Vaccine development for difficult pathogens (e.g., HIV) is being tackled by rational approaches involving identifying functional antibodies, templating immunogens from the antibodies, and then evaluating the immunogens iteratively.
Abstract
Functional antibodies, i.e., those with antipathogen activity in in vitro assays, are generally the best correlate of vaccine protection. Mimics of natural infection, including live attenuated and killed pathogens, which induce such antibodies in vivo, have generated highly successful vaccines. However, pathogens that induce functional antibodies at lower levels or more sporadically have been more refractory to vaccine design. Such pathogens are being tackled by more systematic approaches involving identifying functional antibodies, templating immunogens from the antibodies, and then evaluating the immunogens iteratively. I believe this is a powerful new approach to vaccine design as discussed below.
WHAT ARE AND HOW DO VACCINES WORK?
A description of the most powerful immunogen design vaccine strategies would clearly benefit from understanding how vaccines might work for each individual pathogen. Google defines a vaccine as “a substance used to stimulate the production of antibodies and provide immunity against one or several diseases, prepared from the causative agent of a disease, its products, or a synthetic substitute, treated to act as an antigen without inducing the disease.” The definition is lacking in that it is likely that cellular immune responses induced by immunization contribute, at least in some cases, to vaccine protection. The most successful vaccine strategies for the future will unravel the relative contributions of humoral and cellular immunity to protection for each pathogen and incorporate this knowledge into vaccine design. Nevertheless, a good case can be made that for many vaccines the antibody response is crucial, and antibody induction is the focus here.
GREAT DEBATES.
What are the most interesting topics likely to come up over dinner or drinks with your colleagues? Or, more importantly, what are the topics that don't come up because they are a little too controversial? In Immune Memory and Vaccines: Great Debates, Editors Rafi Ahmed and Shane Crotty have put together a collection of articles on such questions, written by thought leaders in these fields, with the freedom to talk about the issues as they see fit. This short, innovative format aims to bring a fresh perspective by encouraging authors to be opinionated, focus on what is most interesting and current, and avoid restating introductory material covered in many other reviews.
The Editors posed 13 interesting questions critical for our understanding of vaccines and immune memory to a broad group of experts in the field. In each case, several different perspectives are provided. Note that while each author knew that there were additional scientists addressing the same question, they did not know who these authors were, which ensured the independence of the opinions and perspectives expressed in each article. Our hope is that readers enjoy these articles and that they trigger many more conversations on these important topics.
The provision of antibody-based immunity requires memory, which can be conceived in two forms, both generated following contact with antigen: circulating specific high-affinity antibody produced by long-lived plasma cells in the bone marrow and circulating memory B cells expressing surface antibody receptors for antigen so that such cells can expand and differentiate to produce specific high-affinity antibody on new antigen contact. Circulating antibody has the great advantage that it can act immediately against an invading pathogen. B-cell memory will require a longer time to become effective, although plasmablasts generated by reactivated memory B cells could potentially provide protective levels of antibody in a matter of a few days after pathogen contact. Nevertheless, in many scenarios, the most powerful design strategies will seek to induce sustained high levels of circulating functional antibody. A clear-cut example here is HIV, in which the prevention of the establishment of latency likely requires circulating antibody rather than stimulation of B-cell memory. In other instances, the prevention of disease may not require an overwhelming rapid antibody response and B-cell memory may play a greater role. The notion of “functional antibody” is key in thinking about immunogen design strategies. For viruses, functionality is often associated with an in vitro neutralization assay in which antibodies inhibit productive viral entry to target cells. Many pathogens have evolved mechanisms to evade antibody recognition and immunogen design must seek to deal with these mechanisms and elicit potent functional antibodies.
IMMUNOGEN DESIGN
Antibodies evolve through mutation and selection to recognize molecular shapes. In principle, immunogen design problems boil down to creating the appropriate molecular shapes. The stunning successes achieved with whole organism vaccines such as live attenuated and killed pathogens are because of the effective presentation, to the humoral immune system, of the very same molecular shapes that are found on the surface of the virulent pathogen. These pathogens are typically “evasion lite” in that there is little evidence that molecular features have evolved to evade immune recognition. The life cycle of the pathogen may not require any such evasion (e.g., measles or polio viruses). Other pathogens such as HIV, influenza virus, and malaria have life cycles that require survival at some point in an antibody-rich milieu. These pathogens tend to be “evasion strong” in that they have evolved a multitude of mechanisms that result in the elicitation of suboptimal antibody responses with respect to long-term protection against circulating forms of the pathogen. Evasion can occur by different mechanisms, many of which involve the molecular nature of the pathogen surface. Examples include extreme sequence variation of the surface proteins (HIV, influenza virus, and malaria) and dense glycan coating (HIV and hepatitis C virus [HCV]). Powerful immunogen design strategies for these pathogens require wholly different and much more precise approaches for the elicitation of antibodies to the pathogen surface than have typically been applied. Classical vaccine approaches have not worked to date and are unlikely to do so in the future.
STRATEGIES FOR IMMUNOGEN DESIGN: GETTING THE RIGHT SHAPES
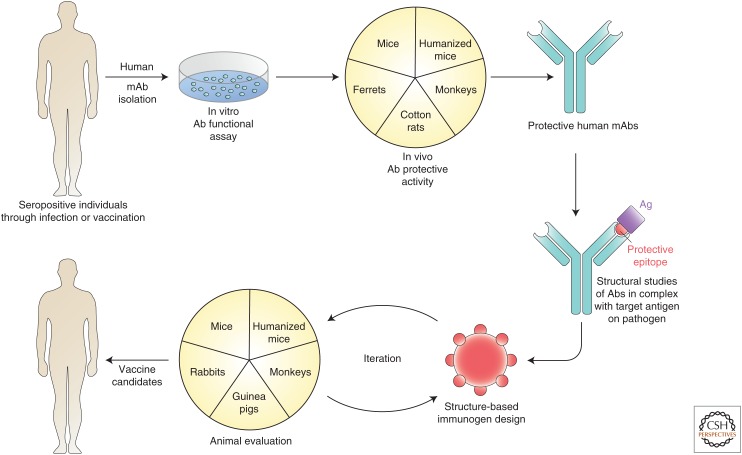
The first problem for immunogen design is to generate molecular shapes that will elicit functional antibodies. How to do this? Here, natural infection can provide valuable leads. In virtually all infections, a proportion of individuals, sometimes quite small, will make highly potent functional antibody responses. Monoclonal antibodies can be isolated from these individuals and used to guide vaccine design in a reversal of the normal flow of vaccines to antibodies in an approach described as reverse vaccinology 2.0 (Fig. 1) (Burton 2002; Rappuoli et al. 2016). (Reverse vaccinology 1.0 is an approach based on bioinformatics and animal immunization and challenge to determine the antigens most appropriate for inclusion in a vaccine. The approach led directly to the meningitis B vaccine.) It is no coincidence that the upsurge of recent interest in rational vaccine design has followed the ability to isolate relatively rare but potent functional antibodies from human infections (Burton et al. 2012). With the appropriate monoclonal antibodies in hand, high-resolution structures from crystallography and cryoelectron microscopy (cryoEM) of antibody-pathogen surface molecules can reveal important details of epitopes to facilitate immunogen design (Ward and Wilson 2015). Ideally, one would wish to have multiple functional antibodies from many donors to single epitope regions to capture the diversity of antibody combining site shapes that can recognize the critical region. The process of collecting many antibodies against the same region has a further advantage in that it may reveal necessary features of such antibodies that can help direct immunogen evaluation. For example, one class of functional (broadly neutralizing) antibodies to HIV that imitate CD4 in binding to the CD4-binding site (CD4bs) of gp120 of the surface Envelope spike (Env) use a particular VH gene segment (VH1-2) (Zhou et al. 2013). Therefore, evaluation of immunogens designed to elicit this type of antibody can look for activation of VH1-2 germline antibodies. However, perhaps the most important facet of having multiple functional antibodies to the same epitope region available is that one has multiple related antibody templates for immunogen design. This can be particularly important when dealing with highly antigenically variable pathogens that may escape from one single monoclonal antibody (mAb) but not several.
Figure 1.
Reverse vaccinology 2.0. Typically memory B cells or plasmablasts from seropositive individuals with high serum functional titers are used as a source of human monoclonal antibodies (mAbs) that are isolated by direct functional screening or antigen selection approaches. The mAbs are investigated for in vivo protective activity in appropriate animal models (e.g., mice and ferrets have been much used for influenza virus, cotton rats for RSV, and monkeys for HIV). Protective mAbs are studied in interaction with their pathogen target (e.g., Env molecules for many viruses and the structural data used to help guide immunogen design). Immunogens are then evaluated in animal models. Typically iterative improvements are expected in the immunogen design before moving forward to vaccine candidates to be tested in humans. (Image adapted from data in Burton 2002 and Rappuoli et al. 2016.)
Armed with an antibody template, or better antibody templates, how can immunogen design proceed most expeditiously? This will depend on the target pathogen. In the case of respiratory syncytial virus (RSV), two approaches have been adopted. A major problem with attempts to induce neutralizing antibodies to RSV has been the metastability of the viral surface F glycoprotein. The prefusion form of the protein readily decays to a postfusion form, and antibodies induced to the postfusion form tend to bind poorly to the prefusion form and do not neutralize effectively. McLellan et al. (2013a,b) developed a strategy based on identifying mAbs that bind to and lock in the prefusion form of F, defining the nature of the corresponding antigenic site (dubbed “0”) structurally, modifying F with disulfide linkages and cavity-filling mutations, and using the stabilized F protein as an immunogen. The immunogen induces excellent RSV-neutralizing titers in animal models and shows great promise as a potential human vaccine (McLellan et al. 2013a). This is a brilliant demonstration of the power of an antibody/structure/rational design approach to vaccine development. A second approach is described by Schief and coworkers in which a helix–turn–helix motif from RSV F and recognized by an RSV-neutralizing antibody was generated as a scaffold using computational design methods (Correia et al. 2014). Immunization of macaques with the scaffold presented multivalently on a particle elicited potent neutralizing antibodies. This approach showed the feasibility of extracting a key molecular shape (epitope) from a pathogen and then using it to elicit a highly directed (epitope-focused) response. One could imagine that such an approach would be very valuable if one had an immunogen with multiple distracting immunodominant epitopes and one sought to focus the response on to a more quiescent but functional epitope.
For human cytomegalovirus (HCMV), Corti et al. (2013) used neutralizing antibodies from natural human infection to identify a new class of antibodies and a new vaccine target. Classically, the fusion protein gB had been a favored target, but neutralizing antibodies that were about 100-fold more potent than gB antibodies were then isolated against a pentameric complex of five different proteins required for viral entry (Macagno et al. 2010). Immunization of mice with the pentameric complex generated extraordinarily high neutralizing titers that were up to 1000-fold higher than those found in acute HCMV infection (Kabanova et al. 2014; Hofmann et al. 2015). This approach, dubbed “analytical vaccinology” (Lanzavecchia et al. 2016), again reveals the power of antibodies isolated from natural infection to guide vaccine strategies.
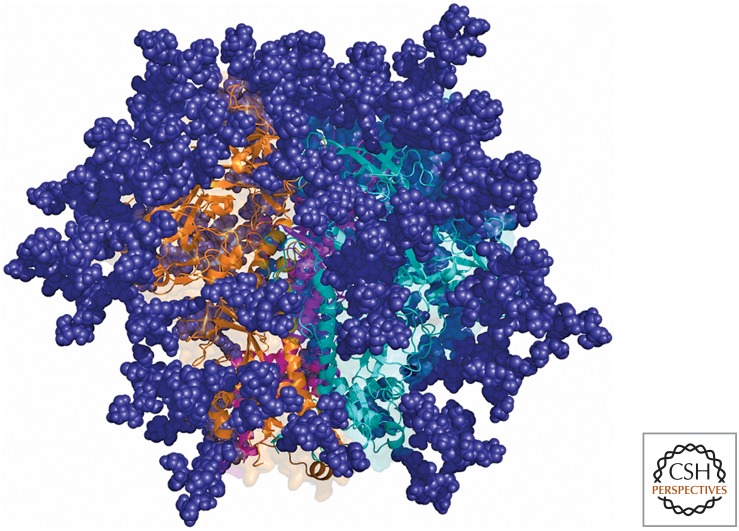
For HIV, if one were to simply examine the highly glycosylated structure of the envelope trimer (Fig. 2), it would be very difficult to determine the relatively conserved targets for broadly neutralizing antibodies (bnAbs). Only the concerted efforts of a number of laboratories that have isolated scores of bnAbs have revealed the full complexity of bnAb epitopes and, therefore, the essential features recognized by these Abs (Burton and Hangartner 2016). For example, bnAbs to the high mannose patch (V3/glycan) bnAb antigenic region recognize elements of the CCR5 coreceptor site, and glycans that camouflage this site from most Ab recognition (Sok et al. 2016b). However, there are notable differences between individual bnAbs in how they recognize this region. Furthermore, in the context of some isolates, bnAbs to this region appear able to interact with multiple glycoforms (Mouquet et al. 2012) and alternate glycans if a given glycan is lost through mutation (Sok et al. 2014), adding extra dimensions to neutralization breadth that an immunogen would ideally capture. Therefore “capturing the right shape” for an HIV immunogen aimed at eliciting HIV bnAbs is complex. This complexity is taken to a new level by the observation that most HIV Env molecules do not generally bind to germline versions of bnAbs (Xiao et al. 2009). This means that, to trigger bnAb responses, one either has to discover Env molecules that were involved in initiating the response in the individual from whom the bnAb was isolated or design germline-targeting molecules de novo. Study of the evolution of bnAbs has allowed for identification of specific germline-targeting Env molecules (Liao et al. 2013; Doria-Rose et al. 2014). At the same time, the use of computational design and library selection using yeast or mammalian display technologies allows for the generation of molecules that target multiple bnAb germlines, which should be advantageous in the context of human translation (Jardine et al. 2015). Once a germline Ab is activated, it is likely that further immunogens will be needed to shepherd the antibody response along toward a bnAb as discussed below.
Figure 2.
The HIV envelope trimer. The trimer is, in many ways, a prototype “evasion-strong” molecule. Much of the protein surface of individual protomers is buried in the interior of the molecule. Glycans are shown in deep blue and individual Env protomers (gp120 and gp41) in orange, teal, and purple. The self-glycan coating is very dense, making antibody access to the protein surface highly restricted. The CD4-binding site is shown in the middle of the molecule. In addition, variable parts of the molecule (not highlighted) are more accessible than more conserved regions that are targeted by bnAbs. The bnAbs have proved invaluable in identifying targets for immunogen design and serves as templates for such design. (Figure courtesy of Sergey Menis, Christina Corbaci, and James Voss.)
Finally, the concepts and successes in HIV bnAbs are being replicated for other highly antigenically variable pathogens, most notably influenza virus (Burton et al. 2012; Krammer and Palese 2015). One can also begin to consider the possibility of designing vaccines based on common or similar epitopes found within a given class of viruses. For instance, an antibody has been described that neutralizes four different paromyxoviruses (Corti et al. 2013) and that might provide clues to the generation of a pan-paromyxovirus vaccine. Yet again, an antibody that could neutralize multiple flaviviruses would be extremely valuable for design of a pan-flavivirus vaccine. Vaccines against a whole class of viruses would have the attraction of potentially offering protection against emerging pathogens related to current prevalent pathogens.
Once the right shapes (immunogens) have been discovered, the next stage is to use them in the most effective combinations and protocols.
IMMUNIZATION STRATEGIES: USING THE SHAPES APPROPRIATELY
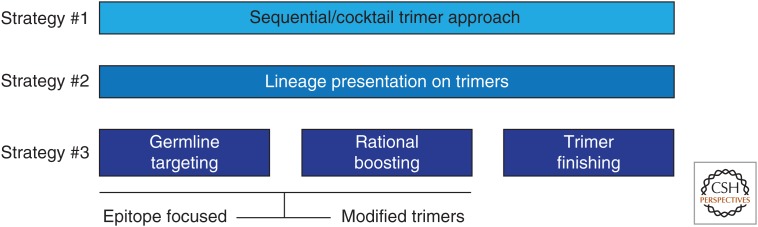
Rationally designed immunogens templated from functional antibodies will typically be in the form of soluble proteins that generally do not induce particularly strong or durable antibody responses (Sundling et al. 2013). One must then seek to present the proteins in a more immunogenic form for the most effective vaccine strategies. This includes the use of adjuvants that are becoming more effective but also the presentation of proteins in a multivalent format on, for example, liposomes, nanoparticles, or virus-like particles. In addition, DNA and RNA vaccination have a number of advantages and limitations. In the particular case of HIV, there is a growing consensus that repeated immunization with the same immunogen will not be effective in inducing bnAbs but rather schemes for sequential and cocktail immunization are under consideration (Fig. 3). Many aspects of the best immunization strategies to take advantage of state-of-the-art immunogen design are still to be worked out. However, it is clear that an important consideration here is the ability to rapidly evaluate immunogens and immunization strategies. As well as classical small animal studies, the use of various strains of mice expressing human antibodies is proving to be of great value (Briney et al. 2016; Escolano et al. 2016; Sok et al. 2016a; Tian et al. 2016).
Figure 3.
Sequential immunization as a strategy to induce broadly neutralizing antibodies to HIV. For “evasion-strong” pathogens such as HIV, it may be necessary to guide affinity maturation in a manner not hereto necessary to thwart evasion mechanisms. This may require the use of multiple different immunogens in a vaccination protocol rather than repeated immunizations with the same immunogen as performed classically. For HIV, examples of possible strategies are shown. Strategy #1 might involve sequential immunization with trimer Env (Fig. 2) molecules from different clades, for example, perhaps presented in cocktails (Wang et al. 2015). Strategy #2 would involve immunization with trimers chosen on the basis of viral evolution in an individual who developed bnAbs (Haynes 2015). Strategy #3 involves the rational design of germline targeting and boosting immunogens that could be based on trimers but also other Env-related molecules followed by a final immunization with native or native-like trimer(s) (Escolano et al. 2016; Steichen et al. 2016).
CONCLUSION
Rational vaccine design based on molecular understanding of the interaction of functional antibodies with their targets on a pathogen is a powerful new approach to vaccine design. There are already successes and indications of successes to come. I believe these successes will usher in a new era of molecular vaccinology.
ACKNOWLEDGMENTS
The author gratefully acknowledges the help and advice from many valued colleagues and the financial support of the National Institute of Allergy and Infectious Diseases (NIAID), the Gates Foundation, the International AIDS Vaccine Initiative (IAVI), and the Ragon Institute.
REFERENCES
- Briney B, Sok D, Jardine JG, Kulp DW, Skog P, Menis S, Jacak R, Kalyuzhniy O, de Val N, Sesterhenn F, et al. 2016. Tailored immunogens direct affinity maturation toward HIV neutralizing antibodies. Cell 166: 1459–1470.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR. 2002. Antibodies, viruses and vaccines. Nat Rev Immunol 2: 706–713. [DOI] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. 2016. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol 34: 635–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. 2012. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337: 183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, et al. 2014. Proof of principle for epitope-focused vaccine design. Nature 507: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. 2013. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature 501: 439–443. [DOI] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. 2014. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Steichen JM, Dosenovic P, Kulp DW, Golijanin J, Sok D, Freund NT, Gitlin AD, Oliveira T, Araki T, et al. 2016. Sequential immunization elicits broadly neutralizing anti-HIV-1 antibodies in Ig knockin mice. Cell 166: 1445–1458.e1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF. 2015. New approaches to HIV vaccine development. Curr Opin Immunol 35: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I, Wen Y, Ciferri C, Schulze A, Fuhner V, Leong M, Gerber A, Gerrein R, Nandi A, Lilja AE, et al. 2015. Expression of the human cytomegalovirus pentamer complex for vaccine use in a CHO system. Biotechnol Bioeng 112: 2505–2515. [DOI] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. 2015. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349: 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabanova A, Perez L, Lilleri D, Marcandalli J, Agatic G, Becattini S, Preite S, Fuschillo D, Percivalle E, Sallusto F, et al. 2014. Antibody-driven design of a human cytomegalovirus gHgLpUL128L subunit vaccine that selectively elicits potent neutralizing antibodies. Proc Natl Acad Sci 111: 17965–17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Palese P. 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14: 167–182. [DOI] [PubMed] [Google Scholar]
- Lanzavecchia A, Fruhwirth A, Perez L, Corti D. 2016. Antibody-guided vaccine design: Identification of protective epitopes. Curr Opin Immunol 41: 62–67. [DOI] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. 2013. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, Gerna G, Sallusto F, Lanzavecchia A. 2010. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol 84: 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, et al. 2013a. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science 342: 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, et al. 2013b. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340: 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci 109: E3268–E3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R, Bottomley MJ, D'Oro U, Finco O, De Gregorio E. 2016. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med 213: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien JP, Menis S, Wickramasinghe L, et al. 2014. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med 6: 236ra263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Briney B, Jardine JG, Kulp DW, Menis S, Pauthner M, Wood A, Lee EC, Le KM, Jones M, et al. 2016a. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353: 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Pauthner M, Briney B, Lee JH, Saye-Francisco KL, Hsueh J, Ramos A, Le KM, Jones M, Jardine JG, et al. 2016b. A prominent site of antibody vulnerability on HIV envelope incorporates a motif associated with CCR5 binding and its camouflaging glycans. Immunity 45: 31–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen JM, Kulp DW, Tokatlian T, Escolano A, Dosenovic P, Stanfield RL, McCoy LE, Ozorowski G, Hu X, Kalyuzhniy O, et al. 2016. HIV vaccine design to target germline precursors of glycan-dependent broadly neutralizing antibodies. Immunity 45: 483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundling C, Martinez P, Soldemo M, Spangberg M, Bengtsson KL, Stertman L, Forsell MN, Karlsson Hedestam GB. 2013. Immunization of macaques with soluble HIV type 1 and influenza virus envelope glycoproteins results in a similarly rapid contraction of peripheral B-cell responses after boosting. J Infect Dis 207: 426–431. [DOI] [PubMed] [Google Scholar]
- Tian M, Cheng C, Chen X, Duan H, Cheng HL, Dao M, Sheng Z, Kimble M, Wang L, Lin S, et al. 2016. Induction of HIV neutralizing antibody lineages in mice with diverse precursor repertoires. Cell 166: 1471–1484.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Mata-Fink J, Kriegsman B, Hanson M, Irvine DJ, Eisen HN, Burton DR, Wittrup KD, Kardar M, Chakraborty AK. 2015. Manipulating the selection forces during affinity maturation to generate cross-reactive HIV antibodies. Cell 160: 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AB, Wilson IA. 2015. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci 40: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun 390: 404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. 2013. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity 39: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]