswitch may also contain a good candidate for a switching sequence.
Even if a riboswitch does not contain a clear candidate for a switching sequence in the native conformation, there are additional steps to consider that may lead to an effective sensor. Critically, the U bases of the Spinach H1 stem can base pair with A or G in RNA, and Spinach tolerates some mutations in the H1 stem, as long of it retains a stem structure, forming A-U, G-C, or G-U base pairs. There is evidence that certain modifications of the 3′ end of this transducer sequence do not impair the function of Spinach-based metabolite sensors, as shown in Figure 3C. Thus, there is flexibility to change the “native” 5′-UCC-3′ of Spinach into other sequences that can act as the transducing sequence that will interact with the switching sequence. Changing the sequence of the riboswitch to adjust transduction of the sensor is also possible, but may impair sensitivity of the sensor for the metabolite target. These principles allow us to design sensors from a wider variety of riboswitches for detection of many different metabolites.
2.2 Structure-guided design of linker sequences
The way in which the riboswitch and Spinach are linked can change the behavior of the sensor. Previous sensors have placed the riboswitch on the 5′ end of Spinach, but in principle the order of domains in the 5′-to-3′ sequence is not restricted. Once putative switching sequences in the riboswitch and transducer sequences in Spinach have been determined, the linker sequence between the riboswitch and Spinach should be considered, which also benefits greatly from a structural approach.
The purpose of adjusting the linker sequence is to optimize positioning of the transducer sequence relative to the switching sequence. The crystal structure of Spinach demonstrates that the distance between its 5′ end and the CUU within the transducer sequence and the base triplet is roughly 3.4 nm (Warner et al., 2014). The distance between the riboswitch’s 3′ end and switching sequence normally cannot be changed (Serganov, Polonskaia, Phan, Breaker, & Patel, 2006), but the linker length can be controlled to fine tune a sensor, as illustrated in Figure 3A. The optimal distance from the linker to the switching sequence should be around this 3.4 nm distance. For example, as shown in the structural model of the optimized thiamine pyrophosphate (TPP) Spinach riboswitch sensor (Figure 3A), the predicted distance from the linker to the switching sequence (3.0 nm) is nearly the same as the distance to the corresponding U base of the transducer sequence (3.4 nm).
Optimized length and flexibility of the linker can also enhance sensor function. Long lengths of single-stranded linkers allow flexibility that lets the sensor explore different orientations between the domains. On the other hand, a long linker may allow “too much flexibility” if the domains of the sensor are already in an ideal position. Less flexible double-stranded linkers that extend from the stem of the riboswitch or Spinach can also be tested. One consideration is that double-stranded linkers will both extend and rotate the Spinach or riboswitch helixes, thereby inducing steric effects that may facilitate or disturb the interaction between the transducer and switching sequences. Once again, structural information is a critical guide for optimizing linker types and the behavior of RNA-based sensors.
3. IDENTIFYING SPINACH RIBOSWITCH SENSORS IN VITRO
Once a series of putative sensors have been conceived, the next step towards imaging cellular metabolites is to characterize these RNA sequences in vitro. As discussed above, there are several structure-guided design approaches that may lead to discovery of an optimized Spinach riboswitch sensor. To identify which designs of switching sequence, transducer sequence, and linker sequence are most successful, one must enzymatically synthesize each RNA and then observe fluorescence changes with different levels of metabolite concentration. It is also important to measure other sensor characteristics, such as kinetic response and specificity of recognition. All of these in vitro tests are critical to identifying potential Spinach riboswitch sensors for intracellular imaging.
3.1 In vitro preparation of RNA-based sensor variants
3.1.1 Materials and equipment
PCR thermal cycler
PCR clean-up kit (e.g., QIAquick PCR purification kit, Qiagen, Cat No. 28104)
Ampliscribe T7-flash transcription kit (Epicentre, Cat No. ASF3507)
Incubator, 37 °C
RNA purification kit (e.g., RNA clean & concentrator, Zymo Research, Cat No. R1018; Bio-Spin columns, BioRad, Cat No. 732-6251)
Nanodrop or spectrophotometer for UV-Vis absorption
3.1.2 Procedures
Once the riboswitch is selected, and the transducer and switching sequences are identified, double-stranded DNA (dsDNA) templates for variants of these sequences should be generated. The T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′) should be added to the 5′ end of the template for successful in vitro transcription.
The protocol for standard PCR can be easily found elsewhere. Here, we briefly introduce the procedure for overlap-extension PCR, a method for generating a long (> 100 nt) dsDNA template that is difficult to synthesize chemically and can be expensive when purchased. During overlap-extension PCR, two equimolar template fragments are assembled together to generate the full dsDNA template. These two template fragments contain an overlap region with melting temperature typically around 60 °C. A typical thermocycler program can be as follows: heat samples to 98 °C for 20 seconds, followed by 15 repeated cycles of: denaturing primers at 98 °C for 5 seconds, annealing at 58 °C for 20 seconds, and extending template primers at 72 °C for 20 seconds. After the repeated cycles, a final extension at 72 °C is held for 5 min, followed by cooling to hold samples stable at 4 °C.
After overlap-extension PCR, standard primers complementary to the ends of the product are added to the template mix. Amplification of the template follows the same thermocycler program but with 30 cycles rather than 15 cycles.
PCR products should be verified for size by mobility on a 2% agarose gel. If the samples are sufficiently pure, then they can be purified using a PCR clean-up kit (e.g., QIAquick PCR purification kit, Qiagen, Cat No. 28104), otherwise they must be separated from impurities by agarose gel electrophoresis, followed by excision and purification from gel.
The reaction for generating RNA from the dsDNA templates consists of using a T7 RNA polymerase recognizing the T7 promoter in the 5′ end of the DNA template. The T7 polymerase transcribes through to the end of the template. Following the manufacturer’s protocol, an in vitro transcription reaction can be carried out using the Ampliscribe T7-flash transcription kit (Epicentre, Cat No. ASF3507) in a 20 μl scale. Allow the reaction to incubate at 37 °C for at least 1 hour. If the maximum RNA yield is desired, the reaction can proceed overnight. Afterwards, treat the reaction with DNase at 37 °C for 30 min.
Before proceeding, RNA must be purified from the transcription reaction components. Purification of transcribed RNA can be carried out by column purification or an ethanol precipitation method. Column purification can be achieved using one of several kits (e.g., RNA clean & concentrator, Zymo Research, Cat No. R1018; Bio-Spin columns, BioRad, Cat No. 732-6251). The elution must be made with RNase-free water or TE buffer to prevent base hydrolysis and should be concentrated enough that quantification of concentration by UV absorbance (e.g. with a Nanodrop instrument) is appropriate (0.1 – 5 μg/μl). It is best to store RNA at −80 °C to reduce degradation by incidental RNases.
3.2 Quantification of fluorescence sensor characteristics
3.2.1 Materials and equipment
Fluorimeter
Incubator, 37 °C
DFHBI or DFHBI-1T fluorophore (Lucerna, SKU: 400-1 mg or SKU: 410-1mg, respectively)
3.2.2 Procedures
Testing the fluorescence characteristics of each sensor begins with measurements of fluorescence response to physiological amounts of metabolite. Sensors that exhibit the highest fold-increase in fluorescence after adding metabolite and lowest fluorescence in the absence of metabolite are tested further for dose response and kinetics. Fluorescent aptamers like Spinach, which activate dye fluorescence should be measured with fluorophore in excess because of the low background of fluorophore, when free in solution. In this way, the fluorescence observed will be determined exclusively by the percent of the RNA sensor that adopts a folded conformation. If the RNA is in excess and the fluorophore is limiting, any unfolded sensor will not be detectable since the maximal fluorescence level would be determined by the concentration of the fluorophore. The suitable fluorophore for these sensors include 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI, Lucerna) or its trifluoroethyl derivative, DFHBI-1T (Lucerna). Other measurement conditions may vary from sensor to sensor, however, when using Spinach as a fluorescing domain, sufficient magnesium concentration is necessary for proper folding and fluorescence. Previous Spinach-based sensors are most effective at 2–5 mM magnesium concentration in vitro, which resembles bacterial levels, with fluorophore concentrations roughly 100-fold higher than RNA concentrations.
A starting sample buffer for testing Spinach-based RNA sensors may contain 40 mM HEPES pH 7.4, 100 mM KCl, and 5 mM MgCl2 with 10 μM DFHBI-1T and 0.1 μM of RNA. Assessment of sensors begins with measurement of fluorescence before and after incubation with physiological concentration of metabolite (e.g., 100 μM TPP) at 37 °C for 30 minutes. Any fluorescence that is detected after incubation should also be compared to fluorescence of Spinach without the target-binding aptamer domain (Figure 3C), and any modifications to the Spinach transducer region must be reflected in this control. Measurement of fluorescence with a fluorimeter should be made with 470 nm excitation and 503 nm emission.
The best candidate sensors normally have the highest fold-change upon adding metabolite, and it is critical to see which candidates have a suitable affinity for the metabolite. A suitable affinity (EC50) should fall well within the physiological concentration range of the target, and preferably be slightly higher than the physiological concentration range. In this case, a doubling of the metabolite concentration should result in approximately a doubling in the binding of the metabolite to the sensor and a corresponding increase in fluorescence. Thus, the sensor will produce a fluorescence in a nearly linear manner compared to the metabolite concentration. Titration curves are measured as the fold-change measurement for separate samples with a range of metabolite concentrations or for a single sample with progressive addition of metabolite. These fluorescence measurements form a curve that can be fit by a nonlinear regression to a model for substrate binding.
Kinetic measurements of high-affinity sensors should be made as well to help pick the best sensor and to get a sense of the time course for sensor function in bacteria. Continuous or time-lapsed measurements of fluorescence should be conducted to identify sequences with the highest rate of response to metabolite. An acceptable sensor will have rapid metabolite-induced fluorescence signal change relative to the timescale of physiological dynamics of metabolite. For example, we developed a TPP-targeting Spinach riboswitch sensor that reaches 90% of its maximal fluorescence signal within 10 min. This time frame was suitable for experiments in which we monitored TPP biosynthesis kinetics that occurred over several hours in cells (You et al., 2015).
Furthermore, the specificity of RNA sensors can be measured by determining the fluorescence response to metabolites that are known to occur in cells and which are molecularly similar to the metabolite, e.g., thiamine and thiamine monophosphate for TPP sensors. For sensors designed from naturally occurring riboswitches, this step is still important, even though riboswitches tend to have high specificity for their intended metabolites (Mandal & Breaker, 2004).
4. LIVE CELL IMAGING OF METABOLITES WITH SPINACH RIBOSWITCH SENSORS
After in vitro characterization of RNA-based sensors, it is critical to validate these sensors in live cells. RNA-based metabolite sensors are still difficult to use in mammalian cells, mainly due to the limited stability and abundance of these short and structured RNAs. Despite this, these RNAs typically maintain a steady state level in cells due to a balance of new transcription and degradation. RNA-based sensors with fast kinetic response and readily detectable fluorescence signal are becoming popular for metabolite analysis in bacterial cells (Kellenberger et al., 2013; You et al., 2015). Shown below is a protocol using Spinach riboswitch sensor for TPP imaging in live E. coli cells.
4.1 Preparation of sensor-expressing E. coli
4.1.1 Materials and equipment
PCR thermal cycler
Water bath or heat block, 42 °C
Shaking incubator, 37 °C
LB agar plate: dissolve 10 g of tryptone, 5 g of yeast extract, 20 g of agar and 10 g of NaCl in a final volume of 1 L with deionized water. Autoclave for 15 min at 121 °C. After cooling, add 50 μg/mL kanamycin, and then pour it into 10-cm Petri dishes.
4.1.2 Procedures
Clone Spinach riboswitch sensor into bacterial expression vector. Traditional restriction site cloning can be used to generate a bacterial expression plasmid containing the in vitro characterized sensor. For example, sensor templates can be PCR amplified with primers containing either BglII or XhoI restriction sites on the 5′ and 3′ ends for insertion into the T7 expression cassette of pET28c. These restriction sites place the sensor upstream of a T7 terminator in pET28c. A pETDuet vector, which expresses the sensor and a far-red fluorescent protein from the same promoter as an expression control can also help to gauge sensor function within E. coli. All the following described steps should be performed in a sterile environment.
Take three tubes of chemically competent E. coli Rosetta2 (DE3) cells thawed on ice. Add 40 ng of pET28c-TPP Spinach riboswitch sensor, pET28c-Spinach2 (positive control) and pET28c (negative control), respectively, to each tube. Mix gently by flicking the tubes, and incubate on ice for 5 min.
Heat shock in a 42 °C water bath for 45 s, then immediately place back on ice to chill for 2 min. After chilling, add 250 μl of SOC medium and incubate at 37 °C for 45 min with shaking.
Plate 50 μl of incubated cells onto a LB agar plate supplemented with 50 μg/mL kanamycin. Incubate at 37 °C overnight. Pick several colonies to isolate plasmids. Sequence or use restriction digestion to confirm correct transformation.
4.2 Imaging dynamics of metabolite concentration in live E. coli (TPP sensor)
4.2.1 Materials and equipment
Shaking incubator, 37 °C
Nanodrop or spectrophotometer for UV-Vis absorption
Inverted wide-field fluorescent microscope
Glass-bottom 24-well imaging plates
Poly-D-lysine or poly-L-lysine
LB medium: dissolve 10 g of tryptone, 5 g of yeast extract and 10 g of NaCl in a final volume of 1 L with deionized water. Autoclave for 15 min at 121 °C. The medium can be stored at room temperature.
M9 medium: prepare 5× M9 salts solution by dissolving 64 g of Na2HPO4, 15 g of KH2PO4, 5 g of NH4Cl and 2.5 g of NaCl in a final volume of 1 L with deionized water. Autoclave for 15 min at 121 °C. Final M9 medium contain 1× M9 salts solution, 2 mM MgSO4, 100 μM CaCl2 and 2mg/ml glucose. Sterilize the medium by vacuum filtration through a 0.22 μm filter.
4.2.2 Procedures
Inoculate a single colony inside a sterile culture tube with 5 ml of LB medium and 50 μg/mL kanamycin. Grow the cultures at 37 °C with shaking overnight.
Measure the optical density of an overnight culture at 600 nm (OD600). Dilute the culture to a 0.05–0.1 OD600 units/mL with LB medium with 50 μg/mL kanamycin, grow at 37 °C with shaking till 0.4 OD600 units/mL. Add 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and continue to grow at 37 °C with shaking for 2 h. During this final induction, 300 μg/mL adenosine, an inhibitor of de novo TPP biosynthesis, may be added together with IPTG.
Prepare a glass-bottomed imaging plate, e.g., 24-well MatTek plate, by adding 300 μl of poly-D-lysine solution to each well and incubating at 37 °C for 3 h. Add 500 μl of sterile water twice to rinse each well and remove excess poly-D-lysine.
Spin down 100 μl of an IPTG-induced culture at 5,000g for 2 min at room temperature to pellet the culture. Resuspend in 1 mL of M9 medium. Take a 200 μl aliquot of resuspended culture to plate on the poly-D-lysine-coated plate. Incubate at 37 °C for 45 min to allow cell adherence.
Wash adherent cells twice with 500 μl of M9 medium to remove unattached cells. Then, add 200 μl of M9 medium supplemented with 200 μM DFHBI. Incubate at 25 °C for 1 h.
Image the plate through a 60× oil objective. An example filter set includes a 470/40 nm excitation filter, a dichroic mirror 495 nm (long pass), and an emission filter 525/50 nm. Start imaging with the cells expressing pET28c-Spinach2 (positive control). While imaging, focus on a field containing 50–100 evenly adhered cells. Proper exposure time is normally in the range of 100 ms to 2 s, which allows the highest signal without saturated pixels.
To image dynamics of intracellular concentration dynamics, for example of TPP, add 10 μM thiamine to each well. Thiamine can be converted to TPP by intracellular thiamine kinase. Acquire images immediately (time zero) and every 15 min afterwards for each well. Between each image acquisition, the shutter should be closed to minimize photobleaching of fluorophore and cell damage. Live fluorescence images were taken for 3 h, as demonstrated in Figure 4A. Proper exposure time is normally in the range of 100 ms to 2 s. Time intervals and the required duration may vary based on the metabolite and the biological process being studied.
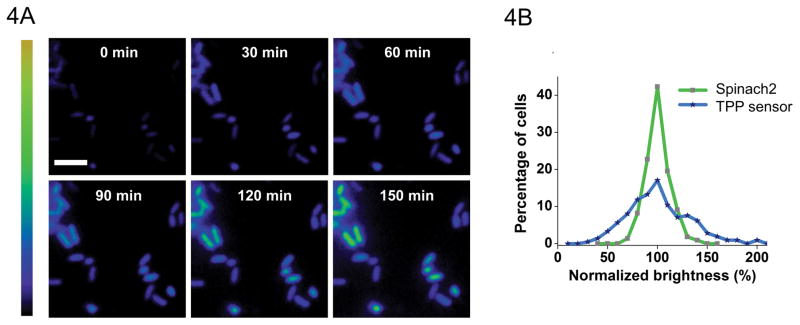
Figure 4.
(A) Representative images of bacteria transformed with TPP Spinach riboswitch. Time course of fluorescence over 3 hours demonstrates intracellular sensor function. (B) Comparison of distributions of fluorescence signal from bacteria expressing TPP Spinach riboswitch and the corresponding Spinach sequence by itself. Demonstrates that variations in cell-to-cell fluorescence are due to differences in TPP concentrations rather than differences in expression from T7 promoter.
4.3 Data analysis of metabolite concentration variations (TPP sensor)
Data analysis can be carried out using image analysis software such as NIS-Elements. To subtract the background fluorescence, which is primarily due to cellular autofluorescence, the image for pET28c-expressing (negative control) cells in media containing DFHBI is used.
Choose cells that are fully adhered and ensure that the field is entirely in focus in each frame. Circle these cells as regions of interest (ROIs). Track their fluorescence intensity through all background-subtracted time-point image frames. Calculate the mean fluorescence intensity per unit volume by dividing the total fluorescence by the calculated volume for each ROI. Volume was estimated using the equation V = πr2 [(4/3)r + a], where V is cell volume, r is radius, and a is the side length of the cell. Collect the mean fluorescence intensity change relative with respect to time for 150–300 cells in each well.
Normalize the sensor fluorescence values to the mean pET28c-Spinach2 (positive control) signal from the corresponding time point. If a vector simultaneously expressing the sensor and a fluorescent protein has been used, the sensor fluorescence can be normalized to the fluorescence protein signal in the same cells. These normalized values represent the fluorescence signal change due to dynamics of TPP concentration.
The fold increase in fluorescence over time is calculated as the ratio of mean intensity at a desired time point to that at time 0. After this value is calculated, cells are binned according to their percentage of total signal relative to the average signal, which is normalized to 100%.
Generate heat maps as a pictorial representation of fold change in fluorescence signal as in Figure 4A. Use the background-subtracted image with the most intense TPP sensor signal to adjust the upper lookup table (LUT) arrow. Assign the brightest cell the red color. Apply the same LUT setting to all other images.
By expressing Spinach alone in the same expression vector, the cell-to-cell variation in RNA expression can be seen. This can help to rule out the possibility that the variations in cellular fluorescence are due to variations in Spinach riboswitch expression level rather than differences in metabolite concentration (Figure 4B).
5. SUMMARY AND CONCLUDING REMARKS
Versatile RNA-based sensor technologies have been developed to monitor dynamics of metabolites inside live cells. Here we describe protocols for designing, optimizing, and implementing modular RNA-based metabolite sensors. We have illustrated a structure-guided strategy to identify riboswitches that can be fashioned into genetically encoded fluorogenic riboswitch sensors, using both their metabolite sensing ability and their structure-switching capacity. Our example of imaging bacterial TPP dynamics based on the thiM TPP riboswitch demonstrates that one can ultimately use a sensor to discern different individual cellular levels of metabolite.
This approach to sensor design could be greatly improved by a number of anticipated advancements in RNA imaging (Filonov, Kam, Song, & Jaffrey, 2015) and new types of metabolite-recognizing riboswitches (Lynch & Gallivan, 2009; Weigand & Suess, 2007). By exploiting the diversity of naturally occurring riboswitches, it should be possible to image a large number of intracellular metabolites. Additionally, improvements in fluorogenic RNA aptamers may lead to enhanced sensitivity and future extension of this technology for dynamic imaging of mammalian metabolites and proteins.
Acknowledgments
We thank Grigory S. Filonov for helpful suggestions and comments. Support was provided by the Tri-Institutional PhD Program in Chemical Biology (MSKCC, Weill-Cornell Medical College, and Rockefeller University). This work was also supported by NIH Grants R01 NS064516 and R01 EB010249 (to S.R.J.)
References
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science (New York, NY) 2012;335(6065):233–5. doi: 10.1126/science.1215063. http://doi.org/10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biology. 2007;8(11):R239. doi: 10.1186/gb-2007-8-11-r239. http://doi.org/10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. Prospects for Riboswitch Discovery and Analysis. Molecular Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. http://doi.org/10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Dodani SC, Chang CJ. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nature Chemistry. 2012;4(12):973–984. doi: 10.1038/nchem.1500. http://doi.org/10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filonov GSS, Kam CWW, Song W, Jaffrey SRR. In-Gel Imaging of RNA Processing Using Broccoli Reveals Optimal Aptamer Expression Strategies. Chemistry & Biology. 2015;22(5):649–660. doi: 10.1016/j.chembiol.2015.04.018. http://doi.org/10.1016/j.chembiol.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. Journal of Biological Chemistry. 2008;283(33):22347–22351. doi: 10.1074/jbc.C800120200. http://doi.org/10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. Structural Insights into Ligand Recognition by a Sensing Domain of the Cooperative Glycine Riboswitch. Molecular Cell. 2010;40(5):774–786. doi: 10.1016/j.molcel.2010.11.026. http://doi.org/10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellenberger Ca, Wilson SC, Sales-Lee J, Hammond MC. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP. Journal of the American Chemical Society. 2013;135:4906–4909. doi: 10.1021/ja311960g. http://doi.org/10.1021/ja311960g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SA, Gallivan JP. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Research. 2009;37(1):184–192. doi: 10.1093/nar/gkn924. http://doi.org/10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgaroli A, Milani D, Meldolesi J, Pozzan T. Fura-2 measurement of cytosolic free Ca2+ in monolayers and suspensions of various types of animal cells. The Journal of Cell Biology. 1987;105(November):2145–2155. doi: 10.1083/jcb.105.5.2145. http://doi.org/10.1083/jcb.105.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nature Reviews Molecular Cell Biology. 2004;5(6):451–63. doi: 10.1038/nrm1403. http://doi.org/10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science (New York, NY) 2012;335(6073):1194. doi: 10.1126/science.1218298. http://doi.org/10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science (New York, NY) 2011;333(6042):642–646. doi: 10.1126/science.1207339. http://doi.org/10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends in Biotechnology. 2011;29(3):144–152. doi: 10.1016/j.tibtech.2010.12.004. http://doi.org/10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nature Reviews Molecular Cell Biology. 2012;13(4):263–9. doi: 10.1038/nrm3314. http://doi.org/10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441(7097):1167–71. doi: 10.1038/nature04740. http://doi.org/10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors. Nature Methods. 2013;10(9):873–5. doi: 10.1038/nmeth.2568. http://doi.org/10.1038/nmeth.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen KA, Malhotra A, Weeks KM. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. Journal of the American Chemical Society. 2010;132(29):9940–9943. doi: 10.1021/ja103781u. http://doi.org/10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack RL, Song W, Jaffrey SR. Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria. Nature Protocols. 2013;9(1):146–155. doi: 10.1038/nprot.2014.001. http://doi.org/10.1038/nprot.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, … Looger LL. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods. 2009;6(12):875–881. doi: 10.1038/nmeth.1398. http://doi.org/10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valeur B. Design principles of fluorescent molecular sensors for cation recognition. Coordination Chemistry Reviews. 2000;205(1):3–40. http://doi.org/10.1016/S0010-8545(00)00246-0. [Google Scholar]
- Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR, Ferré-D’Amaré AR. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nature Structural & Molecular Biology. 2014;21(8):658–663. doi: 10.1038/nsmb.2865. http://doi.org/10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand JE, Suess B. Tetracycline aptamer-controlled regulation of pre-mRNA splicing in yeast. Nucleic Acids Research. 2007;35(12):4179–4185. doi: 10.1093/nar/gkm425. http://doi.org/10.1093/nar/gkm425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biology. 2010;11(3):R31. doi: 10.1186/gb-2010-11-3-r31. http://doi.org/10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419(6910):952–6. doi: 10.1038/nature01145. http://doi.org/10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- You M, Litke JL, Jaffrey SR. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch. Proceedings of the National Academy of Sciences. 2015;2015:201504354. doi: 10.1073/pnas.1504354112. http://doi.org/10.1073/pnas.1504354112. [DOI] [PMC free article] [PubMed] [Google Scholar]