Abstract
Cannabinoid pharmacology has been intensely studied because of cannabis’ pervasive medicinal and non-medicinal uses as well as for the therapeutic potential of cannabinoid-based drugs for the treatment of pain, anxiety, substance abuse, obesity, cancer and neurodegenerative disorders. The identification of allosteric modulators of the cannabinoid receptor 1 (CB1) has given a new direction to the development of cannabinoid-based therapeutics due to the many advantages offered by targeting allosteric site(s). Allosteric receptor modulators hold potential to develop subtype-specific and pathway-specific therapeutics. Here we briefly discuss the first-generation of allosteric modulators of CB1 receptor, their structure-activity relationships, signaling pathways and the allosteric binding site(s) on the CB1 receptor.
Keywords: CB1, allosteric modulator, biased signaling, therapeutic potential
1. Introduction
Cannabinoid receptors are considered to be key regulators of nausea, obesity (Bellocchio et al., 2006; DiMarzo and Després, 2009), pain (Manzanares et al., 2006; Sagar et al., 2009), anxiety, depression, substance use disorders (Mackie et al., 2006) and neurodegenerative disorders such as Alzheimer’s disease (Aso et al., 2014), and Parkinson’s disease (Brotchie et al., 2003). Despite exhaustive research, few cannabis-based therapeutics have reached clinical use, although nabilone (Cesamet) (Frank et al., 2008), dronabinol (Marinol) (Pertwee et al., 2006) and a Δ9-tetrahydrocannabinol (THC)/cannabidiol blend (Sativex) (Blake et al., 2006) are approved for the treatment of spasticity, nausea and pain in various countries. Conventional drug design has primarily targeted the orthosteric site of cannabinoid receptors, creating ligands that compete with the endogenous cannabinoid, anandamide and 2-arachidonoyl-glycerol (2-AG), for binding to this site. However, psychoactive side effects are frequent and often preclude clinical usefulness for ligands that target these sites on the CB1 cannabinoid receptor (CB1) which is expressed in high numbers in many of the regions of the brain (Mackie et al., 2006).It is now well appreciated that allosteric sites exist on many G-protein coupled receptors (GPCRs), including CB1 (Kenakin, 2012; Conn et al., 2014). Targeting these sites offers in principle several advantages such as greater subtype-selectivity (Conn et al., 2009), maintenance of spatial and temporal aspects of receptor activation and consequent attenuation of side effects (Conn et al., 2009; Burford et al., 2013). This manuscript focuses on the allosteric modulation of the CB1 receptor and gives a structural and functional review of its known allosteric modulators and their outcome.
2. Allosteric modulators
Recent research in GPCRs has shifted focus to identifying ligands that bind to a site(s) topographically distinct from where the endogenous ligands bind (orthosteric site). These sites are called allosteric sites and the ligands that bind these sites to modulate receptor activity are called allosteric modulators.
2.1 Types of allosteric modulators
There are four types of allosteric modulators:
Potentiators or positive allosteric modulators (PAMs): ligands that increase receptor function. The positive modulation can be seen by an increase in agonist affinity or efficacy (Gentry et al., 2015). PAMs can also function by blocking desensitization of the receptor (Burford et al., 2013).
Allosteric antagonists or negative allosteric modulators (NAMs): ligands that decrease receptor function through a decrease in agonist affinity or efficacy (Gentry et al., 2015).
Allosteric agonists (ago-allosterics or ago-agonists): allosteric compounds that display positive modulation in the absence of the orthosteric ligand (Gentry et al., 2015).
Neutral allosteric ligands (NALs): ligands that bind at the allosteric site but do not modulate receptor function (Gentry et al., 2015).
2.2 Detection and quantification of allostery
Many different binding or functional assays can be used to detect and quantify allostery at GPCRs. One such assay is equilibrium binding which identifies two important parameters for allostery: KB, the equilibrium dissociation constant which defines the affinity of an allosteric modulator for its receptor; and α, the cooperativity factor, which defines the magnitude and direction of impact the allosteric modulator and orthosteric ligand have on each other when both occupy the receptor. When α>1, the ligand is a positive allosteric modulator whereas when α<1, the ligand is a negative allosteric modulator. α=1 denotes no allosteric modulation. The two parameters are calculated based on an allosteric ternary complex model (Christopoulos et al., 2002; Price et al., 2005), according to the following equation:
where Y represents the fractional specific binding; KA and KB are the equilibrium dissociation constants for the orthosteric and allosteric ligands, respectively; [A] and [B] are the concentrations of the orthosteric and allosteric ligands, and α is the cooperativity factor. Another method to detect the above two parameters through binding assays is to perform kinetic assays to determine the impact of allosteric compounds on the association or dissociation of orthosteric ligands. PAMs increase the association kinetics or decrease the dissociation kinetics of orthosteric agonists. NAMs, on the other hand, increase the dissociation kinetics or decrease the association kinetics of orthosteric agonists (May et al., 2007).
Complementing ligand binding assays, functional assays such cAMP assays and calcium assays can also be performed to identify allosteric interactions of ligands with the orthosteric ligand. Functional assays allow readouts that can easily be translated into therapeutic effects exerted by the allosteric modulator on the signaling pathway of interest (May et al., 2007). To accommodate the impact of allosteric modulators on the efficacy of orthosteric ligands in functional assays, an operational model of allosterism is used which allows quantification of allosteric effects on both affinity and efficacy (Price et al., 2005; May et al., 2007). The model quantifies impact of allostery on binding of orthosteric ligands by a cooperativity factor α and the allosteric impact on efficacy by parameter β. Two other parameters, τA and τB, relate the intrinsic efficacy of the orthosteric ligand and the allosteric ligand, respectively, independent of the other ligand. A composite cooperativity factor logαβ defines the combined modulation effects on affinity and efficacy of orthosteric ligands. The following equation describes the operational model of allosterism (Gregory et al., 2010):]
where E and Em represents the effect and the maximum possible effect respectively and n represents the slope factor that governs the shape of the stimulus-response function. If the allosteric ligand has no intrinsic efficacy, the equation simplifies to the following (May et al., 2007; Gregory et al., 2010):
2.3 Therapeutic potential of allosteric modulators
Allosteric ligands have a number of potential advantages over their orthosteric counterparts as therapeutic agents. First, allosteric modulators may show more subtype specificity than orthosteric ligands do. Since most allosteric sites are not constrained by high evolutionary pressure to maintain endogenous ligand binding, they may not have high sequence identity across species or between subtypes. Thus targeting these sites could help develop subtype-specific drugs (Conn et al., 2009), but also emphasizes the challenges of identifying and characterizing allosteric modulators across species. Subtype specificity of allosteric modulators has been documented for many GPCRs, including adenosine receptors (Goblyos et al., 2011) and metabotropic glutamate receptors (Lindsley et al., 2004). Second, allosteric sites offer an opportunity to develop therapeutics for receptors where targeting the orthosteric site has not yielded successful drugs. For example, for the GLP-1 receptor, no small molecule orthosteric agonists have been identified, yet allosteric agonists have been reported (Knudsen et al., 2007; Burford et al., 2011). Third, unlike orthosteric drugs, “pure” allosteric modulators (not ago-allosteric modulators) may have an effect only when the endogenous ligand is present and thus maintain the temporal and spatial characteristics of endogenous signaling. PAMs, for example, may amplify endogenous signaling without affecting its temporal regulation. This could be of great importance especially with neurotransmitters where signal timing is very important (Conn et al., 2009; Burford et al., 2011). Fourth, orthosteric ligands may cause significant desensitization/downregulation of receptors due to constant receptor activation. Since allosteric activity may not be constant, or may not activate cellular pathways leading to desensitization and down regulation, in some instances these allosteric modulators offer potential for lesser receptor desensitization and/or behavioral tolerance (May et al., 2007). Fifth, allosteric activity often has a ceiling effect, and thus can be used to develop ligands with better adverse effect profiles than efficacious orthosteric ligands, where increasing concentrations may lead to undesirable adverse events (Wild et al., 2013).
3. Allosteric modulators of the CB1 receptor
Several allosteric modulators of CB1 have been identified or proposed. This has opened possibilities to develop subtype-specific ligands since many of the orthosteric ligands for CB1 alsobind CB2 with significant affinities. Some of the known allosteric modulators for CB1 include 5-chloro-3-ethyl-N-(4-(piperidin-1-yl)phenethyl)-1H-indole-2-carboxamide (ORG27569) (Price et al., 2005), 1-(4-chlorophenyl)-3-(3-(6-(pyrrolidin-1-yl)pyridin-2-yl)phenyl)urea (PSNCBAM-1) (Horswill et al., 2010), 3-(4-chlorophenyl)-5-(8-methyl-3-p-tolyl-8-azabicyclo[3.2.1]octan-2-yl)isoxazole (RTI-371) (Navarro et al., 2009), the endogenous ligand (5S,6R,9E,11Z,13E,15S)-5,6,15-trihydroxyicosa-9,11,13-trienoic acid (lipoxin A4) (Pamplona et al., 2012) and 3-(2-nitro-1-phenylethyl)-2-phenyl-1H-indole (GAT211) (as well as its (S-(-)) enantiomer, GAT229), all of which display positive allosteric modulation (PAMs) of CB1 under some conditions. Ligands that display negative allosteric modulation (NAMs) under some conditions have also been reported, including Pepcans (Bauer et al., 2012), cannabidiol (Laprairie et al., 2015), pregnenolone (Vallee et al., 2014), and, likely, GW405833 (Dhopeshwarkar et al., 2017). The most frequently encountered of these allosteric modulators are discussed below and some of the chemical structures are shown in Fig. 1.
Fig. 1. Some well-studied allosteric modulators of the CB1 receptor.
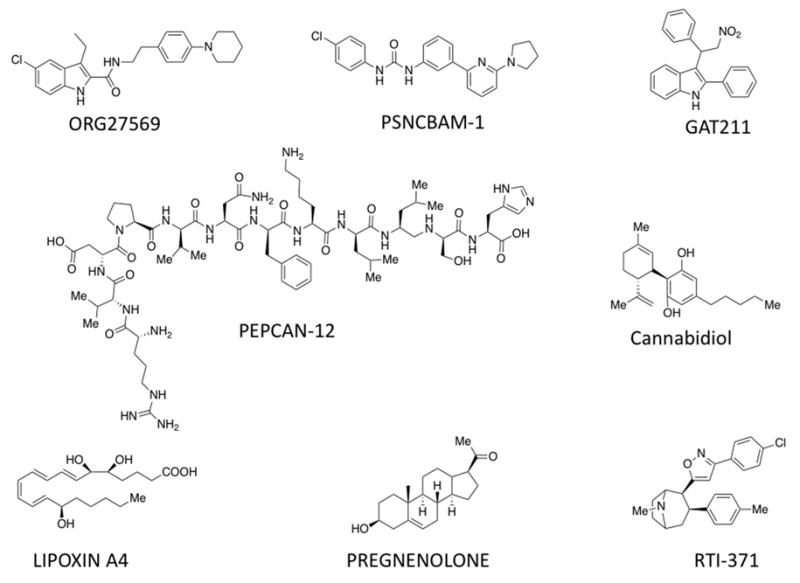
The figure shows the chemical structure of ORG27569, PSNCBAM-1, GAT211, Pepcan-12, Cannabidiol, Lipoxin A4, Pregnenolone and RTI-371. These compounds have been reported to have allosteric modulation of the CB1 receptor for different signaling pathways. The compounds pregnenolone, lipoxin A4 and pepcan-12 represent endogenous compounds; cannabidiol is naturally occurring and ORG27569, PSNCBAM-1, GAT211 and RTI-371 are synthesized compounds.
3.1 ORG27569
The compound ORG27569, developed by Organon (now merged with Merck), was among the first CB1 allosteric modulators to be identified and characterized. Since its discovery, the pharmacological profile of this molecule has been extensively studied. ORG27569 increases the binding of CP55,940, an orthosteric agonist of CB1 and decreases the binding affinity of SR141716A (Rimonabant), an inverse agonist of CB1, indicating that it acts as a PAM for the CB1 receptor (Price et al., 2005; Ahn et al., 2012; Khajehali et al., 2015). Paradoxically, ORG27569 decreased G-protein coupling induced by CP55,940 in membranes expressing the CB1 receptor (Price et al., 2005; Ross et al., 2007; Ahn et al. 2012; Khurana et al., 2014). To resolve this conundrum and address whether ORG27569 promotes an active state of the receptor or not, the Kendall group evaluated the impact of this compound on receptor internalization. Using the inactive mutant T210A CB1 (D’Antona et al., 2006), which primarily expresses CB1 at the cell surface, it was shown that co-treatment of ORG27569 with CP55,940 produced robust CB1 internalization compared to treatment with CP55,940 alone. Furthermore, ORG27569 induced receptor internalization even in the absence of CP55,940. This indicated that although ORG27569 inhibits G-protein coupling, it rapidly promotes a receptor conformation that increases its internalization (Ahn et al. 2012; Ahn et al. 2013).This is a challenging issue to examine since the wild-type CB1 is largely localized on intracellular vesicles in the absence of agonist. It is also consistent with constitutive activities (Leterrier et al., 2004) or in the presence of endogenous agonist (Howlett, A. et al., 2011), all of which may contribute to some of the seemingly confounding results (Cawston et al., 2013; Gamage et al., 2016) including recycling of internalized receptors. For example, Cawston et al. (2013) indicate that treatment with allosteric modulators ORG27569 and PSNCBAM-1 antagonized CP55,940 induced inhibition of cAMP levels and suggest that this allosteric modulator may cause the receptor to favor inactive conformations and traffic accordingly. These results further emphasize the complexity of GPCRs, their allosteric modulation, and the challenge of interpreting assays that may only provide one aspect of signaling rather than more global interconnecting phenomena.
Another important characteristic exhibited by allosteric modulators is ligand (or probe) dependence. The impact of ORG27569 on G-protein coupling activity with three classes of orthosteric agonists i.e. CP55,940 (classical cannabinoid), WIN55,212-2 (aminoalkylindole) and anandamide (endocannabinoid) was examined (Baillie et al., 2013). In GTPγS assays in mouse brain membranes, ORG27569 displayed a concentration-dependent decrease in Emax of G-protein coupling induced by CP55,940 (Price et al.,2005: Ahn et al., 2012; Baillie et al., 2013; Khurana et al., 2014), WIN55,212-2 and anandamide (Baillie et al., 2013). However, a lesser reduction in GTPγS levels was seen with WIN55,212-2-induced G-protein coupling levels than those with CP55,940 and anandamide (Baillie et al., 2013). This issue has practical implications, because while it is often easier to screen potential allosteric modulators by evaluating their impact on binding and signaling of potent synthetic cannabinoids, what matters most for their development as therapeutic agents is how they affect signaling of endogenous cannabinoids.
The effect of ORG27569 on CB1 agonist-mediated inhibition of forskolin-stimulated cAMP production in cell membranes expressing CB1 has also been reported (Baillie et al., 2013). ORG27569 completely abolished the inhibition of CP55,940-mediated inhibition of forskolin-stimulated cAMP production at 10 nM ORG27569 (Baillie et al., 2013; Khajehali et al., 2015). However, it was less effective in reducing WIN55,212-2-mediated inhibition of forskolin-stimulated cAMP production (Baillie et al., 2013).
3.1.1 Impact on signaling pathways by ORG27569
Kendall and colleagues evaluated the impact of ORG27569 on signaling including via phosphorylation of ERK1/2 and JNK1/2, which are members of the MAPK family (Ahn et al., 2012). Typically, activation of the CB1 receptor has been known to cause a G-protein mediated phosphorylation of JNK1/2 (Howlett 2005; Turu & Hunyady 2010). Treatment of CB1-expressing cells with CP55,940 resulted in phosphorylation of JNK1/2, which was abrogated by co-treatment with ORG27569. This is consistent with the G-protein coupling inhibition demonstrated by ORG27569 (Price et al.,2005: Ahn et al., 2012; Baillie et al., 2013; Khurana et al., 2014). Also, ERK1/2 can be phosphorylated by CB1 via G-protein dependent and independent pathways. Treatment of HEK293 cells expressing the CB1 receptor with ORG27569 showed an increase in phosphorylation of ERK1/2. This increase in ERK1/2 phosphorylation was not abrogated by treatment of cells with pertussis-toxin (PTX) indicating that ERK1/2 phosphorylation by ORG27569 is not G-protein mediated. The role of other signaling proteins such as β-arrestins in the G-protein-independent ERK1/2 phosphorylation was evaluated via siRNA co-transfection (Ahn et al., 2013). It was demonstrated that β-arrestin 1 mediates ERK1/2 phosphorylation by ORG27569 but other results have also been reported (Gamadge et al., 2016). This was the first indication that allosteric modulators such as ORG27569 hold promise to develop subtype- and pathway-selective CB1 therapeutics. ORG27569 demonstrates probe dependence in its impact on ERK1/2 phosphorylation since no significant effect was seen on WIN55,212-2 mediated ERK1/2 phosphorylation levels (Baillie et al., 2013). Using the mouse autaptic hippocampal culture model, Straiker and colleagues found that ORG27569 was also a negative allosteric modulator of 2-AG-mediated inhibition of glutamatergic neurotransmission (Straiker et al., 2015). Since ORG27569 may be a biased allosteric modulator (Ahn et al., 2013; Baillie et al., 2013; Khajehali et al., 2015), its analysis is especially challenging; competition due to affinity differences by a G-protein mediated orthosteric agonist and an arrestin–mediated allosteric modulator must be considered. Furthermore, binding of an allosteric modulator to a topological site that precludes G-protein coupling is also possible (Gentry et al., 2015; Stornaiuolo et al., 2015).
3.1.2 Structure-activity relationships of ORG27569
Several groups have investigated the key structural requirements of indole-2-carboxamides for allosteric modulation of CB1 receptors. Critical structural factors include the following:
C3 alkyl chain length is critical. An n-propyl chain substitution enhances the allosteric modulation of orthosteric ligand binding, whereas an n-hexyl chain enhances the affinity of the allosteric modulator for CB1 (Khurana et al., 2014). Substitution with short chain alkyl groups such as methyl improves allostery at CB1 in calcium mobilization assays (Nguyen et al., 2015).
Length of the linker between the amide bond and the phenyl ring is critical. Only an ethylene substitution is tolerated, and any increase or decrease in linker length results in complete loss of allosteric activity (Mahmoud et al., 2013; Khurana et al., 2014).
Electron-withdrawing substituent at the C5 position of the indole ring is important. A fluoro substitution at the C5 position demonstrates improved allostery over a chloro substitution (Nguyen et al., 2015).
Changes in substituents on the phenyl ring influences both the affinity and efficacy of allosteric modulation with a N,N-dimethyl amino group demonstrating improved allostery relative to piperidinyl moiety (Mahmoud et al., 2013; Khurana et al., 2014).
3.2 PSNCBAM-1
PSNCBAM-1 is an allosteric modulator that is structurally distinct from ORG27569. The two compounds show similar profiles for their impact on CP55,940 binding and its G-protein coupling, i.e. increased binding of CP55,940, together with antagonism of CP55,940 induced G-protein coupling (Horswill et al, 2010; Khurana et al., 2017). Similar to ORG27569, PSNCBAM-1 demonstrates biased signaling via β-arrestin 1 as seen in ERK1/2 phosphorylation studies (Khurana et al., 2017). Like ORG25769, PSNCBAM-1 behaves as an inhibitor of 2-AG-mediated inhibition of synaptic transmission (Straiker et al., 2015). In a rat model, PSNCBAM-1 decreased both food intake and body weight gain (Horswill et al., 2010).
3.2.1 Structure-activity relationships of PSNCBAM-1
Structure-activity relationships (SAR) have been established for PSNCBAM-1, which may be modified to improve allostery. German and colleagues (German et al., 2014) utilized calcium mobilization assays whereas Kendall and group utilized radioligand binding assays to generate a SAR for PSNCBAM-1 (Khurana et al., 2017). Key findings of the SAR include (1) Non-cyclic substitutions such as a dimethylamino group at the 2-pyrrolidinylpyridine position of PSNCBAM-1 are more favored at this position (German et al., 2014), and (2) there needs to be an electron-withdrawing substituent at the 4-chorophenyl position with a cyano group being more potent than a chloro group (German et al., 2014; Khurana et al., 2017). However, other electron-withdrawing groups such as COOH, CF3, acetyl or ethoxyacyl groups do not demonstrate high affinity or cooperativity in radioligand binding assays compared to the cyano group (Khurana et al., 2017). (3) The position of the electron-withdrawing group is critical. Change in the cyano group from para to meta position drastically reduced affinity and efficacy of the allosteric modulator (Khurana et al., 2017).
3.3 Pepcans
Pepcans are largely produced and released in the CNS by noradrenergic neurons (Hofer et al. 2015). Bauer and colleagues (Bauer et al., 2012), in their efforts to isolate CB1 binding peptides from mouse brain and mouse and human plasma samples, identified pepcans that demonstrated negative allosteric modulation of the CB1 receptor. A peptide Hpα (α-hemoglobin-derived peptide hemopressin) with the amino acid sequence PVNFKLSH was previously isolated from rat brain membranes and found to exhibit inverse agonist effects at the CB1 receptor (Heimann et al. 2007). Other peptides that were later identified were RVD-Hpα and VD-Hpα (Gomez et al., 2009). Bauer and colleagues generated monoclonal antibodies that identified the C-terminal regions of RVD-Hpα to isolate peptides that included N-terminally extended forms of RVD-Hpα that may bind the CB1 receptor and designated them as Pepcans-12 to 23 depending on the peptide length. In radioligand displacement assays and dissociation kinetic assays, Pepcan-12 demonstrated negative allosteric modulation of CP55,940 and WIN55,212-2 as shown by partial displacement of the orthosteric ligands and an increase in the dissociation rate constants.
3.3.1 Impact on signaling pathways by Pepcan-12
The impact of Pepcan-12 on G-protein-mediated pathways was investigated by GTPγS assays and cAMP assays. Pepcan-12, by itself, did not impact the basal G-protein coupling efficacy in mouse brain membrane preparations. However, the peptide decreases HU-210-induced G-protein coupling efficacy (Emax) by 20% (Bauer et al., 2012). To investigate the impact of Pepcan-12 on endocannabinoid-induced G-protein coupling, the modulation of 2-AG mediated G-protein coupling by Pepcans was investigated. Pepcan-12 completely abrogated the G-protein coupling efficacy of 80 nM 2-AG (Bauer et al., 2012) and substantially attenuated 2-AG mediated inhibition of glutamate release from hippocampal neurons (Straiker et al., 2015), thus demonstrating the negative allosterism of endocannabinoid signaling exhibited by these naturally occurring peptides.
In assays examining Gs-mediated cAMP accumulation by CB1 orthosteric ligands in a recombinant cell system, Pepcan-12 demonstrated concentration-dependent inhibition of cAMP accumulation efficacy by the synthetic agonist WIN55,212-2 and endocannabinoid 2-AG, thus further supporting that Pepcan-12 is a NAM of the CB1 receptor for this signaling pathway as well (Bauer et al., 2012).
3.4 Lipoxin A4
Lipoxin A4 (LXA4) is an endogenously produced CB1 receptor allosteric modulator. LXA4 demonstrated positive allosteric modulation of the CB1 receptor both in vitro and in vivo (Pamplona et al., 2012). It has been previously shown that in mouse models, intracerebroventricular injections of LXA4 elicits cannabimimetic catalepsy, which is blocked by treatment with the CB1 inverse agonist SR141716A (Pamplona at al., 2010). Displacement binding studies with SR141716A and cAMP accumulation assays were performed to evaluate direct binding of LXA4 to the CB1 receptor, which demonstrated that the effects of LXA4 were not consistent with orthosteric agonist activation of CB1 (Pamplona et al., 2012). To evaluate whether allosteric CB1 activation might explain the cannabimimetic effects of LXA4, this compound and subeffective doses of two endocannabinoids, anandamide or 2-AG, were injected in mice. The results showed that anandamide-induced catalepsy was potentiated by LXA4. Decreasing the in vivo synthesis of LXA4 by injecting 5-lipoxygenase (LOX) inhibitors resulted in loss of anandamide activity in vivo. In binding assays, LXA4 potentiated the binding of CP55,940 and WIN55212-2 with a higher modulation of CP55,940 binding than WIN55212-2, thus demonstrating probe dependence (Pamplona et al., 2012).
3.4.1 Impact on signaling pathways by Lipoxin A4
Not only does LXA4 impact orthosteric ligand binding but it also positively modulates Gi/o mediated inhibition of cAMP levels (Pamplona et al., 2012).LXA4 alone did not impact cAMP levels in concentrations between 0.1 nM and 1 μM. However, LXA4 positively modulates the potency of anandamide in decreasing the cAMP levels induced by forskolin. However, in another study, LXA4 failed to augment anandamide-induced suppression of cAMP levels (Khajeheli et al. 2015). 2-AG mediated inhibition of forskolin-induced cAMP was modestly attenuated by LXA4 (Pamplona et al., 2012), while LXA4 did not alter 2-AG mediated inhibition of glutamatergic synaptic transmission in cultured mouse hippocampal neurons (Straiker et al., 2015).
3.5 GAT211 and its enantiomers
GAT211 is an interesting racemic compound that has both direct agonist and positive allosteric properties. Examination of the two enantiomers of GAT211, GAT228 (R) and GAT229 (S), was able to assign allosteric agonism activity to GAT228 and PAM activity to GAT229 (Laprairie et al., 2017). GAT229 appears to be a PAM for a wide range of CB1 agonists, including anandamide, 2-AG, and CP55,940 (Laprairie et al., 2017).
3.6 Pregnenolone
Pregnenolone is best known as an intermediate in sterol synthesis (Miller & Auchus, 2011). However, a very interesting study found that THC increased pregnenolone levels, and that pregnenolone attenuated many of the actions of THC mediated by CB1 cannabinoid receptors. Conversely, preventing pregnenolone synthesis increased the potency and/or efficacy of THC in a range of behavioral tests (Vallee et al., 2014).Pharmacological investigations were consistent with allosteric modulation (i.e., no effect on equilibrium binding of WIN55,212-2 or CP55,940) in a biased fashion (i.e., suppressed THC inhibition of mitochondrial respiration and activation of ERK1/2 phosphorylation, but did not affect THC-mediated inhibition of cAMP accumulation). A subsequent study (Khajehali et al., 2015), also using hCB1-expressing CHO cells found that high concentrations of pregnenolone decreased equilibrium binding of SR141716A, but did not inhibit THC activation of ERK1/2 phosphorylation. Further investigations are needed to resolve this discrepancy.
3.7 Cannabidiol
Cannabidiol (CBD) is a cannabinoid compound present in varying quantities in cannabis preparations (ElSohly et al., 2016) and has attracted substantial recent interest as a possible therapeutic in some forms of pediatric epilepsy (Leo et al., 2016; Reddy 2017) as well as in schizophrenia (Leweke et al., 2012).In addition to its possible therapeutic use in pediatric epilepsy, CBD has long been studied as a modifier of THC actions (Niesink et al., 2013).The combination of CBD with THC was thoroughly explored during the development of nabiximols (Sativex), with the ~1:1 combination of CBD and THC found to be generally better tolerated and more efficacious than equivalent doses of THC alone (Wade et al., 2003; Russo et al., 2008; Johnson et al., 2010). Multiple mechanisms of action of CBD have been reported (see review: Campos et al., 2016), however recent studies suggest that CBD can act as a negative allosteric modulator of CB1 signaling (Laprairie et al., 2015).Negative allosterism of CB1 was observed in multiple systems and ligands at CBD concentrations between 500 nM and 1000 nM. Of note, in pediatric epilepsy patients receiving 20 mg/kg of CBD daily, plasma concentrations reach ~1000 nM (Wong et al., 2016).
4. Biased signaling by CB1 allosteric modulators
It is well appreciated that GPCR signaling is pluridimensional in nature (Fig. 2) and different ligands can evoke different receptor conformations that generate varied pharmacological responses (Luttrell et al., 2014). Some allosteric modulators such as ORG27569 and PSNCBAM-1 increase orthosteric agonist binding, yet decrease G-protein signaling by agonists. For example, ORG27569 is biased for β-arrestin-1 mediated pathways (Ahn et al., 2013). With the recent publication of crystal structures for inverse agonist-bound CB1 (Hua et al., 2016; Shao et al., 2016), it will be intriguing to elucidate the differences in conformation exhibited by CB1, when bound to ORG27569 and how this elicits biased signaling by this allosteric modulator.
Fig. 2. Schematic depiction of allosteric modulation of a GPCR.
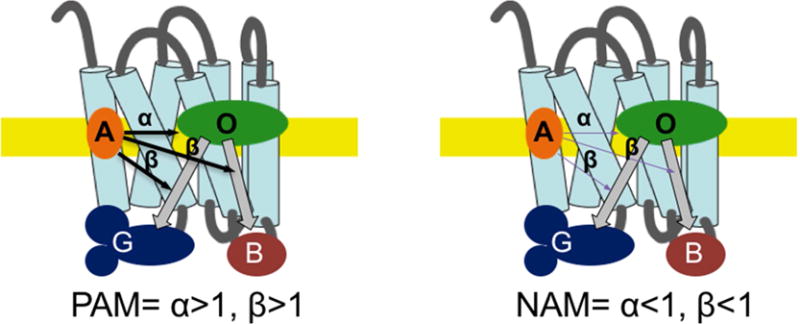
Classically, an orthosteric agonist (O) binds to the GPCR and facilitates signaling through several G protein (only one shown for clarity) and non-G protein pathways (G and B, respectively). The relative strength of signaling often varies among signaling pathways (functional selectivity).An allosteric modulator (A) will bind to the receptor at a site independent from the orthosteric site. The allosteric modulator may positively (PAM) or negatively (NAM) influence orthosteric ligand binding (α) and/or signaling pathways (β), as depicted by thick and thin arrows and discussed in the text.
As one way to address this question, Farrens and colleagues utilized site directed fluorescence labeling techniques to investigate changes in transmembrane helix (TM) movements upon binding of ORG27569 (Fay et al., 2015). They discovered that upon binding to the CB1 receptor, the outward TM6 movements that are necessary to activate the receptor by exposing the binding site crevice for G-protein are blocked by ORG27569, thus explaining why ORG27569 inhibits G-protein coupling (Fay et al., 2012). Further, the group investigated movements in helix 8 (H8) upon ORG27569 binding. Biased signaling by β-arrestins has been proposed previously to involve movements in H8 to allow arrestin binding. Changes in fluorescence upon ORG27569 binding demonstrated increased H8 and TM7 movements, which are not observed with the orthosteric inverse agonist SR141716A. The authors, thus concluded that ORG27569 induces a conformation different from an inactive state conformation with blocked TM6 movements that explain the inhibition of G-protein coupling and high H8 and TM7 movements that may explain the biased β-arrestin signaling by ORG27569 (Fay et al., 2015). It will be interesting to determine if other β-arrestin-biased conformations including those induced by orthosteric ligands (Franks et al., 2014; Laprairie et al., 2016), result in a similar conformation.
5. Allosteric binding site(s) on the CB1 receptor?
With the many allosteric modulators identified for the CB1 receptor so far, one of the most important developments was the identification of the site where some of these allosteric modulators may bind. Identification of the allosteric binding site(s) allows an understanding of the mechanisms by which allosteric modulators such as ORG27569 display a contradictory profile i.e it increases CP55,940 binding while decreasing G-protein coupling associated with CP55,940 (Price et al., 2005; Ahn et al., 2012). Identifying the allosteric binding pockets will also allow structure-based drug design for modulators with improved allostery at the CB1 receptor.
Different research groups have attempted to identify the putative allosteric binding sites of the CB1 receptor. Such sites, however, do not appear to overlap. Fay and colleagues (Fay et al., 2012) investigated how different conformations of the receptor are induced by ORG27569, which could explain the molecular pharmacology of this agent, including receptor activation, enhancement of CP55,940 binding, together with inhibition of G-protein coupling. Site-specific fluorescent labeling of the receptor was done to identify conformational changes in CB1 upon binding of ORG27569. Further investigations by this group identified a disulfide bond in the N-terminus of the receptor that plays an important role in the allosteric effects seen with ORG27569 and PSNCBAM-1. The disulfide bond was formed between Cys98 and Cys107. Interestingly, these authors found that reduction of this amino-terminal disulfide bond enhanced the cooperativity effects seen with ORG27569 and PSNCBAM-1. The authors suggested that allosteric modulators may bind to this region and alter the dissociation of the orthosteric ligand CP55,940, thus enhancing CP55,940 binding (Fay et al., 2013). However, the studies lacked direct evidence to show the impact of the disulfide bridge on binding affinities of the allosteric ligands, suggesting that the disulfide bond may play an indirect effect in the allostery at CB1 receptor.
Shore and colleagues utilized molecular modeling, mutagenesis and G-protein coupling assays to investigate the allosteric binding pocket for ORG27569 (Shore et al., 2013). The approach for the identification of the allosteric site was based on the observation that ORG27569, in ligand displacement assays, decreases the equilibrium binding of SR141716A, an inverse agonist of CB1 receptor. This may be because the receptor active state is enhanced and/or because ORG27569 may have an overlapping binding site with SR141716A. Computational modeling of ORG27569 on the CB1 receptor was done with docking the ligand manually in the TMH3-6-7 region. Modeling data suggested that the binding site for ORG27569 overlaps with the binding of SR141716A. Residues K1923.28, F2003.36, W2795.43 and W3566.48, which have previously been reported to be important for SR141716A binding were mutated to alanine and tested for their impact on G-protein coupling efficacy of CP55,940 with ORG27569. Only the K192A mutant showed an increase in G-protein coupling in presence of the ORG27569 compared to CB1 wild-type, suggesting this amino acid plays an important role in the binding of ORG27569. The computational modeling suggested that the piperidine nitrogen of ORG27569 is involved in forming hydrogen bonding with K192. This was the first attempt at identification of the allosteric site for the CB1 receptor. However, since this residue is conserved between CB1 and CB2, this mutation does not describe why ORG27569 binds to CB1 and not CB2. The computational modeling also predicts F268 from the EC2 loop to form aromatic stacking interactions with the indole-ring of ORG27569. No mutagenesis studies were reported for this residue. Interestingly, other class A GPCRs have also demonstrated the role of extracellular loops in the binding of allosteric modulators. C7/3-phth, for example, a negative allosteric modulator of the orthosteric antagonist N-methylscopolamine (NMS), for M2 muscarinic receptors exhibited cationic-Π interactions between the ammonium groups on the ligand and the aromatic residues in the extracellular ligand-binding vestibule, in molecular dynamic simulations. These simulations suggested two mechanisms for cooperativity between the orthosteric and allosteric ligands. First, electrostatic repulsions between the cationic orthosteric ligand NMS and C7/3-phth decrease the binding affinity of one in the presence of other. Second, the presence of one ligand affects the shape of the binding site of the other, thereby controlling the cooperativity effects. For example, the wide and bulky PAM alcuronium demonstrated that when bound to the receptor, it exhibits a conformation with wide open orthosteric and allosteric binding sites. However, the NAM C7/3-phth demonstrates extensive binding contacts in a closed conformation, causing a closed conformation of the orthosteric site (Dror et al., 2013; Langmead et al., 2014).
Stornaiuolo and colleagues identified an entirely different site of binding for ORG27569 (Stornaiuolo et al., 2015). The group utilized computational modeling techniques using the sphingosine 1-phosphate receptor to build a homology model of the CB1 receptor and to identify five different possible allosteric binding sites. Out of all the sites evaluated, the few residues that were different in CB1 and CB2 were mutated in CB1 to the corresponding residues in CB2 receptor. Fluorescence binding assays were done using a fluorescent tetra-methyl-rhodamine (TAMRA) labeled form of the CB1 inverse agonist, AM251, namely T1117. It has been previously demonstrated by the same group (Bruno et al., 2014) that binding of T1117 to CB1 wild-type is negatively impacted by ORG27569. Loss of T1117 binding would suggest that the mutations are not important for ORG27569 binding. If however, no change in T1117 binding is seen in the presence of ORG27569, the mutation is considered important for ORG27569 binding. Photoactivatable analogs of ORG27569 were also utilized to crosslink with CB1 and residues important for binding were then evaluated based on mass spectrometry. The authors identified an intracellular binding site for ORG27569 (Stornaiuolo et al., 2015). This was in contrast to the previously identified more extracellular site(s) identified. Mutational analysis identified three mutations C1.55Y, H2.41L and F4.46L (P2 in the intracellular regions of TM1, 2 and 4), that demonstrated no loss of T1117 binding upon incubation with ORG27569, suggesting these residues to be important for binding of ORG27569. The mass spectrometry data identified two serines, S2.45 and S3.42, to be important for the binding of ORG27569.
Thus, overall, different researchers have identified different sites of binding of ORG27569 and none agree with each other (Fay et al., 2012; Shore et al., 2013; Stornaiuolo et al., 2015) or perhaps there is more than one site. Also, no investigations into the binding site for PSNCBAM-1 have been reported.
Nonetheless, these studies lay the foundation for the identification of the putative allosteric binding site(s) on the CB1 receptor. With the recently published crystal structures of the CB1 receptor bound to inverse agonists (Hua et al., 2016; Shao et al., 2016), the community awaits for crystal structures of CB1 bound to allosteric modulators and for advanced computational studies (e.g. molecular dynamics simulations) integrating crystal structure information with site-directed mutagenesis which may shed new light on the allosteric binding sites on the CB1 receptor. Alternatively, photoactivatable analogues of indole-2-carboxamides such as ORG27569 with functionalities such as benzophenone, aliphatic or phenyl azide and phenyltrifluoromethyldiazrine have also been reported which may provide novel tools for mapping the allosteric binding site(s) on the CB1 receptor (Qiao et al., 2016).
6. Therapeutic potential of allosteric modulators of the CB1 receptor
It is becoming evident that CB1 allosteric modulators may demonstrate therapeutic potential. PSNCBAM-1, for example, elicits acute hypophagic effects (Horswill et al., 2007) and antagonizes neuronal excitability (Wang et al., 2010), properties which may lead to treatments of obesity and some central nervous system (CNS) disorders. Pregnenolone has demonstrated promise in blocking the psychotic-like symptoms such as THC-impaired cognitive function (Busquets-Garcia et al., 2017).Lipoxin A4 has been shown to protect neuronal cells from β-amyloid-induced neurotoxicity (Pamplona et al., 2012), thus demonstrating possible therapeutic potential for the treatment of Alzheimer’s disease. Moreover, the PAM ZCZ011, enhances agonist binding and attenuates neuropathic pain in a mouse model without cannabimimetic side effects (Ignatowska-Jankowska et al., 2015). Similarly, in a chemotherapy-induced painful neuropathy model, the PAM GAT211 produced CB1-mediated analgesia, without tolerance or CB1-mediated dependence, even with repeated dosing (Slivicki et al., 2016). The potential therapeutic impact of ORG27569 remains unsettled. ORG27569 reduced food consumption in both CB1 (−/−) and CB1 (+/+) mice, questioning the role of CB1 receptors in the hypophagic effects seen with ORG27569. Similarly, the cataleptic, antinociceptive and hypothermic effects associated with THC or CP55,940 were not altered by administration of ORG27569 which raises issues of translation of pharmacologic effects seen at a molecular level to whole animals (Gamage et al., 2014; Ding et al., 2014). On the other hand, ORG27569 resulted in dose-dependent decreases in both cue- and drug-induced reinstatement of cocaine- and methamphetamine-seeking behavior in rat models, thus demonstrating that functional antagonism of CB1 receptors by ORG27569 may hold promise for developing effective therapeutics for drug addiction (Jing et al., 2014).
With increasing evidence of some allosteric modulators of CB1 such as indole-2-carboxamides demonstrating biased signaling, manipulation of selective signaling pathways in a spatially and temporally restricted fashion can be achieved and this may be therapeutically beneficial. An example of this potential is shown by the angiotensin II receptor. This receptor may exhibit G-protein dependent or β-arrestin-dependent signaling. Biased agonism of this receptor to promote β-arrestin mediated effects may provide a beneficial cytoprotective response and obviate a deleterious increase in blood pressure that is observed when this receptor signals in a G-protein manner (Kenakin et al., 2010). This offers tremendous opportunities for developing precision drugs for many disorders that involve CB1 receptors.
7. Future considerations
The identification of allosteric modulators of the CB1 receptor (Fig. 3) has provided new opportunities to develop therapeutics that are subtype-specific and in some cases, pathway-specific. This allows an appreciation of the pluridimensional nature of GPCR signaling and how different ligands can induce different conformations and this differential signaling can be utilized for the treatment of human diseases. Further identification of the allosteric binding site(s) can be pursued as it will further help in the strategic design of small molecules for lead identification and optimization, which is currently hindered by the paucity of reliable models of the CB1 receptor.
Fig. 3. Allosteric compounds for the CB1 receptor and their classification as PAMs or NAMs or agonists based on activity.
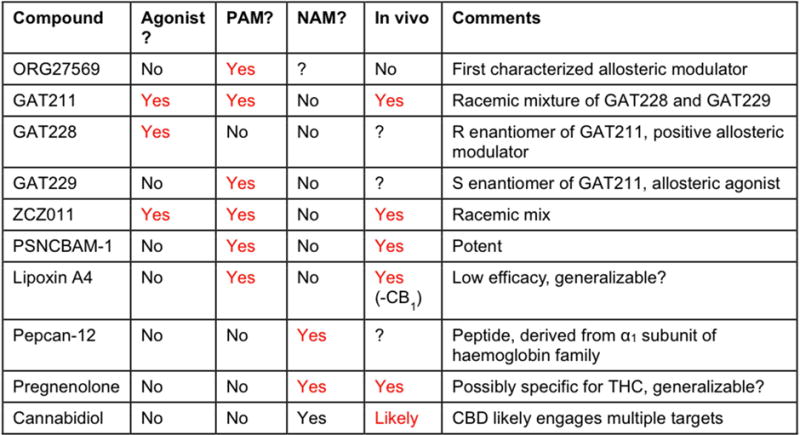
Since a compound can show positive allosterism for one pathway and negative or no allosteric activity for others, defining the status of a compound as a PAM or NAM or agonist is challenging. For example, ORG27569 demonstrated positive allosteric modulation for agonist CP55,940 binding and was internalized as expected for an activated receptor (Price et al., 2005; Ahn et al., 2012) and thus may be classified as a PAM. However, this is an inhibitor of G-protein coupling (Price et al., 2005; Mahmoud et al., 2013) and demonstrates negative modulation of CP55,940 induced calcium mobilization (Nguyen et al., 2015) and thus is also reported as a NAM. Similarly, PSNCBAM-1 demonstrated PAM activity for CP55,940 binding (Horswill et al., 2007) and CP55,940 induced ERK1/2 phosphorylation (Khurana et al., 2017). However, it demonstrates negative modulation of G-protein coupling (Horswill et al., 2007) and CP55,940 induced calcium mobilization (German et al., 2014) and thus acts as inhibitor for these signaling pathways, emphasizing that different signaling pathways can be modulated in opposite directions by an allosteric modulator. Thus, when speaking of an allosteric modulator it is helpful to specify the pathway(s) being discussed. The In Vivo column identifies if the allosteric modulator has in vivo activity consistent with allosteric modulation. The Comments column highlights particularly notable aspects of the allosteric modulator. See text for details. Abbreviations: AM, allosteric modulator.
HIGHLIGHTS.
Modulation of the CB1 receptor by allosteric modulators is an area of intense research.
Receptor subtype and pathway specificity can be achieved by allosteric modulators of the CB1 receptor.
Different allosteric modulators and their impact on signaling pathways is revealed though complexities are apparent.
Putative allosteric binding site(s) on the CB1 receptor are examined.
Acknowledgments
This work was supported in part by NIH Grant DA039942 (to D.A.K.), DA021696 (to K.M.), DA012413 and DP1DA031387 (to D.P.).
Abbreviations
- CB1
cannabinoid receptor one
- GPCR
G-protein coupled receptors
- PAM
positive allosteric modulators
- NAM
negative allosteric modulators
- SAM
silent allosteric modulators
- PTX
pertussis-toxin
- 2-AG
2-arachidonylglycerol
- LXA4
lipoxin A4
- LOX
5-lipoxygenase
- TM
transmembrane helices
- CNS
central nervous system
- THC
Δ9-tetrahydrocannabinol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahn KH, Mahmoud MM, Kendall DA. Allosteric modulator ORG27569 induces CB1 cannabinoid receptor high affinity agonist binding state, receptor internalization, and Gi protein-independent ERK1/2 kinase activation. J Biol Chem. 2012;287:12070–12082. doi: 10.1074/jbc.M111.316463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn KH, Mahmoud MM, Shim JY, Kendall DA. Distinct roles of beta-arrestin 1 and beta-arrestin 2 in ORG27569-induced biased signaling and internalization of the cannabinoid receptor 1 (CB1) J Biol Chem. 2013;288:9790–9800. doi: 10.1074/jbc.M112.438804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett A, Reggio PH, Childers SR, Hampson RE, Ulloa NM, Deutsch DG. Endocannabinoid tone versus constitutive activity of cannabinoid receptors. Br J Pharmacol. 2011;163:1329–1343. doi: 10.1111/j.1476-5381.2011.01364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso E, Ferrer I. Cannabinoids for treatment of Alzheimer’s disease: moving toward the clinic. Front Pharmacol. 2014;5:37. doi: 10.3389/fphar.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie GL, Horswill JG, Anavi-Goffer S, Reggio PH, Bolognini D, Abood ME, McAllister S, Strange PG, Stephens GJ, Pertwee RG, Ross RA. CB(1) receptor allosteric modulators display both agonist and signaling pathway specificity. Mol Pharmacol. 2013;83:322–338. doi: 10.1124/mol.112.080879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M, Chicca A, Tamborrini M, Eisen D, Lerner R, Lutz B, Poetz O, Pluschke G, Gertsch J. Identification and quantification of a new family of peptide endocannabinoids (Pepcans) showing negative allosteric modulation of CB1 receptors. J Biol Chem. 2012;287:36944–36967. doi: 10.1074/jbc.M112.382481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellocchio L, Mancini G, Vicennati V, Pasquali R, Pagotto U. Cannabinoid receptors as therapeutic targets for obesity and metabolic diseases. Curr Opin Pharmacol. 2006;6:586–591. doi: 10.1016/j.coph.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Blake DR. Preliminary assessment of the efficacy, tolerability and safety of a cannabis-based medicine (Sativex) in the treatment of pain caused by rheumatoid arthritis. Rheumatology. 2006;45:50–52. doi: 10.1093/rheumatology/kei183. [DOI] [PubMed] [Google Scholar]
- Brotchie JM. CB1 cannabinoid receptor signalling in Parkinson’s disease. Curr Opin Pharmacol. 2003;3:54–61. doi: 10.1016/s1471-4892(02)00011-5. [DOI] [PubMed] [Google Scholar]
- Burford NT, Clark MJ, Wehrman TS, Gerritz SW, Banks M, O’Connell J, Traynor JR, Alt A. Discovery of positive allosteric modulators and silent allosteric modulators of the μ-opioid receptor. Proc Natl Acad Sci USA. 2013;110:10830–10835. doi: 10.1073/pnas.1300393110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burford NT, Watson J, Bertekap R, Alt A. Strategies for the identification of allosteric modulators of G-protein-coupled receptors. Biochem Pharmacol. 2011;81:691–702. doi: 10.1016/j.bcp.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Busquets-Garcia A, Soria-Gomez E, Redon B, Mackenbach Y, Vallee M, Chaouloff F, Varilh M, Ferriera G, Piazza PV, Marsciano G. Pregnenolone blocks cannabinoid-induced acute psychotic-like states in mice. Mol Psychiatry advanced online publication. 2017 doi: 10.1038/mp.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AC, Foqaca MV, Sonego AB, Guimaraes FS. Cannabidiol, neuroprotection and neuropsychiatric disorders. Pharmaol Res. 2016;112:119–127. doi: 10.1016/j.phrs.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Cawston EE, Redmond WJ, Breen CM, Grimsey NL, Connor M, Glass M. Real-time characterization of cannabinoid receptor 1 (CB1) allosteric modulators reveals novel mechanism of action. Bri J Pharmacol. 2013;170:893–907. doi: 10.1111/bph.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos A, Kenakin T. G protein-coupled receptor allosterism and complexing. Pharmacol Rev. 2002;54:323–374. doi: 10.1124/pr.54.2.323. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discov. 2009;8:41–54. doi: 10.1038/nrd2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PJ, Lindsley CW, Meiler J, Niswender CM. Opportunities and challenges in the discovery of allosteric modulators of GPCRs for treating CNS disorders. Nat Rev Drug Discov. 2014;13:692–708. doi: 10.1038/nrd4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Antona DM, Ahn KH, Kendall DA. Mutations of CB1 T210 produces active and inactive receptor forms: correlations with ligand affinity, receptor stability, and cellular localization. Biochem. 2006;45:5606–5617. doi: 10.1021/bi060067k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhopeshwarkar A, Murataeva N, Makriyannis A, Straiker A, Mackie K. Two Janus Cannabinoids That Are Both CB2 Agonists and CB1 Antagonists. J Pharmacol Exp Ther. 2017;360:300–311. doi: 10.1124/jpet.116.236539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Qiu Y, Jing L, Thorn DA, Zhang Y, Li JX. Behavioral effects of the cannabinoid CB1 receptor allosteric modulator ORG27569 in rats. Pharmacol Res Perspect. 2014;2:e00069. doi: 10.1002/prp2.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMarzo V, Depré s J-P. CB1 antagonisats for obesity—what lessons have we learned from rimonabant? Nat Rev Endocrinol. 2009;5:633–638. doi: 10.1038/nrendo.2009.197. [DOI] [PubMed] [Google Scholar]
- Dror RO, Green HF, Valant C, Borhani DW, Valcourt JR, Pan AC, Arlow DH, Canals M, Lane JR, Rahmani R. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;84:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- ElSohly MA, Mehmedic Z, Foster S, Gon C, Chandra S, Church J. Changes in Cannabis potency over the last 2 decades (1995–2014): Analysis of current data in the United States. Biol Psychiatry. 2016;79:613–619. doi: 10.1016/j.biopsych.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JF, Farrens DL. A key agonist-induced conformational change in the cannabinoid receptor CB1 is blocked by the allosteric ligand Org 27569. J Biol Chem. 2012;287:33873–33882. doi: 10.1074/jbc.M112.352328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JF, Farrens DL. The membrane proximal region of the cannabinoid receptor CB1 N-terminus can allosterically modulate ligand affinity. Biochem. 2013;52:8286–8294. doi: 10.1021/bi400842k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JF, Farrens DL. Structural dynamics and energetics underlying allosteric inactivation of the cannabinoid receptor CB1. Proc Natl Acad Sci USA. 2015;112:8469–8474. doi: 10.1073/pnas.1500895112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank B, Serpell MG, Hughes J, Matthews JNS, Kapur D. Comparison of analgesic effects and patient tolerability of nabilone and dihydrocodeine for chronic neuropathic pain, randomized, crossover, double blind study. BMJ. 2008;336:199–201. doi: 10.1136/bmj.39429.619653.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks LN, Ford BM, Madidi NR, Penthala NR, Crooks PA, Prather PL. Characterization of the intrinsic activity for a novel class of cannabinoid receptor ligands: Indole quinuclidine analogs. Eur J Pharmacol. 2014;737:140–148. doi: 10.1016/j.ejphar.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage TF, Anderson JC, Abood ME. CB1 Allosteric modulaor ORG27569 is an antagonist/inverse agonist of ERK1/2 signaling. Cannabis and Cannabinoid Research. 2016;1:272–280. doi: 10.1089/can.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage TF, Ignatowska-Jankowska BM, Wiley JL, Abdelrahman M, Trembleau L, Greig IR, Thakur GA, Tichkule R, Poklis J, Ross RA, Pertwee RG, Lichtman AH. In-vivo pharmacological evaluation of the CB1-receptor allosteric modulator-ORG27569. Behav Pharmacol. 2014;25:182–185. doi: 10.1097/FBP.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry PR, Sexton PM, Christopoulos A. Novel allosteric modulators of G-protein coupled receptors. J Biol Chem. 2015;290:19478–19488. doi: 10.1074/jbc.R115.662759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German N, Decker AM, Gilmour BP. Diarylureas as Allosteric Modulators of the Cannabinoid CB1 Receptor: Structure–Activity Relationship Studies on 1-(4-Chlorophenyl)-3-{3-[6-(pyrrolidin-1-yl)pyridin-2-yl]phenyl}urea (PSNCBAM-1) J Med Chem. 2014;57:7758–7769. doi: 10.1021/jm501042u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goblysos A, Ijzerman AP. Allosteric modulation of adenosine receptors. Biochim et Biophy Acta. 2011;1808:1309–1318. doi: 10.1016/j.bbamem.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Gomes I, Grushko JS, Golebiewska U, Hoogendoorn S, Gupta A, Heimann AS, Ferro ES, Scarlata S, Fricker LD, Devi LA. Novel endogenous peptide agonists of cannabinoid receptors. FASEB J. 2009;23:3020–3029. doi: 10.1096/fj.09-132142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory KJ, Sexton PM, Christopoulos A. Overview of receptor allosterism. Curr Protoc Pharmacol. 2010;51:1.21.1–1.21.34. doi: 10.1002/0471141755.ph0121s51. [DOI] [PubMed] [Google Scholar]
- Heimann A S, Gomes I, Dale CS, Pagano RL, Gupta A, de Souza LL, Luchessi AD, Castro LM, Giorgi R, Rioli V, Ferro ES, Devi LA. Hemopressin is an inverse agonist of CB1 cannabinoid receptors. Proc Natl Acad Sci USA. 2007;104:20588–20593. doi: 10.1073/pnas.0706980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer SC, Ralvenius WT, Gachet MS, Fritschy J, Zeilhofer HU, Gertsch J. Localization and production of peptide endocannabinoids in the rodent CNS and adrenal medulla. Neuropharmacol. 2015;98:78–89. doi: 10.1016/j.neuropharm.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Horswill JG, Bali U, Shaaban S, Keily JF, Jeevaratnam P, Babbs AJ, Reynet C, Wong Kai In P. PSNCBAM-1, a novel allosteric antagonist at cannabinoid CB1 receptors with hypophagic effects in rats. Br J Pharmacol. 2007;152:805–814. doi: 10.1038/sj.bjp.0707347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett AC. Cannabinoid receptor signaling. Handb Exp Pharmacol. 2005;168:53–79. doi: 10.1007/3-540-26573-2_2. [DOI] [PubMed] [Google Scholar]
- Hua T, Vemuri K, Mengchem P, Qu L, Han GW, Wu Y, Zhao S, Shui W, Li S, Korde A, Laprairie RB, Stahl EL, Ho J-H, Zvonok N, Zhou H, Kufareva I, Wu B, Zhao Q, Hanson MA, Boh LM, Makriyannis A, Stevens RC, Liu Z-J. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell. 2016;167:750–762. doi: 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatowska-Jankowska BM, Baillie GL, Kinsey S, Crowe M, Ghosh S, Owens RA, Damaj IM, Poklis J, Wiley JL, Zanda M, Zanato C, Greig IR, Lichtman AH, Ross RA. A cannabinoid CB1 receptor-posive allosteric modulator reduces neuropathic pain in the mouse with no psychoactive effects. Neuropsychopharmacology. 2015;40:2948–2959. doi: 10.1038/npp.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing L, Qiu Y, Zhang Y, Li J-X. Effects of the cannabinoid CB1 receptor allosteric modulator ORG 27569 on reinstatement of cocaine- and methamphetamine-seeking behavior in rats. Drug Alcohol Depend. 2014;143:251–256. doi: 10.1016/j.drugalcdep.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JR, Burnell-Nugent M, Lossignol D, Ganae-Motan ED, Potts R, Fallon MT. Multicenter, double-blind, randomized, placebo-controlled, parallel-group study of the efficacy, safety, and tolerability of THC:CBD extract and THC extract in patients with intractable cancer-related pain. J Pain Symptom Manage. 2010;39:167–179. doi: 10.1016/j.jpainsymman.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Kenakin TP, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacological reviews. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenakin TP. Biased signaling and allosteric machines: new vistas and challenges for drug discovery. Br J Pharmacol. 2012;165:1659–1669. doi: 10.1111/j.1476-5381.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajehali E, Malone DT, Glass M, Sexton PM, Christopoulos A, Leach K. Biased Agonism and Biased Allosteric Modulation at the CB1 Cannabinoid Receptor. Mol Pharmacol. 2015;88:369–379. doi: 10.1124/mol.115.099192. [DOI] [PubMed] [Google Scholar]
- Khurana L, Ali HI, Olszewska T, Ahn KH, Damaraju A, Kendall DA, Lu D. Optimization of chemical functionalities of indole-2-carboxamides to improve allosteric parameters for the cannabinoid receptor 1 (CB1) J Med Chem. 2014;57:3040–3052. doi: 10.1021/jm5000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana L, Fu B, Duddupudi AL, Liao Y, Immadi SS, Kendall DA, Lu D. Pyrimidinyl biphenylureas: identification of new lead compounds as allosteric modulators of the cannabinoid receptor CB1. J Med Chem. 2017;60:1089–1104. doi: 10.1021/acs.jmedchem.6b01448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen LB, Kiel D, Teng M, Behrens C, Bhumralkar D, Kodra JT. Small-molecule agonists for the glucagon-like peptide 1 receptor. Proc Natl Acad Sci USA. 2007;104:937–942. doi: 10.1073/pnas.0605701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead C, Christopoulos A. Functional and structural perpectives on allosteric modulation of GPCRs. Curr Opin Cell Biol. 2014;27:94–101. doi: 10.1016/j.ceb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Laprairie RB, Bagher AM, Kelly ME, Donovan-Wright EM. Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br J Pharmacol. 2015;172:4790–4805. doi: 10.1111/bph.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprairie RB, Bagher AM, Kelly ME, Donovan-Wright EM. Biased Type 1 cannabinoid receptor signaling influences neuronal viability in a cell culture model of huntington disease. Mol Pharmacol. 2016;89:364–375. doi: 10.1124/mol.115.101980. [DOI] [PubMed] [Google Scholar]
- Laprairie RB, Kulkarnie PM, Deschamps JR, Kelly ME, Janero DR, Cascio MG, Stevenson LA, Pertwee RG, Kenakin T, Denovan-Wright EM, Thakur GA. Enantio-specific allosteric modulation of cannabinoid 1 receptor. Chm Neurosci. 2017 doi: 10.1021/acschemneuro.6b00310. [DOI] [PubMed] [Google Scholar]
- Leo A, Russo E, Elia M. Cannabidiol and epilepsy: Rational and therapeutic potential. Pharmacol Res. 2016;107:85–92. doi: 10.1016/j.phrs.2016.03.005. [DOI] [PubMed] [Google Scholar]
- Leterrier C, Bonnard D, Carrel D, Rossier J, Lenkei Z. Constitutive endocytic cycle of the CB1 cannabinoid receptor. J Biol Chem. 2004;279:36013–36021. doi: 10.1074/jbc.M403990200. [DOI] [PubMed] [Google Scholar]
- Leweke FM, Piomelli D, Pahlisch F, Muhl D, Gerth CW, Hoyer C, Klosterkötter J, Hellmich M, Koethe D. Cannabidiol enhances anandamide signaling and alleviates psychotic symptoms of schizophrenia. Transl Psychiatry. 2012;20 doi: 10.1038/tp.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley CW, Wiscnoski DD, Leister WH, O’Brien JA, Lemaire W, Williams DL, Burno M, Sur C, Kinney GG, Doug JP, Tiller PR, Smith S, Duggan ME, Hartman GD, Conn PJ, Huff JR. Discovery of positive allosteric modulators for the metabotropic glutamate receptor subtype 5 from a series of N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamides that potentiate receptor function in vivo. J Med Chem. 2004;47:5825–5828. doi: 10.1021/jm049400d. [DOI] [PubMed] [Google Scholar]
- Luttrell LM. Minireview: more than just a hammer: ligand “bias” and pharmaceutical discovery. Mol Endocrinol. 2014;28:281–294. doi: 10.1210/me.2013-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie K. Cannabinoid receptors as therapeutic targets. Annu Rev Pharmacol Tox. 2006;46:101–122. doi: 10.1146/annurev.pharmtox.46.120604.141254. [DOI] [PubMed] [Google Scholar]
- Mahmoud MM, Ali HI, Ahn KH, Damaraju A, Samala S, Pulipati VK, Kolluru S, Kendall DA, Lu D. Structure-Activity relationship study of indole-2-carboxamides identifies a potent allosteric modulator for the cannabinoid Receptor 1 (CB1) J Med Chem. 2013;56:7965–7975. doi: 10.1021/jm4009828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares J, Julian M, Carrascosa A. Role of the cannabinoid system in pain control and therapeutic implications for the management of acute and chronic pain episodes. Curr Neuropharmacol. 2006;4:239–257. doi: 10.2174/157015906778019527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May LT, Leach K, Sexton PM, Christopoulos A. Allosteric modulators of G-protein coupled receptors. Annu Rev Pharmacol Toxicol. 2007;47:1–51. doi: 10.1146/annurev.pharmtox.47.120505.105159. [DOI] [PubMed] [Google Scholar]
- Miller WL, Auchus RJ. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro HA, Howard JL, Pollard GT, Carroll FI. Positive allosteric modulation of the human cannabinoid (CB) receptor by RTI-371, a selective inhibitor of the dopamine transporter. Br J Pharmacol. 2009;156:1178–1184. doi: 10.1111/j.1476-5381.2009.00124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisink RJ, van Laar MW. Does cannabidiol protect against adverse psychological effects of THC? Front Psychiatry. 2013;4:130. doi: 10.3389/fpsyt.2013.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, German N, Decker AM. Structure-activity relationships of substituted 1H-indole-2-carboxamides as CB1 receptor allosteric modulators. Bioorg Med Chem. 2015;23:2195–2203. doi: 10.1016/j.bmc.2015.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamplona FA, Menezes-de-Lima O, Jr, Takahashi RN. Aspirin-triggered lipoxin induces CB1-dependent catalepsy in mice. Neurosci Lett. 2010;470:33–37. doi: 10.1016/j.neulet.2009.12.050. [DOI] [PubMed] [Google Scholar]
- Pamplona FA, Ferreira J, Menezes de Lima O, Jr, Duarte FS, Bento AF, Forner S, Villarinho JG, Bellochio L, Wotjak CT, Lerner R, Monory K, Lutz B, Canetti C, Matias I, Calixto JB, Marsicano G, Guimaraes MZ, Takahashi RN. Anti-inflammatory lipoxin A4 is an endogenous allosteric enhancer of CB1 cannabinoid receptor. Proc Natl Acad Sci USA. 2012;109:21134–21139. doi: 10.1073/pnas.1202906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG. Lipoxin A4 is an allosteric endocannabinoid that strengthens anandamide-induced CB1 receptor activation. Proc Natl Acad Sci USA. 2012;109:20781–20782. doi: 10.1073/pnas.1218529110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee RG. The pharmacology of cannabinoid receptors and their ligands: an overview. Int J Obes. 2006;30:S13–18. doi: 10.1038/sj.ijo.0803272. [DOI] [PubMed] [Google Scholar]
- Price MR, Baillie GL, Thomas A, Stevenson LA, Easson M, Goodwin R, McLean A, McIntosh L, Goodwin G, Walker G, Westwood P, Marrs J, Thomson F, Cowley P, Christopoulos A, Pertwee RG, Ross RA. Allosteric modulation of the cannabinoid CB1 receptor. Mol Pharmacol. 2005;68:1484–1495. doi: 10.1124/mol.105.016162. [DOI] [PubMed] [Google Scholar]
- Qiao C, Ali HI, Ahn KH, Kolluru S, Kendall DA, Lu D. Synthesis and biological evaluation of indole-2-carboxamides bearing photoactivatable functionalities as novel allosteric modulators for the cannabinoid CB1 receptor. Eur J Med Chem. 2016;121:517–529. doi: 10.1016/j.ejmech.2016.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy DS. The utility of cannabidiol in the treatment of refractory epilepsy. Clin Pharmacol Ther. 2017;101:182–184. doi: 10.1002/cpt.441. [DOI] [PubMed] [Google Scholar]
- Ross RA. Allosterism and cannabinoid CB1 receptors: the shape of things to come. Trends Pharmacol Sci. 2007;28:567–572. doi: 10.1016/j.tips.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Russo EB. Cannabinoids in the management of difficult to treat pain. Ther Clin Risk Manag. 2008;4:245–259. doi: 10.2147/tcrm.s1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar DR, Gaw AG, Okine BN, Woodhams SG, Wong A, Kendall DA. Dynamic regulation of the endocannabinoid system: implications for analgesia. Mol Pain. 2009;5:59. doi: 10.1186/1744-8069-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Yin J, Chapman K, Grzemska M, Clark L, Wang J, Rosenbaum DW. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature. 2016;540:602–606. doi: 10.1038/nature20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore DM, Baillie GL, Hurst DH, Navas F, Seltzman HH, Marcu JP, Abood ME, Ross RA, Reggio PH. Allosteric modulation of a cannabinoid G protein-coupled receptor: binding site elucidation and relationship to G protein coupling. J Biol Chem. 2014;289:5828–4. doi: 10.1074/jbc.M113.478495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivicki R, Hohmann AG. Society for Neuroscience Meeting. San Diego, CA: 2016. Positive allosteric modulation of cannabinoid CB1 receptor signaling suppresses chemotherapy-induced peripheral neuropathy. [Google Scholar]
- Stornaiuolo M, Bruno A, Botta L, Regina GL, Sandro C, Romano S, Luciana M, Novellino E. Endogenous vs exogenous allosteric modulators in GPCRs: a dispute for shuttling CB1 among different membrane microenvironments. Sci Rep. 2015;5:15453. doi: 10.1038/srep15453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straiker A, Mitjavila J, Yin D, Gibson A, Mackie K. Aiming for allosterism: evaluation of allosteric modulators of CB1 in a neuronal model. Pharmacol Res. 2015;99:370–376. doi: 10.1016/j.phrs.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turu G, Hunyady L. Signal transduction of the CB1 cannabinoid receptor. J Mol Endocrinol. 2010;44:75–85. doi: 10.1677/JME-08-0190. 2010. [DOI] [PubMed] [Google Scholar]
- Vallee M, Vitiello S, Bellocchio L, Hebert-Chatelain E, Monlezun S, Martin-Garcia E, Kasanetz F, Baillie G L, Panin F, Cathala A, Roullot-Lacarriere V, Fabre S, Hurst DP, Lynch DL, Shore DM, Deroche-Garmonet V, Spampinato U, Revest JM, Maldonado R, Reggio PH, Ross RA, Marsicno G, Piazza PV. Pregnenolone can protect the brain from cannabis intoxication. Science. 2014;343:94–98. doi: 10.1126/science.1243985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade DT, Robson P, House H, Makela P, Aram J. A preliminary controlled study to determine whether whole-plant cannabis extracts can improve intractable neurogenic symptoms. Clin Rehabil. 2003;17:21–29. doi: 10.1191/0269215503cr581oa. [DOI] [PubMed] [Google Scholar]
- Wang X, Horsewill JG, Whalley BJ, Stephens GJ. Effects of the allosteric antagonist 1-(4-Chlorophenyl)-3-[3-(6-pyrrolidin-1-ylpyridin-2-yl)phenyl]urea (PSNCBAM-1) on CB1 receptor modulation in the cerebellum. Mol Pharmacol. 2010;79:758–767. doi: 10.1124/mol.110.068197. [DOI] [PubMed] [Google Scholar]
- Wild C, Cunningham KA, Zhou J. Allosteric modulation of G protein-coupled receptors: an emerging approach of drug discovery. J Pharmacol Ther. 2013;2:1. [PMC free article] [PubMed] [Google Scholar]
- Wong M, Devinsky O, Thiele E, Appleton R, Patel A, Harden C, Sommerville K, Greenwood S, Morrison G. A dose ranging safety and pharmacokinetic study of cannabidiol (CBD) in children with Dravet syndrome (GWPCARE1); American Epilepsy Society Annual Meeting; 2016. www.aesnet.org. [Google Scholar]


