Abstract
Background
An uncharacterized histone H2a-coding transcript (E130307C13) has been cloned from a mouse full-length cDNA library. This transcript is encoded on chromosome 6, approximately 4 kb upstream of a histone H4 gene, Hist4h4. The proteins encoded by this transcript and the human H2afj mRNA isoform-2 have the highest amino acid similarity. In this paper, we characterize it from the expression pattern given by quantitative RT-PCR.
Results
Quantitative RT-PCR indicated that the gene that encodes E130307C13 (E130307C13) is regulated in a replication-independent manner, and therefore it is H2afj. Certainly, H2afj transcript lacks a stem-loop structure at the 3'-UTR but contains a poly (A) signal. In addition, its promoter region has a different structure from those of the replication-dependent histone H2a genes.
Conclusion
The bioinformatics imply that E130307C13 is a replication-independent H2a gene. In addition, quantitative RT-PCR analysis shows that it is replication-independent. Thus, it is H2afj, a novel replication-independent H2a gene in mouse.
Background
Replication of the eukaryotic chromosomes requires the synthesis of histones to package the newly replicated DNA into chromatin. Control of the level of histone mRNA accounts for much of the control of histone protein synthesis [1]. Mouse has 18 replication-dependent histone H2a genes [2]. Among these 18 genes, 13 genes are located in the Hist1 cluster on chromosome 13, 4 in the Hist2 cluster on chromosome 3 and 1 in the Hist3 cluster on chromosome 11 [2]. The histone mRNAs that are cell-cycle-regulated increase 35-fold as cells progress from mitosis through G1-phase and into S-phase [3]. The promoters of histone genes contain CCAAT and TATA boxes [4]. The replication-dependent histone H2a genes lack introns and a poly (A) signal. They have a conserved stem-loop structure (5'-GGCTCTTTTCAGAGCC-3') at the 3'-UTR, which plays an important role in mRNA processing and stability [5-7].
Mouse also has two replication-independent histone H2a genes, H2afx on chromosome 9 and H2afz chromosome 3. These two genes encode polyadenylated mRNAs. H2afx mRNA has both a polyadenylated tail and a stem-loop structure [8]. Mouse replication-dependent histone H2a mRNAs and H2afx mRNA have a nuclear export element (5'-ACAACAAGAAGACGCGCATCAT-3') in the protein-coding region that functions to export the mRNA from the nucleus to the cytoplasm [9].
An uncharacterized histone H2a-coding transcript (E130307C13, FANTOM clone ID; NM_177688, Genebank accession number) has been cloned from a mouse full-length cDNA library. E130307C13 lies on chromosome 6, approximately 4 kb upstream of Hist4h4. In this paper, we characterize it from the expression pattern given by quantitative RT-PCR. In addition to this, we compared the structure with the sequences deposited in the international DNA/protein database.
Results and discussion
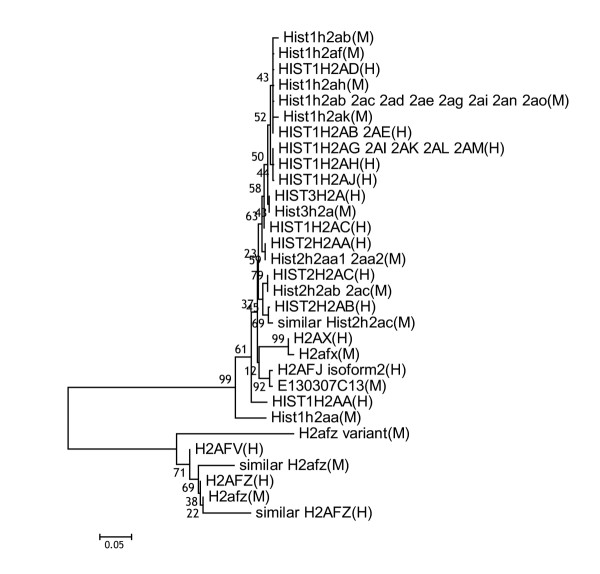
Comparison of the putative amino acid sequence encoded by E130307C13 with the amino acid sequences deposited in the international DNA/protein database showed that it has the highest similarity to that encoded by human H2afj mRNA isoform-2 (NM_177925, Genebank accession number; Figs. 1, 2). Human has two isoforms of H2afj [10,11]. Isoform-1 (NM_018267, Genebank accession number) is produced after splicing of two introns; isoform-2 does not need intron splicing for maturation. Interestingly, H2afj also lies near a histone H4 gene, on human chromosome 12. According to the nomenclature of histone genes [2], if E130307C13 is regulated in a replication-dependent manner, it is recognized as Hist4h2a. But if it is regulated in a replication-independent manner, it is recognized as H2afj.
Figure 1.
Phylogenetic relationships among 44 amino acids sequences from human and mouse histone H2a related proteins. The bar indicates 5% difference of sequence. The numbers at the branches indicate percentage of 1000 bootstrap analyses. (H) and (M) indicate the human sequence and mouse sequence, respectively.
Figure 2.
Alignment of the human and mouse histone H2a proteins in Fig. 1.
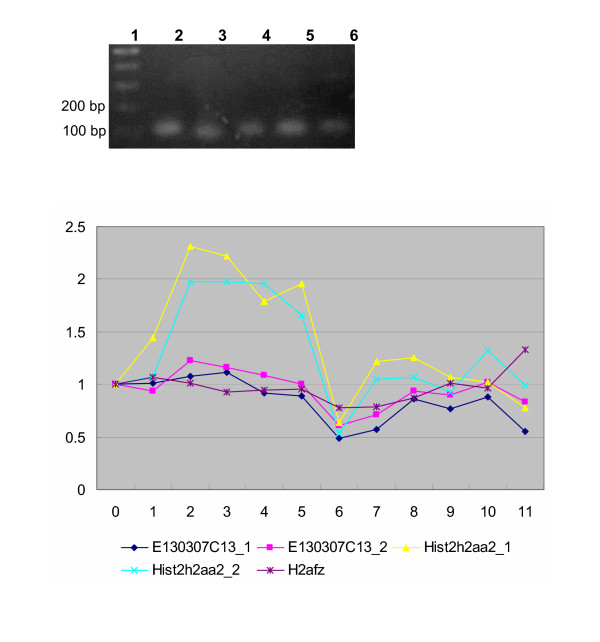
Each product of the quantitative RT-PCR gave a single band on the agarose gel of the expected size (Fig. 3). Observation using the quantitative RT-PCR showed that the expression pattern of E130307C13 is typical of replication-independent histone gene (Table 1, Fig. 3). The expression pattern of E130307C13 is more similar to that of the replication-independent H2afz than to that of the replication-dependent Hist2h2aa2. The expression of Hist2h2aa2 increased along with cell cycle progression from the beginning of S-phase (0 h), peaked at 2 h, and then decreased (Fig. 3). On the other hand, E130307C13 and H2afz were expressed constantly (Fig. 3). These results suggest that E130307C13 is a replication-independent histone H2a gene in mouse. According to the nomenclature of histone genes [2], E130307C13 is recognized not as Hist4h2a but as H2afj.
Figure 3.
RT-PCR products on agarose gel and expression patterns. Lanes 1, 100 bp ladder; 2, RT-PCR product amplified with E130307C13 primer set 2; 3, that with E130307C13 primer set 1; 4, that with Hist2h2aa2 primer set 1; 5, that with Hist2h2aa2 primer set 2; 6, that with H2afz primer set.
Table 1.
CT values, ΔCT, and ΔΔCT
| Time (h) | GAPDH | E130307C13_1 | ΔCT | ΔΔCT | Expression | E130307C13_2 | ΔCT | ΔΔCT | Expression | Hist2h2aa_1 | ΔCt | ΔΔCT | Expression | Hist2h2aa_2 | ΔCT | ΔΔCT | Expression | H2afz | ΔCT | ΔΔCT | Expression |
| 0 | 14.4 | 19.1 | 4.65 | 0 | 1 | 20 | 5.61 | 0 | 1 | 18.1 | 3.63 | 0 | 1 | 19.3 | 4.9 | 0 | 1 | 16.8 | 2.35 | 0 | 1 |
| 1 | 14.4 | 19 | 4.64 | -0.01 | 1.01 | 20.1 | 5.71 | 0.1 | 0.93 | 17.5 | 3.1 | -0.53 | 1.44 | 19.2 | 4.79 | -0.11 | 1.08 | 16.6 | 2.26 | -0.09 | 1.06 |
| 2 | 14.4 | 18.9 | 4.54 | -0.11 | 1.08 | 19.7 | 5.32 | -0.29 | 1.22 | 16.8 | 2.42 | -1.21 | 2.31 | 18.3 | 3.92 | -0.98 | 1.97 | 16.7 | 2.33 | -0.02 | 1.01 |
| 3 | 14.3 | 18.8 | 4.5 | -0.15 | 1.11 | 19.7 | 5.4 | -0.21 | 1.16 | 16.8 | 2.48 | -1.15 | 2.22 | 18.3 | 3.92 | -0.98 | 1.97 | 16.8 | 2.46 | 0.11 | 0.93 |
| 4 | 14.4 | 19.2 | 4.78 | 0.13 | 0.91 | 19.9 | 5.49 | -0.12 | 1.09 | 17.2 | 2.79 | -0.84 | 1.79 | 18.3 | 3.93 | -0.97 | 1.96 | 16.8 | 2.43 | 0.08 | 0.95 |
| 5 | 14.4 | 19.2 | 4.82 | 0.17 | 0.89 | 20 | 5.61 | 0 | 1 | 17.1 | 2.66 | -0.97 | 1.96 | 18.6 | 4.17 | -0.73 | 1.66 | 16.8 | 2.42 | 0.07 | 0.95 |
| 6 | 14.1 | 19.8 | 5.69 | 1.04 | 0.49 | 20.4 | 6.33 | 0.72 | 0.61 | 18.4 | 4.29 | 0.66 | 0.63 | 19.9 | 5.8 | 0.9 | 0.54 | 16.8 | 2.71 | 0.36 | 0.78 |
| 7 | 14.2 | 19.7 | 5.45 | 0.8 | 0.57 | 20.3 | 6.11 | 0.5 | 0.71 | 17.6 | 3.35 | -0.28 | 1.21 | 19 | 4.83 | -0.07 | 1.05 | 16.9 | 2.7 | 0.35 | 0.78 |
| 8 | 14.3 | 19.2 | 4.87 | 0.22 | 0.86 | 20.1 | 5.71 | 0.1 | 0.93 | 17.6 | 3.3 | -0.33 | 1.26 | 19.2 | 4.81 | -0.09 | 1.06 | 16.9 | 2.55 | 0.2 | 0.87 |
| 9 | 14.3 | 19.3 | 5.03 | 0.38 | 0.77 | 20 | 5.76 | 0.15 | 0.9 | 17.8 | 3.54 | -0.09 | 1.06 | 19.3 | 5.01 | 0.11 | 0.93 | 16.6 | 2.34 | -0.01 | 1.01 |
| 10 | 14.2 | 19.1 | 4.83 | 0.18 | 0.88 | 19.8 | 5.58 | -0.03 | 1.02 | 17.8 | 3.6 | -0.03 | 1.02 | 18.7 | 4.5 | -0.4 | 1.32 | 16.6 | 2.4 | 0.05 | 0.97 |
| 11 | 14.3 | 19.8 | 5.5 | 0.85 | 0.55 | 20.2 | 5.87 | 0.26 | 0.84 | 18.3 | 4 | 0.37 | 0.77 | 19.2 | 4.91 | 0.01 | 0.99 | 16.2 | 1.94 | -0.41 | 1.33 |
Upstream of the 5'-end of E130307C13, no TATA box was found. In addition, the first CCAAT box lies 230 bases upstream of the translation start codon (Fig. 4). The other replication-dependent H2a genes have the first CCAAT and TATA boxes within 100 bases upstream of the translation start codon (Fig. 4). The replication-independent genes H2afx and H2afz also have a TATA box (Fig. 4). Thus, histone H2a genes have a TATA box in the promoters, except for E130307C13. Interestingly, the promoter of H2afz lacks CCAAT box but includes TATA box, on the other hand, that of E130307C13 lacks TATA box but includes CCAAT box (Fig. 4).
Figure 4.
Sequences between CCAAT and TATA boxes upstream of the histone H2a and E130307C13 coding regions. Underlines indicate CCAAT and TATA boxes. The ATGs located at the 3'-end indicate translation start codon. Numbers in parentheses represent numbers of bases not shown. The Hist1h2aj is a pseudogene, lacking a start codon.
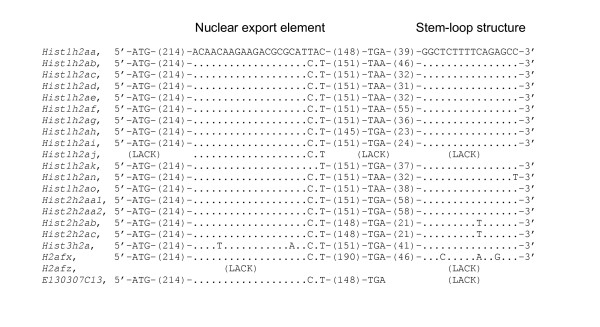
In addition, the 3'-UTR of the E130307C13 mRNA does not include the conserved stem-loop structure (Fig. 5). But the E130307C13 mRNA has two poly (A) signals at the middle and near the 3'-end. It indicates that the E130307C13 gives rise to two differentially polyadenylated mRNA transcripts. Considering the position at the 3'-UTR, it has a possibility that the poly (A) signal near the 3'-end is functional. Except for the pseudogene Hist1h2aj, the replication-dependent H2a mRNAs have the conserved stem-loop structure at the 3'-UTR. H2afx gives rise to a cell-cycle-regulated mRNA ending in the stem-loop when it is transcribed during S-phase, and a polyadenylated mRNA that is present in G1-phase cells [2,8]. H2afz mRNA lacks the stem-loop structure and has poly (A) signals. H2afz contains four introns in the protein-coding region and needs a splicing mechanism to produce the mature mRNA. Interestingly, E130307C13 lacks introns. Comparing the transcript structure of E130307C13 with those of the other histone H2a genes suggests that E130307C13 has replication-independent characteristics.
Figure 5.
Alignment of nuclear export elements and stem-loop structures. ATG indicates translation start codon. TAA or TGA indicates translation stop codon.
Conclusion
The bioinformatics imply that E130307C13 is a replication-independent H2a gene. In addition, quantitative RT-PCR analysis shows that it is replication-independent. Thus, it is H2afj, a novel replication-independent H2a gene in mouse.
Methods
Phylogenetic tree construction
Multiple alignment of 44 amino acids sequences of histone H2a related proteins from human and mouse was created using the CLUSTAL W [12] on the DNA Databank of Japan. The phylogenetic tree by the neighbor-joining method with 1000 bootstrap analyses was constructed based on the multiple alignment using MEGA version 2.1 [13].
Cell cycle synchronization
The cell cycle of Hepa 1–6 cells was synchronized at the end of G1-phase by the addition of thymidine-hydroxyurea. The cell cycle arrest was released by washing out the thymidine-hydroxyurea, then the cells were harvested at intervals of 1 h from 0 h to 11 h.
RNA extraction
Total RNA was extracted by using the RNeasy mini kit (Qiagen) according to the manual for the cell line. After that, each sample was treated with DNase I.
cDNA synthesis
RNA (approximately 0.5 μg) and random hexamer primers were heated to 70°C for 10 min, followed by cooling on ice for 5 min. The cDNA was synthesized using Superscript III First Strand buffer (Invitrogen) according to the manual. The reverse transcriptase was inactivated by a 15-min incubation at 70°C.
Quantitative PCR
The following primers were used: 5'-AACTGTAGCCCGGCCCG-3' and 5'-TTCGTCTGTTTGCGCTTT-3' (primer set 1, product size 100 bp) and 5'-CAACAAGCTGCTGGGCAAA-3' and 5'-TCGCCTTATGGTGGCTCTCC-3' (primer set 2, product size 101 bp) for transcripts of Hist2h2aa2; 5'-ACTCCGGAAAGGCCAAGACA-3' and 5'-GTTGTCCTAGATTTCAGGTG-3' for H2afz, product size 100 bp; 5'-CGTCCTGCCCAATATCCAG-3' and 5'-TCTGCACCCGTCTGTCG-3' (primer set 1, product size 90 bp) and 5'-AAGCAGGGCGGTAAGGTG-3' and 5'-TCCGCGTAGTTGCCCTTC-3' (primer set 2, product size 110 bp) for E130307C13; and 5'-TGTGTCCGTCGTGGATCTGA-3' and 5'-CCTGCTTCACCACCTTCTTGA-3' for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), product size 76 bp. Quantification of GAPDH mRNA was used as a control for data normalization. PCR amplification was performed on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). The PCR conditions were an initial step of 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C. The SYBR premix Ex Taq (Takara) was used according to the manual. Expression was assessed by evaluating threshold cycle (CT) values. The relative amount of expressed RNA was calculated using Livak and Schmittgen's method [14].
Authors' contributions
HN designed this study, carried out the molecular biological studies, and the molecular evolutionary studies. TS and YT carried out synchronization of cells and quantitative RT-PCR. YH participated in the design of this study.
Acknowledgments
Acknowledgements
We thank the two anonymous reviewers for their helpful comments. This work was supported in part by a Research Grant for the RIKEN Genome Exploration Research Project, a Research Grant for Advanced and Innovational Research Program in Life Science, and a Research Grant for the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of the Japanese Government, and in part by a Research Grant for CREST of Japan Science and Technology Corporation to Y.H., and supported in part by grant 15770055 from the MEXT to H.N.
Contributor Information
Hiromi Nishida, Email: hnishida@gsc.riken.jp.
Takahiro Suzuki, Email: t-suzuki@gsc.riken.jp.
Yasuhiro Tomaru, Email: tomaru@gsc.riken.jp.
Yoshihide Hayashizaki, Email: yosihide@gsc.riken.jp.
References
- Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. doi: 10.1016/S0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. doi: 10.1016/S0888-7543(02)96850-3. [DOI] [PubMed] [Google Scholar]
- Harris ME, Böhni R, Schneiderman MH, Ramamurthy L, Schümperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3' end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/S0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- Graves RA, Pandey NB, Chodchoy N, Marzluff WF. Translation is required for regulation of histone mRNA degradation. Cell. 1987;48:615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygum H, Yang X, Duronin RJ, Marzluff WF. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3' processing in vivo. Mol Cell Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannironi C, Bonner WM, Hatch CL. H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3' processing signals. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol Cell. 2001;7:899–905. doi: 10.1016/S1097-2765(01)00233-7. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Musashino K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, Brownstein MJ, Usdin TB, Toshiyuki S, Carninci P, Prange C, Raha SS, Loquellano NA, Peters GJ, Abramson RD, Mullahy SJ, Bosak SA, McEwan PJ, McKernan KJ, Malek JA, Gunaratne PH, Richards S, Worley KC, Hale S, Garcia AM, Gay LJ, Hulyk SW, Villalon DK, Muzny DM, Sodergren EJ, Lu X, Gibbs RA, Fahey J, Helton E, Ketteman M, Madan A, Rodrigues S, Sanchez A, Whiting M, Madan A, Young AC, Shevchenko Y, Bouffard GG, Blakesley RW, Touchman JW, Green ED, Dickson MC, Rodriguez AC, Grimwood J, Schmutz J, Myers RM, Butterfield YS, Krzywinski MI, Skalska U, Smailus DE, Schnerch A, Schein JE, Jones SJ, Marra MA. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment though sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]