Abstract
Subarachnoid hemorrhage (SAH) is a serious clinical condition where leakage of blood into the subarachnoid space causes an acute rise in intracranial pressure and reduces cerebral blood flow, which may lead to delayed cerebral ischemia and poor outcome. In experimental SAH, we have previously shown that the outcome can be significantly improved by early inhibition of the MAPK/ERK kinase/extracellular signal-regulated kinase (MEK/ERK1/2) pathway. The aim of this study was to apply mass spectrometry to investigate the overall late effects of experimental SAH on cerebrovascular protein expression. SAH was induced in rats that were treated with the MEK1/2 inhibitor U0126 or vehicle. Neurological outcome was assessed using a battery of behavioral tests. Specific protein expression of large cerebral arteries was analyzed quantitatively with high-throughput tandem mass spectrometry. SAH resulted in a marked reduction of neurological scores, which was counteracted by U0126 treatment. Mass spectrometry analysis demonstrated regulation of 184 proteins after SAH, regulations that were in part prevented by U0126 treatment. Network analysis identified several protein networks including a strong structural network centered around 14-3-3. Additionally, protein networks with functions in mRNA metabolism and protein folding were identified. Treatment with U0126 inhibited cerebral vessel wall pERK1/2 expression and significantly improved outcome of the rats. In conclusion, we show that SAH induces a broad array of specific changes in the overall protein networks in cerebral artery smooth muscle cells and suggest that this is essential for understanding the vascular pathophysiology after SAH.
Electronic supplementary material
The online version of this article (doi:10.1007/s12031-017-0944-7) contains supplementary material, which is available to authorized users.
Keywords: Animal model, SAH, Proteomics, Mass spectrometry, MEK1/2 inhibition
Introduction
Subarachnoid hemorrhage (SAH) is a condition caused by leakage of blood from a ruptured aneurysm into the subarachnoid space which causes an acute rise in intracranial pressure (ICP) and a diminished cerebral blood flow (CBF). During the acute phase, there is a mortality rate of about 15%. After cessation of the bleeding, patients might suffer from delayed cerebral ischemia (DCI) with pathological constriction of cerebral arteries, also known as cerebral vasospasm, in the days following the insult. The DCI phase is progressive and can result in infarction, neuronal loss, and poor outcome (Edvinsson and Povlsen 2011). Experimental studies have indicated plasticity of contractile cerebrovascular endothelin, 5-hydroxytryptamine 1B (5-HT1B), angiotensin AT1, and thromboxane A2 receptors in the ischemic process (Ansar and Edvinsson 2008; Ansar et al. 2010; Hansen-Schwartz et al. 2003). These receptor alterations are associated with reduced regional CBF and poor outcome (Ansari 2007).
Proteomics, blended from protein and genome, is large-scale studies of proteins. The possibility to obtain a picture of the total protein expression at a given time in a given tissue has greatly improved during the last 10 years with the accessibility of high-throughput mass spectrometry methods (Cottrell 2011). With this technology, thousands of proteins can be identified and quantified in a single study, which has made it possible to investigate, e.g., effects of diseases and treatments on protein expression in various tissues.
In experimental SAH in rats, we have previously shown that the outcome can be significantly improved by inhibition of the MAPK/ERK kinase/extracellular signal-regulated kinase (MEK-ERK) pathway days after the insult (Larsen et al. 2011). We reported that the MEK1/2 inhibitor U0126 prevents upregulation of the contractile receptors ETB and 5-HT1B, and reduces vascular thickening evident 2 days post-SAH (Edvinsson and Povlsen 2011; Parker et al. 2013). Moreover, analysis of acute (0.5–1 h post insult) SAH in vivo events has shown SAH-induced regulation of focal adhesion molecules and actin cytoskeleton dynamics, and in addition a central role of activation of ERK1/2 (Parker et al. 2013).s
The detailed analysis of acute SAH events by Parker et al. (Parker et al. 2013) revealed the proteomic changes immediately associated with the insult. However, no large-scale proteomic analysis of the alterations in proteins 48 h after SAH, a time-point more closely related to the development of DCI, has yet to be performed. Hence, the aim of this study was to apply high-throughput quantitative tandem mass spectrometry to establish later effects of experimental SAH on the protein expression in the cerebral vessels. Moreover, because the inhibition of MEK-ERK1/2 activation has a prophylactic effect in later cerebrovascular changes after SAH, we investigated the effects of U0126 on the SAH-induced proteomic changes (Larsen et al. 2011).
Experimental Procedures
All animal procedures were carried out strictly within national laws and guidelines and were approved by the Danish Animal Experimentation Inspectorate (license no. 2011/561-2025).
Rat Subarachnoid Hemorrhage Model
SAH was induced as described by Prunell et al. (2002). Male Sprague-Dawley rats 350–400 g (Taconic, Denmark) were anesthetized using 3.5% isoflurane (Baxter A/S, Denmark) in atmospheric air and O2 (70:30). Anesthesia was maintained by intubation and artificial ventilation of 1–2% isoflurane in N2O/O2 (70:30) during the surgical procedure. Respiration was monitored by regularly withdrawing blood samples to a blood gas analyzer (Radiometer, Denmark). A temperature probe was rectally inserted to record the body temperature, which was maintained at 37 °C by a heating pad. ICP was measured via a catheter inserted into the basal cisterna via a hole in the atlanto-occipital membrane. The catheter was connected to a pressure transducer and the signal was recorded in the software LabChart via a PowerLab (both from AD Instruments, UK). Mean arterial blood pressure (MABP) was measured via a tail artery catheter connected to a pressure transducer and recorded in LabChart. Cortical CBF was measured with a laser-Doppler fiber optic probe placed directly on the dura mater on the surface of the brain via a hole drilled through the skull, 4 mm anterior from the bregma and 3 mm to the right of the midline, without perforation of the dura. ICP, MABP, and CBF were measured in real time with recordings commencing approximately 30 min before SAH and continuing until 1 h after SAH.
A 27G blunt cannula was stereotactically placed 6.5 mm anterior to the bregma in the midline at an angle of 30° to the vertical plane, it was lowered until it met the base of the scull, and then retracted 1 mm placing the tip of the needle just in front of the chiasma opticum. Rats equilibrated 30 min before 300 μl of blood was withdrawn from the tail catheter and injected manually into the prechiasmatic cisterna at a pressure equal to the MABP (80–100 mmHg). Rats were maintained under anesthesia for another 60 min in order to recover. The ICP catheter was cut and sealed with a removable plug 2 cm from the tip. The tail catheter, the needle, and the laser-Doppler probe were removed and incisions closed. Carprofen (4 mg/kg; Pfizer, Denmark) against pain was administered subcutaneous as well as 15 ml saline for hydration. Rats were then revitalized and extubated. Sham-operated rats went through the same procedure, with the exception that no blood was injected intracisternally.
Experimental Groups
A total of 40 rats underwent surgery for the neurological evaluation and mass spectrometric analysis and 18 rats underwent surgery for Western blot validation. The mass spectrometry was conducted in duplicates and each sample consisted of tissue from four to five rats. These rats also went through neurological assessment, meaning that there were 9–10 biological replicates in each group for neurological evaluation. Western blot was carried out in technical duplicates with four to six biological replicates and each sample consisted of protein purified from one rat. In order to assess whether MEK-ERK1/2 activity was involved in the vascular response in the first 2 days post-SAH, thus, we compared animals treated with either U0126 (1,4-diamino-2,3-dicyano-1,4-bis [2-aminophenylthio] butadiene) (Duncia et al. 1998) or vehicle (DMSO) at 6, 12, 24, and 36 h post-SAH with sham-operated rats that received neither U0126 nor vehicle.
Animals were randomly selected for treatment with either U0126 or vehicle. U0126 was given as 0.05 ml/kg body weight of a 10−5 M solution of U0126 ethanolate (Sigma-Aldrich, MO, USA) in isotonic saline with 0.1% DMSO, yielding a final dose of 0.22 μg /kg body weight. Vehicle consisted of 0.1% DMSO in isotonic saline. Treatment was administered intracisternal through the ICP catheter placed with the tip in the basal cistern.
At 48 h post-surgery, all rats were anesthetized using CO2 and decapitated. In a dissection microscope, the middle cerebral arteries (MCAs), the circle of Willis, and the basilar artery were carefully dissected free of brain tissue and cleaned of connective tissue and blood, and stored at −80 °C.
Neurological Function
Neurological evaluations (rotating pole test and spontaneous behavior observations) were performed by personnel blinded with regard to experimental groups. Tests and observations were performed in a silent room with as few visual disturbing elements as possible.
Rotating Pole Test
Gross sensorimotor function of animals was evaluated by testing their balance and coordination of movements on a rotating pole (45 mm in diameter and 150 cm in length), which was either steady or rotating at different speeds (3 or 10 rpm) (Ohlsson and Johansson 1995). A cage where the floor was covered with bedding material from the home cage of the rat was placed at the end of the pole opposite from the rat to serve as a positive reinforcement for the rat to traverse the pole, and the ability of the rats to traverse the pole was monitored. The performance of the rat was scored according to the following definitions: Score 1, the animal is unable to sit on the pole and falls off immediately. Score 2, the animal has severe difficulties to stay on the pole, but manages to balance for a while and move less than 30 cm on the pole. Score 3, the animal embraces the pole with paws while crossing, but manages to move more than 30 cm. Score 4, the animal manages to traverse the pole and reach the platform but moves with the body close to the pole, embraces the pole, or jumps with hind legs. Score 5, the animal traverse the pole with normal posture but displays one or several of the following deficits: foot slips (less than 10), slightly disturbed pattern of movement, stops along the pole, and has difficulty staying on the pole while standing still. Score 6, the animal traverses the pole perfectly with 0–2 ft slips and no stops. Results were analyzed with one-way ANOVA with Dunn’s multiple comparison post-test. Significance level was set to p < 0.05.
Spontaneous Behavior
Spontaneous activity of the rats was quantified by placing the rats individually in a test cage with fresh bedding and nesting material for 20 min. An observer equipped with a timer recorded all time intervals spent moving around in the cage (locomotion), sitting or lying in the same place (no movement), rearing, or grooming. Results were analyzed with one-way ANOVA and Newman-Keuls multiple comparison post-test. Significance level was set to p < 0.05.
Protein Purification
Tissue from cerebral arteries was dounce homogenized in ice-cold 0.1 M Na2CO3 containing 1 tablet PhosSTOP phosphatase inhibitor cocktail (Roche, France) per 10 ml and 1 ml P8340 protease inhibitor (Sigma-Aldrich, MO, USA) per 20 ml, and then tip-probe sonicated 2 × 20 s on ice. Homogenates were ultra-centrifuged at 150,000×g for 1.5 h. Pellets (membrane fractions) were resuspended in 6 M urea/2 M thiourea. Supernatants (soluble fractions) were precipitated using 20% trichloroacetic acid, and pellets from this precipitation were resuspended in 6 M urea/2 M thiourea. Samples were reduced in 10 mM dithiothreitol, then alkylated using 20 mM iodoacetamide. Lysyl-endopeptidase (Wako, Osaka, Japan) was added for 3 h, and then samples were diluted 6-fold in 50 mM triethyl ammonium bicarbonate. Trypsin (Promega, WI, USA) was added at a ratio of 1:40 (w/w) and left for 18 h to digest. Digests were acidified by addition of formic acid to a final concentration of 2% and the protein contents of samples were quantified using amino acid composition analysis. Equal amounts of protein from each sample were iTRAQ four-plex labeled according to the manufacturer’s instructions (AB Sciex, MA, USA), equal labeling was confirmed on a Bruker UltraFlex MALDI MS/MS instrument (Bruker, MA, USA), and samples were combined to give equal reporter intensity.
Combined peptide samples were cleaned up on a homemade R3 column and then dried down for hydrophilic interaction liquid chromatography (HILIC). Peptide samples were resuspended in 90% acetonitrile (ACN)/0.1% trifluoroacedic acid (TFA) and loaded onto a micro-HILIC HPLC chromatography system. The HILIC resin was TSK-gel Amide 80 (Tosoh Bioscience, Japan). Samples were separated over a gradient of 90–60% organic solvent over 30 min, then 60–0% over 15 min. Fractions were collected every minute at absorbances over 500 mAU and every 5 min otherwise. All fractions were lyophilized.
Mass Spectrometry
Dry peptide fractions were resuspended in 0.1% FA and loaded onto a Thermo Easy-LC system. Peptides were eluted using a 0–34% organic gradient over 70 min, then 34–100% over 20 min. All LC-MS runs were performed on columns with a 75-μm inner diameter, packed with C18 material (Dr. Maisch, Ammerbuch-Entringen, Germany). Mass spectrometry (MS) was performed using higher-energy collisional dissociation (HCD) fragmentation on a Thermo LTQ Velos (Thermo Fisher Scientific, Germany). Quantification was performed on reporter tags at m/z 114, 115, 116, and 117. MS was performed with the following settings: A full MS scan in the mass area of 400–1800 Da was performed in the Orbitrap with a resolution of 30,000 FWHM and a target value of 1 × 106 ions. For each full scan, the seven most intense ions (>+1 charge state) were selected for HCD and detected at a resolution of 7500 FWHM. HCD was performed with the following settings: threshold for ion selection was 20,000, the target value of ions used for HCD was 1 × 105, and activation time was 1 ms, the isolation window was 2.5 Da, and the normalized collision energy was 48.
Analysis of Mass Spectrometry Data
Data was searched using Thermo Proteome Discoverer (version 1.3.0.339) and Mascot (v2.2, Matrix Science Ltd., UK) allowing for variable methionine oxidation. Enzyme specificity was set for trypsin, with two missed cleavages allowed. Precursor mass tolerance was 10 ppm and fragment mass tolerance was set to 0.05 Da. iTRAQ labeling was also set as a variable modification in order to detect lysine modification, while cysteine carbamidomethylation was set as a fixed modification. Data were searched against a Swiss-prot rodent database (UniProtKB/Swiss-Prot 2012_10). Data were filtered to 1% peptide FDR with a decoy approach using percolator and filtered to remove Mascot scores less than 18, as previously described (Engholm-Keller et al. 2012). Data was exported from Proteome Discoverer and manually normalized to protein median values across an entire labeling experiment to correct for protein abundance variation. Protein and peptide regulation was defined based on the log2-transformed iTRAQ ratios and considered regulated when exceeding 2 standard deviations from the median ratio for a given tag. Proteins were classified according to their molecular function using the PANTHER database (www.panther.db) and network analysis was performed using STRING, version 9 (www.string.db) (Szklarczyk et al. 2011).
Western Blot
Each purified protein sample was prepared from MCAs, circle of Willis, and the basilar artery from one animal. Samples were sonicated in RIPA buffer (50 mM Tris-HCl buffer pH 7.5, 150 mM NaCl, 1 mM EDTA pH 8.0, 1% NP-40, 0.1% SDS, 0.5% Triton X-100, 50 mM β-glycerolphosphate, 0.1% deoxycholic acid) with 1 tablet Complete Protease Inhibitor Cocktail (Roche, France) per 50 ml, and 1 tablet PhosSTOP Phosphatase Inhibitor Cocktail (Roche, France) per 10 ml RIPA.
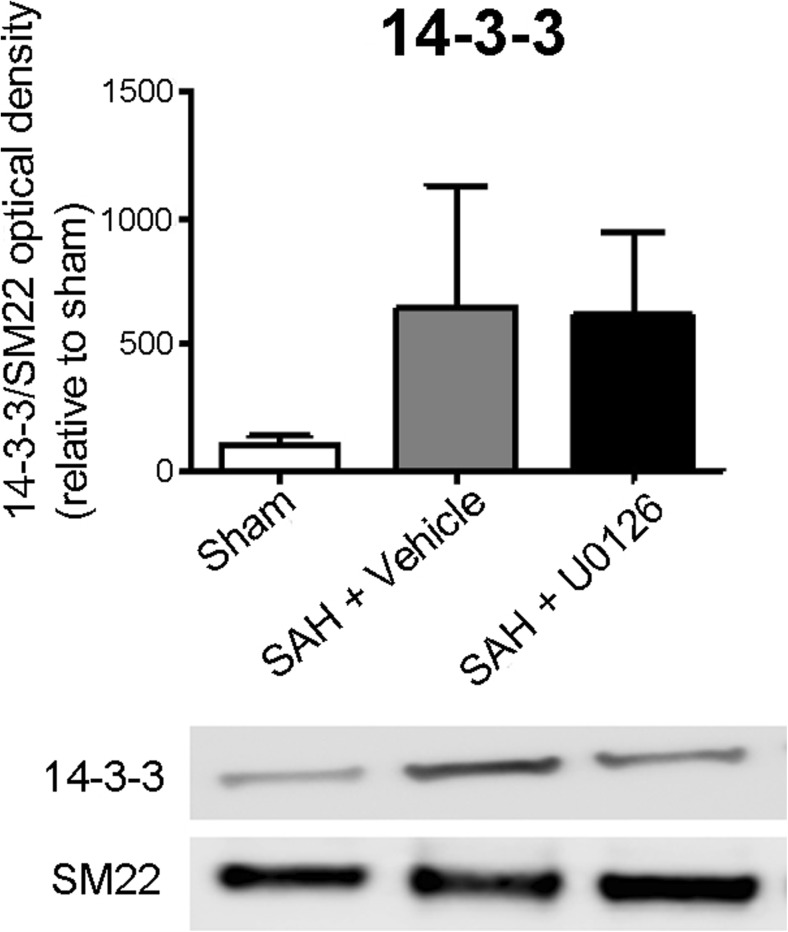
Protein concentrations were determined with a spectrophotometer (Infinite M200, TECAN, Switzerland). Purified protein samples were run on a 4–20% precast SDS gel (Expedeon Inc., USA) and blotted onto a nitrocellulose membrane (GE Healthcare, USA). Membranes were blocked for 1 h in 5% bovine serum albumin (BSA) in Tris-buffered saline (50 mM Tris, 150 mM NaCl, pH 7.6) with 0.05% (v/v) Tween-20 (TBS-T) at room temperature and incubated over night at 4 °C with primary antibodies diluted in 5% BSA TBS-T (Supplementary Table S1). After a 5 min wash, the membranes were incubated 1 h at room temperature with secondary antibody diluted in 5% BSA TBS-T (Supplementary Table S1), washed five times 5 min in TBS-T and developed using ECL Select (GE Healthcare, USA) and a LAS4000 (Fujifilm, Japan). Blots were quantified with Image Gauge V3.2 (Fujifilm, Japan) and intensities for band representing 14-3-3 were normalized to intensities for bands representing SM22 which was used as loading control. Normalized intensities for the U0126-treatment and DMSO-treatment groups, respectively, were compared to normalized intensities for the sham-surgery group using one-way ANOVA and Newman-Keuls test for multiple comparison significance; the level was set to p < 0.05.
Immunohistochemistry
Immunohistochemical staining was performed on cerebral vessel segments (from three rats of the sham group, SAH with vehicle or U0126 group). The tissues were embedded in Tissue-Tek®, frozen on dry ice, and stored at −80 °C until further processing. They were sectioned (10 μm) on a cryostat (Leica, Denmark) and mounted on microscope slides (SuperFrost®, Menzel, Germany). The sections were fixed for 20 min using Stefanini’s fixative (2% paraformaldehyde and 0.2% picric acid in phosphate buffer, pH 7.2) and permeabilized in phosphate-buffered saline (PBS) containing 0.25% Triton X-100 (T-PBS). To prevent nonspecific staining, sections were blocked with 2% donkey serum and 1% BSA in T-PBS for 1 h. Samples were incubated at 4 °C overnight with primary anti-pERK1/2 antibody (1:250, alx-210-506a, Alexis Biochemicals, Nottingham, UK) diluted in T-PBS. The following day, the slides were rinsed three times in T-PBS and incubated with secondary donkey anti-sheep DyLight™ 488 antibody (1:200, 713-485-003, Jackson ImmunoResearch, UK) or secondary donkey anti-mouse FITC (1:100, Jackson Immunoresearch Laboratories) for 1 h in room temperature followed by three washes in T-PBS before mounting with Vectashield (Vector Laboratories, Burlingame, CA, USA). Primary antibody was omitted as a negative control.
Results
Physiological Parameters
Two groups of animals were subjected to SAH and then treated with either U0126 or vehicle at 6, 12, 24, and 36 h post-SAH. A third group of animals were subjected to sham surgery and received neither U0126 nor vehicle (Supplementary Fig. S1).
Measurements of the physiological parameters during SAH surgery showed that ICP reached values of 10–20 mmHg above MABP (80–100 mmHg) and cortical CBF was lowered to 75% compared to flow before insult, and did not return to baseline for a minimum of 10 min. These results (Supplementary Table S2) are comparable with values in our previous study (Povlsen et al. 2013) and values did not differ between groups. The mortality rate after 48 h was 16%.
Neurological Functions
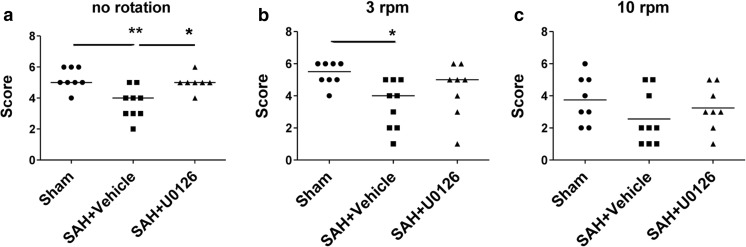
Neurological function was assessed using the rotating pole test and standardized observations of spontaneous behavior by trained personnel blinded to the experimental groups. The ability of the rats to traverse a rotating pole at either no rotation, at 3, or 10 rpm was significantly decreased by induction of SAH. This reduction was restored by U0126 treatment (Fig. 1). The administration of U0126 in SAH animals was well tolerated and did not modify ICP, CBF, or MABP differently as compared to the DMSO-treated SAH group (Larsen et al. 2011).
Fig. 1.
Neurological assessment of rats treated with U0126 or DMSO post-SAH. Rats were tested for their ability to traverse a 1.5-m pole with either no rotation, 3, or 10 rpm. Each data point is the score of one rat; n = 7–9. Horizontal lines are median score. Compared with sham-operated animals, rats that underwent SAH performed significantly poorer at no rotation (a) and 3 rpm (b). At 10 rpm (c), there was no significant difference between sham and SAH. Animals that underwent SAH and were treated with U0126 performed significantly better than animals treated with vehicle at no rotation (a). Data were analyzed with one-way ANOVA and Dunn’s post-test: * = p < 0.05, ** = p < 0.01
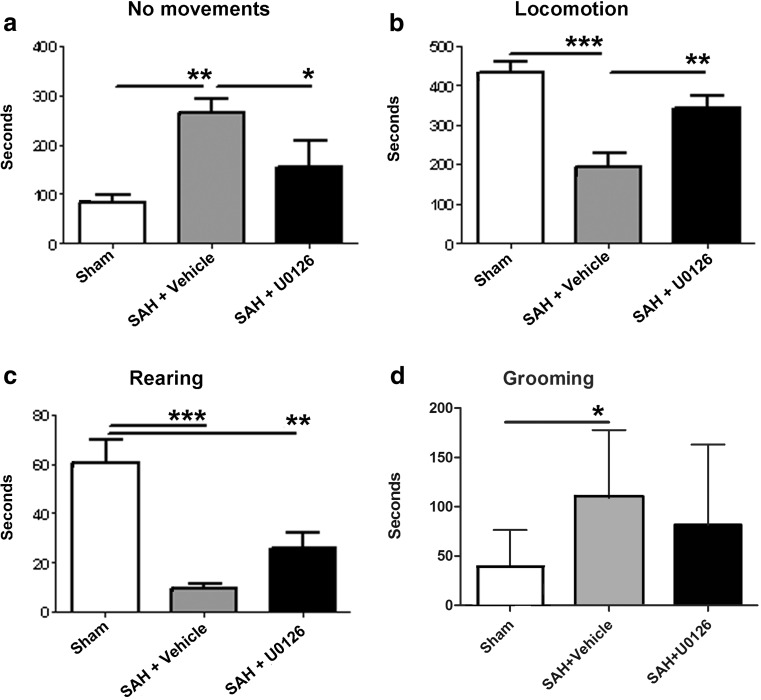
Observations on time spend with no movements, locomotion, rearing, and grooming were recorded using a standardized protocol. It was found that induction of SAH worsened the neurological function of rats as seen by a significant change in time spent with no movement and locomotion (Fig. 2). This change was significantly neutralized by U0126 treatment. In addition, SAH markedly decreased time spent with rearing (p < 0.001) and increased the time of grooming; these effects were partly restored after U0126 treatment (p < 0.05) (Fig. 2).
Fig. 2.
Behavioral observations of rats treated with U0126 or vehicle post-SAH. Rats were observed for 20 min and the duration of their spontaneous activities was quantified. Data are mean time spent on each activity ± S.E.M.; n = 8–9. Compared with sham-operated animals, rats that underwent SAH performed significantly poorer with no movement (a), locomotion (b), rearing (c), and grooming (d). Animals that underwent SAH and were treated with U0126 performed significantly better than animals treated with vehicle with no movement (a) and locomotion (b). Data were analyzed with one-way ANOVA and Newman-Keuls post-test for multiple comparison: * = p < 0.05, ** = p < 0.01, *** = p < 0.001
In conclusion, inhibition of vessel wall pERK1/2 (U0126 treatment) of rats subjected to SAH improved the neurology functional parameters as compared to treatment with vehicle.
Proteome Analysis in SAH in the Absence or Presence of MEK1/2 Inhibition
In order to obtain an overview of protein expression in cerebral vessels after induction of SAH and to assess the effect of treatment with the MEK1/2 inhibitor U0126, tandem mass spectrometry with isobaric tags for relative and absolute quantitation (iTRAQ) was used to generate quantitative total proteomes for the three groups of animals (sham, vehicle-, or U0126 treatment) (Ong and Mann 2005; Ross et al. 2004; Steen and Mann 2004).
The analysis of the cerebral vessel proteome was performed in duplicates, each sample containing a pool of cerebral vessels (MCAs, circle of Willis, and basilar artery) from four to five animals dissected 48 h after rats were subjected to either SAH (and treated with either U0126 or vehicle) or sham surgery. Pilot studies showed no effect of vehicle per se or of U0126 to non-SAH sham surgery animals (data not shown).
The cerebral artery samples were iTRAQ labeled as follows: U0126-treated groups were labeled with iTRAQ 114, vehicle-treated groups were labeled with iTRAQ 115, and sham groups were labeled with iTRAQ 116. Tandem mass spectrometry was performed on a 1:1:1 mixture of all three groups simultaneously. Protein levels in the different groups were determined based on the intensities of the tags compared with sham. Because this was an overall screening proteome study, the results include all identified proteins in the groups.
Core Results
A total of 2677 proteins were identified. The effect of SAH on protein expression in cerebral vessels (without any contamination of brain tissue) was assessed by comparing the regulated proteins of rats subjected to SAH and treated with vehicle with the regulated proteins of rats subjected to sham surgery (this group did not differ in expression from sham surgery with or without U0126; data not shown). When examining the SAH vehicle group, we identified 171 upregulated proteins and 79 downregulated proteins compared with sham. The ten most upregulated and downregulated proteins are shown in Table 1A and B. Using the PANTHER database, we sorted the proteins based on their molecular function.1 Of the 171 upregulated proteins, 159 were recognized in the database with a total of 171 hits.2 Of the allocated proteins, 39.9% (68 proteins) were involved in catalytic activity, 30.4% (52 proteins) in binding of either nucleic acids, proteins, or calcium ions, 17% (29 proteins) in structural activity, 1.8% (3 proteins) in enzyme regulator activity, 2.9% (5 proteins) in transcription factor activity, 3.5% (6 proteins) in receptor activity, 12.3% (4 proteins) in translation, and 2.3% (4 proteins) have transporter functions (Supplementary Fig. S2).
Table 1.
The ten most upregulated and downregulated proteins after SAH. The table shows proteins that are regulated after SAH and the level of regulation is expressed as log2 of the iTRAQ ratios. (A) The ten most upregulated proteins after SAH when compared with sham. (B) The ten most downregulated proteins after SAH when compared with sham
| Accession number | Protein name | Log2 |
|---|---|---|
| A: The ten most upregulated proteins after SAH | ||
| P50116 | Protein S100-A9 | 3.96 |
| 9JJ54 | Heterogeneous nuclear ribonucleoprotein D0 | 3.84 |
| Q9Z1Z3 | Epsin-2 | 3.78 |
| Q925G0 | Putative RNA-binding protein 3 | 3.74 |
| B3EWD2 | Hemoglobin subunit beta | 3.70 |
| Q8VC52 | RNA-binding protein with multiple splicing 2 | 3.70 |
| Q9DBR1 | 5′-3′ Exoribonuclease 2 | 3.58 |
| Q63083 | Nucleobindin-1 | 3.58 |
| P11348 | Dihydropteridine reductase | 3.57 |
| P63281 | SUMO-conjugating enzyme UBC9 | 3.55 |
| B: The ten most downregulated proteins after SAH | ||
| Q62715 | Neutrophil antibiotic peptide NP-2 | −5.38 |
| P04764 | Alpha-enolase | −5.38 |
| D3ZLY9 | Histone H2B type 1-H | −4.82 |
| P05811 | Alpha-crystallin B chain | −4.47 |
| Q5RKG9 | Eukaryotic translation initiation factor 4B | −4.09 |
| Q6LED0 | Histone H3.1 | −3.96 |
| P04646 | 60S ribosomal protein L35a | −3.85 |
| P08081 | Clathrin light chain A | −3.75 |
| P08009 | Glutathione S-transferase Yb-3 | −3.64 |
| Q9JM53 | Apoptosis-inducing factor 1, mitochondrial | −3.42 |
Of the 79 downregulated proteins, 72 were found in the PANTHER database and gave 82 hits. Of these proteins, 30.5% (25 proteins) were involved in binding,3 29.3% (24 proteins) in catalytic activity, 15.3% (11 proteins) in structural activity, 9.8% (8 proteins) have transporter functions, 7.3% (6 proteins) are involved in receptor activity, 6.1% (5 proteins) in enzyme regulator activity, 2.4% (2 proteins) in translation, and 1.2% (1 protein) in transcription factor activity (Supplementary Fig. S2). This demonstrates that the response to SAH in the major cerebral arteries involves both up- and downregulation of mainly proteins with binding, catalytic, and structural functions. The stronger of the two responses seems to be upregulation of proteins rather than downregulation. Importantly, we show that a wide range of proteins were regulated after SAH, hence targeting a single protein is not a viable way to counteract the detrimental effects of SAH.
The Effect of MEK1/2 Inhibition
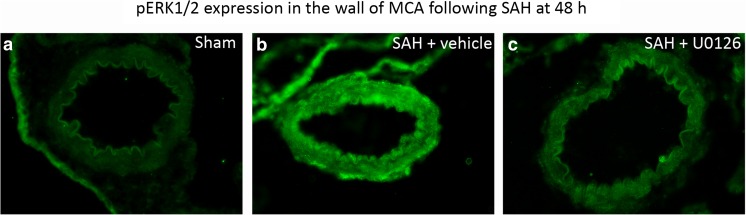
The first part was verification in the circle of Willis arteries that treatment with U0126 had efficacy per se. This has been done in a Western blot quantitative study before using the same SAH method (Ansar and Edvinsson 2008). Here, we confirm this effect using immunohistochemistry. We verified that SAH induced enhanced expression of pERK1/2 in the cerebral vascular smooth muscle cells (VSMC), while this was unaltered in the U0126-treated vessels (Fig. 3). The effect of MEK1/2 inhibition on SAH-induced changes in protein expression was assessed in a two-step procedure where the first step was to compare SAH animals treated with U0126 with sham animals. Hereby, 104 proteins were identified as upregulated and 183 proteins were found to be downregulated, which indicates that MEK1/2 inhibition affects the SAH-induced expressional changes in cerebral vessels. The next step in order to identify downstream targets of MEK1/2 was to search for proteins that fulfilled one of the following criteria: 1, the protein is upregulated in vehicle-treated animals (as compared with sham animals) and either downregulated or not affected in U0126-treated animals (as compared with sham animals). 2, The protein is downregulated in vehicle-treated animals (as compared with sham animals) and either upregulated or not affected in U0126-treated animals (as compared with sham animals). A list of 133 proteins (Table 2) was identified using criteria 1. This group (hereafter denoted group 1) represents proteins with an SAH-induced expressional increase that does not persist after MEK1/2 inhibition. A list of 51 proteins (Table 3) was identified based on criteria 2. This group (hereafter denoted group 2) represents proteins with an SAH-induced expressional decrease that does not persist after MEK1/2 inhibition.
Fig. 3.
pERK immunohistochemistry on middle cerebral artery. a The image shows MCA of a sham-operated animal. No pERK is visible in the vessel. b The image demonstrates specific pERK immunoreactivity in the vessel after induction of SAH. c Treatment with U0126 attenuates the pERK immunoreactivity in the vessel
Table 2.
Proteins that show a SAH-induced upregulation which does not persist after MEK inhibition. The list shows proteins that fulfilled the following criteria: the protein is upregulated in vehicle-treated animals (as compared with sham animals) and either downregulated or not affected in U0126-treated animals (as compared with sham animals). Regulation is expressed as log2 of the iTRAQ ratios. This group of 133 proteins is referred to as group 1 and represents proteins with an SAH-induced expressional increase that does not persist after MEK1/2 inhibition
| Accession number | Protein name | Log2 |
|---|---|---|
| Q9JJ54 | Heterogeneous nuclear ribonucleoprotein D0 | 3.84 |
| Q4FZY0 | EF-hand domain-containing protein D2 | 3.78 |
| Q9Z1Z3 | Epsin-2 | 3.78 |
| Q925G0 | Putative RNA-binding protein 3 | 3.74 |
| B3EWD2 | Hemoglobin subunit beta | 3.70 |
| B5DFF2 | RNA-binding protein with multiple splicing 2 | 3.70 |
| Q9DBR1 | 5′-3′ Exoribonuclease 2 | 3.58 |
| Q63083 | Nucleobindin-1 | 3.58 |
| P11348 | Dihydropteridine reductase | 3.57 |
| P63281 | SUMO-conjugating enzyme UBC9 | 3.55 |
| Q8BQ30 | Phostensin | 3.54 |
| Q6P686 | Osteoclast-stimulating factor 1 | 3.52 |
| Q63797 | Proteasome activator complex subunit 1 | 3.46 |
| Q8K297 | Procollagen galactosyltransferase 1 | 3.45 |
| Q4KM73 | UMP-CMP kinase | 3.41 |
| Q924S5 | Lon protease homolog, mitochondrial | 3.38 |
| Q9QX47 | Protein SON | 3.34 |
| Q5U318 | Astrocytic phosphoprotein PEA-15 | 3.33 |
| P61149 | Fibroblast growth factor 1 | 3.28 |
| Q5BJK8 | Golgi integral membrane protein 4 | 3.28 |
| O08586 | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase | 3.25 |
| D3ZQN7 | Laminin subunit beta-1 | 3.22 |
| P62815 | V-type proton ATPase subunit B, brain isoform | 3.22 |
| Q9DBZ5 | Eukaryotic translation initiation factor 3 subunit K | 3.16 |
| P61972 | Nuclear transport factor 2 | 3.15 |
| Q4FZY0 | EF-hand domain-containing protein D2 | 3.13 |
| Q6PEV3 | WAS/WASL-interacting protein family member 2 | 3.11 |
| P63326 | 40S ribosomal protein S10 | 3.10 |
| Q6AY09 | Heterogeneous nuclear ribonucleoprotein H2 | 3.03 |
| Q63663 | Interferon-induced guanylate-binding protein 2 | 3.02 |
| P52303 | AP-1 complex subunit beta-1 | 2.97 |
| P60868 | 40S ribosomal protein S20 | 2.97 |
| Q64611 | Cysteine sulfinic acid decarboxylase | 2.90 |
| P51583 | Multifunctional protein ADE2 | 2.89 |
| Q63871 | DNA-directed RNA polymerases I, II, and III subunit RPABC4 | 2.89 |
| Q5U211 | Sorting nexin-3 | 2.87 |
| P27615 | Lysosome membrane protein 2 | 2.86 |
| Q4KM65 | Cleavage and polyadenylation specificity factor subunit 6 | 2.86 |
| Q62865 | cGMP-inhibited 3′,5′-cyclic phosphodiesterase A | 2.85 |
| P55260 | Annexin A4 | 2.84 |
| Q8R361 | Rab11 family-interacting protein 5 | 2.81 |
| P97379 | Ras GTPase-activating protein-binding protein 2 | 2.81 |
| Q66H20 | Polypyrimidine tract-binding protein 2 | 2.78 |
| Q6AXS3 | Protein DEK | 2.72 |
| P36876 | Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B alpha isoform | 2.72 |
| B5DFC8 | Eukaryotic translation initiation factor 3 subunit C | 2.72 |
| Q9QWE9 | Gamma-glutamyltransferase 5 | 2.71 |
| Q63433 | Serine/threonine-protein kinase N1 | 2.70 |
| Q4KM74 | Vesicle-trafficking protein SEC22b | 2.69 |
| Q5U2T8 | Corepressor interacting with RBPJ 1 | 2.64 |
| Q9WVA3 | Mitotic checkpoint protein BUB3 | 2.61 |
| Q8CIE6 | Coatomer subunit alpha | 2.59 |
| G3V9C7 | Histone H2B type 1-K | 2.57 |
| Q9Z340 | Partitioning defective 3 homolog | 2.55 |
| P97633 | Casein kinase I isoform alpha | 2.47 |
| P41499 | Tyrosine-protein phosphatase non-receptor type 11 | 2.46 |
| P60711 | Actin, cytoplasmic 1 | 2.45 |
| Q9R1Z8 | Vinexin | 2.45 |
| Q63560 | Microtubule-associated protein 6 | 2.42 |
| Q27W02 | Protein mago nashi homolog | 2.39 |
| Q6AYD6 | PDZ and LIM domain protein 2 | 2.37 |
| Q4AEF8 | Coatomer subunit gamma | 2.37 |
| P52481 | Adenylyl cyclase-associated protein 2 | 2.37 |
| P49138 | MAP kinase-activated protein kinase 2 | 2.36 |
| Q99P96 | Histone deacetylase 7 | 2.36 |
| P36372 | Antigen peptide transporter 2 | 2.35 |
| P05811 | Alpha-crystallin B chain | 2.34 |
| Q5HZV9 | Protein phosphatase 1 regulatory subunit 7 | 2.33 |
| P70615 | Lamin-B1 | 2.32 |
| Q9WUX5 | Protein MRVI1 | 2.32 |
| P16675 | Lysosomal protective protein | 2.31 |
| Q9EPH8 | Polyadenylate-binding protein 1 | 2.29 |
| Q63560 | Microtubule-associated protein 6 | 2.28 |
| Q60436 | Serrate RNA effector molecule homolog (Fragment) | 2.26 |
| Q8R081 | Heterogeneous nuclear ribonucleoprotein L | 2.26 |
| Q561S0 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | 2.26 |
| Q5U2R0 | Methionine adenosyltransferase 2 subunit beta | 2.24 |
| P46664 | Adenylosuccinate synthetase isozyme 2 | 2.21 |
| P0C5W1 | Microtubule-associated protein 1S | 2.19 |
| Q9WTT6 | Guanine deaminase | 2.17 |
| O55125 | Protein NipSnap homolog 1 | 2.17 |
| Q6MG49 | Large proline-rich protein BAG6 | 2.16 |
| P04692 | Tropomyosin alpha-1 chain | 2.16 |
| Q8VC85 | U6 snRNA-associated Sm-like protein LSm1 | 2.16 |
| Q6AYE2 | Endophilin-B1 | 2.16 |
| D3ZU13 | Eukaryotic translation initiation factor 4 gamma 1 | 2.15 |
| Q6MG61 | Chloride intracellular channel protein 1 | 2.14 |
| O55012 | Phosphatidylinositol-binding clathrin assembly protein | 2.13 |
| Q5U301 | A-kinase anchor protein 2 | 2.13 |
| Q9Z2K1 | Keratin, type I cytoskeletal 16 | 2.13 |
| Q9EPL8 | Importin-7 | 2.10 |
| P14426 | H-2 class I histocompatibility antigen, D-K alpha chain | 2.06 |
| Q8BSY0 | Aspartyl/asparaginyl beta-hydroxylase | 2.05 |
| Q6AYQ4 | Transmembrane protein 109 | 2.02 |
| O55096 | Dipeptidyl peptidase 3 | 2.01 |
| P39069 | Adenylate kinase isoenzyme 1 (Fragments) | 1.98 |
| P05982 | NAD(P)H dehydrogenase [quinone] 1 | 1.34 |
| Q9QXQ0 | Alpha-actinin-4 | 1.27 |
| Q62507 | Cochlin | 1.26 |
| P19132 | Ferritin heavy chain | 1.20 |
| P38983 | 40S ribosomal protein SA | 1.17 |
| P52944 | PDZ and LIM domain protein 1 | 1.17 |
| P70619 | Glutathione reductase (Fragment) | 1.14 |
| P24368 | Peptidyl-prolyl cis-trans isomerase B | 1.12 |
| P38652 | Phosphoglucomutase-1 | 1.07 |
| P63326 | 40S ribosomal protein S10 | 1.06 |
| Q5XI28 | Ribonucleoprotein PTB-binding 1 | 1.05 |
| Q9Z1P2 | Alpha-actinin-1 | 1.05 |
| B0BNF1 | Septin-8 | 1.04 |
| P97633 | Casein kinase I isoform alpha | 1.04 |
| P63102 | 14-3-3 protein zeta/delta | 1.03 |
| Q60972 | Histone-binding protein RBBP4 | 0.99 |
| Q9DB16 | Calcium-binding protein 39-like | 0.96 |
| Q497B0 | Omega-amidase NIT2 | 0.96 |
| B0BNA7 | Eukaryotic translation initiation factor 3 subunit I | 0.95 |
| P47853 | Biglycan | 0.94 |
| Q921M3 | Splicing factor 3B subunit 3 | 0.93 |
| O55126 | Protein NipSnap homolog 2 | 0.93 |
| Q8VHI3 | GDP-fucose protein O-fucosyltransferase 2 | 0.92 |
| P61983 | 14-3-3 protein gamma | 0.91 |
| P28661 | Septin-4 | 0.89 |
| P14046 | Alpha-1-inhibitor 3 | 0.88 |
| P15651 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | 0.87 |
| P19804 | Nucleoside diphosphate kinase B | 0.86 |
| P85972 | Vinculin | 0.85 |
| P15865 | Histone H1.2 | 0.84 |
| Q58FK9 | Kynurenine--oxoglutarate transaminase 3 | 0.84 |
| P62260 | 14-3-3 protein epsilon | 0.84 |
| Q5XI78 | 2-Oxoglutarate dehydrogenase, mitochondrial | 0.82 |
| B2GV06 | Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial | 0.82 |
| P31977 | Ezrin | 0.81 |
| Q01129 | Decorin | 0.80 |
| O88483 | [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, mitochondrial | 0.80 |
| P18418 | Calreticulin | 0.80 |
| P12007 | Isovaleryl-CoA dehydrogenase, mitochondrial | 0.79 |
| O35854 | Branched-chain-amino-acid aminotransferase, mitochondrial | 0.78 |
| Q66HS7 | PDZ and LIM domain protein 3 | 0.78 |
| P07335 | Creatine kinase B-type | 0.77 |
| Q9ESW0 | DNA damage-binding protein 1 | 0.77 |
Table 3.
Proteins that show a SAH-induced downregulation which does not persist after MEK inhibition. The list shows proteins that are downregulated in vehicle-treated animals (as compared with sham animals) and either upregulated or not affected in U0126-treated animals (as compared with sham animals). Regulation is expressed as log2 of the iTRAQ ratios. This group of 51 proteins is denoted group 2 and represents proteins with an SAH-induced expressional decrease that does not persist after MEK1/2 inhibition
| Accession number | Protein name | Log2 |
|---|---|---|
| Q64478 | Histone H2B type 1-H | −4.82 |
| P05811 | Alpha-crystallin B chain | −4.47 |
| Q8BGD9 | Eukaryotic translation initiation factor 4B | −4.09 |
| P08009 | Glutathione S-transferase Yb-3 | −3.64 |
| Q5U2R0 | Methionine adenosyltransferase 2 subunit beta | −3.39 |
| P62860 | 40S ribosomal protein S30 | −3.29 |
| P23457 | 3-Alpha-hydroxysteroid dehydrogenase | −3.25 |
| Q9Z2G8 | Nucleosome assembly protein 1-like 1 | −3.14 |
| P55063 | Heat shock 70 kDa protein 1-like | −3.03 |
| Q8VDM6 | Heterogeneous nuclear ribonucleoprotein U-like protein 1 | −2.93 |
| P22057 | Prostaglandin-H2 D-isomerase | −2.87 |
| P04897 | Guanine nucleotide-binding protein G(i) subunit alpha-2 | −2.76 |
| P27321 | Calpastatin | −2.76 |
| Q04940 | Neurogranin | −2.72 |
| O35206 | Collagen alpha-1(XV) chain | −2.72 |
| Q9CSU0 | Regulation of nuclear pre-mRNA domain-containing protein 1B | −2.39 |
| Q63362 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | −2.32 |
| Q63768 | Adapter molecule crk | −2.32 |
| P26043 | Radixin | −2.23 |
| Q63584 | Transmembrane protein Tmp21 | −2.20 |
| P27274 | CD59 glycoprotein | −2.18 |
| Q3T1I4 | Protein PRRC1 | −2.16 |
| P62142 | Serine/threonine-protein phosphatase PP1-beta catalytic subunit | −2.15 |
| P55002 | Microfibrillar-associated protein 2 | −1.39 |
| Q04857 | Collagen alpha-1(VI) chain | −1.36 |
| P04157 | Receptor-type tyrosine-protein phosphatase C | −1.22 |
| P11240 | Cytochrome c oxidase subunit 5A, mitochondrial | −1.22 |
| P15508 | Spectrin beta chain, erythrocyte | −1.20 |
| P31399 | ATP synthase subunit d, mitochondrial | −1.15 |
| Q62737 | Cytochrome b-245 light chain | −1.10 |
| Q02788 | Collagen alpha-2(VI) chain | −1.09 |
| P26051 | CD44 antigen | −1.08 |
| Q8BUR4 | Dedicator of cytokinesis protein 1 | −1.06 |
| P11662 | NADH-ubiquinone oxidoreductase chain 2 | −1.03 |
| P24623 | Alpha-crystallin A chain | −1.03 |
| P61805 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit DAD1 | −0.95 |
| P97700 | Mitochondrial 2-oxoglutarate/malate carrier protein | −0.90 |
| Q9JM51 | Prostaglandin E synthase | −0.88 |
| Q6AXX6 | UPF0765 protein C10orf58 homolog | −0.87 |
| P26151 | High affinity immunoglobulin gamma Fc receptor I | −0.86 |
| Q9R233 | Tapasin | −0.85 |
| Q9CQ91 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3 | −0.84 |
| P29419 | ATP synthase subunit e, mitochondrial | −0.83 |
| Q3B8Q1 | Nucleolar RNA helicase 2 | −0.83 |
| P06687 | Sodium/potassium-transporting ATPase subunit alpha-3 | −0.80 |
| P97544 | Lipid phosphate phosphohydrolase 3 | −0.79 |
| P00697 | Lysozyme C-1 | −0.77 |
| P11247 | Myeloperoxidase | −0.77 |
| P08050 | Gap junction alpha-1 protein | −0.77 |
| Q99P82 | Claudin-11 | −0.77 |
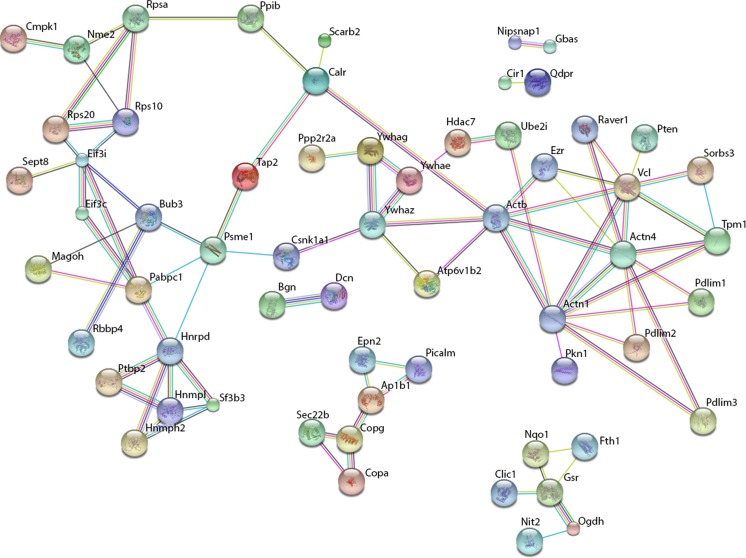
Network analysis of group 1 proteins was performed using the STRING database for protein interactions4 (Figs. 3 and 4). Of the 133 identified proteins, the database recognized 107 and constructed a network of protein interactions (Fig. 4). The analysis showed that many of the proteins were regulated through each other either directly or indirectly. In particular, the analysis revealed a network of proteins with structural functions which includes actinin and vinculin. Actinin is a structural protein and it is involved in cytoskeletal stability. Vinculin is a protein involved in focal adhesions via interaction with actin and other structural proteins (Sun et al. 2008). Another group of highly interconnected proteins includes three isoforms of the adaptor protein 14-3-3 (14-3-3 protein zeta/delta, gamma, and epsilon) (Fig. 6). The 14-3-3 adaptor protein has a large number of reported interaction partners and roles in apoptosis, cell cycle, migration, and differentiation (Mhawech 2005). Networks of proteins involved in translation, mRNA processing, and protein folding such as translational initiation factor 3 subunits, Polyadenylate-binding protein 1 and ribosomal protein subunits S40 isoforms were also identified. Furthermore, a network of proteins responsible for vesicle transport and formation including epsin and phosphatidylinositol-binding clathrin assembly protein and a network of reductases were found including quinone reductase 1 and glutathione reductase.
Fig. 4.
Proteins upregulated after experimental SAH and affected by U0126 treatment (group 1 proteins). Proteins whose upregulation in large cerebral arteries of rats 48 h after SAH did not persist after U0126 treatment were subjected to network analysis performed with STRING (www.string.db) software. The network consists of roughly six clusters, the most predominant containing structural proteins
Fig. 6.
Validation of expression of 14-3-3 in large cerebral arteries. The expression of 14-3-3 in large cerebral arteries of sham animals, and animals subjected to SAH and treated with vehicle or U0126 was investigated by Western blot. 14-3-3 is present in all groups and was upregulated in both the vehicle and the U0126-treated groups (p < 0.05). Data were analyzed with one-way ANOVA and Newman-Keuls test for multiple comparisons
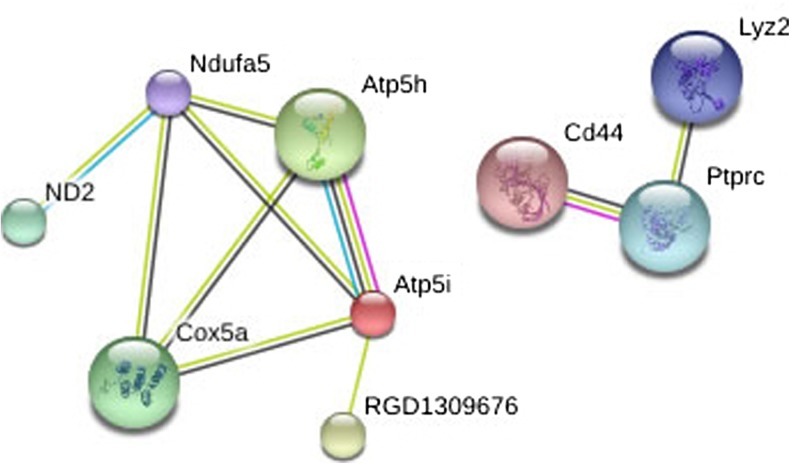
When performing a network analysis of those proteins whose SAH-induced downregulation disappeared after U0126 treatment (group 2 proteins), two networks were suggested (Fig. 5). One network included six proteins involved in metabolism such as ATP synthase and cytocrome C synthase. The other network contained three proteins involved in immune responses.
Fig. 5.
Proteins downregulated in experimental SAH and affected by U0126 treatment (group 2 proteins). Proteins whose downregulation in large cerebral arteries of rats 48 h after SAH did not persist after U0126 treatment were subjected to network analysis performed with STRING (www.string.db) software. The network consists of two individual clusters, the most predominant containing proteins involved in cellular metabolism
Validation of Specific Proteins and Their Regulation
We chose to validate proteins from the strong networks of the proteins that were found upregulated after SAH and normalized by MEK1/2 inhibition (group 1 proteins). We studied the expression of 14-3-3 protein in animals subjected to sham surgery or SAH with or without U0126 treatment with Western blot. This protein was detectable in all three groups, supporting that they are indeed expressed in cerebral vessels. Furthermore, the adaptor protein 14-3-3 showed regulation (one-way ANOVA, p = 0.05), more specifically 14-3-3 tends to be upregulated after SAH, whereas treatment with U0126 did not affect this upregulation (Fig. 6).
Discussion
This study is the first to show on an overall proteomic level the expressional changes that occur in large cerebral arteries at 48 h post-SAH and to address how these proteomic changes are affected by treatment with a MEK1/2 inhibitor. Localization studies demonstrated this to occur early after SAH and to maintain elevated levels in the VSMCs (Ansar and Edvinsson 2008). It is worth noting that much of previous work to find novel ways of therapy in order to reduce cerebral vasospasm and the development of late cerebral ischemia after SAH have centered on single molecular approaches. Our study clearly reveals that a large number of proteins show differential expression in the cerebral vessel walls following an experimental SAH and this supports our vision that therapy must be directed towards a common signal transduction mechanism (Edvinsson and Povlsen 2011).
We have shown that the MEK/ERK pathway is activated in cerebral ischemia and that the activation results in enhanced expression of contractile cerebrovascular receptors such as endothelin type B, 5-hydroxytryptamie type 1B, angiotension II type AT1, and thromboxane receptors in the cerebral arteries associated with the ischemic region. Most interesting the specific inhibitor U0126 not only prevents this receptor upregulation but importantly has a beneficial effect on the outcome after experimental subarachnoid hemorrhage in rats (Larsen et al. 2011). Compared to rats treated with vehicle, the U0126-treated rats perform better on a rotating pole test and showed increased spontaneous activity, rearing, grooming, and locomotion as compared to vehicle-treated animals. These findings are in line with and extend previous work (Edvinsson and Povlsen 2011; Larsen et al. 2011; Parker et al. 2013). The present study thus adds to the evidence pointing to the MEK/ERK pathway as a major player in SAH pathophysiology and outcome.
The proteome of major cerebral arteries after SAH revealed that pathways linked to structural modifications of the cerebral blood vessel wall, mainly the VSMCs, are highly regulated after SAH. In addition, we found that proteins involved in binding of nucleic acids, proteins, or calcium ions, and in catalytic activity are highly regulated.
The main results of this study is the identification of 250 proteins regulated in cerebral vessels after SAH and the finding that 184 of these proteins returned to sham level or were contra regulated after treatment with U0126. This underlines the importance of the MEK/ERK pathway in cerebrovascular pathology after SAH. The proteins identified in this study comprise strong networks of structural proteins, adaptor proteins, and in addition networks of proteins involved in translation, mRNA processing, and protein folding.
Remodeling of cerebral arteries is a well-known phenomenon to occur after SAH (Edvinsson and Povlsen 2011; Zhang and Macdonald 2006) and the high number of structural proteins that were observed to be regulated at 2 days after induction of SAH in this study supports that a structural remodeling takes place after SAH. Among the structural proteins we observed to be upregulated by SAH was actinin, an important structural protein in smooth muscle cells. Actinin acts as a linker between integrins and actin filaments to strengthen the cytoskeleton and mediate contraction of the smooth muscle cell (Sun et al. 2008). Another upregulated structural protein is vinculin, a focal adhesion protein involved in binding integrins and actin in the cytoskeleton. Vinculin plays a role in the connection between integrins and actin filaments in the vasculature (Critchley 2004). The network of structural proteins also included tropomyosin and β-actin, both involved in contractility of VSMCs.
Interestingly, the network analysis in addition highlighted the 14-3-3 protein family, whose members previously have been shown to affect the ability of VSMCs to undergo phenotypic switch in vitro (Chen et al. 2013). The 14-3-3 proteins are a family of conserved regulatory molecules that are expressed in all eukaryotic cells, and we validated their presence in cerebral arteries by Western blot. The 14-3-3 proteins have the ability to bind a multitude of functionally diverse signaling proteins, including kinases, phosphatases, and transmembrane receptors. More than 50 signaling proteins have been reported as 14-3-3 ligands including proteins of the Ras/Raf/MEK/ERK pathway (Fischer et al. 2009; Fu et al. 2000).
Upregulation of proteins involved in the protein synthesis pathways of the VSMCs is in line with the fact that in addition to the structural proteins previously mentioned, several proteins such as receptors, cytokines, and ion channels have been observed to be upregulated (Edvinsson and Povlsen 2011; Kamp et al. 2012; Maddahi et al. 2012). Their upregulation might depend severely on a more efficient protein synthesis and thereby an upregulation of the protein synthesis pathways. However, neither the receptors nor the Ca2+ channels were identified as regulated in this study, but this might very well be explained by their relatively low abundance compared to, e.g., structural proteins.
The downregulation of mitochondrial proteins is difficult to explain. It seems like a paradox that energy-consuming processes such as protein synthesis are upregulated, while energy-generating pathways are downregulated. However, we speculate that the downregulation is not beneficial since it is abolished after treatment with MEK inhibition, which according to our findings is beneficial after SAH (Fig. 1).
We show that targeting the MEK-ERK1/2 pathway improves outcome of animals and normalizes protein changes 48 h after SAH. Previous work on SAH have focused on limited targets, e.g., single receptors, cytokines, or matrix metalloproteinases (Ansar et al. 2007; Maddahi et al. 2012; Wang et al. 2012); however, our approach made it possible to draw a global picture of multiple changes. One example of the “single target” approach is the Clazosentan trial project (an endothelin receptor antagonist). By targeting the endothelin receptors with a specific antagonist Clazosentan, the arterial narrowing of the vessels was reduced, but it did not improve outcome after SAH (Macdonald et al. 2012). Therefore it seems that additional processes play a role in the vascular pathology after SAH and a total quantitative proteome is a promising tool to get a broader view of the events that follow SAH.
In summary, the overall findings of this study suggest that the changes in structural properties of VSMC change after SAH in a very complex way. These changes can be reduced by treatment with U0126 and this reduction is associated with improved neurological outcome. This suggests that MEK1/2 inhibition after SAH might be a possible treatment strategy for SAH patients. Moreover, the data from this study comprise a large dataset that could be very useful for further investigations and validation of other pathways involved in the complex aftermath of SAH with or without treatment with U0126.
Electronic Supplementary Material
(DOC 79 kb)
(DOC 80 kb)
(DOC 49 kb)
(DOC 46 kb)
Acknowledgements
This work was supported by The Swedish Research Council (LE - no. 5958), The Heart and Lung foundation, Sweden (LE), and The Lundbeck Foundation (LE—Center of excellence, M.R.L—Junior Group Leader Fellowship, and A.V.G.E—a project grant in biomedicine).
Author Contributions Statement
AHM, GKP, AVGE, MRL, and LE conceived and designed the experiments. AHM, GKP, KW, and AVGE performed the experiments. AHM, AVGE, and JN analyzed the data. AHM, AVGE, JN, KW, and LE wrote the paper. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflicts of interest.
Footnotes
One protein can have several functions and it might therefore appear in more than one group. Additionally, the database did not allocate all the identified proteins.
Hits are defined as the total number of molecular functions assigned for the recognized proteins.
In the PANTHER classification system, binding is defined as calcium ion binding, calcium-dependent phospholipid binding, chromatin binding, lipid binding, nucleic acid binding, and protein binding (www.pantherdb.org).
STRING did not allocate interactions to all identified proteins. The network analyses only contain proteins where interactions could be allocated.
Electronic supplementary material
The online version of this article (doi:10.1007/s12031-017-0944-7) contains supplementary material, which is available to authorized users.
References
- Ansar S, Edvinsson L. Subtype activation and interaction of protein kinase C and mitogen-activated protein kinase controlling receptor expression in cerebral arteries and microvessels after subarachnoid hemorrhage. Stroke. 2008;39:185–190. doi: 10.1161/STROKEAHA.107.487827. [DOI] [PubMed] [Google Scholar]
- Ansar S, Larsen C, Maddahi A, Edvinsson L. Subarachnoid hemorrhage induces enhanced expression of thromboxane A2 receptors in rat cerebral arteries. Brain Res. 2010;1316:163–172. doi: 10.1016/j.brainres.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Ansar S, Vikman P, Nielsen M, Edvinsson L. Cerebrovascular ETB, 5-HT1B, and AT1 receptor upregulation correlates with reduction in regional CBF after subarachnoid hemorrhage. Am J Physiol Heart Circ Physiol. 2007;293:H3750–H3758. doi: 10.1152/ajpheart.00857.2007. [DOI] [PubMed] [Google Scholar]
- Ansari N. Re: Vitiligo: auto-immunity and immune responses. Int J Dermatol. 2007;46:1314–1315. doi: 10.1111/j.1365-4632.2007.03484.x. [DOI] [PubMed] [Google Scholar]
- Chen YC, Chu LY, Yang SF, Chen HL, Yet SF, Wu KK. Prostacyclin and PPARalpha agonists control vascular smooth muscle cell apoptosis and phenotypic switch through distinct 14-3-3 isoforms. PloS one. 2013;8:e69702. doi: 10.1371/journal.pone.0069702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell JS. Protein identification using MS/MS data. J Proteomics. 2011;74:1842–1851. doi: 10.1016/j.jprot.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Critchley DR. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem Soc Trans. 2004;32:831–836. doi: 10.1042/BST0320831. [DOI] [PubMed] [Google Scholar]
- Duncia JV, et al. MEK inhibitors: the chemistry and biological activity of U0126, its analogs, and cyclization products. Bioorg Med Chem Lett. 1998;8:2839–2844. doi: 10.1016/S0960-894X(98)00522-8. [DOI] [PubMed] [Google Scholar]
- Edvinsson L, Povlsen GK. Late cerebral ischaemia after subarachnoid haemorrhage: is cerebrovascular receptor upregulation the mechanism behind? Acta Physiol. 2011;203:209–224. doi: 10.1111/j.1748-1716.2010.02227.x. [DOI] [PubMed] [Google Scholar]
- Engholm-Keller K, Birck P, Storling J, Pociot F, Mandrup-Poulsen T, Larsen MR. TiSH--a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J Proteomics. 2012;75:5749–5761. doi: 10.1016/j.jprot.2012.08.007. [DOI] [PubMed] [Google Scholar]
- Fischer A, et al. Regulation of RAF activity by 14-3-3 proteins: RAF kinases associate functionally with both homo- and heterodimeric forms of 14-3-3 proteins. J Biol Chem. 2009;284:3183–3194. doi: 10.1074/jbc.M804795200. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Ann Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Hansen-Schwartz J, Hoel NL, Xu CB, Svendgaard NA, Edvinsson L. Subarachnoid hemorrhage-induced upregulation of the 5-HT1B receptor in cerebral arteries in rats. J Neurosurg. 2003;99:115–120. doi: 10.3171/jns.2003.99.1.0115. [DOI] [PubMed] [Google Scholar]
- Kamp MA, Dibue M, Schneider T, Steiger HJ, Hanggi D. Calcium and potassium channels in experimental subarachnoid hemorrhage and transient global ischemia. Stroke Res Treat. 2012;2012:382146. doi: 10.1155/2012/382146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen CC, Povlsen GK, Rasmussen MN, Edvinsson L. Improvement in neurological outcome and abolition of cerebrovascular endothelin B and 5-hydroxytryptamine 1B receptor upregulation through mitogen-activated protein kinase kinase 1/2 inhibition after subarachnoid hemorrhage in rats. J Neurosurg. 2011;114:1143–1153. doi: 10.3171/2010.6.JNS1018. [DOI] [PubMed] [Google Scholar]
- Macdonald RL, et al. Randomized trial of clazosentan in patients with aneurysmal subarachnoid hemorrhage undergoing endovascular coiling. Stroke. 2012;43:1463–1469. doi: 10.1161/STROKEAHA.111.648980. [DOI] [PubMed] [Google Scholar]
- Maddahi A, Povlsen GK, Edvinsson L. Regulation of enhanced cerebrovascular expression of proinflammatory mediators in experimental subarachnoid hemorrhage via the mitogen-activated protein kinase kinase/extracellular signal-regulated kinase pathway. J Neuroinflamm. 2012;9:274. doi: 10.1186/1742-2094-9-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhawech P. 14-3-3 proteins—an update. Cell Res. 2005;15:228–236. doi: 10.1038/sj.cr.7290291. [DOI] [PubMed] [Google Scholar]
- Ohlsson AL, Johansson BB. Environment influences functional outcome of cerebral infarction in rats. Stroke. 1995;26:644–649. doi: 10.1161/01.STR.26.4.644. [DOI] [PubMed] [Google Scholar]
- Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- Parker BL, Larsen MR, Edvinsson LI, Povlsen GK. Signal transduction in cerebral arteries after subarachnoid hemorrhage—a phosphoproteomic approach. J Cereb Blood Flow Metab. 2013;33:1259–1269. doi: 10.1038/jcbfm.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povlsen GK, Johansson SE, Larsen CC, Samraj AK, Edvinsson L. Early events triggering delayed vasoconstrictor receptor upregulation and cerebral ischemia after subarachnoid hemorrhage. BMC Neurosci. 2013;14:34. doi: 10.1186/1471-2202-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell GF, Mathiesen T, Svendgaard NA. A new experimental model in rats for study of the pathophysiology of subarachnoid hemorrhage. Neuroreport. 2002;13:2553–2556. doi: 10.1097/00001756-200212200-00034. [DOI] [PubMed] [Google Scholar]
- Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Steen H, Mann M. The ABC’s (and XYZ’s) of peptide sequencing. Nat Rev Mol Cell Biol. 2004;5:699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- Sun Z, Martinez-Lemus LA, Hill MA, Meininger GA. Extracellular matrix-specific focal adhesions in vascular smooth muscle produce mechanically active adhesion sites. Am J Physiol Cell Physiol. 2008;295:C268–C278. doi: 10.1152/ajpcell.00516.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Potential contribution of matrix metalloproteinase-9 (mmp-9) to cerebral vasospasm after experimental subarachnoid hemorrhage in rats. Ann Clin Lab Sci. 2012;42:14–20. [PubMed] [Google Scholar]
- Zhang ZD, Macdonald RL. Contribution of the remodeling response to cerebral vasospasm. Neurol Res. 2006;28:713–720. doi: 10.1179/016164106X151990. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 79 kb)
(DOC 80 kb)
(DOC 49 kb)
(DOC 46 kb)