Abstract
The isocitrate dehydrogenase IDH2 produces α-ketoglutarate by oxidizing isocitrate, linking glucose metabolism to oxidative phosphorylation. In this study, we report that loss of SIRT3 increases acetylation of IDH2 at lysine 413 (IDH2-K413-Ac), thereby decreasing its enzymatic activity by reducing IDH2 dimer formation. Expressing a genetic acetylation mimetic IDH2 mutant (IDH2K413Q) in cancer cells decreased IDH2 dimerization and enzymatic activity and increased cellular reactive oxygen species (ROS) and glycolysis, suggesting a shift in mitochondrial metabolism. Concurrently, overexpression of IDH2K413Q promoted cell transformation and tumorigenesis in nude mice, resulting in a tumor-permissive phenotype. Immunohistochemical staining showed that IDH2 acetylation was elevated in high-risk luminal B patients relative to low-risk luminal A patients. Overall, these results suggest a potential relationship between SIRT3 enzymatic activity, IDH2-K413 acetylation-determined dimerization, and a cancer-permissive phenotype.
Keywords: Sirtuins, SIRT3, IDH2, Acetylation, Acetylome, Warburg effect, Glycolysis, Metabolism, Detoxification, Aging, Signaling
INTRODUCTION
Isocitrate dehydrogenases (IDHs) catalyze the irreversible oxidation and decarboxylation of isocitrate into α-ketoglutarate and are key enzymes in the Krebs Cycle, regulating energy metabolism and NADH/NADPH production (1). Previous studies in yeast have shown that mutations in IDHs inhibit yeast IDH activity and result in decreases in isocitrate binding (2), AMP affinity (2), NAD+ reduction (2) and yeast respiration (3), suggesting their critical roles in energy production and metabolism. Mammalian IDH2 is a NADP+-dependent homodimer which is responsible for the mitochondrial energy flow in the Krebs Cycle that links glycolysis and the Krebs Cycle to mitochondrial electron transport chain and oxidative phosphorylation. In addition, mammalian IDH2 catalyzes the production of NADPH through reduction of NADP+ to maintain mitochondrial redox homeostasis.
IDH2 enzymatic activity can be inhibited by acetylation at lysine 413 (4). Acetylation of IDH2, at least in some significant part, is directed by SIRT3, the primary mitochondrial deacetylase (5), a member of the Sirtuin family (homolog of yeast Silent Information Regulator 2, (Sir2) family. Sirtuins were initially discovered in yeast and C. elegans, and these genes play a critical role in extending the life cycle by suppressing toxic rDNA formation, suggesting potential anti-aging effects of sirtuins (6). Therefore, it seems reasonable to propose that enzymatic inhibition of IDH2 by acetylation of K413 promotes phenotypes, similar to those observed in yeasts lacking Sir2, which should negatively affect mitochondrial metabolism and energy homeostasis as well as potentially dysregulating aging-related pathways.
SIRT3 functions as a mitochondrial nicotinamide adenine dinucleotide (NAD+)-dependent class III histone deacetylase. In the last five years, numerous downstream deacetylation targets have been identified that direct mitochondrial processes, including β-oxidation, TCA cycle, oxidative phosphorylation, and detoxification (7–9). Loss of SIRT3 enzymatic activity dysregulates mitochondrial energy and detoxification pathways, as well as promotes transformation and/or tumor permissive phenotypes, as observed in cells and/or mice lacking Sirt3. In this regard, it has been shown that Sirt3 KO mice exhibited elevated cellular and mitochondrial reactive oxygen species (ROS) compared with Sirt3 wild-type (WT) mice (9) and this is thought to play a role, at least in some significant part, in the mechanism by which mice lacking Sirt3 exhibit a tumor permissive phenotype. Metabolically, loss of Sirt3 shifts metabolism from oxidative phosphorylation to glycolysis, and overexpression of SIRT3 partially reverses the preferential utilization of glycolysis, referred as the Warburg effect, in cancer cells (8, 10, 11).
It has long been proposed there is a relationship between mitochondrial redox, the Warburg effect, and breast cancer carcinogenesis. SIRT3 has been regarded as a mitochondrial localized tumor suppressor gene by protecting cells from ROS accumulation as well as promoting oxidative phosphorylation as a mechanism to generate ATP (8, 12). In this regard, mice lacking Sirt3 develop estrogen receptor positive (ER+) mammary tumors, and at least one copy of SIRT3 is deleted in 40% of women diagnosed with breast cancer (11). There appears to be a subgroup of human ER+ breast malignancies that exhibit a decrease in SIRT3 levels (13). In this manuscript, we report that loss of SIRT3 activity in mice lacking Sirt3 or cancer cells expressing shSIRT3 increases IDH2-K413-Ac and decreases IDH2 dimerization. Expression of IDH2K413Q decreased IDH2 dimerization and enzymatic activity. Metabolically, expression of IDH2K413Q alters cancer cell metabolism and results in accumulation of ROS. Furthermore, expression of IDH2K413Q promotes tumor permissive phenotypes in vitro and in vivo, finally linking SIRT3 and IDH2 acetylation to breast cancer malignancy risk correlatively.
Materials and Methods
IDH2 activity assay
Animal protocols were approved by Northwestern University IACUC. Sirt3 wild-type and knockout livers were collected at five and eight months and mitochondrial liver samples were used to measure IDH2 activity (Sigma). IDH2 is an NADP+-dependent enzyme, whereas IDH3 is an NAD+-dependent isocitrate dehydrogenase, and only NADP+ is added to the reactions; and only NADP+-dependent IDH2 activity is measured, per manufacturer’s protocol.
Cell culture and stable cell line
HEK-293T, MCF7 and NIH3T3 cells were obtained from ATCC in 2012, authenticated using STR profiling with CellCheck 9 Plus by IDEXX Bioresearch, and tested for mycoplasma (Mycoplasma Detection Kit, InvivoGen, Inc) in April 2016. Early passages of cells were frozen and cells were passaged for fewer than six months in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (Sigma) and Antibiotic Antimycotic solution (Sigma) and incubated in a 37°C chamber with 5% CO2.
Virus production, plasmids, short hairpin RNA constructs, and site-directed mutagenesis
To generate lenti-virus, HEK-293T cells were transfected with 5μg DNA, 5μg psPAX2 packaging plasmid, and 500ng VSV.G envelope plasmid. pLKO.1 human SIRT3 shRNA was purchased from OpenBiosystem. pLKO.1 human IDH2 shRNA oligos (CCTCTCTGGAGGCCTTTCTAG) was purchased (IDT) and cloned into the pLKO.1 vector (Addgene). The human IDH2 expression vector, pCDNA3-Flag-IDH2, was used as the IDH2 wild-type (WT) and for mutagenesis. K413 was converted to Arginine (R: deacetyl mimic) or Glutamine (Q: acetyl mimic) by site-directed mutagenesis (Bioinnovatise). The lenti-viral IDH2 vector was generated by PCR amplification into two PCDH-CMV vectors (Lentiviral vector, System Biosciences). Cells were infected with 5 MOI of lenti-virus and selected in 2 μg/mL puromycin (Invitrogen) for 14 days or 100 μg/mL G418 sulfate (Invitrogen). Two-weeks, selection, cells were grown in standard DMEM. MCF7 cells were grown in high and low serum as a control to determine the relative level of mitochondrial acetylation (Supplemental Fig. S1).
Immunoblotting
Cells and tissues were washed with cold 1X PBS, harvested and lysed for 30 mins in IP buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 5% glycerol) with protease inhibitors (BioTool) and TSA (Trichostatin A, Sigma). Lysates were quantified with Bradford assay (BioRad) and immunoblotted using: SIRT3 (Cell signaling), IDH2 (Proteintech), IDH2-K413-Ac (9), actin (Sigma) and GAPDH (Millipore). For IDH2 dimerization assay, lysates were cross-linked with 0.05% glutaldehyde for 10 minutes at room temperature and immunoblotted with IDH2 antibody. For Native-PAGE analysis, flag-tagged IDH2 was transiently transfected into MCF7 or HEK-293T cells, eluted with flag-beads (Sigma), and separated on a Bis-Tris Native gel with the addition of G-250 sample additive (Invitrogen).
Total glutathione level measurement
One million cells were lysed in 1.34mM Diethylenetriaminepenta-acetic acid (DETAPAC, Sigma) dissolved in 143 mM sodium phosphate (Sigma), 6.3 mM EDTA (Sigma), and 5% 5-sulfosalicylic acid (SSA, Sigma). 50 μL of lysate were mixed with 700 μL 0.298 mM NADPH (Sigma) in sodium phosphate buffer, 100 μL 6mM 5,5′-dithio-bis-2-nitrobenzoic acid (DTNB, Sigma) sodium phosphate buffer, 100 μL water (Corning), and 50 μL 0.023 U/μL glutathione reductase (GR) (Sigma). The kinetic absorbance was read at 412 nm every 15 s for 2.5 min using xMark™ Microplate Absorbance Spectrophotometer (BioRad), and the rates were compared to a standard curve. Tumors were chopped and lysed in DETAPAC buffer before assayed. Protein concentrations were measured for standardization of total glutathione levels.
Mitochondrial ROS measurements using MitoSOX
Cells were trypsinized and resuspended in 1X PBS with 5 mM pyruvate, labeled with MitoSOX™ red (2μM, in 0.1% DMSO, 20 mins), and incubated at 37°C. Samples were analyzed by Fortessa flow cytometer (Becton Dickinson Immunocytometry System, INC., Mountain View, CA) (excitation 488 nm, emission 585 nm band-pass filter). The mean fluorescence intensity of 10,000 cells was analyzed and corrected for autofluorescence from unlabeled cells. The MFI data was normalized to control levels.
Clonogenic cell survival colony assay analysis
For the clonogenic survival analysis, cells were seeded onto a 6-well plate. After 14 days colonies were stained with crystal violet and counted with a Zeiss Stemi dissecting microscope.
Soft agar colony formation assay analysis
Ten thousand cells were plated on 0.3% agar in growth medium over 0.6% base agar foundation layer in growth medium. After 21 days, the colonies were visualized under a 20X microscope (Zeiss) and images were acquired.
Oxygen consumption rate (OCR) and extracellular acidification rate analysis (ECAR)
MCF7 cells were seeded on a XF-24 microplate (Seahorse Biosciences) and incubated with DMEM containing 10% FBS overnight. For OCR analysis, 2.5 μM oligomycin A (Sigma), 10 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP, mitochondrial uncoupler, Sigma), and 2 μM antimycin/rotenone (complex I/III inhibitors, Sigma) were sequentially added into different ports of the same Seahorse cartridge. For ECAR analysis, thirty minutes before ECAR measurement, the medium was switched to DMEM without bicarbonate containing 0.5% FBS (pH=7.5). Twenty-five mM glucose (Corning), 2.5 μM oligomycin A and 50 mM 2-deoxyglucose (2-DG, Sigma) were sequentially added into different ports of the same Seahorse cartridge.
Xenograft in vivo tumorigenesis analysis
Five million cells expressing WT, IDH2K413R, and IDH2K413Q were injected into six weeks old Foxn1nu athymic nude mice (The Jackson Laboratory). Tumor size were examined using a Vernier caliper every two days. Volumes were calculated using V=1/2*W2*L (14).
Immunohistochemistry staining and analysis
Breast cancer arrays (Biomax) were stained with IDH2-K413-Ac and SIRT3 antibodies for 48 hours at 4 °C. After treatment the slides were incubated with secondary antibody (Sigma) for one hour. The slides were treated with VECTASTAIN ABC kit (Vector Laboratories), DAB peroxidase substrate kit (Vector Laboratories), stained in hematoxylin (Sigma) for 10 minutes and destained before dehydration. The intensities were quantified using HistoQuest software (Tissuegnostics).
Statistical analysis
Ordinary one-way ANOVAs, two-tailed unpaired t-tests and correlation analysis were conducted using Prism 6. The statistical significance was reported when p < 0.05.
Results
Loss of Sirt3 affects IDH2 acetylation at lysine 413 and IDH2 activity in mice
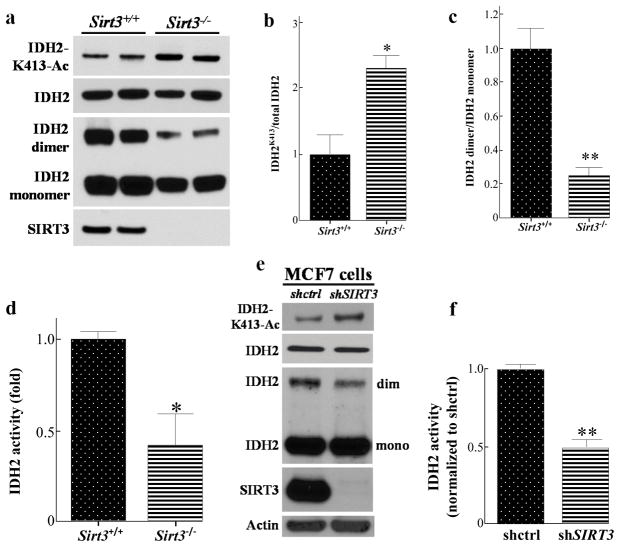
It has been previously shown that loss of Sirt3 results in IDH2-K413-Ac and decreases IDH2 activity (4). To confirm that SIRT3 plays a role in IDH2 enzymatic activity through IDH2 deacetylation in mice, liver samples from Sirt3 wild-type (Sirt3+/+) and Sirt3 knock-out (Sirt3−/−) mice were collected and the levels of total IDH2, IHD2-K413-Ac, and IDH2 dimerization were determined. While no significant difference in the total amount of IDH2 was observed, loss of Sirt3 increases in vivo IHD2-K413-Ac (Fig. 1a, b). In addition, after the liver lysates were cross-linked with glutaldehyde and run on an SDS-PAGE gel, we found that loss of Sirt3 decreased the levels of dimerized IDH2 (Fig. 1a, c). To test mitochondrial NADP+-dependent IDH2 activity in mice, five wild-type and Sirt3−/− mice livers were fractionated, and IDH2 activity was measured. A significant decrease in IDH2 activity was detected (Fig. 1d) in the livers of mitochondria lacking Sirt3, suggesting that loss of Sirt3 decreases IDH2 activity in vivo. The results of these experiments suggest that loss of Sirt3 increased IDH2-K413-Ac, decreased IDH2 dimerization, as well as decreased IDH2 enzymatic activity.
Figure 1. Loss of SIRT3 increases IDH2 K413-Ac, decreases IDH2 dimerization, and its activity.
(a) Wild-type and Sirt3−/− mice livers lysates were SDS-PAGE gel separated and immunoblotted with anti-acetyl IDH2K413 and anti-IDH2 antibodies. For IDH2 dimerization, lysates were treated with 0.05% glutaldehyde and separated by SDS-PAGE. (b–c) IDH2 acetylation and IDH2 dimerization were quantified using ImageJ software, and results were normalized to Sirt3+/+ liver samples (n=5). (d) The mitochondrial extracts from wild-type and Sirt3−/− mice livers (n=5) were collected and mitochondrial NADP+-dependent IDH2 activity was measured. (e) Lysates of MCF7-shctrl and MCF7-shSIRT3 cells were immunoblotted with anti-IHD2-K413-Ac, IDH2, SIRT3 and actin antibodies. (f) MCF7-shctrl or MCF7-shSIRT3 cell mitochondrial extracts (n=3) were collected and IDH2 activity was measured. Representative images are shown. Results are from three separate experiments, and error bars represent one standard deviation. * indicates p<0.05, ** indicates p<0.01, by two-tailed unpaired t test using Prism 6.0.
Loss of SIRT3 directs IDH2 acetylation at lysine 413 and IDH2 dimerization in cancer cells
To extend these results to an in vitro model system, MCF7 breast cancer cells infected with a lenti-shRNA vector targeting the human SIRT3 transcript were used. These experiments showed that MCF7 cells infected with lenti-shSIRT3 (MCF7-shSIRT3 cells) to knock-down SIRT3, as compared with control, MCF7-shctrl cells, exhibited an increase in IDH2-K413-Ac as well as a decrease in the levels of the IDH2 dimerization complex (Fig. 1e). We also collected the mitochondrial lysates from both control and MCF7-shSIRT3 cells, and a significant two-fold decrease of IDH2 activity was observed (Fig. 1f).
IDH2 lysine 413 acetyl mimetic decreases IDH2 dimerization and IDH2 activity in cancer cells
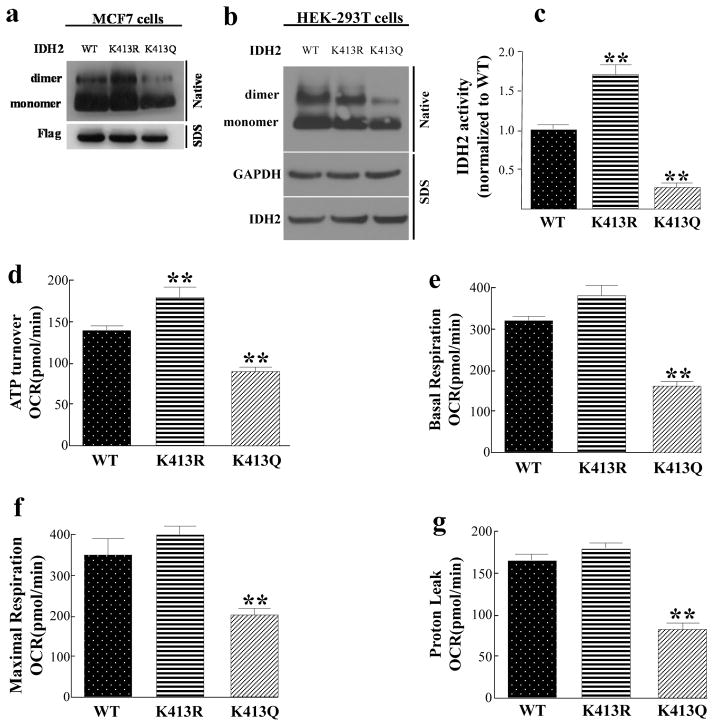
To examine if IDH2-K413-Ac status directs enzymatic activity and dimerization, a series of IDH2-K413 site-directed mutants were constructed. In this regard, it has previously been shown that substitution of a lysine with a glutamine (K413Q) mimics the acetylated lysine state, while substitution with an arginine (K413R) mimics deacetylation (9, 15, 16). Thus, mutating lysine IHD2-K413 to arginine would be predicted to mimic a deacetylated lysine, while substitution with a glutamine would be expected to mimic an acetylated lysine. We transfected flag-tagged wild-type IDH2, IDH2K413R and IDH2K413Q into MCF7 cells. After elution of flag-tagged IDH2 proteins, we ran these samples on a native gel. We found that eluted IDH2K413Q proteins displayed a decreased IDH2 dimerization (Fig. 2a). We confirmed our result using HEK-293T cells, as expression of IDH2K413Q showed decreased IDH2 dimerization as well (Fig. 2b). Furthermore, expression of IDH2K413Q significantly decreased mitochondrial IDH2 activity compared to cells expressing wild-type or IDH2K413R. (Fig. 2c). These experiments suggest that the acetylation status of IDH2, as well as the ability of IDH2 to form a dimerization complex, directs enzymatic activity in MCF7 cancer cells and HEK-293T cells.
Figure 2. Expression of IDH2K413Q in IDH2 knock-downed MCF7 and HEK-293T cells decreases IHD2 dimerization, mitochondrial respiration capacity, and increases glycolytic capacity.
(a–b) MCF7 and HEK-293T cells were transfected with flag-tagged IDH2WT, IDH2K413R (de-acetyl mimetic), or IDH2K413Q (acetyl mimetic). Forty-eight hours after transfection, cells were lysed, eluted, and separated on a native-PAGE gel and immunoblotted with anti-IDH2 antibody. (c) MCF7-shIDH2-IDH2WT, MCF7-shIDH2-IDH2K413R, or MCF7-shIDH2-IDH2K413Q cells (n=3) were measured for IDH2 activity as above. (d–g) MCF7-shIDH2-IDH2WT, MCF7-shIDH2-IDH2K413R, or MCF7-shIDH2-IDH2K413Q cells (n=7) were measured for (d) ATP turnover, (e) mitochondrial basal respiration, (f) mitochondrial respiration capacity, and (g) proton leak. The ATP turnover rate was determined by the difference between the starting OCR and the OCR after adding oligomycin. The basal respiration rate was determined by the difference between the starting OCR and the OCR after adding antimycin /rotenone mixture. The maximal respiration rate was determined by the difference between the OCR after adding CCCP and the OCR after adding antimycin/rotenone. The proton leak rate was determined by the difference between the OCR after adding oligomycin and the OCR after adding antimycin/rotenone mixture. Results are from three separate experiments and error bars represent one standard deviation. * indicates p<0.05, ** indicates p<0.01, comparing to WT by two-tailed unpaired t-test using Prism 6.0.
The IDH2K413 acetylation mimetic impairs mitochondrial respiration and cancer cell metabolism
Our results clearly show that the acetylation status of IDH2 changes IDH2 enzymatic activity, and the dysregulation of this key enzyme in the Krebs Cycle likely disrupts metabolic pathways as well as results in a metabolite imbalance. Thus, cells expressing the IDH2 acetyl mimetic should disrupt the Krebs Cycle, oxidative phosphorylation, as well as mitochondrial metabolism. To examine how expression of the IDH2-K413 site directed mutant alters cellular metabolism, the cell lines used above were measured for mitochondrial function. In this regard, it is shown that MCF7-shIDH2 cells expressing IDH2K413Q (MCF7-shIDH2-IDH2K413Q) exhibited a significant decrease in ATP turnover (Fig. 2d), basal respiration (Fig. 2e), mitochondrial respiration capacity (Fig. 2f), and proton leak (Fig. 2g), as compared with MCF7-shIDH2-IDH2WT and MCF7-shIDH2-IDH2K413R cells, suggesting that the acetylation status IDH2-K413 alters breast cancer cell mitochondrial metabolism.
IDH2K413 acetylation mimetic increases an in vitro and in vivo transformation phenotype
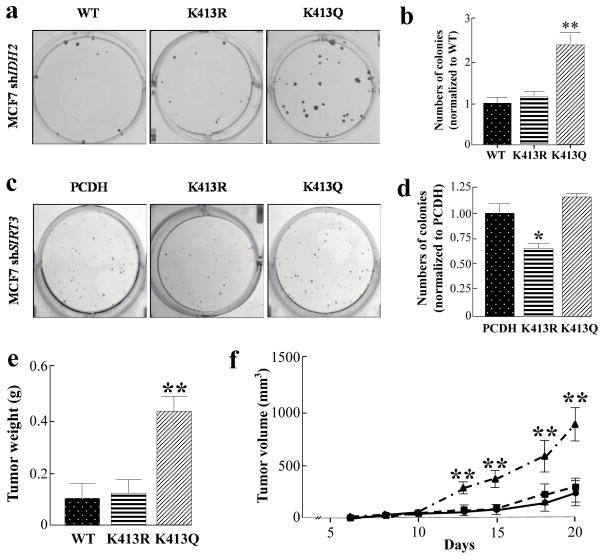
We have previously shown that loss of Sirt3 promotes tumor permissive phenotypes, in vitro and in vivo, suggesting that SIRT3 can function as a tumor suppressor (TS) protein. Therefore, we hypothesized that acetylation of IDH2 at lysine 413, which would be observed with either deletion of Sirt3 or due to a decrease in enzymatic activity that should occur with increased age, might play a role, at least in some part, in this tumor permissive phenotype. In this regard, MCF7-shIDH2-IDH2K413Q cells formed more colonies when grown at low density, as compared with MCF7 cells expressing wild-type IDH2 (MCF7-shIDH2-IDH2WT) (Fig. 3a, b). In addition, fewer colonies were observed in the MCF7-shSIRT3-IDH2K413R cells, suggesting that re-expression of active, de-acetylated IDH2 mimetic gene can, at least in some part; partially reverse the in vitro tumor permissive phenotype (Fig. 3c, d).
Figure 3. Expression of IDH2K413Q promotes a transformation permissive phenotype.
(a) Fifty MCF7-shIDH2-IDH2WT, MCF7-shIDH2-IDH2K413R, or MCF7-shIDH2-IDH2K413Q cells were plated on a 6-well cell culture plate for 14 days before colonies were stained using crystal violet. (b) Quantification of colony numbers (n=4). (c) Two hundred and fifty MCF7-shSIRT3-PCDH, MCF7-shSIRT3-IDH2K413R and MCF7-shSIRT3-IDH2K413Q cells were plated on a 6-well cell culture plate for 14 days before colonies were stained using crystal violet. (d) Quantification of colony numbers (n=4). (e) Tumors from immunodeficient mice injected with five million MCF7-shIDH2-IDH2WT, MCF7-shIDH2-IDH2K413R, or MCF7-shIDH2-IDH2K413Q cells were harvested and weighed (n=5). (f) Tumor growth was measured starting at day six of tumor incidence (n=5). Results are the average of five biological replicates, and error bars represent one standard deviation. * indicates p<0.05, ** indicates p<0.01, comparing to WT by two-tailed unpaired t-test using Prism 6.0.
MCF7-shIDH2-IDH2K413Q, MCF7-shIDH2-IDH2K413R, and MCF7-shIDH2-IDH2WT cells were subsequently injected into immunodeficient mice and examined for tumor growth / proliferation, as measured by tumor size and weight (Table 1). When the tumors were harvested, the sizes and the weight of the tumors expressing IDH2K413Q were significantly larger and heavier than the tumors expressing wild-type IDH2 and IDH2K413R (Fig. 3e, Supplemental Fig. S2). Finally, the tumors expressing IDH2K413Q grew faster than the tumors expressing wild-type IDH2 and IDH2K413R when the tumor volumes were examined (Fig. 3f).
Table 1.
Xenograft tumor number, weight, and size
| Group i.e., gene | Number of mice | Mice with tumors | Ave tumor weight (g) | Ave tumorsize (mm3) |
|---|---|---|---|---|
| Wild-type | 5 | 4 | 0.14 | 243 |
| K413R | 5 | 4 | 0.16 | 263 |
| K413Q | 5 | 5 | 0.52 | 908 |
Based on the results above, it seemed reasonable to determine if enforced expression of IDH2K413R (de-Ac-mimic) and/or IDH2K413Q (Ac-mimic) would alter the transformative properties of immortalized NIH3T3 cells infected with virus expressing Myc or oncogenic Ras (RasG12D) (17, 18). When we examined NIH3T3 cell growth in soft agar, we found that cells expressing Myc and/or oncogenic RasG12D were transformed, as they were able to form colonies in soft agar. Surprisingly, simultaneous expression of IDH2K413R with Myc or RasG12D significantly decreased the sizes of colonies, or even abolished colony formation in soft agar. In contrast, simultaneous expression of IDH2K413Q with Myc or RasG12D significantly increased the size of colonies that grew in soft agar, suggesting the invasiveness and transformative properties of NIH3T3 cells expressing IDH2K413Q and Myc/RasG12D (Supplemental Fig. S3, S4).
To determine whether adding metabolites from the Krebs Cycle that mimics an increase of IDH2 activity (cell-permeable dimethyl-alpha-ketoglutarate (α-KG)) or a decrease of IDH2 activity (isocitrate) affects in vitro phenotypes, we analyzed clonogenic survival and growth of MCF7-shIDH2-IDH2K413Q cells treated with α-KG or isocitrate. These experiments showed that MCF7-shIDH2-IDH2K413Q cells grown in α-KG exhibited a significant decrease in clonogenic survival, as compared to controls (Supplemental Fig. S5a–b). In contrast, MCF7 cells grown in isocitrate exhibited an increase in clonogenic growth (Supplemental Fig. S5c–d). These results suggest that low IDH2 activity promotes tumor aggressiveness, whereas high IDH2 activity partially reverses the proliferative phenotype. To analyze if the addition of α-KG and/or isocitrate alters cancer cell metabolism, mitochondrial respiration and cellular glycolytic rate was measured. In this regard, the addition of α-KG increased mitochondrial respiration capacity (Supplemental Fig. S5e); in contrast, the addition of isocitrate increased glycolysis (Supplemental Fig. S5f).
IDH2K413 acetylation mimetic directs glycolysis, impairs mitochondrial detoxification and increases ROS levels
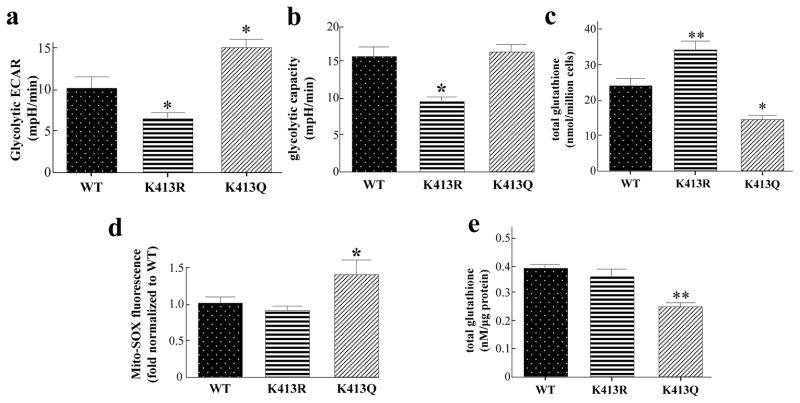
In contrast to cells with active IDH2, cells with inactive IDH2 should rely on specific compensatory mechanisms responding to the dysregulation of the Krebs Cycle and oxidative phosphorylation. Thus, it is proposed that the accumulation of metabolites upstream of IDH2 and the Krebs Cycle in cancer cells expressing IDH2 acetyl mimetic may reprogram to preferentially use glucose for glycolytic metabolism. Since the enforced expression of the IHD2-K413-Ac mutant alters metabolism, it seemed reasonable to determine any potential changes in glycolysis and/or mitochondrial redox balance (19–21). In this regard, we found that MCF7-shIDH2-IDH2K413Q cells displayed an increase in glycolysis (Fig. 4a), while MCF7-shIDH2-IDH2K413R cells showed a decrease in glycolytic capacity (Fig. 4b), suggesting that acetyl mimetic of IDH2K413 directs glycolysis activity. To directly examine whether the IHD2-K413-Ac directs mitochondrial redox balance in vitro, MCF7-shIDH2 cells were used to determine the levels of glutathione, a critical reducing equivalent of regulating redox balance in MCF7 cells. MCF7-shIDH2-IDH2K413Q cells displayed a decrease in total glutathione levels while in contrast; MCF7-shIDH2-IDH2K413R cells showed an increase in total glutathione, as compared with wild-type MCF7-shIDH2-IDH2WT cells (Fig. 4c). In addition, MCF7-shIDH2-IDH2K413Q cells have increased ROS levels, as compared with control cells, as determined by MitoSOXTM fluorescence (Fig. 4d).
Figure 4. MCF7 cells expressing the acetylation mimic mutants exhibit altered mitochondrial energy and detoxification metabolism.
(a–b) Glucose, oligomycin, and 2-deoxyglucose (2-DG) were sequentially added to measure extracellular acidification rates (ECAR) in the XF24 analyzer from Seahorse Bioscience (n=7). Glycolytic ECAR was determined by the difference between the ECAR after adding glucose and the ECAR after adding 2-DG. Glycolytic capacity was determined by the difference between the ECAR after adding oligomycin and the ECAR after adding 2-DG. (c) Total glutathione levels were measured in MCF7-shIDH2-IDH2WT, MCF7-shIDH2-IDH2K413R, or MCF7-shIDH2-IDH2K413Q cells (n=4). (d) MCF7 cells were transfected with IDH2WT, IDH2K413R and IDH2K413Q plasmids, trypsinized and resuspended before MitoSOXTM (2 μM) was added to the cells. The fluorescence representing mitochondrial superoxide was measured via flow cytometry (n=3). (e) Total glutathione levels of tumors from the immunodeficient mice were measured (n=4). Results are from three separate experiments. Error bars represent one standard deviation. * indicates p<0.05, ** indicates p<0.01, comparing to WT by two-tailed unpaired t-test using Prism 6.0.
To examine if the acetylation status of IDH2-K413 altered mitochondrial detoxification of ROS in vivo, MCF7-shIDH2-IDH2WT, MCF7-shIDH2-IDH2K413R, and MCF7-shIDH2-IDH2K413Q cells were injected into immunodeficient mice. The examination of the total glutathione levels showed that the expression of IDH2K413Q in xenograft tumors displayed significantly decreased total glutathione levels, as compared with tumors expressing either the wild-type IDH2 or IDH2K413R (Fig. 4e). To study how the chemical inhibition of ROS production will potentially reverse the tumor permissive phenotypes, we added mitoTEMPO to the MCF7-shIDH2-IDH2K413Q cells, and we found a significant decrease of clonogenic survival when grown with 25μM or 50μM mitoTEMPO (Supplementary Fig. S6). These results suggest that the chemical removal of mitochondrial ROS in these cells decreased colony formation, similar to cells expressing lenti- IDH2K413R.
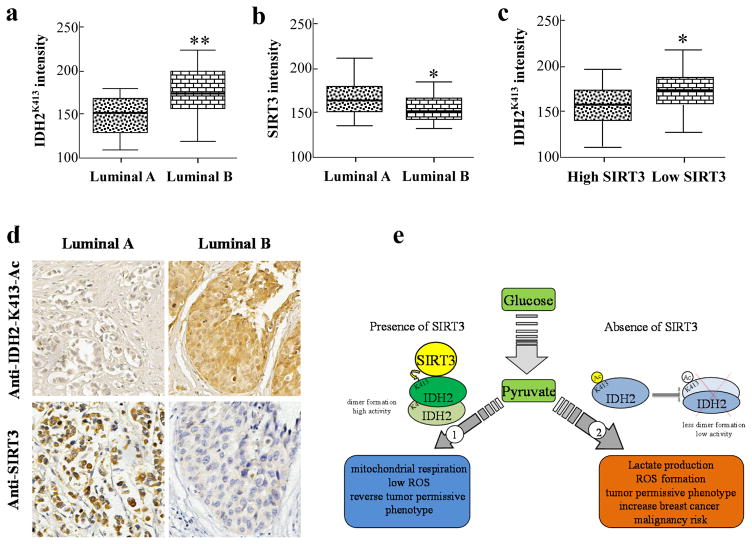
Acetylation of IDH2 at lysine 413 correlates with Luminal B breast cancer risk
To establish a potential correlative clinical relationship between IDH2 acetylation and breast cancer risk, we used breast cancer patient tissue array slides consisting of different subtypes of breast malignancies and examined the levels of IDH2-K413-Ac and SIRT3 by immunohistochemistry (IHC) using an anti-IDH2-K413-Ac and anti-SIRT3 antibody. We found that the levels of IDH2-K413-Ac in Luminal B breast patient samples were significantly higher than Luminal A breast cancer patient samples. In addition, the levels of SIRT3 in Luminal B patient samples were significantly lower than Luminal A breast cancer patient samples (Fig. 5a, b, d). Finally, patients with low SIRT3 and high SIRT3 levels were grouped based on their IHC levels, and we found that the subgroup with lower SIRT3 levels showed higher IDH2 acetylation levels than the group with higher SIRT3 levels (Fig. 5c). In addition, a more rigorous correlation analysis showed that IDH2 acetylation levels negatively correlate with SIRT3 protein levels (Supplemental Fig. S7). These results suggested a strong negative correlation between acetylation of IDH2 at lysine 413, as a result of a loss and/or decrease in SIRT3 enzymatic activity, and breast cancer risk (Fig. 5e).
Figure 5. IDH2-K413-Ac is correlated with breast cancer risk.
(a–b) Slides containing Luminal A (n=17) or Luminal B (n=38) patient samples (Biomax) were stained using anti-IDH2-K413-Ac and anti-SIRT3 antibody. (c) Samples were separated into high SIRT3 (n=25) and low SIRT3 (n=30) groups, and IDH2K413 levels. (d) A representative image demonstrating IDH2K413 and SIRT3 staining in Luminal A and Luminal B cancer samples. (e) Summarized mechanism. The shaded boxes represent the interquartile range; whiskers represent min and max; bars represent the median. * indicates p<0.05, ** indicates p<0.01 by two-tailed unpaired t-test using Prism 6.0.
DISCUSSION
Mammalian SIRT3 is regarded as the primary mitochondrial protein deacetylase that directs the acetylation status of mitochondrial proteins. Loss of SIRT3 enzymatic activity results in aberrant hyperacetylation of multiple mitochondrial proteins (22), including ATP synthase F1 (23), MnSOD (9), and IDH2 (4), resulting in abnormal mitochondrial homeostasis, aberrantly elevated ROS levels, and metabolic reprogramming in tumor cells that exhibit a preferential use of glucose consumption (8) that is similar to that observed in the Warburg effect (24–26). The NADP+-dependent, dimeric form of IDH2 oxidizes isocitrate into α-ketoglutarate and is a key enzyme in the Krebs Cycle. It has been shown that inhibiting IDH2 enzymatic activity by acetylation or knocking it down negatively affects mitochondrial redox balance (4). In our study, we suggest an additional mechanism linking IDH2 acetylation at K413, due to loss of SIRT3, a protein regarded as a mitochondrial tumor suppressor, to this enzymatic function by altering its homodimeric protein-protein interaction.
Based on what we have observed in this project and in other unpublished projects, we found that when we measure parameters like ROS production and glycolytic capacity, the difference between wild-type and de-acetyl mimetic (the R form) of different mitochondrial SIRT3 deacetylation target proteins was not always significant. In contrast, it seems that the acetyl mimetic (the Q form) of SIRT3 downstream targets tends to decrease activity. In this regard, we speculate that de-acetylated IDH2 is already present in significant amounts within cells of our model systems and hence, depending on the cell type, adding additional IDH2 de-acetyl mimetic (the R form) will not always significantly alter either the phenotype or biochemical biology. However, the overexpression of IDH2K413R in aerobically glycolytic (Warburg) cancer cells has the ability to alter the aberrant metabolic biochemical phenotype.
The alteration of acetylation stoichiometry, due to loss of sirtuin enzymatic activity, ranges from less than 1% to about 98%, suggesting that changes in mitochondrial acetylation status can have significant effects in cellular metabolism and signaling pathways (27). Therefore, it seems likely that the overall dysregulation of mitochondrial metabolism, due to an increase in protein acetylation, contribute to the tumor permissive phenotype observed in cells lacking SIRT3. As such, SIRT3 is proposed to be a tumor suppressor protein that connects mitochondrial energy generation, metabolism, and carcinogenesis (12, 21, 28).
Currently, several new metabolic properties related to IDHs have been discovered. First, mutations of IDH1 and IDH2 caused a neomorphic enzymatic activity, resulting in the production of an onco-metabolite named 2-hydroxyglutarate (2HG) (29). It has been shown that 2HG can impair histone demethylation and switch cell differentiation pathways (30). In addition, a reversible reduction of α-KG to isocitrate has also been discovered under hypoxic conditions with wild-type IDH1 and IDH2, and an elevation of 2HG levels was also detected in this process (31). The NAD+-dependent IDH3 has long been regarded as the main enzyme for the conversion from isocitrate to α-KG (32). However, simultaneous loss of IDH2 and IDH3 inhibits ATP formation in pancreatic β-cells, suggesting that NADP+-dependent IDH2 is also involved in mitochondrial metabolism (33). In addition, knocking down IDH2 results in a 25% decrease of NADPH levels in cells, suggesting that IDH2 plays a significant role in mitochondrial NADPH production (4). Furthermore, the main function of IDH2 includes producing NADPH in the mitochondria, regenerating reduced glutathione and defending against elevated ROS levels, suggesting its potential role as an anti-oxidant enzyme by protecting cells against oxidative damage. Previous discoveries have shown that loss of SIRT3 increases ROS levels, activating HIF and oncogenic pathways (10). It has also been shown that expression of active IDH2 and SIRT3 prevents hearing loss in mice by production of NADPH. This regenerates reduced glutathione, decreases GSSG:GSH ratio, and inhibits ROS production. Furthermore, both IDH2 and SIRT3 have potential effects in protecting against acute hemorrhagic leukoencephalitis (AHL), suggesting the beneficial roles of IDH2 and SIRT3 in providing a redox homeostasis environment for cells (34). Therefore, it is suggested that acetylation of IDH2 at K413, due to loss of SIRT3 enzymatic activity, poses risks that lead to pro-oncogenic properties in cells.
Our data suggests that MCF7 cells expressing IDH2K413Q exhibited altered metabolism and elevated oxidative stress. When these cells were injected into nude mice, these tumors also exhibited similar dysregulated biochemical properties (increased oxidative stress) that were previously observed in Sirt3 knock-out mouse model (12). In addition, our previous discovery in Sirt3 knock-out mammary tumors and our observations in human Luminal A and Luminal B breast cancer patient samples have provided a correlative explanation between IDH2 acetylation, SIRT3 expression and breast cancer malignancy risk (13). Furthermore, studies by the Denu group have also discovered that decreased SIRT3 mRNA levels, decreased SIRT3 enzymatic activity, and hyper-acetylation of SIRT3 target proteins (IDH2 and MnSOD) can result in B cell malignancies due to increased ROS levels, suggesting that acetylation of IDH2 can promote malignancy risks in different types of cancers (35).
Overall, our data has also provided an additional mechanistic explanation of how IHD2-K413-Ac negatively alters its enzymatic activity and promotes transformation-permissive phenotypes in vitro and in vivo. Therefore, it seems reasonable to propose that activators of SIRT3 and/or inhibition of mitochondrial ROS production may be used to protect cells from mitochondrial redox imbalance and transformation (21, 36).
Supplementary Material
Acknowledgments
Financial Support
D. Gius is supported by 2R01CA152601-A1, 1R01CA152799-01A1, 1R01CA168292-01A1, 1R01CA214025-01, the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust, Zell Family Foundation, and the Avon Foundation for Breast Cancer Research. Y. Zhu is supported by a Robert H. Lurie Comprehensive Cancer Center Translation Bridge Fellowship Award.
Footnotes
The authors have no conflict of interest
References
- 1.Xu X, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279(32):33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 2.Cupp JR, McAlister-Henn L. Kinetic analysis of NAD(+)-isocitrate dehydrogenase with altered isocitrate binding sites: contribution of IDH1 and IDH2 subunits to regulation and catalysis. Biochemistry. 1993;32(36):9323–9328. doi: 10.1021/bi00087a010. [DOI] [PubMed] [Google Scholar]
- 3.Haselbeck RJ, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J Biol Chem. 1993;268(16):12116–12122. [PubMed] [Google Scholar]
- 4.Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. 2012;287(17):14078–14086. doi: 10.1074/jbc.M112.355206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombard DB, Zwaans BM. SIRT3: As Simple As It Seems. Gerontology. 2013 doi: 10.1159/000354382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarente L. Sirtuins in aging and disease. Cold Spring Harbor symposia on quantitative biology. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 7.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41(2):139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozden O, et al. SIRT3 deacetylates and increases pyruvate dehydrogenase activity in cancer cells. Free Radic Biol Med. 2014;76:163–172. doi: 10.1016/j.freeradbiomed.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tao R, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40(6):893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigis MC, Deng CX, Finley LW, Kim HS, Gius D. SIRT3 is a mitochondrial tumor suppressor: a scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 2012;72(10):2468–2472. doi: 10.1158/0008-5472.CAN-11-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finley LW, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desouki MM, Doubinskaia I, Gius D, Abdulkadir SA. Decreased mitochondrial SIRT3 expression is a potential molecular biomarker associated with poor outcome in breast cancer. Hum Pathol. 2014;45(5):1071–1077. doi: 10.1016/j.humpath.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24(3):148–154. doi: 10.1007/BF00300234. [DOI] [PubMed] [Google Scholar]
- 15.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103(27):10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316(5827):1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- 17.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304(5927):596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 18.Land H, Chen AC, Morgenstern JP, Parada LF, Weinberg RA. Behavior of myc and ras oncogenes in transformation of rat embryo fibroblasts. Mol Cell Biol. 1986;6(6):1917–1925. doi: 10.1128/mcb.6.6.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vassilopoulos A, et al. SIRT3 Deacetylates ATP Synthase F Complex Proteins in Response to Nutrient- and Exercise-Induced Stress. Antioxid Redox Signal. 2014 doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao R, Vassilopoulos A, Parisiadou L, Yan Y, Gius D. Regulation of MnSOD enzymatic activity by Sirt3 connects the mitochondrial acetylome signaling networks to aging and carcinogenesis. Antioxid Redox Signal. 2014;20(10):1646–1654. doi: 10.1089/ars.2013.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou X, Santa-Maria CA, O’Brien J, Gius D, Zhu Y. Manganese Superoxide Dismutase Acetylation and Dysregulation, Due to Loss of SIRT3 Activity, Promote a Luminal B-Like Breast Carcinogenic-Permissive Phenotype. Antioxid Redox Signal. 2016 doi: 10.1089/ars.2016.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lombard DB, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vassilopoulos A, et al. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21(4):551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 25.Warburg O. Iron, the Oxygen-Carrier of Respiration-Ferment. Science. 1925;61(1588):575–582. doi: 10.1126/science.61.1588.575. [DOI] [PubMed] [Google Scholar]
- 26.Warburg O, Wind F, Negelein E. The Metabolism of Tumors in the Body. The Journal of general physiology. 1927;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baeza J, et al. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289(31):21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, et al. SIRT3 and SIRT4 are mitochondrial tumor suppressor proteins that connect mitochondrial metabolism and carcinogenesis. Cancer Metab. 2014;2:15. doi: 10.1186/2049-3002-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108(49):19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiaffino S, Reggiani C, Kostrominova TY, Mann M, Murgia M. Mitochondrial specialization revealed by single muscle fiber proteomics: focus on the Krebs cycle. Scand J Med Sci Sports. 2015;25(Suppl 4):41–48. doi: 10.1111/sms.12606. [DOI] [PubMed] [Google Scholar]
- 33.MacDonald MJ, et al. Knockdown of both mitochondrial isocitrate dehydrogenase enzymes in pancreatic beta cells inhibits insulin secretion. Biochim Biophys Acta. 2013;1830(11):5104–5111. doi: 10.1016/j.bbagen.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu W, et al. Loss of SIRT3 Provides Growth Advantage for B Cell Malignancies. J Biol Chem. 2016;291(7):3268–3279. doi: 10.1074/jbc.M115.702076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillai VB, et al. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat Commun. 2015;6:6656. doi: 10.1038/ncomms7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.