Abstract
The deubiquitylase (DUB) USP37 is a component of the ubiquitin system and controls cell proliferation by regulating the stability of the cyclin-dependent kinase inhibitor 1B, (CDKN1B/p27Kip1). The expression of USP37 is down-regulated in human medulloblastoma tumor specimens. In the current study we show that USP37 prevents medulloblastoma growth in mouse orthotopic models, suggesting that it has tumor suppressive properties in this neural cancer. Here, we also report on the mechanism underlying USP37 loss in medulloblastoma. Previously, we observed that the expression of USP37 is transcriptionally repressed by the RE1 Silencing Transcription Factor (REST), which requires chromatin remodeling factors for its activity. Genetic and pharmacological approaches were employed to identify a specific role for G9a, a histone methyltransferase (HMT), in promoting methylation of histone H3 lysine-9 (H3K9) mono- and di-methylation, and surprisingly tri-methylation, at the USP37 promoter to repress its gene expression. G9a inhibition also blocked the tumorigenic potential of medulloblastoma cells in vivo. Using isogenic low- and high-REST medulloblastoma cells, we further showed a REST-dependent elevation in G9a activity, which further increased mono- and tri-methylation of histone H3K9, accompanied by down regulation of USP37 expression. Together, these findings reveal a role for REST-associated G9a and histone H3K9 methylation in the repression of USP37 expression in medulloblastoma.
Keywords: USP37-deubiquitylase, G9a-histone methylation, Medulloblastoma
Introduction
Medulloblastoma is a malignant pediatric brain tumor. It frequently arises in the cerebellum and is characterized by poor neuronal differentiation and hyper-proliferation (1–3). Current therapies while efficacious against primary tumors, cause severe neurologic deficits in children with brain tumors (4). Therapies targeting tumor-specific molecular events are needed to minimize treatment-related toxicities (4).
The RE1 Silencing Transcription Factor (REST), a repressor of neuronal specification, has been implicated in medulloblastoma development. Its expression is elevated in human medulloblastoma samples and is associated with poor prognostic significance for patients (5–7). Previous work showed that REST knockdown in human medulloblastoma cells abrogated its tumorigenic potential in murine orthotopic models, suggesting that it is required for tumor maintenance (8). REST is also co-expressed with c-Myc or N-Myc in human medulloblastoma samples and co-overexpression of REST and v-Myc in mouse neural progenitors promoted cerebellar tumors that resembled human medulloblastomas (6). These observations are consistent with a function for REST in tumor progression. Finally, REST-positive tumors were poorly differentiated, which is in agreement with its known function as a repressor of neuronal differentiation genes (9–11).
In more recent studies, we observed that REST elevation also contributed to deregulation of medulloblastoma cell proliferation (7,12). We determined that REST-dependent maintenance of tumor cell proliferation was mediated by an ubiquitin specific protease (USP) called USP37, which we identified as a novel transcriptional target of REST’s repressive activity (5). USP37 is a member of the USP family of deubiquitylases (DUBs), and is one of a large number of DUBs that have been described in the literature based on sequence comparison (13–17). DUBs are enzymes that catalyze the proteolytic removal of ubiquitin moieties from proteins and oppose the coordinated activities of ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2 and ubiquitin ligase E3, which catalyze the addition of ubiquitin residues to proteins (18,19). Thus, these two processes maintain cellular homeostasis and is perturbed in a number of cancers (20). Because ubiquitylation is a reversible process and can be targeted pharmacologically, DUBs are also gaining traction as druggable targets for cancer (13,21).
We had previously shown that USP37 is required for the stabilization of the cyclin-dependent kinase inhibitor (CDKI), p27 (5). The failure to stabilize p27 in REST-high and USP37-low medulloblastoma cell lines was shown to directly promote deregulated cell proliferation (5). However, the requirement for USP37 in medulloblastoma growth in vivo, and the specific mechanism(s) utilized by the REST complex in repressing USP37 in tumor cells are not known. Here, we demonstrate that constitutive USP37 expression suppresses medulloblastoma growth in mouse orthotopic models. We also provide genetic and pharmacological evidence in support of a role for the HMT G9a and histone H3K9 methylation in silencing USP37 expression and show that pharmacological inhibition of G9a blocks the growth of medulloblastoma cells in vitro and suppresses their tumorigenic potential in vivo. Finally, using isogenic low-REST and high-REST medulloblastoma cell lines, we show that G9a promotes repression of USP37 expression in a REST-dependent manner. Thus, our findings not only suggest a tumor suppressive role for USP37 in medulloblastoma development, but also provide data to support the further exploration of G9a inhibitors for therapeutic intervention in REST-positive medulloblastomas.
Materials and Methods
Cell Culture/Transfections
DAOY (American Type Cell Culture), DAOY-REST, D283, and UW426 were cultivated as previously described (7). Cell identity was confirmed using short tandem repeat (STR) fingerprinting (Characterized Cell Line Core, M.D. Anderson Cancer Center, Houston, TX). Mycoplasma testing is performed every six months using MycoAlert Mycoplasma Detection Kit (Lonza, Allendale, NJ). Cells are passaged at least twice after thawing prior to experiments. All experiments are performed within two-three weeks after cells are thawed and passaged no more than ten times.
Ectopic expression of mutant G9a was achieved by transient transfection of 2μg of plasmid pcDNA3.1(+)-G9aH1113K (a gift of Dr. Xiaobing Shi) or vector alone using Lipofectamine 2000 Reagent (Thermo Fischer Scientific, Waltham, MA). Cells were harvested after 24 hours (hrs) and processed for RNA and protein analyses.
MTT Assay
E67 was synthesized in accordance to the methods described by Chang et al (22). UNC 0638 was purchased from the Cayman Chemical Company (Ann Arbor, MI) (23). Both drugs were dissolved in DMSO and stored at −20°C. DAOY, DAOY-REST cells (3×104), UW426 (4×104) and D283 (7.5×104) were seeded at the indicated densities in 96-well plates containing 100 μl of media. After overnight incubation, cells were treated for 24, 48, 72 hrs with E67, UNC 0638 or DMSO at concentrations ranging from 0.125 μM-10 μM and 0.125 μM -4 μM, respectively. MTT assays were performed as described previously (5,7,24). Absorbance at 570 nm and background at 650 nm were read on a multiwell spectrophotometer. Results were expressed as a percentage (%) of the control (DMSO).
Live cell nuclear staining assay
DAOY or DAOY-REST cells were seeded at a density of 90,000 cells/well in 6 well plates and incubated overnight. Cells were treated with their respective 48hrs-IC50 concentrations of UNC 0638 (1.1 μM and 2.5μM) in a final volume of 3ml media, incubated for 12, 24, or 48 hrs, prior to harvesting for analysis by live cell nuclear staining assay. 1×105 cells were resuspended in 200 μl PBS and cells were incubated 2 μl of DRAQ5 for 3 minutes at 37°C and then 5–10 minutes at room temperature. Flow cytometry was performed using a FACSCalibur (Becton Dickinson, Franklin Lakes, New Jersey)
Q-RT-PCR Analyses
DAOY, D283 and or UW426 cells were treated with either 5 μmol/L SAHA (Cayman Chemical Company, Ann Arbor, MI) for 6, 12 and 24 hrs, or 2.5 μmol/L MS-275 (Enzo Life Sciences, Farmingdale, NY) for 6, 16 and 24 hrs, or 1.1μM UNC 0638 (IC25 at 48 hrs) for 4 or 12 hrs. Cells were harvested for RNA extraction using Quick RNA kit (Genesee Scientific, San Diego, CA). RNA recovery and purity were determined by NanoDrop Spectrophotometer ND-1000 (Thermo Fischer Scientific, Waltham, MA). cDNA libraries were synthesized using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA) and Q-RT-PCR was performed using Roche Lightcycler 96 (Roche, Indianapolis, IN) according to manufacturer’s instructions. For primers see supplementary data.
Chromatin immunoprecipitation (ChIP) assays
DAOY or DAOY-REST cells were treated with 1.1μM UNC 0638 or DMSO for 12 hrs prior to fixation, cross-linking, and processing for ChIP analyses as described previously (5). Cross-linked cells were resuspended in sonication buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA [pH 8.0], 1% SDS, and protease inhibitors) and sonicated, and 10% of this material was saved as input DNA. The remainder of the samples were diluted 5-fold with ChIP dilution buffer [16.7 mM Tris-HCl [pH 8.0], 167 mM NaCl, 1.2 mM EDTA (pH 8.0), 1.1% Triton X-100, and protease inhibitors], precleared, and incubated with various antibodies for 12 hrs at 4°C. Following incubation with protein-A beads, washing, and elution, the cross-linking was reversed and DNA was purified with a QiaQuick PCR Purification Kit (Qiagen, Valencia, CA). Bound DNA was quantified by SYBRGreen qPCR using Roche Lightcycler 96. Data were analyzed using the comparative 2-ΔΔCT method (7). For antibody and primer information see supplementary data.
Westerns Blot Analysis
Cells were harvested in RIPA lysis buffer containing a cocktail of protease inhibitors (Thermo Fischer Scientific, Waltham, MA) and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO). Equal amount of protein were electrophoresed and subjected to Western blot analyses. For antibody list see supplementary data. Immunoreactive bands were detected using Supersignal West Dura Extended Duration Substrate (Pierce, Rockford, IL).
In vivo measurement of tumor growth
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC). Orthotopic cerebellar injections of isogenic DAOY and DAOY-REST cells, pre-treated with UNC 0638 (IC25 for DAOY cells at 48 hrs, 1.1 μM) for 12 hrs, were performed using 4–6 week old NOD scid gamma null (NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ JAX) mice (Jackson Laboratories, Bar Harbor, Maine) (24). Bioluminescence imaging (BLI) was performed to monitor tumor growth using an IVIS Spectrum (Caliper), following intraperitoneal injection (IP) injection of D-Luciferin. Data were analyzed using Living Image (Xenogen) software. Mice were sacrificed after 13 days and brains were formalin fixed for histopathology assessment.
Immunohistochemistry
IHC staining was performed on paraffin-embedded brain sections (4-μm). Paraffin-embedded brain sections were deparaffinized, rehydrated and treated to inactivate the endogenous peroxidase activity. Antigen retrieval was then performed using a pressure cooker (Aroma) using a sodium citrate buffer (10mM, pH 6.0). Samples were washed and blocked in Tris-Buffered Saline – Tween (TBST) containing 1% bovine serum albumin (BSA) and 5% normal goat serum. Incubation with primary antibody was then carried out overnight at 4°C. For antibody list see supplementary data. Following incubation with the corresponding secondary antibody, slides were washed with TBST and developed using 3,3′-diaminobenzidine (DAB) (Vector Laboratories; Burlingame, Ca) and counterstained with Hematoxylin. Stained slides were viewed using a Nikon ECLIPSE E200 microscope and images were captured under 4X, 10X, and 40X magnification with an OLYMPUS SC100 camera.
Results
Constitutive USP37 expression suppresses medulloblastoma growth
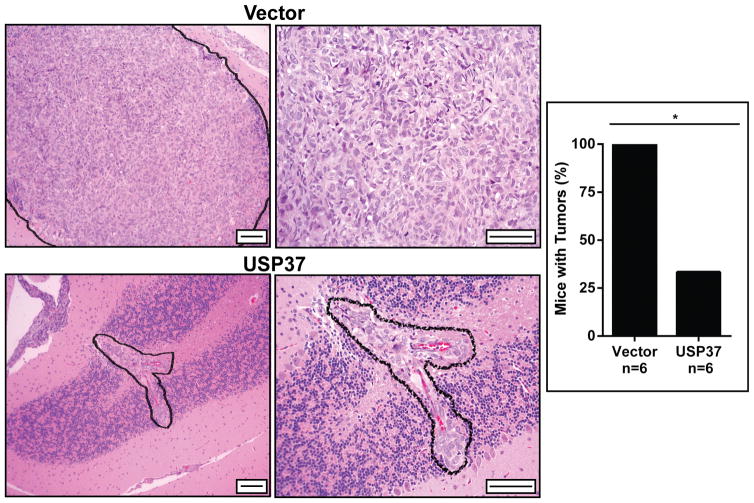
We had previously identified the deubiquitylase USP37 as a novel target of the transcriptional repressor REST in medulloblastoma cell lines (5). Constitutive USP37 expression in medulloblastoma cells with high REST expression led to stabilization of p27 and cessation of cell proliferation in vitro, suggesting that it may have tumor suppressive properties (5). To test this possibility, in vivo experiments were performed wherein DAOY medulloblastoma cells were transiently transfected with a vector expressing USP37 and injected into the cerebella of immunodeficient mice (n=6). Control animals were injected with cells transfected with vector plasmid. Animals were monitored periodically for signs of morbidity including head tilt and hemi-paresis. All animals were sacrificed at 3 weeks when control mice receiving injections of vector transfected cells showed signs of morbidity. Brains were harvested and sectioned for tumor analysis. Sections were stained with hematoxylin and eosin (H&E), which revealed a significant reduction in tumor burden in mice receiving injections of DAOY cells expressing USP37 transgene compared to animals injected with vector transfected cells (Fig. 1, right panel). Although tumors where seen in 2 of the 6 mice, the size of tumors was considerably smaller than those in the control group (Fig. 1, left panel). These observations suggest that USP37 has tumor suppressive properties.
Figure 1. Constitutive USP37 expression suppresses the growth of medulloblastoma cells in mouse orthotopic models.
DAOY cells were transiently transfected with plasmid pDEST26-Flag-HA-USP37 or a vector and implanted into mice cerebella (n=6 each). Animals were sacrificed and brains harvested for H&E staining. Scale bars = 50μM (left) and 100μm (right). The bar graph demonstrates the percentage of mice affected with tumor, vector (6/6) and USP37 WT (2/6). Wilcoxon-Rank Sum non-parametric analysis was performed to determine significance (*=p<0.05).
G9a inhibition blocks medulloblastoma growth in vitro and in vivo
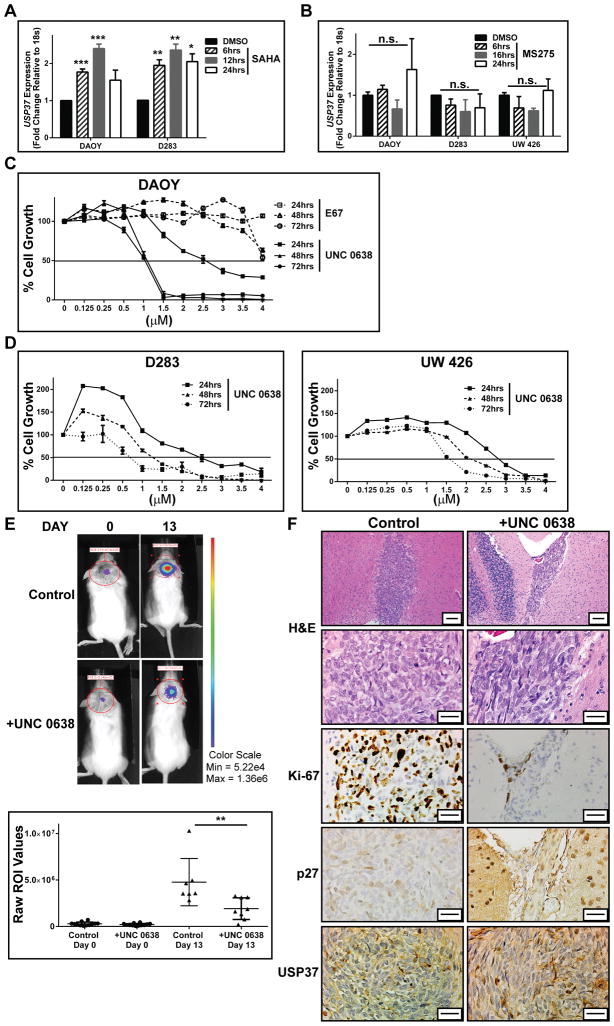
REST is associated with the mSin3A- histone deacetylase 1/2 (HDAC 1/2) complex at its amino (N) and carboxy (C) termini (25). The carboxy (C) terminus of REST binds to the Co-REST complex, which contains the histone H3K9 HMT-G9a, the histone H3K9 and K4 lysine specific demethylase 1 (LSD1) as well as HDAC1/2 (25) To identify the specific REST-associated activity that is required for regulation of USP37 expression, REST-expressing human medulloblastoma cells were treated with drugs that target the N- and C-terminal co-repressor complexes. We had previously shown that the pan HDAC inhibitor SAHA (Vorinostat) as well as the Group 1 HDAC inhibitor MS-275 (Entinostat) blocked the growth of medulloblastoma cells in vitro and promoted the upregulation of REST-target differentiation genes (7). To ask if HDAC inhibition is sufficient for USP37 upregulation, DAOY, D283 and UW426 cells were treated with either SAHA or MS-275 for 6–24 hrs and USP37 gene expression was measured by Q-RT-PCR. As shown in Fig. 2A treatment of DAOY and D283 cells with SAHA, which promotes REST degradation, resulted in a significant upregulation of USP37 gene expression (7). Although MS-275 treatment inhibited REST activity as measured by upregulation of its target neuronal differentiation genes, it did not upregulate USP37 gene expression (Fig. 2B). These results indicate that targeting HDACs 1/2 activity alone is insufficient to de-repress USP37.
Figure 2. REST regulates USP37 expression through H3K9 methylation.
A. DAOY and D283 cells were treated with 5 μmol/L SAHA (Vorinostat) and B. DAOY, D283 and UW426 cells were treated with 2.5 μmol/L MS-275 (Entinostat) or vehicle (DMSO) for various times followed by Q-RT-PCR analyses to measure changes in USP37 gene expression in drug-treated cells compared to untreated cells. C. MTT assays were used to measure the effect of G9a inhibitors E67 and UNC 0638 on mitochondrial activity and cell growth. Solid black line indicates the IC50 value, which is also tabulated. D. D283 and UW426 cells were treated with UNC 0638 as described in C. and MTT assays were performed to obtain IC50 values indicated by dotted line. E. DAOY cells stably expressing firefly luciferase were pre-treated with 1.1 μM (IC50 at 48 hrs) for 12 hrs and implanted into NSG mice cerebella. Tumor growth was measured by BLI on the day of injection (day 0) and every 4 days thereafter. BLI on Days 0 and 13 of representative animals receiving drug-treated (+UNC 0638) and untreated cells (control) is shown (top). Relative flux for the cohort receiving control and drug-treated cells is shown in the bottom graph. F. Representative sections obtained from control and UNC 0638-treated cerebellar sections were processed for H&E staining and IHC with antibodies as indicated. Scale bars = 50 μm (top) and 20 μm (bottom). (*=p<0.05, **=p<0.01 and ***p=<0.001, n.s., not significant)
As a first step in assessing the involvement of the Co-REST complex in regulating USP37 expression, the response of DAOY cells to pharmacological inhibition of G9a was measured as a function of drug concentration and time of exposure to the drug. Treatment of DAOY cells with the G9a inhibitors, E67 and UNC 0638, lead to a decline in cell growth as determined by MTT assays (Fig. 2C). Although the entire range of drug concentrations tested for E67 are not shown, a temporal comparison of IC50 values (7.4 μM, 6.5 μM, 4.2 μM versus 2.7 μM, 1.1 μM and 1.1 μM at 24, 48 and 72 hrs respectively) confirmed that UNC 0638 had a more potent effect on cell growth compared to E67 (Table 1). The decrease in cell growth in response to UNC 0638 treatment was also confirmed in medulloblastoma cell lines with higher (D283) and lower (UW426) REST levels than DAOY cells (7) (Fig. 2D). The effect of G9a inhibition on tumorigenic potential of DAOY cells was also examined in mouse orthotopic models. DAOY cells (50,000/animal, n=7) were treated ex vivo with 1.1μM UNC 0638 (IC50 dose at 48 hrs) for 12 hrs prior to implantation into the cerebellum of immunodeficient NOD SCID-γ (NSG) animals. A 6.25% decrease in cell viability was expected with these pre-implantation treatment parameters. A control group of mice (n=8) were implanted with similar number of cells that were treated with DMSO. Mice were monitored for tumor growth every 4 days by bioluminescence imaging (BLI). Animals were sacrificed 13 days post tumor cell implantation when control mice began displaying signs of morbidity. Shown in Fig. 2E (left panel) are representative images of BLI signals obtained with mice in control (top row) and treatment (bottom row) groups at days 0 and 13. In contrast to the treatment group, a significant increase in bioluminescence intensity was noted within regions of interest (ROI) in the control cohort at day 13 relative to day 0 (Fig. 2E). H&E staining of brain sections confirmed the decrease in tumor burden in animals receiving UNC 0638 treated cells compared to the control cohort that was seen by BLI analyses (Figs. 2E and F). Immunohistochemical (IHC) analyses of brain sections from animals implanted with cells exposed to UNC 0638 also revealed a substantial reduction in Ki-67 positivity, which was associated with an increase in p27 and USP37 staining when compared to the control brains (Fig. 2F). These results suggest that the potential for growth of medulloblastoma cells in vitro and in vivo can be blocked by G9a inhibition.
Table 1.
IC50 values for G9a inhibitors in DAOY cells
| IC50 Values | |||
|---|---|---|---|
| Time (hrs) | 24 | 48 | 72 |
| E67 (μM) | 7.4 | 6.5 | 4.2 |
| UNC 0638 (μM) | 2.7 | 1.1 | 1.1 |
G9a represses USP37 expression by histone H3K9 methylation
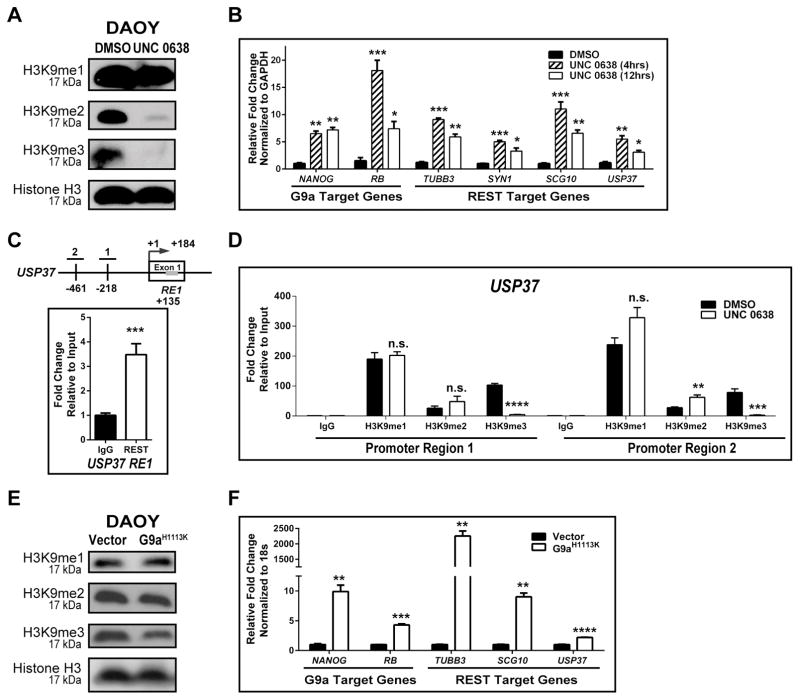
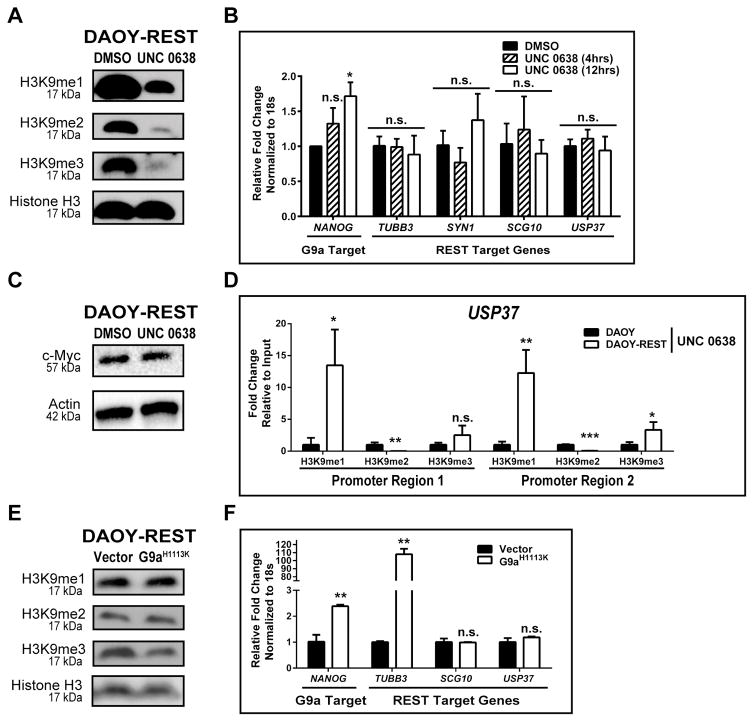
Since USP37 elevation was seen by IHC following UNC 0638 treatment, we asked if G9a-dependent histone H3K9 methylation is involved in the regulation of USP37 gene expression in medulloblastoma cells. First, inhibition of G9a activity following UNC 0638 exposure was studied by Western blotting using lysates of drug or vehicle (DMSO)-treated DAOY cells, which revealed a global decrease in H3K9-me2 and -me3, but not –me1 (Fig. 3A). Further confirmation of these results was obtained through Q-RT-PCR analyses where a 6.5–7.2-fold and 18.1–7.4-fold increase in the expression of the known G9a target genes, NANOG and RB, following exposure of DAOY cells to UNC 0638 for 4 and 12 hrs, respectively (Fig. 3B). REST target genes, TUBB3 (10.1- and 5.9-fold), SYN1 (5- and 3.3-fold), SCG10 (11- and 6.6-fold) and USP37 (5.5- and 3.1-fold) were also upregulated under these conditions, indicating G9a inhibition affected the activity of the REST complex (Fig. 3B).
Figure 3. G9a regulates USP37 expression.
A. Western blot analysis was performed with extracts prepared from DAOY cells that were treated with 1.1 μM UNC 0638 or DMSO for 12 hrs. Global changes in histone H3K9-me1, -me2, and -me3 were determined. Total histone H3 was used as a loading control. B. Changes in the expression of G9a and REST-target genes were established by Q-RT-PCR analyses following treatment with UNC 0638 for 4 or 12 hrs. DMSO treated cells were used as controls. C. Schematic representation of the USP37 promoter region with locations of the RE1 site (gray bar, +135 bp), and CpG island proximal region-1 (-218bp) and region-2 (-461bp), is shown. Specific REST binding to the RE1 site (normalized to IgG) was demonstrated by ChIP assay. Data is represented as fold change relative to input. D. Changes in histone H3K9 -me1, -me2, and -me3 at promoter regions-1 and -2 following DMSO or UNC 0638 treatment (12 hrs), were evaluated by ChIP assay. Data is shown as percent fold change relative to input. DAOY cells transduced with a vector control or a plasmid expressing the G9a mutant-G9aH1113K were analyzed by (E) Western blotting to detect global changes in histone H3K9 -me1, -me2, and -me3, with histone H3 serving as a loading control, and (F) Q-RT-PCR to assess expression of known G9a and REST-target genes in DAOY cells ectopically expressing mutant G9aH1113K. (*=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.000, and n.s. = not significant)
ChIP assays were next performed to first measure REST binding to the RE1 consensus sequence in exon 1 of the USP37 coding region and then to assess the status of histone H3K9 -me1, -me2 and -me3 within the USP37 promoter in DAOY cells. Q-PCR analyses using a pair of primers in the upstream regulatory region of USP37 proximal to the RE1 sites, showed a 3.4-fold increase in amplification of DNA pulled down by anti-REST antibody relative to control IgG, confirming REST occupancy of the site within exon1 of USP37 (Fig. 3C). Antibodies specific to histone H3K9 -me1, -me2 and -me3 marks was also used to demonstrate a 190-fold, 25.7-fold and 103.1-fold increase in pull-down of DNA relative to control IgG around position −218 (region-1) and a 238-fold, 27.6-fold and 78.5- increase in pull-down of DNA relative to control IgG around position −461 bp (region-2) of the USP37 promoter (Fig. 3D). Interestingly, UNC 0638 treatment did not alter the levels of histone H3K9-me1 relative to untreated controls at region-1, but did cause a 1.4-fold increase in histone H3K9-me1 at region-2 (Fig. 3D). A 20- and 30-fold decrease in H3K9-trimethylation and a 2 to 2.4-fold increase in histone H3K9-me2 were also observed upon UNC 0638 treatment at regions-1 and -2 of the USP37 promoter, respectively (Fig. 3D). A role for G9a in the regulation of USP37 gene expression was also confirmed using genetic approaches. DAOY cells were transfected with the plasmid pcDNA3.1-G9a-H1113K, which expresses a catalytically inactive mutant of G9a with a histidine to lysine change at position 1113 (H1113K) of the protein (26). As shown in Fig. 3E, a global decrease in histone H3K9 -me2 and me3 was seen upon expression of G9a-H1113K mutant, confirming interference with endogenous G9a activity in these cells. A 2.2-fold increase in USP37 gene expression was observed in cells expressing mutant G9a compared to vector-transfected cells (Fig. 3F). The expression of SCG10 and TUBB3 underwent 9.9- and 2255-fold increases, whereas expression of the G9a target gene, NANOG, was elevated 9.9-fold under these conditions (Fig. 3F). Together, the above results validate our hypothesis that USP37 gene expression is regulated by G9a.
REST-associated G9a activity represses USP37 gene expression
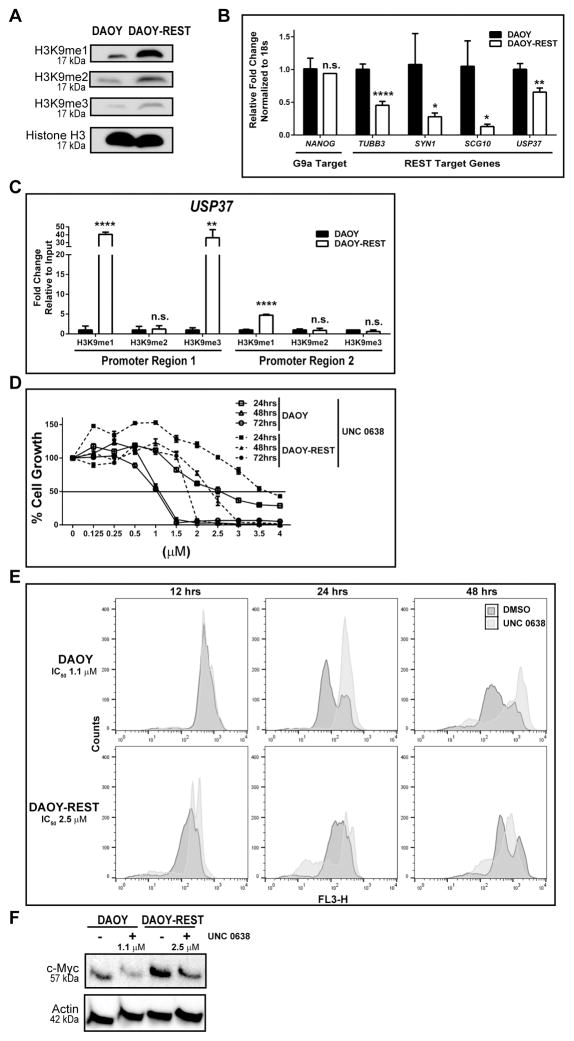
G9a is associated with a number of chromatin remodeling complexes (27,28). To assess if REST associated G9a regulates USP37 gene expression, we first asked if G9a activity was altered in a REST-dependent manner. Western blot analysis of lysates from DAOY cells expressing endogenous REST or human REST transgene (DAOY-REST) showed REST levels to correlate with global increases in histone H3K9 -me1, -me2, and me3 (Fig. 4A). Q-RT-PCR analyses identified significant decreases in the expression of the REST target genes, TUBB3, SYN1, SCG10 and USP37 (Fig. 4B). However, expression of the G9a target gene, NANOG, was unaffected in REST-high cells, indicating that the effect was specific to REST target genes (Fig. 4B). Consistent with these observations, ChIP experiments revealed a 40.5-fold increase in histone H3K9-me1 and a 36.2-fold increase in histone H3K9-me1 in region-1 of the USP37 promoter in DAOY-REST cells compared to DAOY cells (Figs. 3C-top panel and 4C). A smaller 4.7-fold increase in histone H3K9-me1 was noted at region-2 of USP37 promoter in DAOY-REST cells compared to DAOY cells (Fig. 4C). Additionally, growth of DAOY-REST cells was less affected by UNC 0638 treatment compared to DAOY cells at all drug doses and exposure times, which was reflected in higher IC50 values of UNC 0638 for DAOY-REST cells relative to DAOY cells (Fig. 4D and Table 2). These observations indicate that elevation of REST protein increases the activity of G9a in complex with REST.
Figure 4. Elevated REST expression increases G9a-dependent histone methylation and target gene repression.
A. Western blotting analysis was performed with extracts from isogenic DAOY and DAOY-REST cells to determine the effect of REST elevation on global histone H3K9-me1, -me2, and -me3. Total histone H3 served as the loading control. B. Changes in G9a and REST-target gene expression were evaluated by Q-RT-PCR. C. ChIP assay were performed to demonstrate an increase in histone H3K9 -me1 and -me3 at USP37 promoter region-1 and -me1 at region-2 in DAOY-REST cells relative to DAOY cells. D. MTT assays were used to examine the effect of REST elevation in DAOY-REST cells on its response to G9a inhibition compared to DAOY cells. Solid black line indicates the IC50 value, which is also tabulated. (*=p<0.05, **=p<0.01, ****=p<0.0001, and n.s. = not significant). E. Live-cell nuclear staining assay was performed using DRAQ5 to distinguish between cytotoxic and cytostatic effects of UNC 0638 treatment on DAOY and DAOY-REST cells. F. Western blotting analysis was performed to measure the levels of the GLP target protein c-Myc following treatment of DAOY and DAOY-REST cells with 48hrs-IC50 doses of UNC 0638. β-actin was used as a loading control.
Table 2.
Comparison of IC50 values of UNC 0638 in low- and high-REST isogenic cells
| IC50 Values | |||
|---|---|---|---|
| Time (hrs) | 24 | 48 | 72 |
| DAOY (μM) | 2.7 | 1.1 | 1.1 |
| DAOY-REST (μM) | 3.6 | 2.5 | 1.8 |
Since MTT assay measures relative cell growth, the contribution of cell proliferation, differentiation or apoptosis to this reduction in cell growth could not be studied. Because USP37 de-repression and p27 stabilization would be expected to lead to terminal G1 cell cycle exit/arrest prior to neurogenesis, we also used a DRAQ5 live-cell nuclear staining assay to investigate the effect of G9a inhibition on the cell cycle transit of DAOY and DAOY-REST cells. Therefore, both cells were treated with DMSO or their respective 48hrs-IC50 doses of UNC 0638 for 12, 24 or 48 hrs and changes in their DNA contents were followed by flow cytometry. First, while DAOY cells displayed an S-G2/M-G1/G0 cell cycle profile at 12, 24 and 48 hrs respectively, DAOY-REST cells exhibited a G0/G1/S-S-S/G2/M/G1 shift at these time points, suggesting that REST elevation causes a delay in cell cycle transit (Fig. 4E, top and bottom panels). Second, when compared to control DMSO-treated DAOY cells, UNC 0638-treated cells accumulated with S and G2/M/sub-G1 DNA contents at 24 and 48 hrs respectively, suggesting a cell cycle delay and possibly arrest at G2/M, followed by induction of apoptosis in a small fraction of cells (Fig. 4E, top panel). In contrast, UNC 0638 treatment of DAOY-REST cells revealed transition of cells with a predominantly G1/G0/S DNA content at 12 hrs to a sub-G1/G1/S profile at 24 hrs and 48 hrs. A striking difference was the substantial increase in the number of DAOY-REST cells with sub-G1 DNA content, suggesting that REST elevation sensitizes these cells to apoptosis by UNC 0638 treatment. However, an arrest phenotype seen with a significant number of DAOY and DAOY-REST cells would suggest that the effect of G9a is cytostatic, and is consistent with a role for REST in controlling neuronal differentiation.
We next asked if UNC 0638 treatment affected the activity of G9a-like-protein (GLP), a heterodimeric partner for G9a, by measuring levels of its target protein c-Myc by Western blotting. As shown in Fig. 4F, c-Myc levels declined in both DAOY and DAOY-REST cells following exposure to their respective 48hrs-IC50 concentrations of the drug for 12 hrs, indicating that G9a inhibition also affected the activity of GLP in a REST-dependent manner.
Finally, to understand the molecular basis of resistance of DAOY-REST cells to growth inhibition by UNC 0638, these cells were treated with the drug under the same conditions that elicited a de-repression of REST target genes in DAOY cells. Despite the global decrease in histone H3K9 -me1, -me2 and -me3, treatment with UNC 0638 failed to upregulate the expression of REST target genes involved in neuronal differentiation (SYN1, TUBB3 and SCG10) and cell cycle control (USP37), although NANOG expression was elevated under these conditions (Figs. 5A and 5B). A decline in c-Myc levels was also not observed following treatment with 1.1 μM UNC 0638 (Fig. 5C). An analysis of histone H3K9 methylation at regions-1 and -2 of USP37 promoter following drug treatment showed an elevated retention (13.5- to 12.3-fold) of histone H3K9-me1 at regions-1 and -2 in DAOY cells compared to DAOY-REST cells. A 3-fold increased retention of histone H3K9-me3 was also seen at region-2 of the USP37 promoter, while the relative histone H3K9-me2 at regions-1 and -2 were significantly lower in DAOY cells when compared to DAOY-REST cells (Fig. 5D). Ectopic expression of the G9a-H1113K mutant in DAOY-REST cells resulted in decreased global histone H3K9-me3 only (Fig. 5E). Under these conditions, expression of NANOG and TUBB3 expression was de-repressed, whereas USP37 and SCG10 transcripts were unaffected relative to control vector transfected cells (Fig. 5F). Treatment of DAOY-REST cells with the IC50 dose of UNC 0638 used in previous experiments with DAOY cells did not significantly block tumor growth of DAOY-REST cells in vivo relative to animals implanted with untreated cells, suggesting that REST elevation associated increase in G9a activity may contribute to the decreased response of cells to UNC 0638 (Fig. 2E and data not shown).
Figure 5. REST elevation counters de-repression of target genes by G9a inhibition.
A. DAOY-REST cell extracts were subjected to Western blot analyses to measure levels of histone H3K9-me1, -me2, and -me3 following exposure to 1.1 μM UNC 0638 (or DMSO) for 12 hrs. Total histone H3 served as the loading control. B. Changes in the expression of G9a and REST-target genes in DAOY-REST cells treated with 1.1 μM UNC 0638 was shown by Q-RT-PCR. C. Western blotting analysis was performed to measure the levels of the GLP target protein c-Myc following treatment of DAOY and DAOY-REST cells with a 48hrs-IC50 dose of UNC 0638 (1.1 μM) for DAOY cells. β-actin was used as a loading control. D. ChIP assays were performed to demonstrate residual histone H3K9-me1, -me2, and -me3 within USP37 promoter regions-1 and -2 in DAOY-REST cells relative to DAOY cells following drug treatment. Data shown as fold change relative to input. E. Western blotting was performed to assess global changes in G9a-dependent histone H3K9-me1, -me2, and -me3 in DAOY-REST cells transduced with G9aH1113K or vector (control). Histone H3 was used as loading control. F. Q-RT-PCR to assess expression of known G9a and REST-target genes in DAOY-REST cells ectopically expressing mutant G9aH1113K. (*=p<0.05, **=p<0.01, ***=p<0.001, and n.s. = not significant).
Discussion
The application of next generation sequencing technologies to the analysis of human medulloblastoma samples has led to the identification of genomic aberrations such as copy number variations and recurrent mutations in these tumors (30–32). This has provided the basis for not only a new molecular classification of medulloblastomas, but also the foundation for functional studies in animal models (33,34). An interesting outcome of these high throughput studies is the uncovering of somatic mutations, amplifications and loss of chromatin modifying enzymes, implicating epigenetic perturbations in medulloblastoma genesis (35). However, this is an emerging field and the biological consequences of these changes, or how these events can be targeted for clinical application remain to be elucidated. Aberrations in post-translational modifications in medulloblastoma and their contribution to tumor development are even less understood, although work in many cancers has highlighted their critical role in oncology. Here, we have focused on dissecting the role of protein ubiquitination and its deregulation in medulloblastoma biology.
In our previous work, we showed that the DUB, USP37, plays a significant role in controlling medulloblastoma cell proliferation and onset of terminal differentiation (5). Work from previous groups has underscored the importance of the CDKI-p27, and its spatial and temporal elevation in granule cell progenitors (GCPs) in coordinating the switch between cell cycle exit and initiation of neuronal differentiation during normal cerebellar development (36–38). In human medulloblastoma samples, p27 is either expressed at low levels or absent when compared to normal cerebella. The regulation of p27 during normal post-natal cerebellar development and in pathological conditions such as medulloblastoma is not well understood. Our discovery that the downregulation or loss of USP37 in medulloblastoma cells and the consequent failure to stabilize p27 served to introduce the involvement of DUBs in medulloblastoma etiology (5).
Downregulation of USP37 and p27 observed in human medulloblastoma tumors and cell lines, as well as the consequent deregulation of proliferation in cell lines, suggest that USP37 may have a tumor suppressive function in this neural tumor (5). Consistent with this prediction, orthotopic transplantation assays revealed a loss of tumorigenic potential and Ki-67 staining in REST-positive medulloblastoma cells with ectopic USP37 expression. A similar tumor suppressive role for USP37 has been suggested by a recent report by Hernandez-Perez et al, who identified the licensing factor Cdt1 as a target of USP37 (39). Cdt1 in concert with Cdc18/CDC6 monitors origin firing and regulates DNA ploidy, and genomic integrity (40,41) Cdt1 levels are therefore tightly regulated and its stabilization by USP37 in late G1 and its degradation in S phase likely prevents re-initiation of DNA synthesis from the same origin and within a single cell cycle. USP37 was also recently identified as a facilitator of sister chromatid separation and mitotic progression in an RNAi screen (42). The assumption here would be that its loss during mitosis will lead to improper chromosome segregation and genomic instability.
However, the following studies ascribe an oncogenic role for USP37. USP37 is known to be necessary for the deubiquitination and stabilization of the cell cycle regulatory protein, cyclin A, which is critical for G1/S transition in HeLa and U2OS cells (17). The oncogene, c-Myc, is overexpressed in many cancers including lung cancer and this elevation occurs in part due its USP37-dependent stabilization (15,43). The retinoic acid receptor alpha (RARA) and the promyelocytic leukemia zinc finger (PLZF) fusion protein is a target of USP37 and the stabilization of PLZF and oncogenic transformation of acute promyelocytic leukemia cells has been attributed to USP37 (16). USP37 activity is believed to underlie the overexpression of 14-3-3γ protein and the consequent increase in proliferation of leukemia cells, providing support for an oncogenic role for USP37 (44).
The regulation of USP37 expression and activity involves transcriptional and post-transcriptional mechanisms in non-neural cells such as Hela (17,45). Our findings described here serve to highlight the contribution of epigenetics to USP37 regulation in neural cells. USP37 was identified by our group as a novel target of REST, a major regulator of brain development and neurogenesis, and whose expression is aberrantly elevated in human medulloblastoma tumors (5). REST controls the expression of target genes through one or both of its co-repressor complexes and in a context-dependent manner. The failure of the HDAC inhibitor, MS-275, to upregulate USP37 expression suggests that either HDAC1/2 activities are not necessary or are alone insufficient to de-repress USP37 expression (Fig. 2B).
However, our pharmacological and genetic data do implicate the Co-REST complex, and specifically G9a, in epigenetic regulation of USP37 expression. G9a catalyzes mono- and dimethylation of histone H3K9 and its HMT activity correlates with gene repression, which is in agreement with our observations with respect to USP37 (28,46,47). Interestingly, the USP37 promoter exhibited a significant level of tri-methylation of histone H3K9, which was considerably diminished following inhibition of G9a activity by UNC 0638 (Fig. 3D). In support of this, a decrease in histone H3K9 tri-methylation has been demonstrated at a subset of G9a target genes in cells treated with the G9a inhibitor, BIX-01294 (48). Although a direct role for G9a in promoting histone H3K9 tri-methylation cannot be excluded in our studies, G9a is known to exist in a complex with histone H3K9 tri-methylases Suv39H1 and SETDB1 and its heterodimeric partner GLP, in the context of specific genes (49). It is also well established that the activity of these tri-methylases can affect the expression of G9a target genes (49). Thus, a function link between G9a and these proteins at the USP37 promoter could potentially explain the observed elevation in histone H3K9 tri-methylation.
A second point of interest stems from our findings of a REST-dependent global increase in G9a-dependent histone H3K9 mono- and di- and tri-methylation as well as an augmentation of histone H3K9 mono- and tri-methylation at the USP37 promoter. A role for the zinc finger binding protein, WIZ, in the retention of G9a on chromatin has been suggested by a recent study by Simon and colleagues (50). Whether REST, also a zinc finger protein, promotes a similar retention of G9a at genes with RE1 elements remains to be evaluated. In this regard, it is important to note that WIZ isoforms are highly expressed in the cerebellum, and medulloblastoma is a disease of cerebellar origins (51). The G9a/GLP complex has been implicated in the regulation of c-Myc protein (29). The REST-dependent increase in GLP activity as measured by a decline in c-Myc protein levels in DAOY cells (Figs. 4F and 5C), not only connects REST to the G9a partner-GLP, but also provides the first link between REST and c-Myc, which are often co-elevated in human medulloblastoma (6).
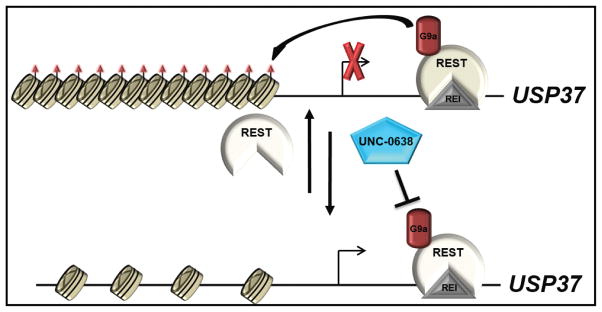
In summary, our study is the first to implicate G9a-dependent chromatin remodeling in the control of USP37 expression (Fig. 6). Blockade of G9a activity reactivates USP37 expression, which we previously showed as being necessary for stabilization of p27 and cessation of medulloblastoma cell proliferation, and impeded their tumorigenic potential in vivo (Fig. 6). We therefore suggest that G9a inhibitors may have therapeutic application, which warrants further evaluation in patient derived xenograft models.
Figure 6. Model to summarize the effect of G9a inhibition on USP37 gene expression.
REST and its associated chromatin remodelers including G9a, binds to a RE1 binding site in exon 1 of USP37 gene and represses its expression by di- and tri-methylation of histone H3K9 (orange triangles). From previous studies, USP37 is a p27 specific deubiquitylase, and its loss in REST-expressing medulloblastoma cells leads to p27 degradation and sustained proliferation. Inhibition of REST-associated G9a enzymatic activity by UNC 0638 de-represses USP37 gene expression by blocking histone H3K9 methylation at its promoter, causes an increase in p27 levels and promotes a blockade of tumor growth in mouse intracranial models.
Supplementary Material
Implications.
Reactivation of USP37 by G9a inhibition has the potential for therapeutic applications in REST-expressing medulloblastomas.
Acknowledgments
Financial Support: This work was supported by Grants from the National Institutes of Health (5R01-NS-079715-01), American Cancer Society (RSG-09-273-01-DDC) and Cancer Prevention Research Institute of Texas (CPRIT-RP150301) to V. Gopalakrishnan, The UT MD Anderson Cancer Center-CCE Scholar Program to T.H.W. Dobson, and Cancer Center Support Grant NCI # CA16672 to the Characterized Cell Line Core.
We thank Dr. Xiaobing Shi for providing reagents and Dr. Mark Bedford for helpful comments.
References
- 1.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta neuropathologica. 2012;123(4):473–84. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rusert JM, Wu X, Eberhart CG, Taylor MD, Wechsler-Reya RJ. SnapShot: Medulloblastoma. Cancer cell. 2014;26(6):940–e1. doi: 10.1016/j.ccell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta neuropathologica. 2012;123(4):465–72. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coluccia D, Figuereido C, Isik S, Smith C, Rutka JT. Medulloblastoma: Tumor Biology and Relevance to Treatment and Prognosis Paradigm. Current neurology and neuroscience reports. 2016;16(5):43. doi: 10.1007/s11910-016-0644-7. [DOI] [PubMed] [Google Scholar]
- 5.Das CM, Taylor P, Gireud M, Singh A, Lee D, Fuller G, et al. The deubiquitylase USP37 links REST to the control of p27 stability and cell proliferation. Oncogene. 2013;32(13):1691–701. doi: 10.1038/onc.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Molecular and cellular biology. 2006;26(5):1666–78. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor P, Fangusaro J, Rajaram V, Goldman S, Helenowski IB, MacDonald T, et al. REST is a novel prognostic factor and therapeutic target for medulloblastoma. Molecular cancer therapeutics. 2012;11(8):1713–23. doi: 10.1158/1535-7163.MCT-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nature medicine. 2000;6(7):826–31. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 9.Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nature genetics. 1998;20(2):136–42. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- 10.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80(6):949–57. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 11.McGann JC, Oyer JA, Garg S, Yao H, Liu J, Feng X, et al. Polycomb- and REST-associated histone deacetylases are independent pathways toward a mature neuronal phenotype. eLife. 2014;3:e04235. doi: 10.7554/eLife.04235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canzonetta C, Mulligan C, Deutsch S, Ruf S, O’Doherty A, Lyle R, et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. American journal of human genetics. 2008;83(3):388–400. doi: 10.1016/j.ajhg.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases from genes to organism. Physiological reviews. 2013;93(3):1289–315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 14.Jara JH, Frank DD, Ozdinler PH. Could dysregulation of UPS be a common underlying mechanism for cancer and neurodegeneration? Lessons from UCHL1. Cell biochemistry and biophysics. 2013;67(1):45–53. doi: 10.1007/s12013-013-9631-7. [DOI] [PubMed] [Google Scholar]
- 15.Pan J, Deng Q, Jiang C, Wang X, Niu T, Li H, et al. USP37 directly deubiquitinates and stabilizes c-Myc in lung cancer. Oncogene. 2015;34(30):3957–67. doi: 10.1038/onc.2014.327. [DOI] [PubMed] [Google Scholar]
- 16.Yang WC, Shih HM. The deubiquitinating enzyme USP37 regulates the oncogenic fusion protein PLZF/RARA stability. Oncogene. 2013;32(43):5167–75. doi: 10.1038/onc.2012.537. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Molecular cell. 2011;42(4):511–23. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 19.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Barakat K, Tuszynski J. Relaxed complex scheme suggests novel inhibitors for the lyase activity of DNA polymerase beta. Journal of molecular graphics & modelling. 2011;29(5):702–16. doi: 10.1016/j.jmgm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 21.D’Arcy P, Linder S. Proteasome deubiquitinases as novel targets for cancer therapy. The international journal of biochemistry & cell biology. 2012;44(11):1729–38. doi: 10.1016/j.biocel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Chang Y, Ganesh T, Horton JR, Spannhoff A, Liu J, Sun A, et al. Adding a lysine mimic in the design of potent inhibitors of histone lysine methyltransferases. Journal of molecular biology. 2010;400(1):1–7. doi: 10.1016/j.jmb.2010.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature chemical biology. 2011;7(8):566–74. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. Journal of neurosurgery. 2000;92(2):326–33. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]
- 25.Nomura M, Uda-Tochio H, Murai K, Mori N, Nishimura Y. The neural repressor NRSF/REST binds the PAH1 domain of the Sin3 corepressor by using its distinct short hydrophobic helix. Journal of molecular biology. 2005;354(4):903–15. doi: 10.1016/j.jmb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Stewart MD, Li J, Wong J. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Molecular and cellular biology. 2005;25(7):2525–38. doi: 10.1128/MCB.25.7.2525-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. The Journal of biological chemistry. 2006;281(13):8476–85. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. The Journal of biological chemistry. 2001;276(27):25309–17. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Yu Y, Liu F, Wei X, Wrobel JA, Gunawardena HP, et al. A chromatin activity-based chemoproteomic approach reveals a transcriptional repressome for gene-specific silencing. Nat Commun. 2014;5:5733. doi: 10.1038/ncomms6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23(34):8853–62. doi: 10.1200/JCO.2005.02.8589. [DOI] [PubMed] [Google Scholar]
- 31.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nature genetics. 2009;41(4):465–72. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ecke I, Petry F, Rosenberger A, Tauber S, Monkemeyer S, Hess I, et al. Antitumor effects of a combined 5-aza-2′deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer research. 2009;69(3):887–95. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 34.Patties I, Kortmann RD, Glasow A. Inhibitory effects of epigenetic modulators and differentiation inducers on human medulloblastoma cell lines. Journal of experimental & clinical cancer research : CR. 2013;32:27. doi: 10.1186/1756-9966-32-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones DT, Northcott PA, Kool M, Pfister SM. The role of chromatin remodeling in medulloblastoma. Brain pathology. 2013;23(2):193–9. doi: 10.1111/bpa.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayrault O, Zindy F, Rehg J, Sherr CJ, Roussel MF. Two tumor suppressors, p27Kip1 and patched-1, collaborate to prevent medulloblastoma. Molecular cancer research : MCR. 2009;7(1):33–40. doi: 10.1158/1541-7786.MCR-08-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatia B, Northcott PA, Hambardzumyan D, Govindarajan B, Brat DJ, Arbiser JL, et al. Tuberous sclerosis complex suppression in cerebellar development and medulloblastoma: separate regulation of mammalian target of rapamycin activity and p27 Kip1 localization. Cancer research. 2009;69(18):7224–34. doi: 10.1158/0008-5472.CAN-09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(31):11579–83. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez-Perez S, Cabrera E, Amoedo H, Rodriguez-Acebes S, Koundrioukoff S, Debatisse M, et al. USP37 deubiquitinates Cdt1 and contributes to regulate DNA replication. Molecular oncology. 2016;10(8):1196–206. doi: 10.1016/j.molonc.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown GW, Jallepalli PV, Huneycutt BJ, Kelly TJ. Interaction of the S phase regulator cdc18 with cyclin-dependent kinase in fission yeast. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(12):6142–7. doi: 10.1073/pnas.94.12.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gopalakrishnan V, Simancek P, Houchens C, Snaith HA, Frattini MG, Sazer S, et al. Redundant control of rereplication in fission yeast. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(23):13114–9. doi: 10.1073/pnas.221467598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeh C, Coyaud E, Bashkurov M, van der Lelij P, Cheung SW, Peters JM, et al. The Deubiquitinase USP37 Regulates Chromosome Cohesion and Mitotic Progression. Current biology : CB. 2015;25(17):2290–9. doi: 10.1016/j.cub.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 43.Sun XX, Sears RC, Dai MS. Deubiquitinating c-Myc: USP36 steps up in the nucleolus. Cell cycle. 2015;14(24):3786–93. doi: 10.1080/15384101.2015.1093713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JO, Kim SR, Lim KH, Kim JH, Ajjappala B, Lee HJ, et al. Deubiquitinating enzyme USP37 regulating oncogenic function of 14-3-3gamma. Oncotarget. 2015;6(34):36551–76. doi: 10.18632/oncotarget.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burrows AC, Prokop J, Summers MK. Skp1-Cul1-F-box ubiquitin ligase (SCF(betaTrCP))-mediated destruction of the ubiquitin-specific protease USP37 during G2-phase promotes mitotic entry. The Journal of biological chemistry. 2012;287(46):39021–9. doi: 10.1074/jbc.M112.390328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mozzetta C, Boyarchuk E, Pontis J, Ait-Si-Ali S. Sound of silence: the properties and functions of repressive Lys methyltransferases. Nature reviews Molecular cell biology. 2015;16(8):499–513. doi: 10.1038/nrm4029. [DOI] [PubMed] [Google Scholar]
- 47.Shankar SR, Bahirvani AG, Rao VK, Bharathy N, Ow JR, Taneja R. G9a, a multipotent regulator of gene expression. Epigenetics. 2013;8(1):16–22. doi: 10.4161/epi.23331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25(3):473–81. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37(1):46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 50.Simon JM, Parker JS, Liu F, Rothbart SB, Ait-Si-Ali S, Strahl BD, et al. A Role for Widely Interspaced Zinc Finger (WIZ) in Retention of the G9a Methyltransferase on Chromatin. The Journal of biological chemistry. 2015;290(43):26088–102. doi: 10.1074/jbc.M115.654459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matsumoto K, Ishii N, Yoshida S, Shiosaka S, Wanaka A, Tohyama M. Molecular cloning and distinct developmental expression pattern of spliced forms of a novel zinc finger gene wiz in the mouse cerebellum. Brain research Molecular brain research. 1998;61(1–2):179–89. doi: 10.1016/s0169-328x(98)00216-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.