Abstract
Introduction
Despite the recognized importance of atrophy in multiple sclerosis (MS), methods for its quantification have been mostly restricted to the research domain. Recently, a CE labelled and FDA approved MS-specific atrophy quantification method, MSmetrix, has become commercially available. Here we perform a validation of MSmetrix against established methods in simulated and in vivo MRI data.
Methods
Whole-brain and gray matter (GM) volume were measured with the cross-sectional pipeline of MSmetrix and compared to the outcomes of FreeSurfer (cross-sectional pipeline), SIENAX and SPM. For this comparison we investigated 20 simulated brain images, as well as in vivo data from 100 MS patients and 20 matched healthy controls. In fifty of the MS patients a second time point was available. In this subgroup, we additionally analyzed the whole-brain and GM volume change using the longitudinal pipeline of MSmetrix and compared the results with those of FreeSurfer (longitudinal pipeline) and SIENA.
Results
In the simulated data, SIENAX displayed the smallest average deviation compared with the reference whole-brain volume (+ 19.56 ± 10.34 mL), followed by MSmetrix (− 38.15 ± 17.77 mL), SPM (− 42.99 ± 17.12 mL) and FreeSurfer (− 78.51 ± 12.68 mL). A similar pattern was seen in vivo. Among the cross-sectional methods, Deming regression analyses revealed proportional errors particularly in MSmetrix and SPM. The mean difference percentage brain volume change (PBVC) was lowest between longitudinal MSmetrix and SIENA (+ 0.16 ± 0.91%). A strong proportional error was present between longitudinal percentage gray matter volume change (PGVC) measures of MSmetrix and FreeSurfer (slope = 2.48). All longitudinal methods were sensitive to the MRI hardware upgrade that occurred during the time of the study.
Conclusion
MSmetrix, FreeSurfer, FSL and SPM show differences in atrophy measurements, even at the whole-brain level, that are large compared to typical atrophy rates observed in MS. Especially striking are the proportional errors between methods. Cross-sectional MSmetrix behaved similarly to SPM, both in terms of mean volume difference as well as proportional error. Longitudinal MSmetrix behaved most similar to SIENA. Our results indicate that brain volume measurement and normalization from T1-weighted images remains an unsolved problem that requires much more attention.
Keywords: Multiple sclerosis, MRI, Neurodegeneration, Gray matter, Atrophy
Highlights
-
•
MSmetrix is validated against established atrophy quantification methods.
-
•
MSmetrix, FreeSurfer, SIENA(x) and SPM provide different results.
-
•
Most striking are the proportional errors between methods.
-
•
Cross-sectional MSmetrix behaves most similar to SPM.
-
•
Longitudinal MSmetrix behaves most similar to SIENA.
1. Introduction
In the past decade, brain atrophy in multiple sclerosis (MS) has been recognized as a crucial component of the disease (Bermel and Bakshi, 2006). Especially gray matter (GM) atrophy has attracted a lot of attention because (i) it could be present in early phases of the disease (Calabrese et al., 2015), (ii) it shows a different course across clinical subtypes (Fisher et al., 2008), and (iii) it shows stronger associations with clinical (especially cognitive) dysfunction than the well-known focal white matter pathology (Bermel and Bakshi, 2006). Although the exact mechanism underlying GM tissue loss in MS remains to be elucidated (Chard and Miller, 2016), atrophy measures are now widely accepted as a surrogate marker for neurodegeneration and disease progression in research. The importance of brain atrophy in MS is further illustrated by the increasing number of recent clinical trials that used brain atrophy measures as outcome measures (Cohen et al., 2010, Cohen et al., 2015, Comi et al., 2012, Kappos et al., 2007, Mikol et al., 2008, O'Connor et al., 2011, Olsson et al., 2014).
Although atrophy in MS is substantially faster than in normal ageing (Fisher et al., 2008), accurate and sensitive quantification of atrophy in MS remains challenging (Vrenken et al., 2013). Firstly, the presence of MS pathology other than atrophy (e.g., gray and white matter MS lesions and diffuse abnormalities) severely complicates reliable measurement of atrophy in MS patients using “standard” image analysis techniques. Secondly, reliable atrophy measurements require 3D imaging, while most institutions and clinical trials still tend to use traditional 2D sequences. Finally, and most importantly, due to the lack of standardized benchmark datasets, only a few studies have systematically evaluated the accuracy and reproducibility of the atrophy measurement techniques that are widely available and generally used in research, trial and clinical settings (Derakhshan et al., 2010, Eggert et al., 2012, Klauschen et al., 2009).
Recently, the MSmetrix software package (Icometrix, Flanders, Belgium) has received CE labeling and FDA approval for the automatic quantification of cerebral (and WM lesion) volumes from FLAIR and T1-weighted images in patients with MS (Jain et al., 2015, Lysandropoulos et al., 2016, Smeets et al., 2016). MSmetrix accepts any of these clinical sequences (FLAIR with a maximum slice thickness of 3 mm) and generates an online report with the measured white matter lesion, whole-brain and gray matter volumes. MSmetrix's approval gives patients and physicians access to a new generation of disease monitoring tools, which are specifically developed to measure atrophy at an individual level in a clinical setting, as well as being tailored to MS patients. However, a thorough evaluation of MSmetrix has not been performed yet. Therefore, the aim of the current study was to compare MSmetrix with three established atrophy measurement techniques, i.e. FreeSurfer, SIENA(x) and SPM. We firstly compared the cross-sectional MSmetrix results with known reference values and the results of the established methods in simulated data. Secondly, we compared the cross-sectional and longitudinal MSmetrix results with the established methods in ‘real’, in vivo MR images of MS patients and healthy controls.
2. Materials and methods
2.1. Data
To evaluate the agreement of the different atrophy measurement methods, we used i) 20 simulated single time-point T1-weighted images of healthy controls provided by the Simulated Brain Database (http://brainweb.bic.mni.mcgill.ca/brainweb/) and ii) in vivo MR images of 100 MS patients and 20 healthy controls, of whom 50 MS patients and 8 healthy controls also had longitudinal follow-up data.
2.1.1. Simulated MRI data
The BrainWeb data set consists of twenty simulated MR images based on the anatomical models of healthy controls (Aubert-Broche et al., 2006a, Aubert-Broche et al., 2006b). For this study, we used the 3D T1-weighted spoiled FLASH sequence (1.5 Tesla, TR = 22 ms, TE = 9.2 ms, FA = 30°, 1 mm isotropic voxels) simulated with 3% noise and 0% intensity-inhomogeneity. The anatomical subject of each model consists of 12 volumes that each describe the voxelwise fuzzy membership (comparable with partial volume) for the corresponding tissue class (background, CSF, GM, WM, fat, muscle, muscle/skin, skull, vessels, ‘tissue around fat’, dura mater and bone marrow). For each subject, we estimated the reference total brain volume from these anatomical models adding up the voxelwise GM and WM memberships across the entire image. As the FLAIR image contrast is not available for the BrainWeb images, all atrophy measurements on simulated data were performed on the T1-weighted images only.
2.1.2. In vivo MRI data
The in vivo data set was selected from two ongoing study cohorts acquired at the MS Center Amsterdam, VU University medical center Amsterdam, consisting of patients with clinically-definite MS and matched healthy controls (Schoonheim et al., 2012, Steenwijk et al., 2014). The institutional ethics review board approved the study protocol and subjects gave written informed consent prior to participation. We randomly selected a subset of 100 MS patients of whom 50 had longitudinal follow-up. Disease severity was measured on the day of scanning using the expanded disability status scale (EDSS) (Kurtzke, 1983). The baseline cohort consisted of 100 MS patients (65% female) with an average (± standard deviation) age of 43.49 ± 10.19 years, disease duration of 9.95 ± 6.93 years, and median EDSS score of 3 (range: 0–8). The matched control group consisted of 20 healthy volunteers, with an average age of 43.34 (± 10.21) years of which 15 (75%) were female. The subset of fifty MS patients (64% female) who had longitudinal follow-up were on average 43.81 ± 10.53 years old at baseline, had a baseline disease duration of 9.98 ± 7.41 years, a median baseline EDSS of 3 (range: 0–6.5), and a mean follow-up time of 4.92 ± 0.95 years (range: [3.03, 6.33] years). The 7 healthy controls (43% female) with follow-up were at baseline on average 38.72 ± 7.45 years old, and had a mean follow-up time of 6.20 ± 0.62 years (range: [5.02, 6.33] years).
MRI was performed using a clinical 3.0 T whole body scanner (GE, Milwaukee, WI, USA). The system underwent a hardware upgrade between the baseline (Signa HDxt) and the follow-up acquisition (Discovery MR750). At both time points, the same eight-channel head coil was used. The imaging protocol was identical at baseline and follow-up, containing among others a 3D T1-weighted fast spoiled gradient echo (FSPGR) sequence (TR = 7.8 ms, TE = 3 ms, FA = 12°, 240 × 240 mm2 field of view (FOV), 176 sagittal slices of 1 mm, 0.94 × 0.94 mm2 in-plane resolution) for anatomical information and a fat-saturated 3D fluid attenuated inversion recovery (FLAIR) sequence (TR = 8000 ms, TE = 125 ms, TI = 2350 ms, 250 × 250 mm2 FOV, 132 sagittal slices of 1.2 mm, 0.98 × 0.98 mm2 in-plane resolution) for MS lesion detection. All baseline measurements were taken before the upgrade and all follow-up measurements were taken after the upgrade.
2.2. Image analysis
In this study, we assessed the agreement of cross-sectional atrophy measurement methods (MSmetrix, FreeSurfer, SIENAX, and SPM) as well as the agreement of longitudinal atrophy measurement methods (longitudinal MSmetrix, longitudinal FreeSurfer, and SIENA). Prior to measuring atrophy, all image data was anonymized, corrected for geometric distortions due to gradient non-linearities, and converted to nifti file format. Icometrix (Leuven, Flanders, Belgium) performed the MSmetrix analyses while being blinded to subject characteristics, except that they were informed whether the subject was an MS patient or healthy control: lesion segmentation was actively removed from the MSmetrix processing pipeline when the data of a healthy control was analyzed. The other analyses were performed at VU University medical center.
2.2.1. MSmetrix: cross-sectional and longitudinal analysis
MSmetrix has a separate cross-sectional and longitudinal pipeline of which the details are published elsewhere (Jain et al., 2015, Lysandropoulos et al., 2016, Smeets et al., 2016). In short, the cross-sectional pipeline combines lesion segmentation, lesion filling and tissue segmentation in an iterative algorithm that converges to segmentations that allow for measurement of lesion volume, whole-brain volume (BV), and gray matter volume (GMV). Since lesion filling is embedded in the segmentation pipeline, it is not necessary to perform lesion filling prior to applying MSmetrix. MSmetrix normalizes for head size by computing a factor from a matrix that linearly transforms the subjects' T1-weighted image to MNI152 standard space. The longitudinal pipeline analyzes two time points: it takes the cross-sectional segmentations of both time points and uses a non-linear registration approach to derive the percentage change of whole-brain (PBVC) and gray matter (PGVC) volume. Icometrix did not have access to the segmentation results of the established atrophy measurement methods, which were created at the VU University medical center.
2.2.2. Established methods: preprocessing
In contrast to MSmetrix, the established atrophy measurement methods did not include lesion filling by default. Therefore, the images were preprocessed to reduce the impact of hypo-intense MS lesions on the segmentations. This involved segmentation of the WM lesions from the 3D FLAIR images using k-Nearest Neighbor classification with Tissue Type Priors (kNN-TTP) (Steenwijk et al., 2013) and lesion filling on the 3D T1-weighted images using Lesion Automated Processing (LEAP) (Chard et al., 2010). A detailed explanation of the pipeline can be found elsewhere (Steenwijk et al., 2014).
2.2.3. Established cross-sectional methods: FreeSurfer, SIENAX and SPM
-
-
FreeSurfer 5.3 (http://surfer.nmr.mgh.harvard.edu) was used to derive FreeSurfer estimated total intracranial volume (eTIV), brain volume (BV), gray matter volume (GMV) and white matter volume (WMV). Although FreeSurfer is specifically optimized for cortical thickness measurements, the pipeline has also been used for quantification of whole-brain atrophy. Normalization for differences in head size was done by computing the ratio of tissue volumes compared to eTIV, resulting in brain parenchymal fraction (BPF), gray matter fraction (GMF), and white matter fraction (WMF).
-
-
SIENAX (part of FSL 5.0.4, http://www.fmrib.ox.ac.uk/fsl), a software package commonly used for cross-sectional atrophy measurements in MS, was also used to obtain BV, GMV and WMV. To ensure optimal brain extraction, the brain extraction tool was employed using optimized parameter settings as recommended previously, yielding bias field correction and a liberal -f (i.e., fractional intensity) threshold of 0.2 (Popescu et al., 2012). We did not optimize the brain extraction settings for each subject individually, as it is known that differences in CSF layer thickness may affect the SIENAX GM/WM segmentation results (Popescu et al., 2011). SIENAX performs normalization for head size by obtaining a scaling factor that normalizes the skull of each subject to the skull in the MNI standard space to obtain NBV, NGMV and NWMV.
-
-
SPM12 (http://www.fil.ion.ucl.ac.uk/spm/software/spm12), another software package commonly used for atrophy measurements in MS, was also used to compute BV, GMV, WMV and total intracranial volume (TIV = GMV + WMV + CSF volume). Normalization for differences in head size was done by computing the fraction of tissue with respect to TIV, resulting in BPF, GMF, and WMF. The default processing parameters were used.
2.2.4. Established longitudinal methods: FreeSurfer and SIENA
-
-
The longitudinal FreeSurfer 5.3 pipeline was used to derive percentage brain volume change (PBVC) and percentage gray matter volume change (PGVC). The longitudinal FreeSurfer pipeline aims to reduce measurement errors that can be induced by different initialization of the algorithm at different time points.
-
-
SIENA (part of FSL), another software package commonly used for longitudinal whole-brain atrophy measurement in MS and even being used as secondary outcome parameter in clinical trials, was also used to compute PBVC (Smith et al., 2002). Optimal brain extraction was ensured by using optimized parameter settings (Popescu et al., 2012).
2.3. Impact of the hardware upgrade
To investigate the impact of the hardware upgrade on the cross-sectional and longitudinal atrophy measurement methods, we also investigated an independent validation cohort of 13 healthy males who were scanned just before and directly after the upgrade, which took about six months to complete. The mean age of this sample was 33.96 ± 8.63 years at baseline, and the mean time to follow-up time was 6.22 ± 0.70 months. The impact of the upgrade was measured for all abovementioned methods.
We evaluated the impact of the scanner upgrade on cross-sectional methods by computing the percentage volume change from baseline (before the upgrade) to follow-up (after the upgrade) in the validation cohort. A paired t-test was used to investigate differences between the time points. The impact of the scanner upgrade on longitudinal methods was evaluated by computing the longitudinal percentage volume change between the two time points. Since the validation cohort consisted of relatively young healthy controls, and the follow-up time was approximately 6 months, close to zero brain volume changes should be expected (Fjell et al., 2009, Raz et al., 2005, Resnick et al., 2003).
2.4. Statistical analysis
Statistical analyses were performed in IBM Corp. Released, 2011 and Matlab R2011a (Mathworks, Natick, MA).
The between-method agreement of normalized cross-sectional whole-brain and gray matter atrophy measures was assessed by computing Spearman-rank correlations. The rationale for using Spearman-rank correlations is that the units of the normalized atrophy measures were different (i.e., ratio versus ratio ∗ volume for BPF and NBV respectively) and a non-linear relationship can be expected.
Mean volumetric was used to investigate the between method agreement of the non-normalized (i.e., raw) atrophy measures. Deming regression analysis was used to investigate the presence of fixed and proportional differences. In contrast to standard regression analysis, Deming regression analysis computes the residue orthogonally to the regression line itself. Thereby, the technique distributes the measurement error over both the dependent and independent variable in the fit. This is necessary when two measurement techniques, each having a measurement error, are compared.
To enhance the sensitivity to differences, we report uncorrected P-values: in all analyses, P-values < 0.05 were considered as statistically significant.
3. Results
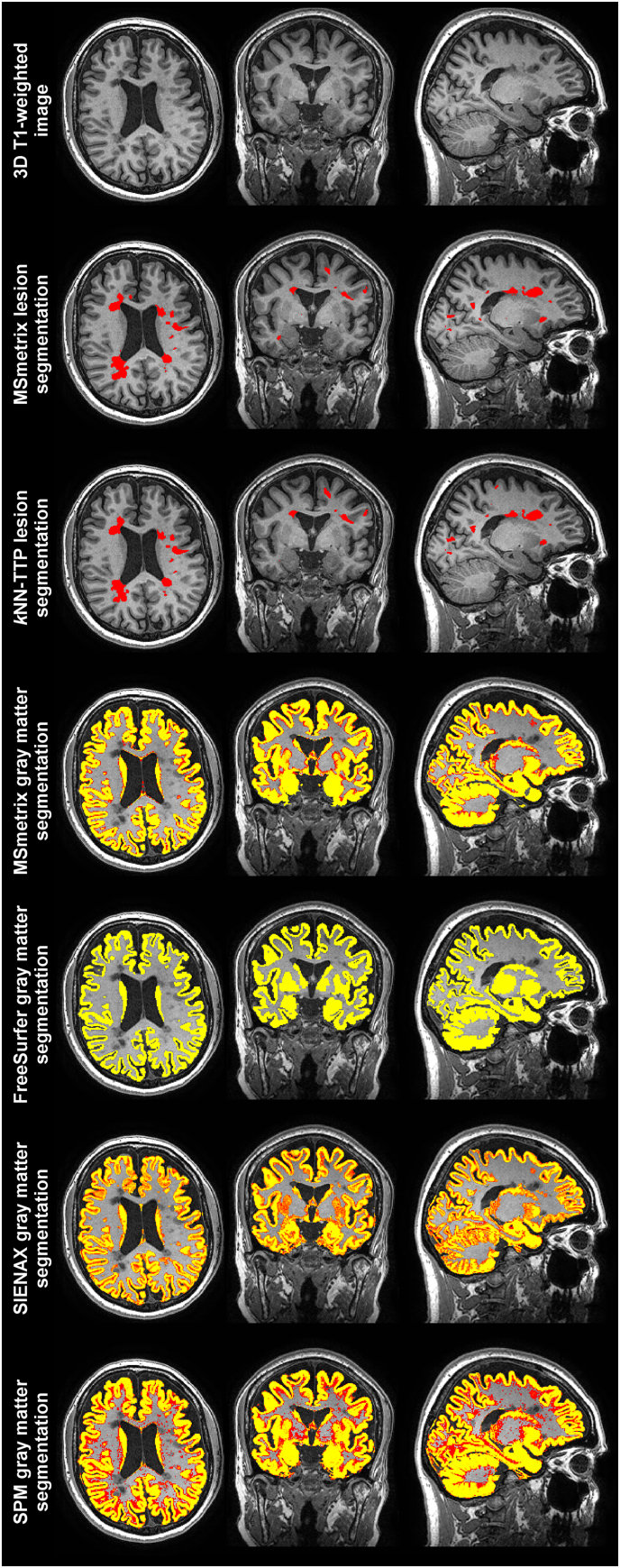
The average measurement results for each method and group are presented in Supplementary Table 1 (cross-sectional methods) and Supplementary Table 2 (longitudinal methods). A typical example of the cross-sectional (lesion and gray matter) segmentation results in an MS patient is displayed in Fig. 1.
Fig. 1.
An example of the cross-sectional segmentation results in a typical MS patient drawn from the in vivo MRI dataset overlaid on the 3D T1-weighted image. The first row displays the raw 3D T1-weighted image. Red areas in the second and third row indicate the lesions found by MSmetrix and kNN-TTP respectively. The lower four rows indicate the voxelwise gray matter partial volume estimates (PVE; red: PVE = 0.01; yellow: PVE = 1) of the different cross-sectional tissue segmentation methods. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.1. Simulated MRI data: agreement of cross-sectional methods
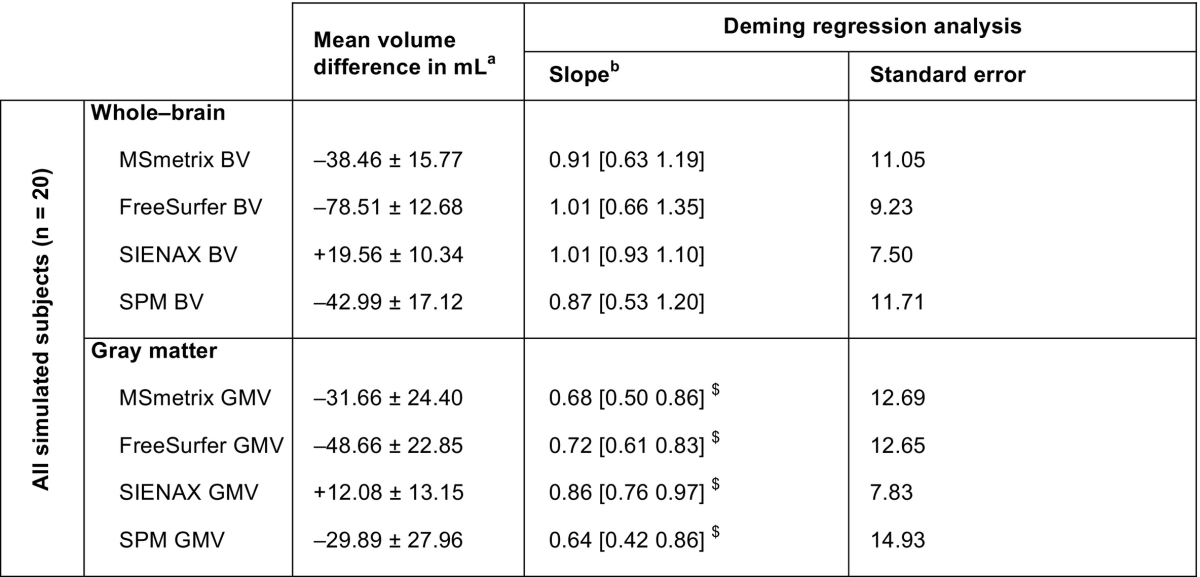
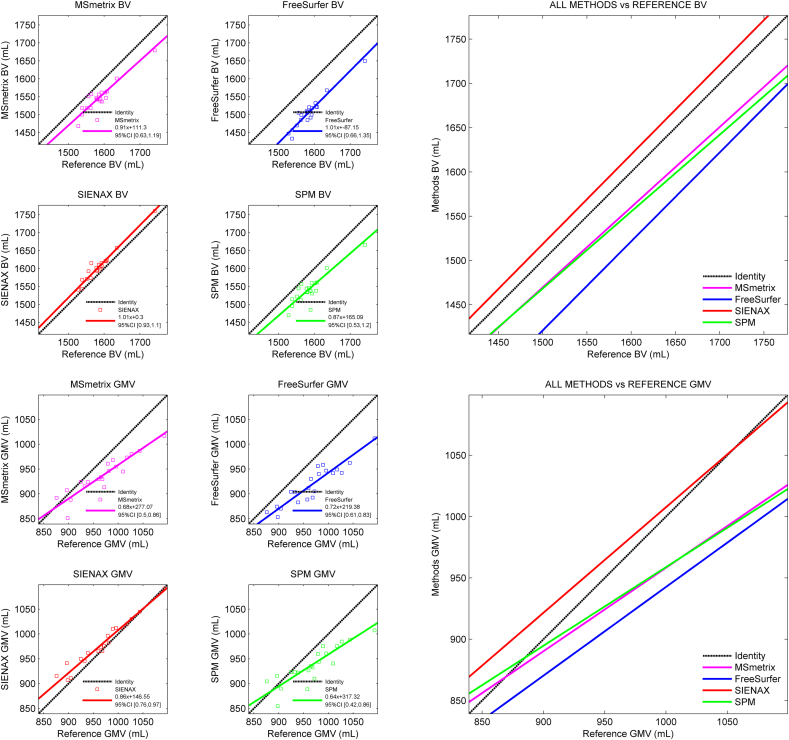
The agreement of the raw (i.e., non-normalized) cross-sectional atrophy measurement methods with the estimated reference volumes and each other is presented in Table 1 and Fig. 2. SIENAX displayed on average the smallest deviation from the whole-brain reference volume (mean difference: + 19.56 ± 10.34 mL), while MSmetrix and SPM showed larger average deviations (mean difference: − 38.46 ± 15.77 mL and − 42.99 ± 17.12 mL, respectively). FreeSurfer showed the largest deviation from the whole-brain reference volume (mean difference: − 78.51 ± 12.68 mL). The regression analyses revealed trends towards proportional errors in the brain volumes measured by MSmetrix and SPM: both methods tend to increasingly underestimate brain volume with increasing reference brain volumes (see Fig. 2A and D). The FreeSurfer and SIENAX whole-brain results did not show such a trend.
Table 1.
The raw whole-brain and gray matter volumes obtained by the cross-sectional methods compared with the reference volume in the simulated MRI data.
Fig. 2.
Scatter plots displaying the agreement of the raw whole-brain and gray matter volumes obtained by cross-sectional methods with the reference volumes in simulated MRI data.
The top row displays the measured whole brain volumes and corresponding fits with respect to the reference whole brain volume. The bottom row displays the measured gray matter volume fits with respect to the reference gray matter volume.
Similar characteristics were observed in the gray matter: SIENAX displayed the smallest average deviation from the reference gray matter volumes (mean difference: + 12.08 ± 13.15 mL), while MSmetrix and SPM showed larger deviations (mean difference: − 31.66 ± 24.40 mL and − 29.89 ± 27.96 mL, respectively). Again, FreeSurfer showed the largest average deviation, with a mean difference of − 48.66 ± 22.85 mL. All methods showed significant proportional errors with respect to the reference gray matter volume (i.e., the slope of the regression line between reference and method-wise gray matter volume differed significantly from 1), reporting lower gray matter volumes for subjects with more true gray matter (see Table 1). Proportional errors were smallest in SIENAX and FreeSurfer (slope = 0.86 and slope = 0.72, respectively), while SPM and MSmetrix showed the largest proportional errors (slope = 0.64 and slope = 0.68, respectively).
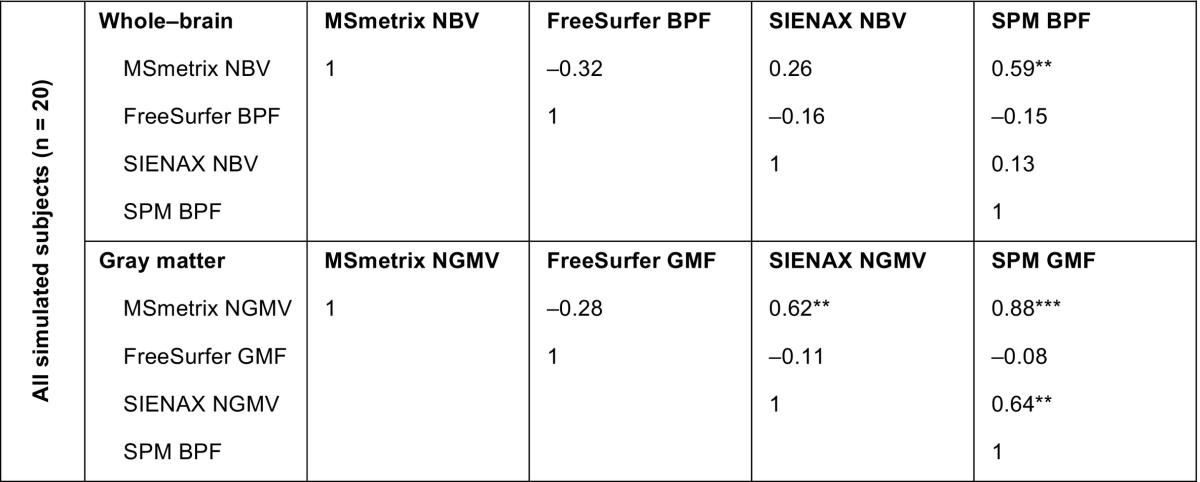
The agreement of the normalized cross-sectional atrophy measurement results in the simulated MRI data is displayed in Table 2. At the whole-brain level, a significant correlation was only observed between MSmetrix and SPM (ρ = 0.588, p = 0.006). For the gray matter, correlations were observed between MSmetrix NGMV, SIENAX NGMV and SPM GPF (all ρ > 0.60). FreeSurfer's normalized whole-brain and gray matter atrophy measurements did not show a correlation with the other methods at all.
Table 2.
Spearman rank correlations between normalized cross-sectional atrophy measurement results in simulated MRI dataa.
Spearman's ρ.
3.2. In vivo MRI data: agreement of cross-sectional methods at baseline
Cross-sectional MSmetrix failed in two MS patients (both female) as a result of a conversion error. These subjects were excluded from all further analyses. One of them was originally included in the longitudinal cohort but was excluded due to this failure at baseline.
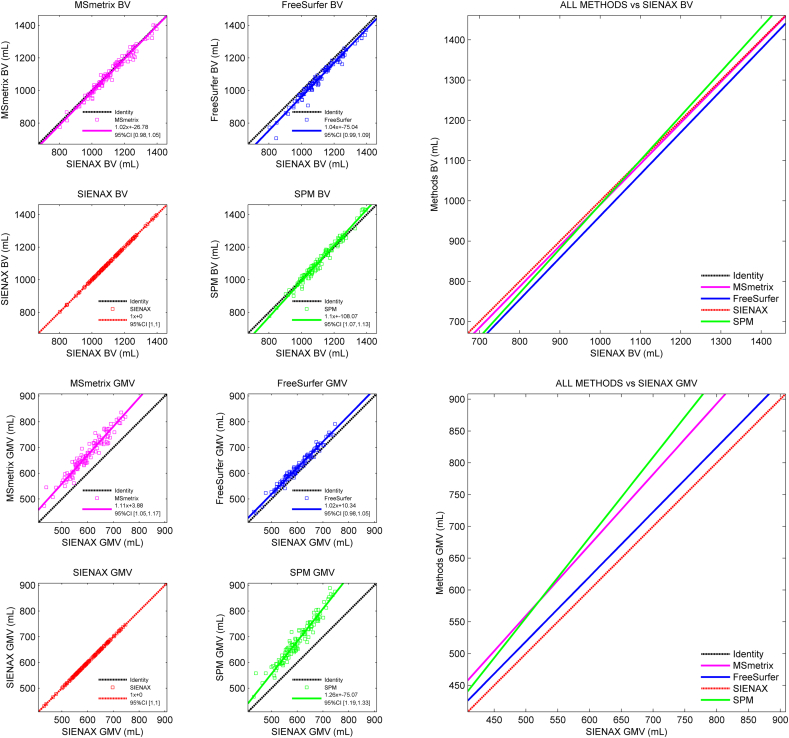
The in vivo agreement between the raw (i.e., non-normalized) cross-sectional atrophy measurement methods at baseline was analyzed by computing the mean difference (see Supplementary Table 3), performing Deming regression analyses (see Supplementary Tables 4 and 5), and constructing scatter plots (see Fig. 3). When constructing the scatter plots, SIENAX was selected as the reference because this method showed the smallest deviation from the true reference volume in the simulated MRI data.
Fig. 3.
Scatter plots displaying the agreement of the raw whole-brain and gray matter volumes obtained by the cross-sectional methods with the SIENAX volumes in in vivo MRI data.
The top row displays the measured whole brain volumes and corresponding fits with respect to the SIENAX volume. The bottom row displays the measured gray matter volume fits with respect to the reference SIENAX volume.
In the pairwise comparison, differences at the whole-brain level between methods were on average smaller than 10 mL. FreeSurfer deviated most from the other methods and showed the largest discrepancy with SPM (mean difference: − 35.57 ± 24.93 mL). Deming regression analyses revealed a proportional error in brain volumes measured by SPM compared to the other methods, overestimating the brain volume for subjects with larger (true) brain volumes.
In the gray matter, MSmetrix and SPM showed the smallest mean volumetric difference (12.15 ± 25.31 mL) closely followed by FreeSurfer and SIENAX (− 20.95 ± 10.72 mL). As can be seen in Fig. 3, the MSmetrix-SPM and FreeSurfer-SIENAX pairs also showed similar behaviour in terms of proportional errors. Of all gray matter method pairs, the FreeSurfer-SIENAX combination was the only one without a proportional error of one method with respect to the other (see Supplementary Table 4). Most severe proportional errors were observed in the gray matter volumes measured by SPM versus the other methods, with a slope coefficient of 1.26 (95% CI [1.19, 1.33]) for SPM versus SIENAX.
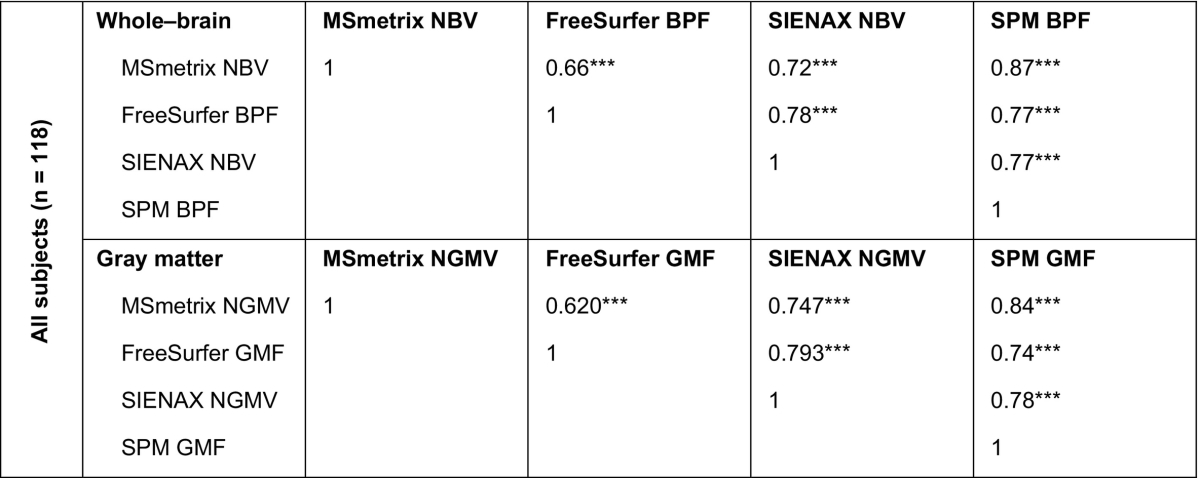
The agreement of the normalized cross-sectional atrophy measurements results in the in vivo MRI data at baseline is displayed in Table 3. SPM was on average most consistent with the other methods. The highest correlation at the whole-brain level was observed between MSmetrix and SPM (ρ = 0.870, p < 0.001). FreeSurfer showed the weakest average correlation with the other methods: the lowest correlation was observed between MSmetrix and FreeSurfer (ρ = 0.664, p < 0.001). Similar trends were seen for the gray matter, with SPM showing the highest correlations with other methods and FreeSurfer the lowest correlations. Also in the gray matter, MSmetrix and SPM showed the highest correlation (ρ = 0.840, p < 0.001).
Table 3.
Spearman rank correlations between normalized cross-sectional atrophy measurement results in in vivo MRI data at baselinea.
Spearman's ρ.
3.3. In vivo MRI data: agreement of longitudinal methods
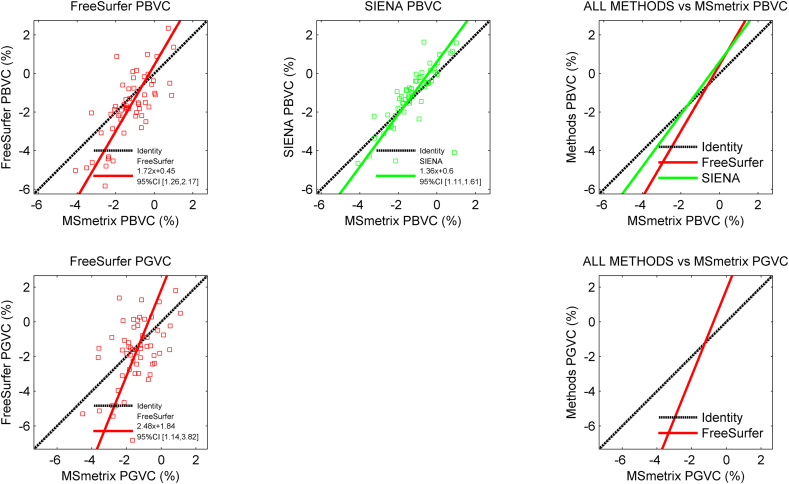
For the longitudinal data, the mean differences between the methods are displayed in Supplementary Table 6, and the results of the Deming analyses are displayed in Supplementary Tables 7 and 8. A visual impression is provided in Fig. 4. MSmetrix and SIENA showed the smallest mean PBVC difference with respect to each other (mean PBVC difference: − 0.16 ± 0.91%), while the FreeSurfer-SIENA pair showed the largest discrepancies (mean PBVC difference: − 0.60 ± 1.26%). All method-pairs displayed significant proportional errors with respect to each other, except for the FreeSurfer-SIENA pair (regression slope: 1.29, 95% CI [0.90, 1.67]).
Fig. 4.
Scatter plots displaying the agreement of the longitudinal measurements with MSmetrix in the in vivo MRI data.
The top row displays the measured percentage whole brain volume change of FreeSurfer and SIENA with respect the percentage gray matter volume change measured by MSmetrix. The bottom row displays the measured percentage gray matter volume change measured by FreeSurfer with respect to the percentage gray matter volume change measured by MSmetrix.
Because the other longitudinal methods could not evaluate the gray matter, MSmetrix PGVC was only compared with FreeSurfer PGVC. The mean difference between these methods was relatively small (mean PGVC difference: − 0.20 ± 1.64%), but a strong proportional error was observed between both methods (regression slope: 2.48, 95% CI [1.14, 3.82]), indicating that the percentage change measured by FreeSurfer is much larger than change found with MSmetrix.
3.4. In vivo MRI data: agreement of lesion segmentation
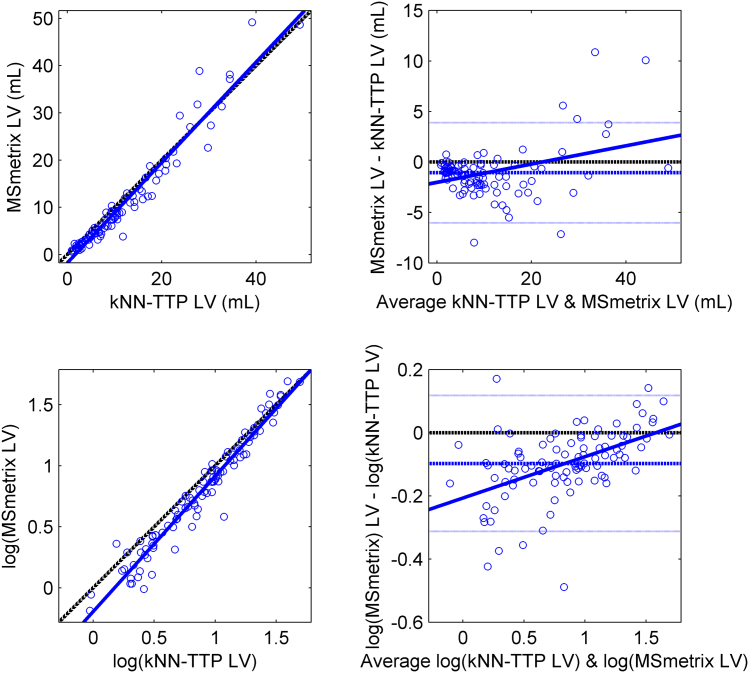
Because the different approaches in lesion segmentation may have affected the in vivo atrophy measurements, we compared the lesion volumes obtained by MSmetrix and kNN-TTP at baseline. This comparison revealed a high volumetric agreement between both methods (ICC consistency = 0.969), but a Bland-Altman plot (see Fig. 5) and Deming regression analysis revealed a proportional error between the methods. MSmetrix reported significantly higher lesion volumes than kNN-TTP in patients with a high lesion load (raw data regression slope = 1.06, 95% CI [1.01, 1.12]; log-transformed regression slope = 1.11, 95% CI [1.06, 1.16]).
Fig. 5.
Scatter (left) and Bland-Altman (right) plot displaying the agreement of MSmetrix and kNN-TTP measuring lesion volumes in the MS patients at baseline.
Because WM lesion volume is not normally distributed, a log-converted version of the raw volumes is displayed in the bottom panels.
3.5. In vivo MRI data: effect of hardware upgrade
The effects of the hardware upgrade on the (cross-sectional and) longitudinal measurements that were analyzed using the 13 healthy controls in the validation cohort are discussed in the Supplementary Materials. In short, the results of all longitudinal methods were affected by the upgrade. Some of the methods reported a mean percentage brain volume change up to approximately 0.5%, while a volume change close to zero was expected.
4. Discussion
In the current study, we compared atrophy measures obtained by MSmetrix with those of established research techniques. Even at the whole-brain level, our results demonstrated differences between methods that were large compared to typical atrophy rates in MS and were marked by strong proportional errors. Our results suggest that cross-sectional MSmetrix behaves much like SPM, while the longitudinal MSmetrix results were reasonably consistent with those of SIENA. In addition, our results indicate that brain volume measurement and normalization from T1-weighted images remains an unsolved problem that requires improvement both on the acquisition and the analysis front.
4.1. Agreement of cross-sectional methods
In the simulated MRI data, our study confirmed the findings of several previous studies that performed a volumetric evaluation of earlier versions of established cross-sectional atrophy measurement techniques (Eggert et al., 2012, Klauschen et al., 2009). FreeSurfer and SPM underestimated brain volume, while SIENAX overestimated the volume but to a much smaller extent. In addition, our results indicate strong proportional errors of the automatic methods compared to the reference volume (i.e., they increasingly over- or underestimate volumes for larger true volumes). In the simulated MRI data, MSmetrix behaved similarly to SPM, in terms of mean difference with the ground truth and in exhibiting a similar proportional error. The similarity between both methods is probably best explained by the underlying method, as both methods use an iterative approach to segment the brain tissue, which is restricted by spatial, registration-based, regularization. A difference between both methods is the approach to perform lesion filling: while SPM was run from lesion filled images, MSmetrix includes WM lesion segmentation and lesion filling in the main segmentation loop. The latter may be advantageous because the segmentation is iteratively refined using this approach. However, a deep investigation of both methods at code level is required to unravel the exact cause of the similar behaviour of both methods, which could be part of future studies. Taking the results of the simulated raw volumes together, our data suggested that SIENAX was most consistent with the reference volume. Therefore, SIENAX results were taken as a reference in the in vivo analyses. However, it should be acknowledged that people have debated the accuracy of BrainWeb to model partial volume effects in a realistic fashion (Bromiley, 2008). This lack in accuracy may explain discrepancies between methods in the simulated data to a certain extent, and forces us to be cautious making statements on whether SIENAX also performs best.
Large differences between methods appeared when comparing the atrophy measures of the simulated MRI data between methods after normalization for head size. While MSmetrix and SPM volume measurements showed acceptable correlations, other method pairs showed severely different (and even inverted) associations. Given the fact that raw volumes did not show such a bad correspondence between methods, the discrepancies after normalization are most likely explained by differences in normalization approaches themselves. We are not aware of other studies that compared the normalization approaches between methods. Future studies should further investigate these differences, as they are likely to highly disturb atrophy measurements.
The in vivo results corroborated our findings in the simulated data. Again, at the whole brain level, the average difference was smallest between MSmetrix and SPM. However, the SPM volumes had a clear proportional error with respect the other methods, measuring lower brain volumes for smaller brains and larger brain volumes for larger brains (see Fig. 2). FreeSurfer deviated most from the other methods, underestimating the whole-brain volume on average by up to 3%, but it should be acknowledged that FreeSurfer was originally not specifically designed for measuring whole brain volumes. Instead, FreeSurfer was specifically optimized for measuring cortical thickness, which we did not evaluate in the current study.
In the gray matter, we observed similar results, MSmetrix behaving similar to SPM. However, in turn, SPM gray matter volume deviated most from the SIENAX results which were closest to the reference in the simulated data. In addition, the SPM gray matter volumes displayed the strongest proportional errors with the other methods (regression slopes down to 0.8). Taken together, this raises the question whether the volumes measured by MSmetrix (and SPM) are accurate in terms of measuring the true brain volumes. The similar behaviour of MSmetrix and SPM may be again be explained by the segmentation approach underlying both methods, but it turns out that the precision of this approach needs to be proven.
After normalization for head size, the correlations between methods in the in vivo MRI data were much stronger than those observed in the simulated MRI data. The highest correlations were observed between MSmetrix and SPM. No clear differences in correlations were observed between MS patients and healthy controls (data not shown). The stronger correlations between methods in vivo may be explained by the presence of more evident atrophy (i.e., inclusion of MS patients) and the larger number of subjects included. Moreover, it could be that the skull boundaries used to determine head size (or TIV) may appear slightly artificial in the simulated data, each of the methods responding differently. Future studies are necessary to further investigate this difference.
4.2. Agreement of longitudinal methods
We additionally assessed the agreement of longitudinal atrophy measurement methods. Given the absence of simulated longitudinal data, we restricted the evaluation to the validation dataset (i.e., 13 right-handed male healthy controls scanned directly before and after the hardware upgrade) and the in vivo data set. From the validation dataset, it is clear that all longitudinal methods are sensitive to the hardware upgrade. This upgrade is a clear limitation of our study, but simultaneously reflects the importance of using identical hardware and sequences for performing longitudinal atrophy measurements (Vrenken et al., 2013). A recent study investigated the sensitivity of MSmetrix and SIENA to changes in contrast by computing the PBVC between 1.5 T and 3.0 T data of 18 MS patients measured by both scanners at the same day (Lysandropoulos et al., 2016). Here, the authors concluded that MSmetrix is more robust to contrast changes than SIENA measuring a ten-fold lower PBVC error when using MSmetrix. The better robustness of longitudinal MSmetrix to the hardware upgrade in terms of whole-brain volume change compared to SIENA was replicated in our study, however we only detected a 2.5-fold improvement. When considering the measurement of gray matter volume change, FreeSurfer showed superior robustness compared to MSmetrix.
Aware of the effect of the hardware upgrade, the results of the longitudinal atrophy measurement in the current work have to be interpreted with great care. One may suggest to post hoc correct the longitudinal measurements with the values obtained in the validation cohort, but we decided to not do so, since the validation cohort was small and it is not known whether the hardware upgrade effects have a systematic and/or proportional nature. Given this limitation, the whole-brain level changes measured by MSmetrix were on average closest to those of SIENA, however, a substantial proportional error was present between both methods. Changes in gray matter volume measured by MSmetrix could only be compared to FreeSurfer: although the mean difference between both methods was very small, again, a substantial proportional error was present between the methods.
4.3. Atrophy measurement methods disagree and better evaluation is crucial
Our results indicate that brain volume measurement and normalization of whole brain volumes on T1-weighted scans are still unsolved problems that require much improvement on both the acquisition and analysis fronts. Moreover, our data highlights the fact that proper validation of atrophy measurement methods deserves much more attention, both in MS as well as in neurodegenerative diseases in general. Another recent study indicated that regional atrophy measurement techniques tend to disagree when assessing correlations between regional atrophy and clinical variables (Popescu et al., 2016). It is alarming that, even when looking at the largest possible scale (i.e., the whole-brain level), established atrophy measurement techniques hardly agree on the actual brain volume that is present – even after lesion filling and without normalization for head size. Some of the methods even show proportional errors that depend on actual brain volume, which may even become worse after normalizing for head size.
In the current work we show that group-averaged whole brain volumes may differ up to 3% (around 35 mL) and gray matter volumes may differ up to 12% (around 83 mL) depending on the measurement techniques used. Longitudinal atrophy measurements show more than a two-fold difference in rate. These are unacceptable disagreements, given the typical annual atrophy rates observed in MS (Fisher et al., 2008) of whole brain (around 0.4%/y, which would be around 62 mL/y at 1.5 L baseline volume) and of gray matter volume (around 0.4%/y, which would be around 38 mL/y at a baseline GM volume of 0.95 L).
This raises questions as to the reliability of MRI studies that have used atrophy measurements in MS, especially those studying regional or local differences. It is clear that there is wide window for better algorithms, yielding more accurate results and less subjects to be included in clinical studies. Future work should address these issues by developing better algorithms and providing better proofs of their validity. Improved benchmark dataset may help to achieve some of these goals.
In this regard, several characteristics of MSmetrix should also be improved. First, the current study shows that cross-sectional atrophy MSmetrix behaves much like SPM. Both methods show strong proportional errors with respect to the true brain volumes, which may be a serious drawback. Secondly, longitudinal MSmetrix suffered from the hardware upgrade, although all methods did. This may be problematic when intending to use the method in clinical practice, where data from the same scanner is typically not available. Third, it is not clear to what extent the estimates of MSmetrix' longitudinal gray matter atrophy are accurate. Our results showed large discrepancies with FreeSurfer's results, which is considered as the gold standard in this field. Lastly, the MSmetrix lesion segmentation routine should be improved. Although the MSmetrix lesion segmentation algorithm performs comparable to other methods available in the public domain, Jain et al. acknowledged room for improvement given the fact that kNN-TTP performed better in a direct comparison (Jain et al., 2015). Because of the potential impact of these methodological uncertainties, it can be rightly questioned whether the current version of MSmetrix is able to fulfil its marketing promises (i.e., reliable individual atrophy measurement, robust to scanner differences, use in clinical practice, etc.). Moreover, little data exists on the value of atrophy measures in clinical decision making (which is clearly a different question and was not within the scope of this study). All aspects together tend to suggest that better algorithms and more thorough validations are necessary before the use atrophy measurement methods can be considered in clinical practice.
4.4. Limitations
Some limitations apply to this work. First, unblinding of MSmetrix to the clinical characteristics of the subjects may be seen as a limitation. We included healthy controls to provide a ‘connection’ between the in vivo and simulated data. However, Icometrix applied a slightly different pipeline in the healthy controls (i.e., not including lesion segmentation and filling) compared to the MS patients, which may have affected our results. To overcome this in the future, future studies are recommended to perform blinded analyses. Second, people have debated the accuracy of BrainWeb to model partial volume effects in a realistic fashion (Bromiley, 2008), which may explain discrepancies between methods in the simulated data to a certain extent. However, discrepancies between methods were also observed in the in vivo data suggesting that the imperfect PVE model of BrainWeb particularly affected the comparison with the reference volume. More realistic simulations (including well-characterized artefacts such as noise and inhomogeneity) and reference datasets should be developed to truly solve this problem. In addition, longitudinal methods may be investigated using methods that simulate longitudinal atrophy (Sharma et al., 2013). Third, we have not corrected P-values for multiple-testing which may have increased the risk that some observed differences are actually chance findings. However, in this validation paper we have considered it more appropriate to display uncorrected P-values allowing the reader to make their own assessment than to apply a formal correction for multiple comparisons. Another limitation is the hardware upgrade that was performed between baseline and follow-up. Although this hardware upgrade was part of regular maintenance, the upgrade interfered substantially with the longitudinal atrophy measurements and hampered reliable comparison of the longitudinal methods. The effect was even worse for the cross-sectional methods, in which the scanner upgrade had a large influence on the normalization for head size. This forced us to be very cautious on making statements on the reliability and accuracy of the longitudinal methods and simultaneously stressed the crucial need to always use data from identical scanners and sequences, also when using MSmetrix. Future research with data that satisfies this requirement is the only way to overcome this limitation. Furthermore, the size of the longitudinal healthy control group was limited. Although a comparison between clinical groups was not a purpose of this study, the heterogeneous group and limited size of this group may explain the absence of a difference in atrophy rates between controls and patients in this study. Finally, the lesion segmentation and filling approach was similar for all established atrophy measurement methods, while MSmetrix used its own implementation. Although it was not reflected by our results and the differences in lesion volume were small compared to the differences in lesion volume, we cannot rule out that this may have caused a slight advantage in terms of agreement between the established methods, compared to MSmetrix. As a post-hoc analysis, we therefore also explored the agreement between methods when running the MSmetrix atrophy measurement algorithm on kNN-TTP/LEAP preprocessed data, and running the established atrophy measurement methods on data preprocessed by using MSmetrix. We did not find evident differences in the results (data not shown).
4.5. Conclusions
In conclusion, our results demonstrated differences between MSmetrix, FreeSurfer, FSL and SPM that were large compared to typical atrophy rates in MS. Especially striking are the proportional errors that were observed. Cross-sectional MSmetrix behaved much like SPM, both in terms of mean difference from the reference (and other methods) as well as proportional error. Longitudinal MSmetrix was (as the other longitudinal methods) sensitive to the hardware upgrade that occurred during the time of the study and behaved most similar to SIENA. Our results indicate that brain volume measurement and normalization from T1-weighted images remains an unsolved problem that requires improvement both on the acquisition and the analysis front.
Funding
Funding for this project was provided by Novartis Pharma B.V. under Research Grant Agreement SP 037.15/432282. The research was conducted independently by the authors and without any intervention by Novartis or Icometrix.
Disclosures
Dr. Steenwijk reports no disclosures.
Dr. Amiri reports no disclosures.
Dr. Schoonheim receives research support from the Dutch MS Research Foundation, grant number 13-820, and has received compensation for consulting services or speaker honoraria from ExceMed, Genzyme, Novartis and Biogen.
Alexandra de Sitter reports no disclosures.
Dr. Barkhof serves on the editorial boards of Brain, European Radiology, Neuroradiology, Multiple Sclerosis Journal and Radiology and serves as a consultant for Bayer-Schering Pharma, Sanofi-Aventis, Biogen-Idec, Teva, Novartis, Roche, Synthon BV, Jansen Research.
Dr. Vrenken has received research support from the Dutch MS Research Foundation, grant numbers 05-358c, 09-358d, 10-718MS and 14-876MS, and has received research support from Pfizer, Novartis, MerckSerono and Teva; speaker honoraria from Novartis; and consulting fees from MerckSerono. All funds were paid directly to his institution.
Dr. Pouwels has received research support from the Dutch MS Research foundation, grant number 14-876MS.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2017.06.034.
Contributor Information
Martijn D. Steenwijk, Email: m.steenwijk@vumc.nl.
Houshang Amiri, Email: h.amiri@vumc.nl.
Menno M. Schoonheim, Email: m.schoonheim@vumc.nl.
Frederik Barkhof, Email: f.barkhof@vumc.nl.
Petra J.W. Pouwels, Email: pjw.pouwels@vumc.nl.
Hugo Vrenken, Email: h.vrenken@vumc.nl.
Appendix A. Supplementary data
Supplementary material
References
- Aubert-Broche B., Evans A.C., Collins L. A new improved version of the realistic digital brain phantom. NeuroImage. 2006;32:138–145. doi: 10.1016/j.neuroimage.2006.03.052. [DOI] [PubMed] [Google Scholar]
- Aubert-Broche B., Griffin M., Pike G.B., Evans A.C., Collins D.L. Twenty new digital brain phantoms for creation of validation image data bases. IEEE Trans. Med. Imaging. 2006;25:1410–1416. doi: 10.1109/TMI.2006.883453. [DOI] [PubMed] [Google Scholar]
- Bermel R.A., Bakshi R. The measurement and clinical relevance of brain atrophy in multiple sclerosis. Lancet Neurol. 2006;5:158–170. doi: 10.1016/S1474-4422(06)70349-0. [DOI] [PubMed] [Google Scholar]
- Bromiley P. Tina-Vision.Net; 2008. Problems with the Brainweb MRI Simulator in the Evaluation of Medical Image Segmentation Algorithms, and an Alternative Methodology. [Google Scholar]
- Calabrese M., Magliozzi R., Ciccarelli O., Geurts J.J.G., Reynolds R., Martin R. Exploring the origins of grey matter damage in multiple sclerosis. Nat. Rev. Neurosci. 2015;16:147–158. doi: 10.1038/nrn3900. [DOI] [PubMed] [Google Scholar]
- Chard D.T., Miller D.H. What lies beneath grey matter atrophy in multiple sclerosis? Brain. 2016;139:7–10. doi: 10.1093/brain/awv354. [DOI] [PubMed] [Google Scholar]
- Chard D.T., Jackson J.S., Miller D.H., Wheeler-Kingshott C.A.M. Reducing the impact of white matter lesions on automated measures of brain gray and white matter volumes. J. Magn. Reson. Imaging. 2010;32:223–228. doi: 10.1002/jmri.22214. [DOI] [PubMed] [Google Scholar]
- Cohen J.A., Barkhof F., Comi G., Hartung H., Khatri B.O., Montalban X., Pelletier J., Capra R., Gallo P., Izquierdo G., Tiel-Wilck K., de Vera A., Jin J., Stites T., Wu S., Aradhye S., Kappos L. Oral fingolimod or intramuscular interferon for relapsing multiple sclerosis. N. Engl. J. Med. 2010;362:402–415. doi: 10.1056/NEJMoa0907839. [DOI] [PubMed] [Google Scholar]
- Cohen J.A., Khatri B., Barkhof F., Comi G., Hartung H.-P., Montalban X., Pelletier J., Stites T., Ritter S., von Rosenstiel P., Tomic D., Kappos L. Long-term (up to 4.5 years) treatment with fingolimod in multiple sclerosis: results from the extension of the randomised TRANSFORMS study. J. Neurol. Neurosurg. Psychiatry. 2015:1–8. doi: 10.1136/jnnp-2015-310597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi G., Jeffery D., Kappos L., Montalban X., Boyko A., Rocca M.A., Filippi M. Placebo-controlled trial of oral laquinimod for multiple sclerosis. N. Engl. J. Med. 2012;366:1000–1009. doi: 10.1056/NEJMoa1104318. [DOI] [PubMed] [Google Scholar]
- Derakhshan M., Caramanos Z., Giacomini P.S., Narayanan S., Francis S.J., Arnold D.L., Collins D.L. Vol. 52. 2010. NeuroImage Evaluation of Automated Techniques for the Quantification of Grey Matter Atrophy in Patients With Multiple Sclerosis; pp. 1261–1267. [DOI] [PubMed] [Google Scholar]
- Eggert L.D., Sommer J., Jansen A., Kircher T., Konrad C. Accuracy and reliability of automated gray matter segmentation pathways on real and simulated structural magnetic resonance images of the human brain. PLoS One. 2012;7 doi: 10.1371/journal.pone.0045081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher E., Lee J.-C., Nakamura K., Rudick R.A. Gray matter atrophy in multiple sclerosis: a longitudinal study. Ann. Neurol. 2008;64:255–265. doi: 10.1002/ana.21436. [DOI] [PubMed] [Google Scholar]
- Fjell A.M., Walhovd K.B., Fennema-notestine C., Mcevoy L.K., Hagler D.J., Holland D., Brewer J.B., Dale A.M. Vol. 29. 2009. One-year Brain Atrophy Evident in Healthy Aging; pp. 15223–15231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IBM Corp. Released . IBM Corp; Armonk, NY: 2011. IBM SPSS Statistics for Windows, Version 20.0. [Google Scholar]
- Jain S., Sima D.M., Ribbens A., Cambron M., Maertens A., Van Hecke W., De Mey J., Barkhof F., Steenwijk M.D., Daams M., Maes F., Van Huffel S., Vrenken H., Smeets D. Automatic segmentation and volumetry of multiple sclerosis brain lesions from MR images. NeuroImage Clin. 2015;8:367–375. doi: 10.1016/j.nicl.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappos L., Freedman M.S., Polman C.H., Edan G., Hartung H.P., Miller D.H., Montalbán X., Barkhof F., Radü E.-W., Bauer L., Dahms S., Lanius V., Pohl C., Sandbrink R. Effect of early versus delayed interferon beta-1b treatment on disability after a first clinical event suggestive of multiple sclerosis: a 3-year follow-up analysis of the BENEFIT study. Lancet. 2007;370:389–397. doi: 10.1016/S0140-6736(07)61194-5. [DOI] [PubMed] [Google Scholar]
- Klauschen F., Goldman A., Barra V., Meyer-Lindenberg A., Lundervold A. Evaluation of automated brain MR image segmentation and volumetry methods. Hum. Brain Mapp. 2009;30:1310–1327. doi: 10.1002/hbm.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- Lysandropoulos A.P., Absil J., Metens T., Mavroudakis N., Vlierberghe E. Van, Smeets D., David P., Maertens A., Hecke W. Van. Quantifying brain volumes for multiple sclerosis patients follow-up in clinical practice – comparison of 1.5 and 3 Tesla magnetic resonance imaging. Brain Behav. 2016;422:1–8. doi: 10.1002/brb3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikol D.D., Barkhof F., Chang P., Coyle P.K., Jeffery D.R., Schwid S.R., Stubinski B., Uitdehaag B.M.J. Comparison of subcutaneous interferon beta-1a with glatiramer acetate in patients with relapsing multiple sclerosis (the REbif vs Glatiramer Acetate in Relapsing MS Disease [REGARD] study): a multicentre, randomised, parallel, open-label trial. Lancet Neurol. 2008;7:903–914. doi: 10.1016/S1474-4422(08)70200-X. [DOI] [PubMed] [Google Scholar]
- O'Connor P., Wolinsky J.S., Confavreux C., Comi G., Kappos L., Olsson T.P., Benzerdjeb H., Truffinet P., Wang L., Miller A., Freedman M.S. Randomized trial of oral teriflunomide for relapsing multiple sclerosis. N. Engl. J. Med. 2011;365:1293–1303. doi: 10.1056/NEJMoa1014656. [DOI] [PubMed] [Google Scholar]
- Olsson T., Boster A., Fernández Ó., Freedman M.S., Pozzilli C., Bach D., Berkani O., Mueller M.S., Sidorenko T., Radue E.-W., Melanson M. Oral ponesimod in relapsing-remitting multiple sclerosis: a randomised phase II trial. J. Neurol. Neurosurg. Psychiatry. 2014;85:1198–1208. doi: 10.1136/jnnp-2013-307282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu V., Hoogstrate W.S., van Schijndel R.A., Barkhof F., Vrenken H. The amount of peripheral CSF affects brain tissue type segmentation in MS. Mult. Scler. J. 2011;17:S53–S276. [Google Scholar]
- Popescu V., Battaglini M., Hoogstrate W.S., Verfaillie S.C.J., Sluimer I.C., van Schijndel R.A., van Dijk B.W., Cover K.S., Knol D.L., Jenkinson M., Barkhof F., de Stefano N., Vrenken H. Optimizing parameter choice for FSL-Brain Extraction Tool (BET) on 3D T1 images in multiple sclerosis. NeuroImage. 2012;61:1484–1494. doi: 10.1016/j.neuroimage.2012.03.074. [DOI] [PubMed] [Google Scholar]
- Popescu V., Schoonheim M.M., Versteeg A., Chaturvedi N., Jonker M., Xavier de Menezes R., Gallindo Garre F., Uitdehaag B.M.J., Barkhof F., Vrenken H. Grey matter atrophy in multiple sclerosis: clinical interpretation depends on choice of analysis method. PLoS One. 2016;11 doi: 10.1371/journal.pone.0143942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz N., Lindenberger U., Rodrigue K.M., Kennedy K.M., Head D., Williamson A., Dahle C., Gerstorf D., Acker J.D. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb. Cortex. 2005 doi: 10.1093/cercor/bhi044. [DOI] [PubMed] [Google Scholar]
- Resnick S.M., Pham D.L., Kraut M.A., Zonderman A.B., Davatzikos C. Vol. 23. 2003. Longitudinal Magnetic Resonance Imaging Studies of Older Adults: A Shrinking Brain; pp. 3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonheim M.M., Popescu V., Rueda Lopes F.C., Wiebenga O.T., Vrenken H., Douw L., Polman C.H., Geurts J.J.G., Barkhof F. Subcortical atrophy and cognition: sex effects in multiple sclerosis. Neurology. 2012;79:1754–1761. doi: 10.1212/WNL.0b013e3182703f46. [DOI] [PubMed] [Google Scholar]
- Sharma S., Rousseau F., Heitz F., Rumbach L., Armspach J.-P. On the estimation and correction of bias in local atrophy estimations using example atrophy simulations. Comput. Med. Imaging Graph. 2013;37:538–551. doi: 10.1016/j.compmedimag.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Smeets D., Ribbens A., Sima D.M., Cambron M., Horakova D., Jain S., Maertens A., Van Vlierberghe E., Terzopoulos V., Van Binst A.-M., Vaneckova M., Krasensky J., Uher T., Seidl Z., De Keyser J., Nagels G., De Mey J., Havrdova E., Van Hecke W. Reliable measurements of brain atrophy in individual patients with multiple sclerosis. Brain Behav. 2016;6 doi: 10.1002/brb3.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Zhang Y., Jenkinson M., Chen J., Matthews P.M., Federico A., De Stefano N. Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. NeuroImage. 2002;17:479–489. doi: 10.1006/nimg.2002.1040. [DOI] [PubMed] [Google Scholar]
- Steenwijk M.D., Pouwels P.J.W., Daams M., van Dalen J.W., Caan M.W.A., Richard E., Barkhof F., Vrenken H. Accurate white matter lesion segmentation by k nearest neighbor classification with tissue type priors (kNN-TTPs) NeuroImage Clin. 2013;3:462–469. doi: 10.1016/j.nicl.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwijk M.D., Daams M., Pouwels P.J.W., Balk L.J., Tewarie P.K., Killestein J., Uitdehaag B.M.J., Geurts J.J.G., Barkhof F., Vrenken H. What explains gray matter atrophy in long-standing multiple sclerosis? Radiology. 2014;272:832–842. doi: 10.1148/radiol.14132708. [DOI] [PubMed] [Google Scholar]
- Vrenken H., Jenkinson M., Horsfield M.A., Battaglini M., van Schijndel R.A., Rostrup E., Geurts J.J.G., Fisher E., Zijdenbos A., Ashburner J., Miller D.H., Filippi M., Fazekas F., Rovaris M., Rovira A., Barkhof F., de Stefano N. Recommendations to improve imaging and analysis of brain lesion load and atrophy in longitudinal studies of multiple sclerosis. J. Neurol. 2013;260:2458–2471. doi: 10.1007/s00415-012-6762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material