Abstract
Poly(ADP-ribose) polymerases (PARPs) are a family of enzymes that modulate diverse biological processes through covalent transfer of ADP-ribose from NAD+ onto substrate proteins. Here, we report a robust NAD+ analog-sensitive approach for PARPs, which allows PARP-specific ADP-ribosylation of substrates that is suitable for subsequent copper-catalyzed azide-alkyne cycloaddition reactions. Using this approach, we mapped hundreds of sites of ADP-ribosylation for PARPs 1, 2, and 3 across the proteome, as well as thousands of PARP-1-mediated ADP-ribosylation sites across the genome. We found that PARP-1 ADP-ribosylates and inhibits NELF, a protein complex that regulates promoter-proximal pausing by RNA polymerase II (Pol II). Depletion or inhibition of PARP-1, or mutation of the ADP-ribosylation sites on NELF-E, promotes Pol II pausing, providing a clear functional link between PARP-1, ADP-ribosylation, and NELF. This analog-sensitive approach should be broadly applicable across the PARP family, and has the potential to illuminate the ADP-ribosylated proteome and the molecular mechanisms used by individual PARPs to mediate their responses to cellular signals.
ADP-ribosylation of proteins is an important regulator of cellular processes, from the regulation of chromatin and transcription to protein translation and stability (1). Most of the 17 PARP family members encoded in the human genome are enzymes with either mono- or poly(ADP-ribosyl) transferase activities, which covalently link ADP-ribose derived from NAD+ to their target proteins, primarily at glutamate, aspartate, and lysine residues (2). PARPs 1, 2, and 3, collectively referred to as the DNA-dependent PARPs, are a group of nuclear proteins with DNA-dependent mono (PARP-3) or poly (PARPs 1 and 2) ADP-ribosyl transferase activities involved in DNA repair, chromosome maintenance, chromatin regulation, and gene expression (2, 3). Previous studies using immune-based enrichment, various affinity resins, and protein microarrays to identify the targets of ADP-ribosylation lacked specificity for individual PARP family members (4). A recent chemical genetics approach targeting a conserved residue in the nicotinamide binding site of the PARP catalytic domain was a technological advance, but unfortunately ablates the poly(ADP-ribosyl) transferase activity of PARP enzymes, while preserving mono(ADP-ribosyl) transferase activity (5, 6). A single chemical genetics approach that preserves the natural mono- and poly(ADP-ribosyl) transferase activities of PARP enzymes and is broadly applicable across the PARP family should be of great utility.
An analog-sensitive PARP (asPARP) approach targeting the adenine moiety of NAD+
Our previous studies indicated that the adenine moiety of NAD+ is a useful target for chemical modification to alter the catalytic activity and chemistry of PARP family members (7). In this regard, we developed an adenine-focused, NAD+ analog-sensitive approach for PARPs that preserves their poly(ADP-ribosyl) transferase activity (Figs. 1A, S1A) and is capable of identifying the specific targets of individual PARP family members. Analog-sensitivity is achieved by mutation of a large “gatekeeper” amino acid in the active site of a protein to a smaller residue, creating a pocket that fits a bulky R group on an engineered substrate whereas interaction of the bulky R group with the wild-type enzyme would have been sterically blocked (8). We initially focused on PARP-1, an abundant and ubiquitously expressed PARP protein in metazoans. To identify a “gatekeeper” residue in PARP-1, we changed 10 large residues buried within the active site and facing the adenine ring of NAD+ to glycine or alanine based on a molecular model (Figs. 1A, S1B, S2). We selected the 8 position of the adenine ring of NAD+ as the site for R group addition since its modification precludes ADP-ribosylation with wild-type PARP-1 (wtPARP-1) or other PARPs (7), a feature critical to the analog-sensitive approach. We then synthesized a library of 11 NAD+ analogs, each with a different R group at position 8, from 8-methylamino-NAD+ to 8-benzylamino-NAD+ (Fig. 1B). In a screen of the 20 PARP-1 mutants versus the 11 NAD+ analogs, we identified two different gatekeeper residues, leucine 877 and isoleucine 895, whose mutation to alanine results in analog-sensitive activity in a PARP-1 automodification assay (Figs. 1C, S1C, S3). Critically, both of these residues are buried within the active site of the enzyme, thus their mutation is unlikely to affect protein-protein interactions that might affect substrate selectivity. Although L877 and I895 are 18 amino acids from one another in the PARP-1 linear sequence, they are adjacent to one another and proximal to the 8 position of the adenine ring in three-dimensional space (Fig. 1D). These results support our molecular model of PARP-1 interactions with NAD+ (Figs. S1B, S2), as well as the structural basis for our asPARP approach.
Figure 1. Structure-based engineering of an NAD+ analog-sensitive PARP-1 (asPARP-1) mutant.
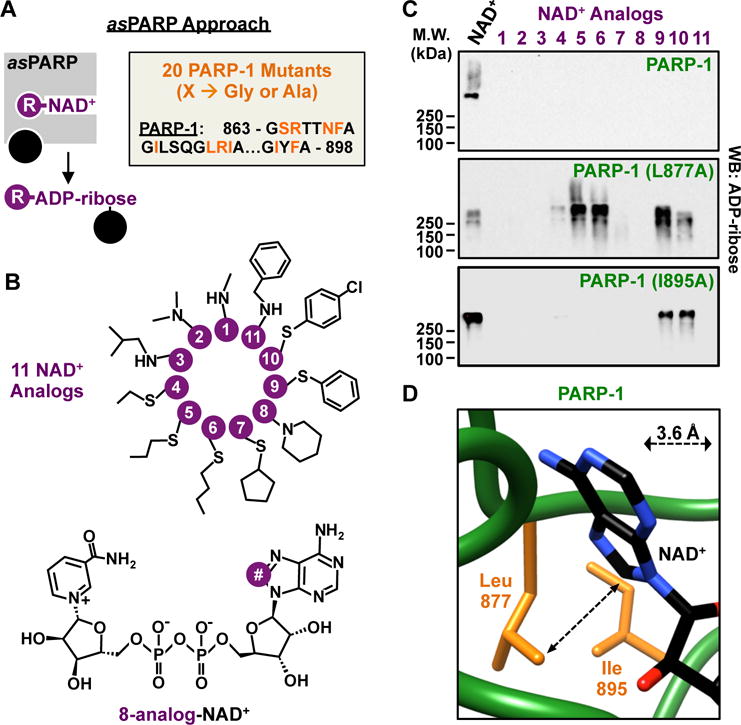
(A) (Left) Schematic illustrating NAD+ analog-sensitivity in PARP proteins. (Right) Residues in PARP-1 selected for mutation to glycine or alanine for discovery of gatekeeper residues.
(B) Chemical structures of the 11 NAD+ analogs used for screening for asPARP-1.
(C) Western blot for ADP-ribose from automodification reactions containing PARP-1 or PARP-1 mutants (L877A and I895A) and NAD+ or NAD+ analogs.
(D) Depiction of the spatial relationship between position 8 of the adenine ring in NAD+ and the gatekeeper residues.
To extend the utility of our asPARP approach, we functionalized the R group 8-Butylthio-NAD+ (Fig. 1B, NAD+ analog 6), with an alkyne moiety to generate 8-Bu(3-yne)T-NAD+ (Fig. 2A). 8-Bu(3-yne)T-NAD+ is an NAD+ analog with a single bi-functional R group at position 8, which facilitates asPARP-selective ADP-ribosylation. The R group also allows incorporation of alkyne into the post-translationally modified substrates for subsequent use in azide-alkyne cycloaddition (‘click’ chemistry) reactions to label or purify the PARP targets (Fig. 2B). asPARP-1 (L877A) with 8-Bu(3-yne)T-NAD+ yielded similar activity as the previously screened analogs, nearing wild-type enzyme kinetics when compared to PARP-1 and NAD+ (Fig. S4). Critically, this clickable NAD+ analog also supports activity with asPARP-2 and asPARP-3 mutants (L443A and L394A, respectively, homologous to L877 of PARP-1) (Figs. 2C, S5A, S5B). The ability to transfer this analog-sensitive activity to the other PARPs by mutation of the conserved gatekeeper residue (Figs. 2C, S5C–G) suggests broad utility of this approach across the PARP family, for both mono- and poly(ADP-ribosyl) transferases.
Figure 2. Activity of asPARPs 1, 2, and 3 with a clickable NAD+ analog.
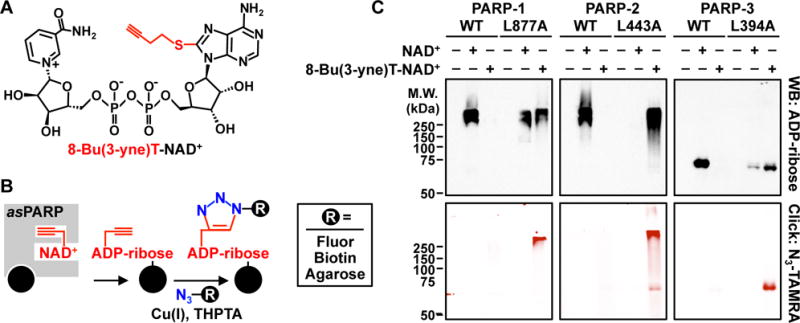
(A) Chemical structure of the bi-functional NAD+ analog 8-Bu(3-yne)T-NAD+ with the clickable analog sensitivity-inducing, alkyne-containing R group highlighted in red.
(B) Schematic illustrating asPARP activity-dependent, click chemistry-mediated covalent attachment of fluorophores, biotin, or agarose resin to 8-Bu(3-yne)T-ADP-ribosylated proteins.
(C) Automodification reactions with wild-type or analog-sensitive PARP-1, PARP-2, and PARP-3 analyzed by Western blotting for ADP-ribose (top) or click chemistry-based in-gel fluorescence (bottom).
Identification of site-specific nuclear substrates of PARPs 1, 2, and 3
We used the asPARP approach to identify site-specific nuclear substrates of PARPs 1, 2, and 3 with an approach that focuses on glutamate and aspartate residues (9). We incubated purified recombinant asPARPs 1, 2, or 3 with HeLa cell nuclear extract in the presence of 8-Bu(3-yne)T-NAD+, which resulted in PARP-specific labeling of extract proteins (Fig. 3A). We then clicked the 8-Bu(3-yne)T-ADP-ribose-labeled proteins to azide-agarose resulting in their covalent attachment to the agarose resin, allowing extensive washing with denaturants, strong detergents, and organic solvents. We performed trypsin-based peptide identification of the ADP-ribosylated proteins by LC-MS/MS (Peptide ID), washed extensively again, and eluted the ADP-ribosylated peptides using hydroxylamine to identify the sites of ADP-ribosylation by LC-MS/MS (Site ID) (9) (Fig. 3B). This approach revealed unique, as well as overlapping, sites of PARP-1-, 2-, and 3-mediated ADP-ribosylation (Figs. 3C, S6A). Ontological analyses of the target proteins revealed enrichment of terms related to transcription and DNA-repair, consistent with the known biology of PARPs 1, 2, and 3, as well as additional terms suggesting previously unknown functions (Figs. 3D, S6B).
Figure 3. Using analog-sensitive PARP-1 mutants to unambiguously identify the ADP-ribosylation targets of DNA-dependent PARPs.
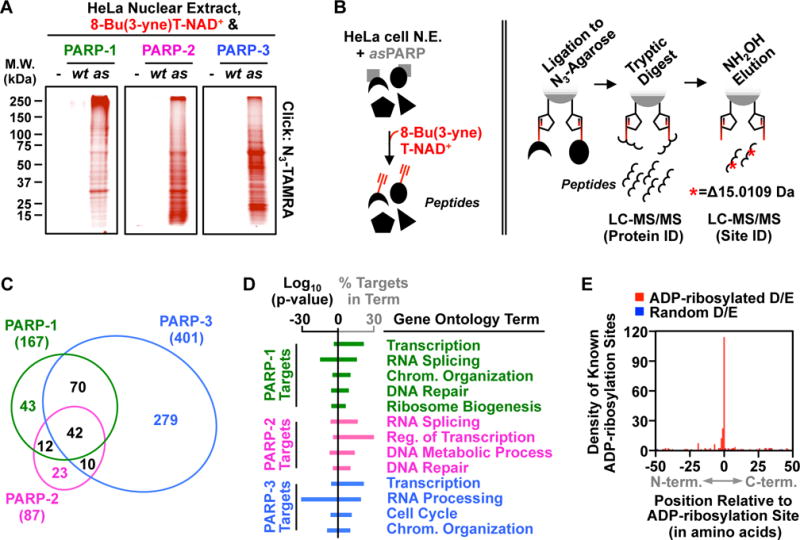
(A) In-gel fluorescence of HeLa cell nuclear extract proteins conjugated to azido-TAMRA following reactions with 8-Bu(3-yne)T-NAD+ in the presence of wild-type (wt) or analog-sensitive (as) PARP-1, PARP-2, or PARP-3.
(B) Depiction of the strategy for LC-MS/MS detection of PARP-specific ADP-ribosylation sites. (Left) asPARP-dependent labeling of HeLa cell nuclear extract (N.E.) proteins (represented by various shapes) using 8-Bu(3-yne)T-NAD+ (red). (Right) Post-labeling sample processing for LC-MS/MS. The 8-Bu(3-yne)T-ADP-ribosylated proteins are covalently linked to azide-agarose by copper-catalyzed cycloaddition (‘click’ chemistry; represented by pentagons), washed, and digested with trypsin to release peptides for protein identification. The remaining covalently linked peptides are eluted using hydroxyl amine (NH2OH) with a mass shift of 15.0109 Da, which allows for identification of ADP-ribosylation sites.
(C) Venn diagram depicting the overlap of the protein targets of PARP-1, PARP-2, and PARP-3.
(D) Gene ontology terms enriched for the sets of PARP-1, PARP-2 and PARP-3 targets.
(E) Histogram of the two-dimensional relationship between previously identified ADP-ribosylation sites (9) with those identified herein.
Motif analyses at the sites of PARP-1-, 2-, and 3-mediated ADP-ribosylation indicate some similarities in sequence preference among the three PARPs (e.g., glutamate proximal to the site of modification), but differences as well (Fig. S6C). The sites of PARP-1-, 2-, and 3-mediated ADP-ribosylation that we identified herein partially overlapped and were more numerous than sites of ADP-ribosylation identified using other approaches (Fig. S7), with excellent agreement for the specific sites of ADP-ribosylation in common targets when compared to a previous cell-based bulk ADP-ribosylation assay (9) (Fig. 3E). We also observed considerable overlap with an asPARP-1 data set that we generated from intact MEF nuclei (Fig. S8). Collectively, these results show that our asPARP approach robustly and faithfully identifies sites of ADP-ribosylation mediated by a specific PARP family member.
Negative Elongation Factor (NELF) is ADP-ribosylated in a P-TEFb-dependent Manner
Previous reports implicating the D. melanogaster homolog of PARP-1 as a key modulator of Pol II pause release at heat shock loci (10, 11), together with the identification of NELF-A and E as ADP-ribosylated proteins (Figs. 4A, S6B), led us to explore whether PARP-1 activity and ADP-ribosylation of the NELF complex might play a role in the control of transcription elongation. The “negative elongation factor” complex (NELF-A, B, C/D, E) functions to restrict transcriptional elongation and stimulate promoter-proximal pausing by RNA polymerase II (Pol II) (12). Immunoaffinity purification of NELF from mammalian cells expressing FLAG epitope-tagged NELF-E demonstrated that PARP-1 interacts with the NELF complex (Fig. 4B), and that NELF-E and NELF-A are ADP-ribosylated in mammalian cells (Fig. 4C). Mutation of the four glutamate residues in NELF-E that we identified in our proteomic screen (E122, E151, E172, and E374; Fig. 4A) to glutamines, a structurally similar residue refractory to ADP-ribosylation, resulted in a substantial reduction in NELF-E modification by PARP-1 (Fig. 4D). Finally, using an electrophoretic mobility shift assay with a model NELF-E-interacting RNA (i.e., HIV TAR), we found that ADP-ribosylation of NELF-E ablates its ability to bind RNA, a function of NELF-E necessary for the establishment paused Pol II (13) (Figs. 4E, S9).
Figure 4. P-TEFb-dependent ADP-ribosylation of NELF by PARP-1.
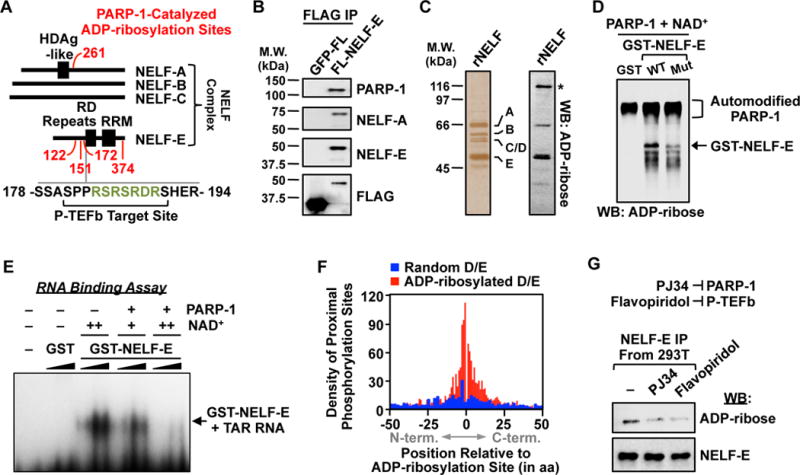
(A) Schematic showing the distribution of PARP-1 ADP-ribosylation sites (red), P-TEFb phosphorylation sites, and a PARP target-enriched 7-mer RSRSRDR (green) on proteins in the NELF complex.
(B) Western blot analysis of immunoprecipitated FLAG-tagged NELF-E or GFP from 293T cells.
(C) Silver stained SDS-PAGE gel (left) and ADP-ribose Western blot (right) of immunopurified NELF complex. Asterisk = automodified PARP-1.
(D) Western blot for ADP-ribose of in vitro modification reactions containing GST, GST-tagged wild-type NELF-E, or GST-tagged ADP-ribosylation site point mutant NELF-E, PARP-1, and NAD+ as indicated.
(E) NELF-E/TAR RNA electrophoretic mobility shift assay with or without PARP-1-mediated ADP-ribosylation. GST or GST-NELF-E was titrated between 0.1 to 1.0 μM and NAD+ was added at 25 μM (+) or 100 μM (++) during the ADP-ribosylation reaction.
(F) Histogram of the relationship between ADP-ribosylation sites identified herein and the nearest incidence of known phosphorylation modifications on PARP target proteins.
(G) Western blot analysis of immunoprecipitated FLAG-tagged NELF-E from 293T cells treated with vehicle, the PARP inhibitor PJ34, or the P-TEFb/CDK9 inhibitor flavopiridol.
We found that phosphorylation sites (Fig. 4F) and, to a lesser extent, sites of other post-translational modifications (Fig. S10A), are frequently found at or near ADP-ribosylation sites across the proteome. This suggests a broad role for ADP-ribosylation as a modulator at hubs of regulatory activity, as well as a more specific regulatory role for ADP-ribosylation (and PARPs) in cooperation with phosphorylation (and kinases) across the proteome. In fact, using a PARP inhibitor (i.e., PJ34) and a CDK9 inhibitor (i.e., flavopiridol, FP), we observed that ADP-ribosylation of NELF-E in mammalian cells is dependent on phosphorylation by CDK9/P-TEFb (Fig. 4G), a kinase that phosphorylates Pol II, DSIF, and NELF-E (14). Furthermore, both inhibitors reduced the extent of serine 2 phosphorylation (Ser2P) in the CTD heptapeptide repeat of the Pol II RPB1 subunit in cells (Fig. S10B). Since elevated Ser2P is associated with actively elongating Pol II (14), these results indicate a reduction in elongating Pol II upon inhibition of PARP-1 and CDK9. Interestingly, a 7-mer amino acid sequence (RSRSRDR) enriched in targets of PARP-1 (Fig. S10C) is located within the previously identified phosphorylation target site for P-TEFb in NELF-E, near a cluster of PARP-1-mediated ADP-ribosylation sites (Fig. 4A). Together, these results highlight the functional links between PARP-1-mediated ADP-ribosylation and transcription-related phosphorylation.
Identifying sites of PARP-1-mediated ADP-ribosylation across the genome
Although recent genomic approaches have facilitated the detection of sites of ADP-ribosylation genome-wide in the context of DNA damage (15), they have not allowed unambiguous assignment of genomic ADP-ribosylation events to a specific PARP family member in unstimulated cells. To unambiguously define sites of PARP-1-mediated ADP-ribosylation across the genome, we developed an assay, which we call “Click-ChIP-seq” (click chemistry-based chromatin isolation and precipitation with deep sequencing), using the asPARP-1 approach in nuclei. We expressed GFP (as a control), wtPARP-1, or asPARP-1 in Parp1−/− mouse embryo fibroblasts (MEFs) (Fig. S11A). ADP-ribosylation following addition of 8-Bu(3-yne)T-NAD+ was clearly evident in the nuclei of Parp1−/− MEFs expressing asPARP-1, but not wtPARP-1 (Fig. S11B). We then collected 8-Bu(3-yne)T-NAD+-treated nuclei, crosslinked them with formaldehyde, clicked the 8-Bu(3-yne)T-ADP-ribose to biotin, and sheared the chromatin by sonication, finally subjecting the material to an assay analogous to chromatin immunoprecipitation (Figs. 5A, S11C). A qPCR-based assay of the enriched genomic DNA revealed asPARP-1-specific ADP-ribosylation at gene promoters in nuclei isolated from MEFs (Fig. S11D).
Figure 5. Click-ChIP-seq, an asPARP-1-based method for identifying the genome-wide distribution of ADP-ribosylation events catalyzed by a specific PARP protein.
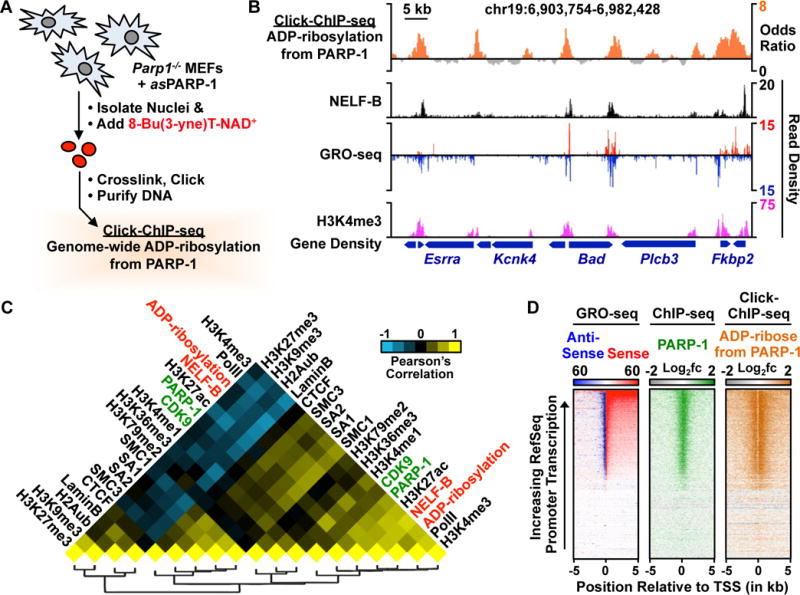
(A) Schematic representation of Click-ChIP-seq. Nuclei are isolated from Parp−/− MEFs expressing asPARP-1, labeled with 8-Bu(3-yne)T-NAD+, subjected to crosslinking with formaldehyde, and then processed for chromatin immunoprecipitation. The enriched DNA is subjected to deep sequencing.
(B) Genome browser view of a multi-gene locus of the mouse genome showing PARP-1-catalyzed ADP-ribosylation (from Click-ChIP-seq) with other genomic features.
(C) Heat map showing pairwise clustered correlations between genomic features and PARP-1-mediated ADP-ribosylation from click-ChIP-seq.
(D) Heat map representations showing PARP-1-catalyzed ADP-ribosylation (from Click-ChIP-seq) in comparison to PARP-1 (from ChIP-seq) and transcription (from GRO-seq) at the promoters of all RefSeq genes.
Click-ChIP-seq revealed robust enrichment of PARP-1-mediated ADP-ribosylation at the promoters of transcriptionally active genes, which were defined by an enrichment of histone H3 lysine 4 trimethylation (H3K4me3, a mark of active promoters, from ChIP-seq) and actively transcribing Pol II (from GRO-seq) (Fig. 5B). This assay also revealed genomic loci with significant peaks of PARP-1 lacking enrichment of PARP-1-mediated ADP-ribosylation located in regions of repressed chromatin (Fig. S12), which are difficult to discern in the absence of the specificity provided by the asPARP approach. Genome-wide correlation analyses between PARP-1-mediated ADP-ribosylation and a variety of other histone modifications and chromatin/transcription-related factors revealed strong clustering, as well as positive correlations with PARP-1 (0.606) and NELF-B (0.754) (Fig. 5C). Heat map representations of the genomic data highlight the relationships at gene promoters among PARP-1-mediated ADP-ribosylation, Pol II accumulation, and H3K4me3, NELF-B, and PARP-1 enrichment (Figs. 5D, S13A). PARP-1-mediated ADP-ribosylation and CDK9 occupancy at promoters strongly correlated with low levels of Pol II pausing (Fig. S13B). These results suggested that PARP-1-mediated ADP-ribosylation may act in a similar manner as CDK9/P-TEFb-mediated phosphorylation to promote the release of paused Pol II into productive elongation.
Role of PARP-1-mediated ADP-ribosylation of NELF in the release of paused Pol II
To determine the role of PARP-1 in the release of paused Pol II, we performed GRO-seq in MCF-7 breast cancer cells to monitor the effects of shRNA-mediated PARP-1 knockdown or treatment with the PARP inhibitor (PARPi) PJ34 on Pol II pausing. GRO-seq is a genomic assay that reveals the location of transcriptionally engaged RNA polymerases globally (16). We observed an accumulation of reads in the peaks of paused Pol II upon PARP-1 knockdown or treatment with PARPi (compared to an untreated luciferase knockdown control; Luc) at gene promoters (Fig. S14). This effect was evident genome-wide (Fig. 6A), with a clear increase in global Pol II pausing indices upon PARP-1 depletion or inhibition (Fig. 6B). At active promoters with a significant accumulation of GRO-seq reads in the paused Pol II peak upon PARP-1 knockdown, we observed decreased GRO-seq reads in the gene bodies (Fig. S15; see panel B for the gene body effects), suggesting that PARP-1 activity is necessary to achieve an efficient release of Pol II into productive elongation.
Figure 6. Functional links between PARP-1-catalyzed ADP-ribosylation, NELF binding, and RNA polymerase II pausing genome-wide.
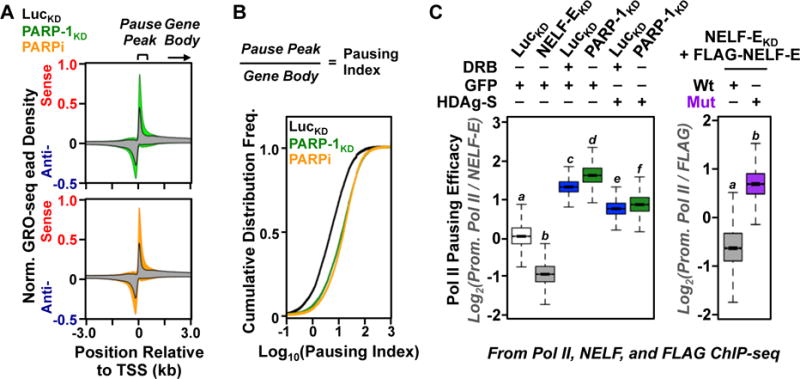
(A) Metagenes of GRO-seq read density at the promoters of all expressed RefSeq genes from MCF-7 cells subjected to shRNA-mediated knockdown with either control/luciferase or PARP-1 shRNAs (top) or treatment with the PARP inhibitor (PARPi) PJ34 (bottom).
(B) RNA polymerase II pausing indices at the promoters of all transcribed RefSeq genes from MCF-7 cells subjected to shRNA-mediated knockdown with either control/luciferase or PARP-1 shRNAs or treatment with PJ34.
(C) Boxplots of promoter proximal Pol II “pausing efficacy” determined by Pol II and NELF ChIP-seq in MCF-7 cells under the different experimental conditions indicated for the top quartile of expressed RefSeq genes. Bars marked with different letters are significantly different (p < 2.16 × 10−16; t-test).
We showed that a large fraction of PARP-1-regulated genes (determined by PARP-1 knockdown) are also regulated by P-TEFb (determined by treatment with DRB) and NELF (determined by NELF knockdown), with respect to expression (by RNA-seq; Fig. S16A, B) and Pol II pausing (by Pol II ChIP-seq; Fig. S16C). In addition, we found that the viral NELF inhibitor, HDAg-S (17), reverses the inhibitory effects of both PARP-1 knockdown and DRB treatment on Pol II pausing, as determined using a ChIP-seq-based “Pol II pausing efficacy” assay (i.e., promoter proximal Pol II enrichment divided by NELF-E enrichment) (Figs. 6C, S17, S18). Finally, we showed that a NELF-E ADP-ribosylation site mutant (Mut) produces a NELF complex that is resistant to the inhibitory effects of PARP-1 and is a more potent inducer of Pol II pausing that NELF complex containing wild-type NELF-E, in spite of lower expression in cells (Figs. 6C, S18, S19). Collectively, our data point to a functional link between CDK9-mediated phosphorylation, PARP-1-mediated ADP-ribosylation, and NELF-mediated Pol II pausing (Fig. S20). Our results indicate that PARP-1-dependent ADP-ribosylation of NELF-E reinforces P-TEFb-mediated Pol II pause release and productive elongation for a subset of NELF-regulated genes, especially those with elevated NELF and Pol II loading, as well as H3K4me3 enrichment (Fig. S21). Finally, these mechanisms are functional even at promoters where PARP-1 serves non-catalytic functions, such as the expulsion of the linker histone H1 from nucleosomes (18) (Fig. S22).
Conclusions and Perspectives
PARP proteins have gained considerable attention as therapeutic targets for the treatment of cancers and other diseases (19), although the broader biology of the PARP family remains largely unexplored. Understanding the biology of PARP proteins requires an understanding of the protein substrates that they modify. In this regard, we have developed an asPARP approach that preserves the natural mono- and poly(ADP-ribosyl) transferase activities of PARP enzymes, which can be coupled with protein mass spectrometry to identify the targets of specific PARP family members. We have also repurposed this asPARP technology for use in genomic assays to identify the genome-wide distribution of ADP-ribosylation events catalyzed by a specific PARP protein. Our studies focusing on NELF illustrate how an integrated approach based on asPARP technology can further the exploration of the biological functions for ADP-ribosylation. Our asPARP approach, which uses a conserved residue in the PARP catalytic domain, should be broadly applicable across the PARP family. This technique will facilitate the rapid, robust, and systematic identification of the molecular targets and mechanisms of action of the entire PARP family, with the potential to transform our understanding of PARP protein functions in physiology and disease.
Supplementary Material
Acknowledgments
The authors thank M. Chae, Q. Liang, J. DeBrabander, U. Havemann, and D. Imren for technical assistance and members of the Kraus Laboratory for their helpful discussions about this project. The asPARP expression constructs and the NAD+ analogs can be obtained from UT Southwestern Medical Center and Biolog Life Science Institute, respectively, under a material transfer agreement. W.L.K., B.A.G., F.S., and H.L. are inventors on patent application U.S. No. 62/144,711 filed by UT Southwestern Medical Center related to the asPARP technology. W.L.K. and B.A.G. are inventors on patent applications No. U.S. 62/009,955 and No. PCT/US2015/034852 filed by UT Southwestern Medical Center related to the ADP-ribose detection reagents. YY is an inventor on patent No. U.S. 8828672 B2 filed by UT Southwestern Medical Center related to technology for the determination of D/E-ADP-ribosylation sites.
This work was supported by a predoctoral fellowship from the American Heart Association to B.A.G., grants from Cancer Prevention and Research Institute of Texas (CPRIT R1103), the Welch Foundation (I-1800), and the UT Southwestern Endowed Scholars Program to Y.Y., who is the Virginia Murchison Linthicum Scholar in Medical Research and a CPRIT Scholar in Cancer Research (a grant from the NIH/NIGMS (GM086703) to H.L., and (4) a grant from the NIH/NIDDK (DK069710) and support from the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowments to W.L.K. W.L.K. is a founder and consultant for Ribon Therapeutics, Inc. The GRO-seq, ChIP-seq, and RNA-seq data sets from this study are available from the NCBI’s GEO database using accession numbers GSE74141 and GSE74142. The proteomic data sets generated for these studies are available in Table S1.
Footnotes
Author Contributions
B.A.G. conceived the asPARP concept with input from W.L.K. based on ideas initially explored in Jiang et al., 2010 (7). W.L.K. conceived the Click-ChIP-seq method, which was further developed by B.A.G. B.A.G. performed all experiments and computational analyses, except as follows. H.J., J.S., H.L., and F.S. synthesized all of precursors and NAD+ analogs used in this study. B.A.G. and H.J. performed the enzyme kinetics assays. B.A.G., Y.Z., and Y.Y. prepared the samples and ran the LC-MS/MS. K.M.H. made the PARP-1 knockdown MCF-7 cells and prepared the GRO-seq samples. W.L.K. secured funding to support this project and provided intellectual support for all aspects of the work. B.A.G. and W.L.K. prepared the figures and wrote the paper.
Supplementary Information
Materials and Methods
Supplementary Figures S1 through S22
Supplementary Tables S1 and S2
Supplementary References
References
- 1.Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 2.Vyas S, et al. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hottiger MO. Nuclear ADP-ribosylation and its role in chromatin plasticity, cell differentiation, and epigenetics. Annu Rev Biochem. 2015;84:227. doi: 10.1146/annurev-biochem-060614-034506. [DOI] [PubMed] [Google Scholar]
- 4.Daniels CM, Ong SE, Leung AK. The Promise of Proteomics for the Study of ADP-Ribosylation. Mol Cell. 2015;58:911. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter-O’Connell I, Jin H, Morgan RK, David LL, Cohen MS. Engineering the substrate specificity of ADP-ribosyltransferases for identifying direct protein targets. J Am Chem Soc. 2014;136:5201. doi: 10.1021/ja412897a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter-O’Connell I, et al. Identifying family-member-specific targets of mono-ARTDs by using a chemical genetics approach. Cell Rep. 2016;14:621. doi: 10.1016/j.celrep.2015.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, Kim JH, Frizzell KM, Kraus WL, Lin H. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. J Am Chem Soc. 2010;132:9363. doi: 10.1021/ja101588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Specht KM, Shokat KM. The emerging power of chemical genetics. Curr Opin Cell Biol. 2002;14:155. doi: 10.1016/s0955-0674(02)00317-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
- 10.Petesch SJ, Lis JT. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 12.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol Cell Biol. 2002;22:2918. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo J, Price DH. RNA polymerase II transcription elongation control. Chem Rev. 2013;113:8583. doi: 10.1021/cr400105n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartolomei G, Leutert M, Manzo M, Baubec T, Hottiger MO. Analysis of chromatin ADP-ribosylation at the genome-wide level and at specific loci by ADPr-ChAP. Mol Cell. 2016;61:474. doi: 10.1016/j.molcel.2015.12.025. [DOI] [PubMed] [Google Scholar]
- 16.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi Y, et al. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science. 2001;293:124. doi: 10.1126/science.1057925. [DOI] [PubMed] [Google Scholar]
- 18.Krishnakumar R, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- 19.Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors: anticancer therapy and beyond. Mol Aspects Med. 2013;34:1217. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


