Abstract
Aquaporins (AQPs) are a class of integral membrane proteins involved in the transport of water and many other small solutes. The AQPs have been extensively studied in many land species obtaining water and nutrients from the soil, but their distribution and evolution have never been investigated in aquatic plant species, where solute assimilation is mostly through the leaves. In this regard, identification of AQPs in the genome of Zostera marina L. (eelgrass), an aquatic ecological model species could reveal important differences underlying solute uptake between land and aquatic species. In the present study, genome-wide analysis led to the identification of 25 AQPs belonging to four subfamilies, plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs) in eelgrass. As in other monocots, the XIP subfamily was found to be absent from the eelgrass genome. Further classification of subfamilies revealed a unique distribution pattern, namely the loss of the NIP2 (NIP-III) subgroup, which is known for silicon (Si) transport activity and ubiquitously present in monocot species. This finding has great importance, since the eelgrass population stability in natural niche is reported to be associated with Si concentrations in water. In addition, analysis of available RNA-seq data showed evidence of expression in 24 out of the 25 AQPs across four different tissues such as root, vegetative tissue, male flower and female flower. In contrast to land plants, higher expression of PIPs was observed in shoot compared to root tissues. This is likely explained by the unique plant architecture of eelgrass where most of the nutrients and water are absorbed by shoot rather than root tissues. Similarly, higher expression of the TIP1 and TIP5 families was observed specifically in male flowers suggesting a role in pollen maturation. This genome-wide analysis of AQP distribution, evolution and expression dynamics can find relevance in understanding the adaptation of aquatic and land species to their respective environments.
Keywords: solute transport, aquaporin evolution, comparative genomics, nodulin 26-like intrinsic proteins, silicon transporter
Introduction
Seagrasses are a group of monocotyledonous angiosperms that diverged from the terrestrial monocots about 130 MYA and subsequently adapted to completely submerged conditions of the marine environment (Janssen and Bremer, 2004). Eelgrass (Zostera marina L.) is an important aquatic weed found in the Atlantic and Pacific oceans as far as the Arctic circle. It provides habitat for several species of fish and invertebrates. Eelgrass improves water quality by absorbing pollutants, and prevents erosion by binding sediments (Moore, 2004). Considering its structural and functional role and importance in many coastal ecosystems, it was recently fully sequenced (Olsen et al., 2016). Genome analysis revealed loss and gain of multiple genes in Z. marina compared to terrestrial or floating aquatic plants, changes assumed to facilitate its adaptation to marine life (Olsen et al., 2016). These adaptations include morphological, physiological and breeding pattern modifications along with the ability to tolerate high salt levels of marine environments.
Notwithstanding the benefits it provides to marine ecosystems, worldwide estimation of eelgrass population suggests a 30% reduction over the past 30 years (Waycott et al., 2009). This is mostly associated with human disturbances (overfishing, eutrophication) and climate-change factors such as increased temperature, changes in mean sea level and biochemical composition of the sea water (Jarvis et al., 2014; Thom et al., 2014). Lower levels of silicon (Si) in marine water is also considered as one of the main reasons for reduced eelgrass population (Kamermans et al., 1999; Disney et al., 2014). In this context, a better understanding of the molecular mechanisms involved in the transport/acquisition of Si and other solutes in eelgrass could find relevance in explaining the alarming reduction in eelgrass populations.
The adverse effect of lower concentrations of dissolved Si on the growth of diatoms has been frequently reported. The diatoms are an important component of ecosystems, and changes in diatom populations substantially affect the marine food web (Ragueneau et al., 2006). Significant changes in the biogeochemistry and aquatic food webs of coastal marine environments have been observed with reduction in dissolved Si content in the Black and Baltic Seas (Humborg et al., 2000). A decreasing proportion of diatoms associated with an increase of flagellates was one of the interesting observations thought to explain the reduction in dissolved Si. The role of Si in aquatic ecosystems has been mostly studied with observational methods, so molecular experiments would contribute to a better definition of this role (Schoelynck and Struyf, 2016). More specifically, a better understanding of Si-transporting aquaporins in aquatic species would adequately support ecological experiments.
Aquaporins are small (21–34 kD) integral proteins, which form transmembrane channels to facilitate movement of water and many other solutes across the cell membrane. The topology of AQPs resembles that of an hourglass structure formed by six transmembranes (TM) α helices (H1 to H6) connected with five inter-helical loops (A to E). At the center of the pore formed by the six TM domains, two distinct constricts are formed, one with highly conserved NPA (Asn-Pro-Ala) motifs and another with four amino acid aromatic arginine (ar/R) region in the channel. These two constrictions determine solute permeability of the AQPs (Lee et al., 2005; Törnroth-Horsefield et al., 2006; Deshmukh et al., 2015).
Aquaporins are found in most living organisms including microbes, animals, and plants. However, AQP’s are comparatively more abundant and diverse in plants than in any other organisms. Based on sequence similarity, plant AQPs were grouped into five subfamilies: plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), nodulin 26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs) and uncategorized intrinsic protein (XIPs) (Chaumont et al., 2001; Johanson et al., 2001; Quigley et al., 2002; Kaldenhoff and Fischer, 2006). Among these, the PIP, TIP, and NIP subfamilies are well-characterized with regards to their localization and function. In addition to water, members of these subfamilies are known to transport urea (Li and Wang, 2014), lactic acid (Bienert et al., 2013), glycerol (Biela et al., 1999), metalloids like boron and silicon (Ampah-Korsah et al., 2016; Song et al., 2016; Ouellette et al., 2017), and gasses like ammonia (NH3) (Jahn et al., 2004), carbon dioxide (CO2) (Kaldenhoff et al., 2014) and hydrogen peroxide (H2O2) (Tian et al., 2016). All AQP subfamilies are widely distributed in different plant species including primitive land plants, with the exception of XIPs that are absent in some higher plants such as Brassicaceae and monocots (Gupta and Sankararamakrishnan, 2009; Deshmukh et al., 2015; Sonah et al., 2017). With the availability of complete genome sequences, the genes encoding AQPs have been characterized in many land plants such as Arabidopsis, rice, Populus, soybean, canola and tomato (Deshmukh et al., 2016a). Surprisingly enough, no systematic study of AQPs has been carried out in aquatic plant species. The recent availability of the annotated genome sequences of eelgrass provides an opportunity to investigate and compare the role of AQPs in plants adapted to aquatic environments.
In the present study, we have performed a genome-wide identification of AQPs in Z. marina. Subsequently, characterization of AQPs was conducted based on phylogenetic analysis, gene structure organization, conserved motifs, ar/R selectivity filters, and homology-based 3D protein structure. Finally, AQP expression profiling in different tissues was studied using available transcriptomic data.
Materials and Methods
Genome-Wide Identification and Distribution of AQPs in Zostera marina
The genome sequence of Z. marina V2.2 was retrieved from the Phytozome database1. A local database of the predicted protein sequences from Z. marina genome was created using BioEdit ver. 7.2.5 (Hall, 1999). Aquaporin homologs were identified by BLASTp search performed against the local database using query sequences of 141 AQPs from rice, Arabidopsis, and soybean (Supplementary Data Sheet 1). An e-value of 10-5 was kept as an initial cut-off to identify high scoring pairs (HSPs). The blast output was tabulated, and the HSPs with >100-bit score was selected. Finally, redundant hits were removed to select unique sequences for further analysis.
Structural Characterization of Zostera marina Aquaporins
The genomic and cDNA sequences of AQPs identified in Z. marina were retrieved from Phytozome database. Structural annotations of the gene models (in gff3 format) were also retrieved from Phytozome. The gene structure of Z. marina AQPs was analyzed using Gene Structure Display Server (GSDS) ver. 2.0 (Hu et al., 2015).
Identification of Functional Motif and Transmembrane Domains
The NPA motifs were identified in protein sequences using conserved domain database at NCBI (CDD). Aquaporins with missing NPA motifs were confirmed with a manual examination. Transmembrane domains in the genes were identified using TMHMM and SOSUI software tools2,3. The TM domains were manually evaluated to confirm alterations or complete loss.
Phylogenetic Analysis of Zostera marina AQPs
The AQP sequences were aligned using CLUSTALW alignment function in MEGA6 (Kumar et al., 2008). The phylogenetic tree was constructed by using maximum likelihood method, and the stability of the branch node was measured by performing 1000 bootstraps. The subfamilies PIP, SIP, TIP, NIP, and XIPs were classified in accordance with the nomenclature used for Arabidopsis, rice and poplar (Quigley et al., 2002; Deshmukh et al., 2015).
Expression Profiling of Zostera marina Aquaporins
The RNA-Seq dataset available at SRA database under the accession SRP056873 was used to analyze the expression of AQPs. A heat map for expression of AQPs was constructed using TIGR Multi Experiment Viewer (MeV4). Hierarchical clustering with average linkage method was performed to cluster the genes (Sonah et al., 2016).
Results
Genome-Wide Identification and Distribution of AQPs in Zostera marina
Genome-wide analysis of Z. marina led to the identification of 25 genes encoding AQPs (Supplementary Table 1). Conserved domain analysis confirmed candidate AQPs as members of the MIP (Major Intrinsic Protein) family (Supplementary Table 2). Prediction of transmembrane helices based on a hidden Markov model revealed the presence of six signature transmembrane domains in 21 out of the 25 identified AQPs (Supplementary Table 3). Furthermore, homology based tertiary protein structure of the AQPs confirmed the typical hourglass-like structure for all 25 proteins.
The Z. marina AQPs were found to be distributed among 22 scaffolds. Out of the 22 scaffolds, 19 contained only one AQP while three scaffolds, 31, 132, and 231, contained two (Table 1). Analysis of genomic distribution of Z. marina AQPs revealed a tandem duplication of ZmNIP4 family members, ZmNIP4-1 and ZmNIP4-2 located on scaffold_231.
Table 1.
Description and distribution of aquaporins identified in Zostera marina genome.
| Scaffold |
Protein |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sl. No. | Gene | Phytozome ID | Gene Length (bp) | Location | Start | End | Transcript length (bp) | CDS length (bp) | Length (aa) | mw (kDa) | pI |
| 1 | ZmNIP1-1 | Zosma431g00060 | 1224 | Scaffold_431 | 34873 | 36096 | 906 | 837 | 278 | 29.52 | 8.62 |
| 2 | ZmNIP1-2 | Zosma7531g00010 | 642 | Scaffold_7531 | 636 | 1277 | 642 | 642 | 213 | 22.52 | 9.99 |
| 3 | ZmNIP1-3 | Zosma84g00450 | 1202 | Scaffold_84 | 363618 | 364819 | 879 | 660 | 219 | 23.81 | 5.9 |
| 4 | ZmNIP4-1 | Zosma231g00020 | 964 | Scaffold_231 | 17959 | 18922 | 801 | 801 | 266 | 29.09 | 8.3 |
| 5 | ZmNIP4-2 | Zosma231g00030 | 752 | Scaffold_231 | 29752 | 30503 | 666 | 666 | 221 | 24.43 | 8.59 |
| 6 | ZmNIP5-1 | Zosma22g01220 | 1447 | Scaffold_22 | 939187 | 940633 | 1294 | 765 | 254 | 26.49 | 6.78 |
| 7 | ZmNIP5-2 | Zosma2446g00010 | 1044 | Scaffold_2446 | 1 | 1044 | 891 | 576 | 192 | 19.96 | 9.04 |
| 8 | ZmNIP5-3 | Zosma26g01390 | 873 | Scaffold_26 | 859679 | 860551 | 720 | 720 | 239 | 25.21 | 9.68 |
| 9 | ZmPIP1-1 | Zosma129g00200 | 1252 | Scaffold_129 | 359101 | 360352 | 1172 | 876 | 291 | 31.39 | 8.3 |
| 10 | ZmPIP1-2 | Zosma16g00380 | 1100 | Scaffold_16 | 235799 | 236898 | 708 | 708 | 235 | 25.55 | 8.22 |
| 11 | ZmPIP2-1 | Zosma21g01120 | 1348 | Scaffold_21 | 945011 | 946358 | 1268 | 843 | 280 | 29.97 | 9.54 |
| 12 | ZmPIP2-2 | Zosma49g00340 | 892 | Scaffold_49 | 249623 | 250514 | 819 | 819 | 272 | 28.97 | 8.45 |
| 13 | ZmSIP1-1 | Zosma12g00280 | 2031 | Scaffold_12 | 480219 | 482249 | 1014 | 774 | 257 | 27.63 | 9.63 |
| 14 | ZmSIP1-2 | Zosma221g00210 | 1785 | Scaffold_221 | 86562 | 88346 | 1160 | 726 | 241 | 26.15 | 7.56 |
| 15 | ZmSIP2-1 | Zosma132g00610 | 1209 | Scaffold_132 | 414430 | 415638 | 1115 | 765 | 254 | 28.17 | 10.38 |
| 16 | ZmSIP2-2 | Zosma132g00640 | 940 | Scaffold_132 | 420728 | 421667 | 768 | 768 | 255 | 28.36 | 10.08 |
| 17 | ZmSIP2-3 | Zosma29g00960 | 1005 | Scaffold_29 | 581320 | 582324 | 866 | 816 | 271 | 29.57 | 9.55 |
| 18 | ZmTIP1-1 | Zosma24g00710 | 979 | Scaffold_24 | 469609 | 470587 | 759 | 759 | 252 | 26.39 | 8.65 |
| 19 | ZmTIP1-2 | Zosma31g00130 | 846 | Scaffold_31 | 53817 | 54662 | 756 | 756 | 251 | 25.66 | 5.09 |
| 20 | ZmTIP1-3 | Zosma43g00730 | 907 | Scaffold_43 | 543521 | 544427 | 753 | 753 | 250 | 26.21 | 7.23 |
| 21 | ZmTIP1-4 | Zosma470g00020 | 1553 | Scaffold_470 | 48092 | 49644 | 1397 | 807 | 268 | 28.24 | 6.96 |
| 22 | ZmTIP1-5 | Zosma50g01000 | 1332 | Scaffold_50 | 651788 | 653119 | 1117 | 750 | 249 | 25.98 | 6.68 |
| 23 | ZmTIP1-6 | Zosma127g00340 | 839 | Scaffold_127 | 475276 | 476114 | 759 | 759 | 252 | 25.88 | 6.13 |
| 24 | ZmTIP3-1 | Zosma4g01350 | 950 | Scaffold_4 | 1086053 | 1087002 | 720 | 720 | 239 | 25.44 | 6.97 |
| 25 | ZmTIP5-1 | Zosma31g01330 | 884 | Scaffold_31 | 757025 | 757908 | 813 | 813 | 270 | 28.24 | 6.93 |
Phylogenetic Distribution of AQPs in Zostera marina
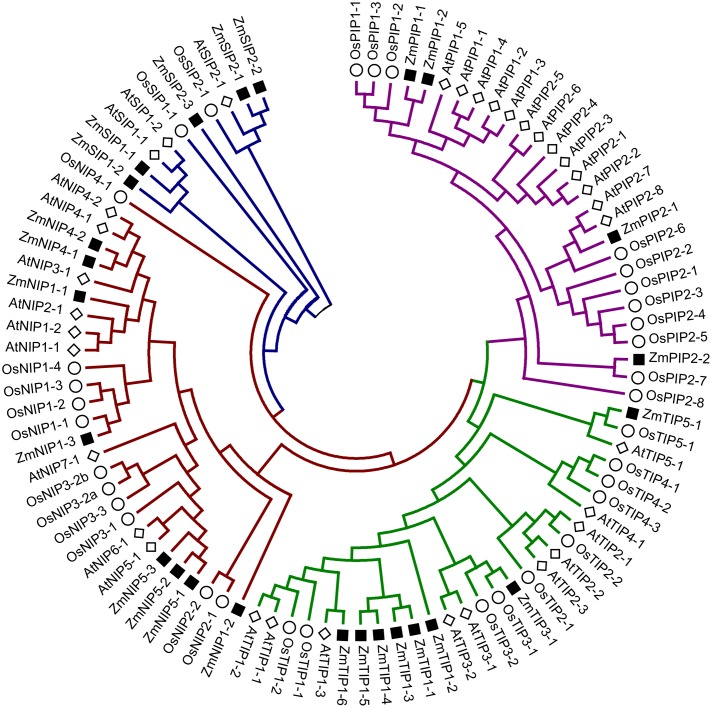
Phylogenetic tree of Z. marina AQPs with the known AQPs from Arabidopsis thaliana, and Oryza sativa showed four distinct clusters representing different subfamilies of AQPs (Figure 1). The Z. marina AQPs were named according to their grouping with the known AQPs, which showed 4 PIPs, 8 TIPs, 8 NIPs, and 5 SIPs. Members of the XIP family are absent from Z. marina genome. Within the groups formed by Z. marina AQPs, two major subgroups were found in ZmPIPs: ZmPIP1 and ZmPIP2. Both ZmPIP1 and ZmPIP2 are comprised of two members each. Similarly, the ZmTIPs formed five subgroups with ZmTIP1 having six members, and ZmTIP3 and ZmTIP5 having one member each. The ZmNIPs formed two distinct groups, ZmNIP1 and ZmNIP5, which are comprised of three members each, and ZmNIP4 containing two members. The NIP2 subgroup (commonly classified as NIP-III) was not found in Z. marina genome, a rather surprising result since the NIP-III subfamily is involved in silicon transport and has been ubiquitously reported in different monocots (Figure 2). ZmSIPs formed two subgroups, ZmSIP1 and ZmSIP2, represented by two and three members, respectively. BLAST search performed against non-redundant Z. marina nucleotide sequences at NCBI further confirmed the loss of XIPs and NIP-IIIs.
FIGURE 1.
Phylogenetic tree of Zostera marina aquaporins (AQPs) along with rice and Arabidopsis AQPs representing five different groups. The genes from Z. marina, rice and Arabidopsis are preceded by the prefixes Zm, Os, and At, respectively.
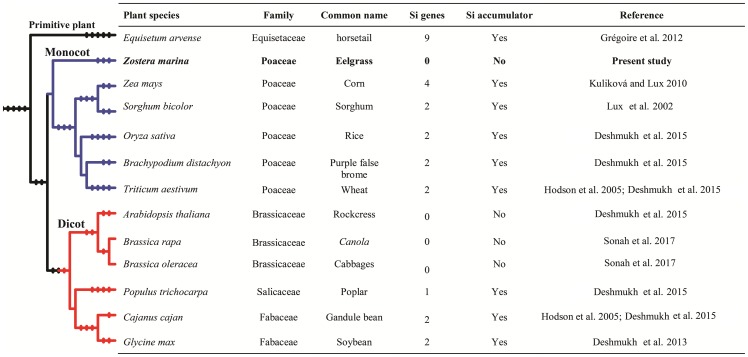
FIGURE 2.
Details of silicon accumulation, and identified silicon influx transporter genes in different monocotyledon and dicotyledon plant species.
Gene Structure, Organization and Evolution of Zostera marina AQPs
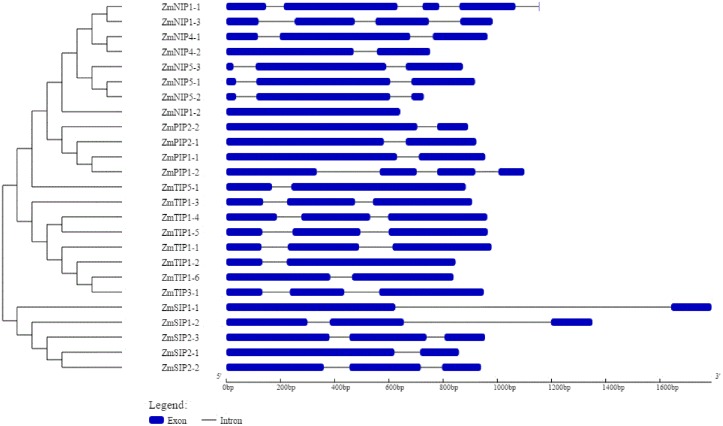
The Z. marina AQPs showed variation in transcript (ranging from 642 to 774 bp) and gene length (ranging from 642 to 2031 bp). Exon–intron structure analysis revealed intron number variation among the AQPs contributing to the variation in gene length (Figure 3). The number of introns in Z. marina AQPs ranged from zero (ZmNIP1-2) to four (ZmNIP1-1). The lowest number of introns were observed in TIPs and SIPs with 1–2 introns followed by PIPs with 1–3, and NIPs with 0–4. Among the eight TIPs, five homologs contained two introns while three homologs harbored a single intron each. Among the NIPs, most of them (4) harbored two introns, and NIP1-2 was found to be intron-free. The SIP family members showed either one or two introns. The identified AQPs from Z. marina predicted proteins ranging from 192 (ZmNIP5-2) to 291 (ZmPIP1-1) amino acids.
FIGURE 3.
Exon–intron organization of 25 aquaporin (AQP) genes identified in Zostera marina genome. Graphical representation of the gene model was obtained with Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). Exons are shown as blue boxes and introns are shown as black lines. The scale at the bottom indicates length in base pairs.
Characterization of NPA Motif, Transmembrane Domains and Sub-Cellular Localization – of Z. marina AQPs
The Z. marina AQPs showed amino acid differences at NPA motifs, ar/R selectivity filters and Froger’s positions (Table 2). Most of the AQPs contained the expected dual NPA motifs except for ZmPIP1-2, and ZmNIP1-3, which contained a single NPA motif. All members from the TIP sub-family showed conserved NPAs, while those from the SIP sub-family showed wide variation in NPA motifs compared to AQP counterparts in Arabidopsis. In the PIP sub-family, ZmPIP2-1 showed the substitution alanine to serine in the first NPA motif. ZmNIP1-2 showed a similar substitution. The NIP family members (ZmPIP5-1, ZmPIP5-2, and ZmPIP5-3) harbored serine in lieu of alanine in the first NPA motif and valine in lieu of alanine in the second one.
Table 2.
Details of conserved domains, aromatic/arginine (ar/R) selectivity filter, and Froger’s residue of aquaporins in Zostera marina genome.
| Loci | NPA (LB) | NPA (LE) | ar/R filters |
Frogers residue |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H2 | H5 | LE1 | LE2 | P1 | P2 | P3 | P4 | P5 | |||||
| Plasma membrane intrinsic proteins (PIPs) | |||||||||||||
| ZmPIP1-1 | NPA | NPA | F | H | T | R | M | S | A | F | W | ||
| ZmPIP1-2 | NPA | F | H | T | R | S | A | F | W | ||||
| ZmPIP2-1 | NPS | NPA | F | H | T | R | M | S | A | F | W | ||
| ZmPIP2-2 | NPA | NPA | F | H | T | R | S | S | A | F | V | ||
| Nodulin 26-like intrinsic proteins (NIPs) | |||||||||||||
| ZmNIP1-1 | NPA | NPA | W | V | A | R | F | T | A | Y | M | ||
| ZmNIP1-2 | NPS | NPA | F | I | G | R | F | S | A | Y | L | ||
| ZmNIP1-3 | NPA | W | V | F | H | F | |||||||
| ZmNIP4-1 | NPA | NPA | W | V | A | R | L | S | S | Y | I | ||
| ZmNIP4-2 | NPA | NPA | W | I | A | R | L | S | S | Y | M | ||
| ZmNIP5-1 | NPS | NPV | A | I | G | R | Y | T | A | Y | L | ||
| ZmNIP5-2 | NPS | NPV | A | I | G | R | Y | T | |||||
| ZmNIP5-3 | NPS | NPV | A | I | G | R | Y | T | A | Y | M | ||
| Tonoplast intrinsic proteins (TIPs) | |||||||||||||
| ZmTIP1-1 | NPA | NPA | H | I | A | V | T | A | S | Y | W | ||
| ZmTIP1-2 | NPA | NPA | Y | I | G | A | T | S | A | Y | W | ||
| ZmTIP1-3 | NPA | NPA | H | I | A | V | T | A | S | Y | W | ||
| ZmTIP1-4 | NPA | NPA | H | I | A | M | T | S | A | Y | W | ||
| ZmTIP1-5 | NPA | NPA | H | I | A | V | T | S | A | Y | W | ||
| ZmTIP1-6 | NPA | NPA | H | I | A | V | T | A | A | Y | W | ||
| ZmTIP3-1 | NPA | NPA | S | I | A | R | T | A | Y | W | |||
| ZmTIP5-1 | NPA | NPA | Q | V | G | R | T | S | A | Y | W | ||
| Small basic intrinsic proteins (SIPs) | |||||||||||||
| ZmSIP1-1 | NPT | NPA | L | V | P | N | M | A | A | Y | W | ||
| ZmSIP1-2 | SLA | NPA | I | I | P | N | M | A | A | Y | W | ||
| ZmSIP2-1 | TTL | SPA | S | D | G | K | V | F | T | N | F | ||
| ZmSIP2-2 | HPL | NPA | Y | D | S | E | F | A | A | Y | V | ||
| ZmSIP2-3 | NPL | NPA | A | H | G | T | F | A | A | Y | W | ||
All the PIP sub-family members showed conserved ar/R filter residues with phenylalanine in H2, histidine at H5, threonine at LE1, and arginine at LE2. In the TIP sub-family, the H2 position of the ar/R filter consisted of histidine/tyrosine/serine/glutamine and the H5 position was comprised of isoleucine, with the exception of valine in ZmTIP5-1. The LE1 position of TIPs was occupied by glycine/alanine, while the LE2 position had four possible residues (alanine/valine/methionine/arginine). The NIP sub-family members contained phenylalanine/tryptophan/alanine (H2), valine/isoleucine (H5) alanine/glycine (LE1) and arginine (LE2). Finally, the SIP family members harbored serine/leucine/isoleucine/alanine/tyrosine (H2), isoleucine/valine/aspartic acid/histidine (H5), glycine/proline/serine (LE1), asparagine/glutamic acid/lysine/threonine (LE2).
To ascertain the expression of Z marina AQPs at different cellular/organellar levels, their sub-cellular localizations were predicted (Supplementary Table 4). The majority of Z marina PIPs were predicted to be localized in the plasma membrane. Most of NIPs (five) were targeted to the vacuole and the remaining three were predicted to be located in the plasma membrane. Among the eight TIPs, four are predicted to be located in the cytoplasm and the other ones in the plasma membrane/nucleus/chloroplast. Among SIPs, SIP1s are predicted to be in the vacuole and SIP2s in either the plasma membrane or the mitochondria.
AQP Expression Profiling in Zostera marina
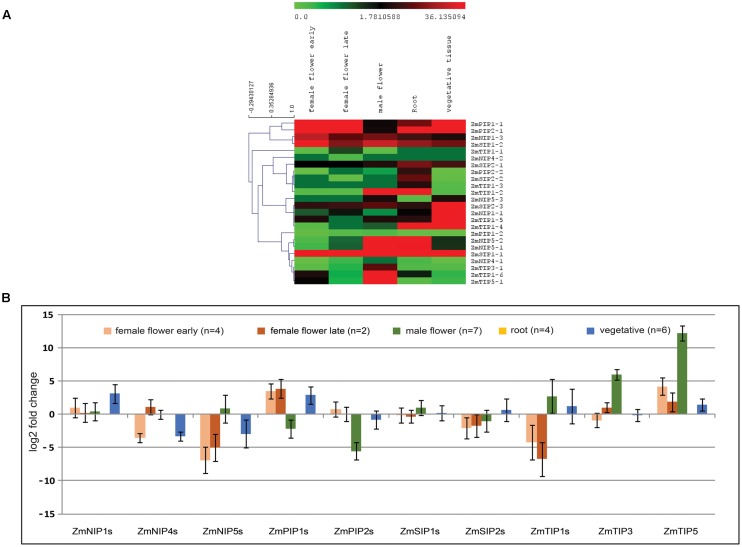
Analysis of reported RNA-seq data showed evidence of expression in 24 out of the 25 predicted Z. marina AQPs, ZmNIP1-2 being the sole absent. Family specific expression of AQPs in different tissues was calculated in terms of fold change in comparison to root tissues. The majority of AQPs showed similar or higher expression in vegetative compared to root tissues (Figure 4A). Among different AQPs, PIPs showed the highest expression across the different tissues analyzed. TIPs showed a particularly high expression in male flowers (Figure 4B).
FIGURE 4.
Analysis of Zostera marina aquaporins (AQPs) expression in different tissues using RNA-seq data (SRP056873, SRA database). (A) Normalized expression of AQPs in terms of reads per kilobase of transcript per million mapped reads (RPKM) showing higher levels of TIP1 and TIP5 in male flowers. (B) Fold change expression of aquaporin (AQP) genes in different tissues compared to roots showing similar or higher levels of expression in vegetative tissues in most AQPs, except for NIP4s and NIP5s. Bars represent standard error from the mean.
Discussion
Aquaporins have gained increased attention recently because of their reported pivotal role in biotic and abiotic stress tolerance (Hu et al., 2012; Liu et al., 2013; Shivaraj et al., 2017; Sonah et al., 2017). Considering the importance of eelgrass in marine environments, and its declining population associated with different stressful conditions, we were therefore interested in identifying all potential aquaporins in the species with the objective that it would become an invaluable resource to better understand its ecology and predicament.
Our genome-wide analyses led to the identification of 25 putative AQPs in Z. marina genome, a number inferior to reference land plants such as rice (34) (Sakurai et al., 2005; Nguyen et al., 2013) and Arabidopsis (35) (Johanson et al., 2001). Lineage of Z. marina diverged from the terrestrial monocots about 130 MYA. Its adaption to submerged conditions occasioned a loss and gain of multiple genes compared to terrestrial or floating aquatic plants (Janssen and Bremer, 2004; Olsen et al., 2016). The reduced number of AQPs in Z. marina compared to terrestrial plants represents likely an adaptive strategy during its evolution to a new environment. More specifically, the aquatic environment may call for a lesser need in proteins involved in water and solute acquisition through the roots.
Phylogenetic analysis of the Z. marina AQPs identified only four different sub-families, PIPs, TIPs, NIPs, and SIP, as observed in monocots and Brassicaceae, with the notable absence of XIP subfamily members. Interestingly, most of the dicots harbor five AQP subfamilies including XIPs (See Supplementary Table 5), which suggests a clear conservation of the absence of XIPs in monocots even following the divergence between terrestrial and aquatic plants. On the other hand, members of NIP2 (NIP-III), NIP3, TIP2, and TIP4 were not observed in Z. marina genome, while they are systematically found in monocots. The loss of the TIP subfamily is rather surprising since TIPs are relatively more conserved across diverse species compared to the rest of AQP subfamilies (Deshmukh et al., 2015). This likely means that TIPs have specific functions in terrestrial plants that are obsolete or redundant in eelgrass. Notwithstanding the differences in the members and number of AQPs in eelgrass, the gene structure and exon–intron organization were found to be quite similar compared to other terrestrial monocots (Danielson and Johanson, 2008). The conserved exon-intron organization is suggestive of functional redundancy across species. Since the exon–intron organization is known to be affected by gene duplication, diversification and changes in exon–intron organization may lead to changes in gene expression profile (Roy and Penny, 2007; Deshmukh et al., 2016b).
The substrate specificity of AQPs is determined by the size and hydrophobic nature of the amino acids forming the pores (Lee et al., 2005; Törnroth-Horsefield et al., 2006). All PIP family members from Z. marina contained a very hydrophilic ar/R selectivity filter, FHTR, a hallmark of water-transporting aquaporins, in contrast to other families. A similar ar/R selectivity filter is also observed in PIP family of aquaporins from other plant species such as O. sativa, A. thaliana, Brassica rapa, Glycine Max, and Ricinus communis (Johanson et al., 2001; Deshmukh et al., 2013; Zhang et al., 2013; Diehn et al., 2015; Zou et al., 2015). Among Z. marina TIP sub family members, TIP1s were found to have residues HIAV, YIGA, and HIAM forming a more hydrophobic ar/R filter compared to the one found in ZmTIP3 and ZmTIP5, which contained SIAR and QVGR residues, respectively. The residues present in ar/R selectivity filter of ZmTIPs were found to be similar to the TIPs from other plant species. TIPs act as functional water transporters and facilitate transport of small solutes such as NH4+, H2O2, and urea (Liu et al., 2003; Holm et al., 2005; Bienert et al., 2007). Conserved NPA motifs and ar/R filter in TIPs suggests their involvement in transport of water as well as solutes in Z. marina.
Members of NIP-III subfamily are known to be involved in transport of metalloids like boron (Takano et al., 2006) and silicon (Ma et al., 2006). Several studies have reported beneficial effects of Si on monocotyledonous plants. Different plant species accumulate a wide range of Si, from 0.2% or less to 10% Si on a dry weight basis (Lux et al., 2002; Hodson et al., 2005; Kuliková and Lux, 2010; Grégoire et al., 2012; Deshmukh et al., 2015). Very few studies have reported Si levels in Z. marina leaves; Herman et al. (1996) found a range between 0.02 and 0.66%, with higher concentrations in plant associated with higher dissolved Si in water. Interestingly, a tight positive correlation was also observed between dissolved Si levels and Z. marina population (Herman et al., 1996; Disney et al., 2014). On the other hand, Kamermans et al. (1999) could not establish if increase in dissolved Si levels resulted in increased biomass of Z. marina. This is an important feature to clarify in light of the association of declining populations of eelgrass with Si concentrations. If lower Si is indeed responsible for this situation, one must be able to establish that eelgrass benefits from Si through its uptake. Indeed, in terrestrial plants, the benefits of Si are directly correlated with a plant’s ability to absorb silicic acid. Recent studies have clearly shown that uptake of Si is mediated by aquaporins, and more specifically by NIP-IIIs containing a GSGR selectivity filter (Deshmukh and Bélanger, 2015). In the present study, we found that NIP-IIIs were completely absent from the Z. marina genome, which would indicate that the species is unable to uptake Si, a conclusion supported by the low Si levels found in the tissue of eelgrass. Under these conditions, it is difficult to rationalize how dissolved Si levels in water would influence eelgrass populations. Better controlled studies are thus needed to determine the role of Si, if any, on eelgrass. In any event, results from this study, namely the absence of NIP-IIIs, offer new avenues to better understand the ecology of eelgrass in the context of Si fluxes in marine environments.
Eelgrass has been extensively studied to understand the plant physiology particularly under aquatic and high saline condition (Yu, 2015; Yang et al., 2017). However, very limited attention was paid to study the involvement of AQPs in physiological processes. In Posidonia oceanica (Seagrass), significant role of PIPs have been observed in salinity tolerance and maintaining water balance in the leaves (Serra et al., 2013). Similarly, expression profiling performed by Cozza and Pangaro (2009) also suggested the role of AQPs in salinity and water balance in seagrass. Apart from such few reports, no significant study evaluating physiological role of AQPs has been performed in aquatic plants.
Analysis of RNA-seq data revealed higher or similar expression of different aquaporins in vegetative compared to root tissues. In terrestrial plants, in addition to providing anchorage, roots play an important role in the absorption of water and nutrients from the soil. However, in submerged aquatic plants, vegetative parts are also actively involved in uptake of water and nutrient from the surrounding environment (Barko and Smart, 1986). PIPs are known to play a central role in transport of water; additionally PIPs are known to facilitate CO2 diffusion in mesophyll tissue of A. thaliana and Nicotiana tabacum affecting photosynthesis (Flexas et al., 2006; Heckwolf et al., 2011). Our expression analysis showed higher expression of PIPs in different plant parts analyzed suggesting a possible role of PIPs in water transport and CO2 diffusion in Z. marina. The members of TIP1 and TIP5 family showed higher expression specifically in male flowers. Different studies in A. thaliana have shown pollen specific accumulation of TIP family members (TIP1;3 and TIP5;1), which are expected to be involved in pollen maturation and germination (Soto et al., 2008; Wudick et al., 2014). Similar expression pattern of homologs of these TIP members in Z. marina indicates their conserved role in pollen development affecting reproduction.
Conclusion
Genome-wide analysis of AQPs performed in the first fully sequenced marine angiosperm, Z. marina revealed several salient features. It has shown loss of AQPs in Z. marina compared to reference land plants such as rice and Arabidopsis. The reduced number of AQPs in Z. marina compared to terrestrial plants represents likely an adaptive strategy during its evolution to a new environment. The Z. marina AQPs formed only four different sub-families as observed in monocots and Brassicaceae, with the notable absence of XIP subfamily members. We also found absence of NIP-III members from the Z. marina genome, which would indicate that the species is unable to uptake Si, a conclusion supported by the low Si levels reported in the tissue of eelgrass. The absence of NIP-IIIs in Z. marina, offer new avenues to understand the ecology of eelgrass in the context of the role of Si in marine environments. The higher expression for most of the AQPs in shoots compared to roots observed with RNA-seq data suggests, as one could expect, a predominant role of vegetative tissues in uptake of water and nutrient from the surrounding environment. The members of TIP1 and TIP5 family showed higher expression specifically in male flowers as observed in A. thaliana indicating their probable role in pollen development affecting reproduction. The identification, classification, and expression of AQPs performed in the present study will be helpful for enhancing our knowledge of distribution and evolution of AQPs in aquatic plant species.
Author Contributions
SS, RD, and HS compiled the data, performed analysis, and wrote first draft of the MS. HS and JB performed expression analysis. RB planned the study, drew the conclusions and contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The project was funded by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC), the Agri-Innovation program Growing Forward 2, SaskCanola and Agriculture and Agri-Food Canada and the Canada Research Chairs Program to RB.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01334/full#supplementary-material
References
- Ampah-Korsah H., Anderberg H. I., Engfors A., Kirscht A., Norden K., Kjellstrom S., et al. (2016). The aquaporin splice variant NbXIP1; 1α is permeable to boric acid and is phosphorylated in the N-terminal domain. Front. Plant Sci. 7:862 10.3389/fpls.2016.00862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barko J. W., Smart R. M. (1986). Sediment-related mechanisms of growth limitation in submersed macrophytes. Ecology 67 1328–1340. 10.2307/1938689 [DOI] [Google Scholar]
- Biela A., Grote K., Otto B., Hoth S., Hedrich R., Kaldenhoff R. (1999). The Nicotiana tabacum plasma membrane aquaporin NtAQP1 is mercury-insensitive and permeable for glycerol. Plant J. 18 565–570. 10.1046/j.1365-313X.1999.00474.x [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Desguin B., Chaumont F., Hols P. (2013). Channel-mediated lactic acid transport: a novel function for aquaglyceroporins in bacteria. Biochem. J. 454 559–570. 10.1042/BJ20130388 [DOI] [PubMed] [Google Scholar]
- Bienert G. P., Møller A. L., Kristiansen K. A., Schulz A., Møller I. M., Schjoerring J. K., et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282 1183–1192. 10.1074/jbc.M603761200 [DOI] [PubMed] [Google Scholar]
- Chaumont F., Barrieu F., Wojcik E., Chrispeels M. J., Jung R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125 1206–1215. 10.1104/pp.125.3.1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozza R., Pangaro T. (2009). Tissue expression pattern of two aquaporin-encoding genes in different organs of the seagrass Posidonia oceanica. Aquat. Bot. 91 117–121. 10.1016/j.aquabot.2009.03.007 [DOI] [Google Scholar]
- Danielson J. A., Johanson U. (2008). Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 8:45 10.1186/1471-2229-8-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R., Bélanger R. R. (2015). Molecular evolution of aquaporins and silicon influx in plants. Funct. Ecol. 30 1277–1285. 10.1111/1365-2435.12570 [DOI] [Google Scholar]
- Deshmukh R. K., Sonah H., Belanger R. (2016a). Plant Aquaporins: genome-wide identification, transcriptomics, proteomics, and advanced analytical tools. Front. Plant Sci. 7:1896 10.3389/fpls.2016.01896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R. K., Sonah H., Singh N. K. (2016b). Intron gain, a dominant evolutionary process supporting high levels of gene expression in rice. J. Plant Biochem. Biotechnol. 25 142–146. 10.1007/s13562-015-0319-5 [DOI] [Google Scholar]
- Deshmukh R. K., Vivancos J., Guerin V., Sonah H., Labbe C., Belzile F., et al. (2013). Identification and functional characterization of silicon transporters in soybean using comparative genomics of major intrinsic proteins in Arabidopsis and rice. Plant Mol. Biol. 83 303–315. 10.1007/s11103-013-0087-3 [DOI] [PubMed] [Google Scholar]
- Deshmukh R. K., Vivancos J., Ramakrishnan G., Guérin V., Carpentier G., Sonah H., et al. (2015). A precise spacing between the NPA domains of aquaporins is essential for silicon permeability in plants. Plant J. 83 489–500. 10.1111/tpj.12904 [DOI] [PubMed] [Google Scholar]
- Diehn T. A., Pommerrenig B., Bernhardt N., Hartmann A., Bienert G. P. (2015). Genome-wide identification of aquaporin encoding genes in Brassica oleracea and their phylogenetic sequence comparison to Brassica crops and Arabidopsis. Front. Plant Sci. 6:166 10.3389/fpls.2015.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney J. E., Thorburn L., Iii G. W. K. (2014). Possible causes of eelgrass (Zostera marina L.) loss in Frenchman Bay, Maine. Bull. Mount Desert Island Biol. Lab. 53 26–28. [Google Scholar]
- Flexas J., Ribas-Carbó M., Hanson D. T., Bota J., Otto B., Cifre J., et al. (2006). Tobacco aquaporin NtAQP1 is involved in mesophyll conductance to CO2 in vivo. Plant J. 48 427–439. 10.1111/j.1365-313X.2006.02879.x [DOI] [PubMed] [Google Scholar]
- Grégoire C., Rémus-Borel W., Vivancos J., Labbé C., Belzile F., Bélanger R. R. (2012). Discovery of a multigene family of aquaporin silicon transporters in the primitive plant Equisetum arvense. Plant J. 72 320–330. 10.1111/j.1365-313X.2012.05082.x [DOI] [PubMed] [Google Scholar]
- Gupta A. B., Sankararamakrishnan R. (2009). Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 9:134 10.1186/1471-2229-9-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41 95–98. [Google Scholar]
- Heckwolf M., Pater D., Hanson D. T., Kaldenhoff R. (2011). The Arabidopsis thaliana aquaporin AtPIP1; 2 is a physiologically relevant CO2 transport facilitator. Plant J. 67 795–804. 10.1111/j.1365-313X.2011.04634.x [DOI] [PubMed] [Google Scholar]
- Herman P., Hemminga M., Nienhuis P., Verschuure J., Wessel E. (1996). Wax and wane of eelgrass Zostera marina and water column silicon levels. Mar. Ecol. Prog. Ser. 144 303–307. 10.3354/meps144303 [DOI] [Google Scholar]
- Hodson M., White P. J., Mead A., Broadley M. (2005). Phylogenetic variation in the silicon composition of plants. Ann. Bot. 96 1027–1046. 10.1093/aob/mci255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. M., Jahn T. P., Møller A. L., Schjoerring J. K., Ferri D., Klaerke D. A., et al. (2005). NH3 and NH4+ permeability in aquaporin-expressing Xenopus oocytes. Pflügers Archiv 450 415–428. 10.1007/s00424-005-1399-1 [DOI] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A.-Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Yuan Q., Wang Y., Cai R., Deng X., Wang J., et al. (2012). Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 53 2127–2141. 10.1093/pcp/pcs154 [DOI] [PubMed] [Google Scholar]
- Humborg C., Conley D. J., Rahm L., Wulff F., Cociasu A., Ittekkot V. (2000). Silicon retention in river basins: far-reaching effects on biogeochemistry and aquatic food webs in coastal marine environments. Ambio 29 45–50. 10.1579/0044-7447-29.1.45 [DOI] [Google Scholar]
- Jahn T. P., Møller A. L., Zeuthen T., Holm L. M., Klærke D. A., Mohsin B., et al. (2004). Aquaporin homologues in plants and mammals transport ammonia. FEBS Lett. 574 31–36. 10.1016/j.febslet.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Janssen T., Bremer K. (2004). The age of major monocot groups inferred from 800+ rbcL sequences. Bot. J. Linn. Soc. 146 385–398. 10.1111/j.1095-8339.2004.00345.x [DOI] [Google Scholar]
- Jarvis J. C., Brush M. J., Moore K. A. (2014). Modeling loss and recovery of Zostera marina beds in the Chesapeake Bay: the role of seedlings and seed-bank viability. Aquat. Bot. 113 32–45. 10.1016/j.aquabot.2013.10.010 [DOI] [Google Scholar]
- Johanson U., Karlsson M., Johansson I., Gustavsson S., Sjövall S., Fraysse L., et al. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126 1358–1369. 10.1104/pp.126.4.1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldenhoff R., Fischer M. (2006). Functional aquaporin diversity in plants. Biochim. Biophys. Acta 1758 1134–1141. 10.1016/j.bbamem.2006.03.012 [DOI] [PubMed] [Google Scholar]
- Kaldenhoff R., Kai L., Uehlein N. (2014). Aquaporins and membrane diffusion of CO2 in living organisms. Biochim. Biophys. Acta 1840 1592–1595. 10.1016/j.bbagen.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Kamermans P., Hemminga M. A., De Jong D. J. (1999). Significance of salinity and silicon levels for growth of a formerly estuarine eelgrass (Zostera marina) population (Lake Grevelingen, The Netherlands). Mar. Biol. 133 527–539. 10.1007/s002270050493 [DOI] [Google Scholar]
- Kuliková Z. L., Lux A. (2010). Silicon influence on maize, Zea mays L., hybrids exposed to cadmium treatment. Bull. Environ. Contam. Toxicol. 85 243–250. 10.1007/s00128-010-0046-5 [DOI] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., Tamura K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9 299–306. 10.1093/bib/bbn017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Kozono D., Remis J., Kitagawa Y., Agre P., Stroud R. M. (2005). Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 Å. Proc. Natl. Acad. Sci. U.S.A. 102 18932–18937. 10.1073/pnas.0509469102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Wang W. (2014). “Urea transport mediated by aquaporin water channel proteins,” in Urea Transporters eds Yang B., Sands J. M. (Dordrecht: Springer; ), 227–265. [DOI] [PubMed] [Google Scholar]
- Liu C., Fukumoto T., Matsumoto T., Gena P., Frascaria D., Kaneko T., et al. (2013). Aquaporin OsPIP1; 1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 63 151–158. 10.1016/j.plaphy.2012.11.018 [DOI] [PubMed] [Google Scholar]
- Liu L.-H., Ludewig U., Gassert B., Frommer W. B., Von Wirén N. (2003). Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol. 133 1220–1228. 10.1104/pp.103.027409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux A., Luxová M., Hattori T., Inanaga S., Sugimoto Y. (2002). Silicification in sorghum (Sorghum bicolor) cultivars with different drought tolerance. Physiol. Plant. 115 87–92. 10.1034/j.1399-3054.2002.1150110.x [DOI] [PubMed] [Google Scholar]
- Ma J. F., Tamai K., Yamaji N., Mitani N., Konishi S., Katsuhara M., et al. (2006). A silicon transporter in rice. Nature 440 688–691. 10.1038/nature04590 [DOI] [PubMed] [Google Scholar]
- Moore K. A. (2004). Influence of seagrasses on water quality in shallow regions of the lower Chesapeake Bay. J. Coast. Res. 20 162–178. 10.2112/SI45-162.1 [DOI] [Google Scholar]
- Nguyen M. X., Moon S., Jung K.-H. (2013). Genome-wide expression analysis of rice aquaporin genes and development of a functional gene network mediated by aquaporin expression in roots. Planta 238 669–681. 10.1007/s00425-013-1918-9 [DOI] [PubMed] [Google Scholar]
- Olsen J. L., Rouzé P., Verhelst B., Lin Y.-C., Bayer T., Collen J., et al. (2016). The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530 331–335. 10.1038/nature16548 [DOI] [PubMed] [Google Scholar]
- Ouellette S., Goyette M. H., Labbe C., Laur J., Gaudreau L., Gosselin A., et al. (2017). Silicon transporters and effects of silicon amendments in strawberry under high tunnel and field conditions. Front. Plant Sci. 8:949 10.3389/fpls.2017.00949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley F., Rosenberg J. M., Shachar-Hill Y., Bohnert H. J. (2002). From genome to function: the Arabidopsis aquaporins. Genome Biol. 3: research0001.1–research0001.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragueneau O., Schultes S., Bidle K., Claquin P., Moriceau B. (2006). Si and C interactions in the world ocean: Importance of ecological processes and implications for the role of diatoms in the biological pump. Glob. Biogeochem. Cycles 20:GB4S02 10.1029/2006GB002688 [DOI] [Google Scholar]
- Roy S. W., Penny D. (2007). Patterns of intron loss and gain in plants: intron loss–dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 24 171–181. 10.1093/molbev/msl159 [DOI] [PubMed] [Google Scholar]
- Sakurai J., Ishikawa F., Yamaguchi T., Uemura M., Maeshima M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46 1568–1577. 10.1093/pcp/pci172 [DOI] [PubMed] [Google Scholar]
- Schoelynck J., Struyf E. (2016). Silicon in aquatic vegetation. Funct. Ecol. 30 1323–1330. 10.1111/1365-2435.12614 [DOI] [Google Scholar]
- Serra I., Nicastro S., Mazzuca S., Natali L., Cavallini A., Innocenti A. (2013). Response to salt stress in seagrasses: PIP1; 1 aquaporin antibody localization in Posidonia oceanica leaves. Aquat. Bot. 104 213–219. 10.1016/j.aquabot.2011.05.008 [DOI] [Google Scholar]
- Shivaraj S., Deshmukh R. K., Rai R., Bélanger R., Agrawal P. K., Dash P. K. (2017). Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 7:46137 10.1038/srep46137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonah H., Deshmukh R., Labbé C., Belanger R. (2017). Analysis of aquaporins in Brassicaceae species reveals high-level of conservation and dynamic role against biotic and abiotic stress in canola. Sci. Rep. 7:2771 10.1038/s41598-017-02877-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonah H., Zhang X., Deshmukh R. K., Fernando D., Belanger R. (2016). Comparative transcriptomic analysis of virulence factors in Leptosphaeria maculans during compatible and incompatible interactions with canola. Front. Plant Sci. 7:1784 10.3389/fpls.2016.01784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Nguyen N., Deshmukh R. K., Patil G. B., Prince S. J., Valliyodan B., et al. (2016). Soybean TIP gene family analysis and characterization of GmTIP1;5 and GmTIP2;5 water transport activity. Front. Plant Sci. 7:1564 10.3389/fpls.2016.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G., Alleva K., Mazzella M. A., Amodeo G., Muschietti J. P. (2008). AtTIP1; 3 and AtTIP5; 1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 582 4077–4082. 10.1016/j.febslet.2008.11.002 [DOI] [PubMed] [Google Scholar]
- Takano J., Wada M., Ludewig U., Schaaf G., Von Wirén N., Fujiwara T. (2006). The Arabidopsis major intrinsic protein NIP5; 1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18 1498–1509. 10.1105/tpc.106.041640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom R., Southard S., Borde A. (2014). Climate-linked mechanisms driving spatial and temporal variation in eelgrass (Zostera marina L.) Growth and assemblage structure in Pacific Northwest estuaries, USA. J. Coast. Res. 68 1–11. 10.2112/SI68-001.1 [DOI] [Google Scholar]
- Tian S., Wang X., Li P., Wang H., Ji H., Xie J., et al. (2016). Plant aquaporin AtPIP1; 4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 171 1635–1650. 10.1104/pp.15.01237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törnroth-Horsefield S., Wang Y., Hedfalk K., Johanson U., Karlsson M., Tajkhorshid E., et al. (2006). Structural mechanism of plant aquaporin gating. Nature 439 688–694. 10.1038/nature04316 [DOI] [PubMed] [Google Scholar]
- Waycott M., Duarte C. M., Carruthers T. J., Orth R. J., Dennison W. C., Olyarnik S., et al. (2009). Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proc. Natl. Acad. Sci. U.S.A. 106 12377–12381. 10.1073/pnas.0905620106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudick M. M., Luu D.-T., Tournaire-Roux C., Sakamoto W., Maurel C. (2014). Vegetative and sperm cell-specific aquaporins of Arabidopsis highlight the vacuolar equipment of pollen and contribute to plant reproduction. Plant Physiol. 164 1697–1706. 10.1104/pp.113.228700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. Q., Zhang Q. S., Zhang D., Sheng Z. T. (2017). Light intensity dependent photosynthetic electron transport in eelgrass (Zostera marina L.). Plant Physiol. Biochem. 113 168–176. 10.1016/j.plaphy.2017.02.011 [DOI] [PubMed] [Google Scholar]
- Yu J. (2015). Studies on physiological response to salt stress of eelgrass (Zostera marina L.). Aquat. Sci. Technol. 4 1–7. 10.5296/ast.v4i1.8696 [DOI] [Google Scholar]
- Zhang D. Y., Ali Z., Wang C. B., Xu L., Yi J. X., Xu Z. L., et al. (2013). Genome-wide sequence characterization and expression analysis of major intrinsic proteins in soybean (Glycine max L.). PLoS ONE 8:e56312 10.1371/journal.pone.0056312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Gong J., Huang Q., Mo Y., Yang L., Xie G. (2015). Gene structures, evolution, classification and expression profiles of the aquaporin gene family in Castor Bean (Ricinus communis L.). PLoS ONE 10:e0141022 10.1371/journal.pone.0141022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.