Abstract
Interpreting the intricate bacterial transcriptomics implies understanding the dynamic relationship established between de novo transcription and the degradation of transcripts. Here, we performed a comparative overview of gene expression levels and mRNA decay rates for different-biovar (trachoma and lymphogranuloma venereum) strains of the obligate intracellular bacterium Chlamydia trachomatis. By using RNA-sequencing to measure gene expression levels at mid developmental stage and mRNA decay rates upon rifampicin-based transcription blockage, we observed that: i) 60–70% of the top-50 expressed genes encode proteins with unknown function and proteins involved in “Translation, ribosomal structure and biogenesis” for all strains; ii) the expression ranking by genes' functional categories was in general concordant among different-biovar strains; iii) the median of the half-life time (t1/2) values of transcripts were 15–17 min, indicating that the degree of transcripts’ stability seems to correlate with the bacterial intracellular life-style, as these values are considerably higher than the ones observed in other studies for facultative intracellular and free-living bacteria; iv) transcript decay rates were highly heterogeneous within each C. trachomatis strain and did not correlate with steady-state expression levels; v) only at very few instances (essentially at gene functional category level) was possible to unveil dissimilarities potentially underlying phenotypic differences between biovars. In summary, the unveiled transcriptomic scenario, marked by a general lack of correlation between transcript production and degradation and a huge inter-transcript heterogeneity in decay rates, likely reflects the challenges underlying the unique biphasic developmental cycle of C. trachomatis and its intricate interactions with the human host, which probably exacerbate the complexity of the bacterial transcription regulation.
Keywords: Biological sciences, Infectious disease, Biochemistry, Genetics
1. Introduction
For the past few years, several studies have shown that bacteria present complex transcriptional activity, showing intricate gene expression regulation and envolving multiple classes of transcripts (including sRNAs) (Sorek and Cossart, 2010; Thomason and Storz, 2010; Georg and Hess, 2011; Lasa et al., 2012). In this context, understanding such transcriptomic scenery is essential to get insights on the pathways of a bacterial physiology, metabolism, and adaptation to changing environments. To address this issue, it is vital to assess the expression profile of each transcript, which in turn is the result of the balance established between de novo transcription and its degradation at a particular growth context. A multitude of studies have focused on the regulation and mechanisms of transcription initiation and/or on measuring steady-state transcript levels in different bacteria, but only a handful of them explored the global trends of transcripts’ molecular stability (some examples: Wang et al., 2002; Bernstein et al., 2002; Hambraeus et al., 2003; Barnett et al., 2007; Kristoffersen et al., 2012; Rustad et al., 2013; Liu et al., 2014; Chen et al., 2015). Although most of these studies revealed a lack of association between several characteristics of a transcript and its decay rate over time (e.g., secondary structures, G + C content, codon composition and transcript length), they further pointed that transcripts’ stability could be species-specific and/or be influenced by bacterial growth rates (Rustad et al., 2013) and by a variety of cell signals and external stimuli (reviewed in Wang et al., 2002). mRNA decay studies have been essentially performed only in extracellular bacteria, which are more undemanding to handle, resulting in a general lack of information for obligate intracellular bacteria.
The high-throughput RNA sequencing (RNA-seq) technology (Goodwin et al., 2016) has been applied for complex analyses of the bacterial transcriptome, including gene expression quantification, identification of operons, detection of antisense transcription, characterization of small RNAs and non-coding RNAs, and even the determination of mRNA decay rates (Croucher and Thomson, 2010; Sharma et al., 2010; Ozsolak and Milos, 2011; Sharma and Vogel, 2014; Creecy and Conway, 2015). For the obligate intracellular bacteria C. trachomatis, the RNA-seq technology has been successfully applied, for instance, to map all TSSs and to identify novel sRNAs (Albrecht et al., 2010), to simultaneously assess the gene’s expression of both the bacterium and the host in response to an infection scenario (Humphrys et al., 2013), and also to evaluate the impact of culture-derived mutations on the transcriptome (Borges et al., 2015). The understanding of the transcriptome of C. trachomatis holds great interest as this is a highly prevalent human pathogen and a major public health concern. In fact, it is the etiologic agent of the blindness trachoma and constitutes the major bacterial cause of sexually transmitted infections worldwide (Burton, 2007; Wright et al., 2008). As a member of Chlamydiaceae, this bacterium displays a unique biphasic developmental cycle of about 36–96 h (Miyairi et al., 2006) during which, the extracellular and infectious form, the EB, enters the host cell and differentiates into the intracellular and replicative form, the RB. After several rounds of binary fission within a membrane-bound vacuole known as the inclusion, RBs differentiate back into EBs, which are released from the host cell, by lysis or inclusion extrusion, to carry on new infections in neighbouring cells.
Overall, the strains of C. trachomatis can be classified into 2 biovars and 15 major serovars according to the polymorphism of the Major Outer Membrane Protein (MOMP, encoded by ompA). The trachoma biovar include strains from serovars A-C and D-K, which preferentially infect epithelial cells of the ocular conjunctivae and the ano-urogenital tract, respectively, causing organ-specific diseases (Peipert, 2003; Miyairi et al., 2006). On the other hand, the LGV biovar include strains from the serovars L1-L3, which also colonize the host through the ano-urogenital tract, but are able to disseminate to the regional draining lymph nodes, via infection of the macrophages (Schachter, 1978). To date, investigators are struggling to understand which factors underlie the phenotypic differences, namely, discrepancies in growth rates, routes of infection, cell tropism and disease-outcomes (Gomes et al., 2006; Thomson et al., 2008; Dehoux et al., 2011; Borges et al., 2012; Harris et al., 2012; Lutter et al., 2012; Abdelsamed et al., 2013; da Cunha et al., 2014; Ferreira et al., 2015; and reviewed in Nunes et al., 2013). Considering that the genetic dissimilarity among the C. trachomatis strains is less than 2%, it has been speculated that differences in the regulation of gene expression also contribute to the above mentioned phenotypic discrepancies. Some studies aiming the evaluation of gene expression differences among C. trachomatis strains have been performed, namely, whole transcriptomic analysis focused on a single strain (Belland et al., 2003; Albrecht et al., 2010; Humphrys et al., 2013), or multi-strain analyses focused on a restricted set of genes (Nunes et al., 2009; Borges et al., 2010; Almeida et al., 2012; Song et al., 2013; Ferreira et al., 2013). Still, intra-strain gene expression dissimilarities observed so far reflect transcripts abundance at discrete moments, and this abundance is completely dependent on the joint action of both the transcription initiation and decay rates (Grunberg-Manago, 1999; Steege, 2000; Pérez-Ortín et al., 2007; Keene, 2010). To give the first steps towards the understanding of these dual transcriptional dynamics in C. trachomatis, we carried out an exploratory global survey regarding the expression level and the decay rate of transcripts in strains representing the two biovars, in an attempt to unveil putative differences that may underlie biovar-specific phenotypes.
2. Materials and methods
2.1. Cell culture, rifampicin treatment and harvesting
C. trachomatis E/CS1025-11 and L2b/CS19-08 clinical strains (subjected to minimal culture passages) were independently inoculated into 15 T25 flasks of confluent HeLa229 cells monolayers by centrifuging at 2200 rpm for 1 h (34 °C), followed by an incubation stage of 1 h at 37 °C, in a 5% CO2 atmosphere. Cell culture medium was then discarded and fresh enriched medium (MEM 10% foetal bovine serum, vitamins, non-essential aminoacids, glucose and 0.5 μg/ml cycloheximide) was added to the cultures. Bacterial cells were allowed to grow at 37 °C with 5% CO2, until the mid-phase of the developmental cycle was achieved (20 h and 24 h pi for L2b/CS19-08 and E/CS1025-11, respectively), because the majority of C. trachomatis genes are being actively transcribed at this point (Belland et al., 2003; Humphrys et al., 2013) and also because, at this stage carry over mRNA, which would be quickly degraded biasing the decay rate calculations, was found to be no longer present (Humphrys et al., 2013). As the estimation of the mid-phase was roughly assessed by microscopic monitoring of inclusions development, we opted by a careful approach by essentially focusing the expression and decay comparative analysis between strains on groups of genes. In fact, a gene-based analysis was solely performed on a discrete manner.
Three sets of five flasks, for each strain, were submitted to different periods of rifampicin treatment (10 μg/ml) − 0 min (T0, no treatment), 10 min (T10) and 30 min (T30) − in order to stop de novo RNA synthesis (Hartmann et al., 1967; Levin and Hatfull, 1993), and thus, allow the evaluation of the mRNA decay. One T25 flask of T0, T10 and T30 sets was immediately harvested by using glass beads, the cell suspension was sonicated for 7 min (Vibra Cell, Bioblock Scientific), for host cells disruption and chlamydial release, submitted to a centrifugation of 7 min at 700 rpm, for cell debris deposit, and finally the supernatant was stored at −20 °C for further DNA extraction. The remaining four T25 flasks of T0, T10 and T30 culture sets, were treated with the rifampicin solution, as previously mentioned. Immediately after the respective treatment period, the solution was replaced with a 2:1 solution of RNAprotect™ Bacteria Reagent (Qiagen) and PBS buffer and left for 2 min, to ensure diffusion into the cells for the bacterial RNA preservation during subsequent handling. Again, cultures were harvested, sonicated and centrifuged as previously referred, and the supernadant was immediately submitted to RNA extraction. Note that we intentionally did not treat the cell culture from which DNA would be extracted, because a preliminary assay showed that the RNAprotect™ Bacteria Reagent (Qiagen) degrades DNA (data not shown). We performed two biological replicates for both strains.
2.2. DNA and RNA extraction
Both nucleic acids were extracted as previously referred (Ferreira et al., 2013). Briefly, DNA was extracted by centrifuging the stored (–20 °C) supernatant at 14,000 rpm for 10 min (at 4 °C) and the pellet was resuspended in 200 μl PBS for QIAamp® DNA Mini Kit (Qiagen) extraction, according to manufacturer’s instructions. DNA was eluted in 50 μl of AE buffer and stored at −20 °C after A260 nm quantification in a NanoDrop 1000 spectrophotometer (Thermo Scientific). For total RNA extraction, the RNeasy® Mini Kit (Qiagen, CA, USA) was used according to manufacturer’s instructions. Briefly, the culture supernatants were subject to a high-speed centrifugation (14,000 rpm) for 10 min at 4 °C, the pellets were suspended in lysozyme-containing TE buffer and treated with RLT buffer with 1% β-mercaptoethanol for cell lysis. An on-column DNase treatment (RNase-free DNase, Qiagen) was included in the procedure to remove contaminant DNA, and RNA was eluted in 40 μl of RNase-free water. RNA yield and purity were determined by absorbance measurement at 260 nm and 280 nm using the NanoDrop 1000 spectrophotometer (Thermo Scientific). For a parallel qPCR quantification of the expression levels (and decay kinetics) along the time frame used, 5 μl of this total RNA was further used for cDNA generation (see below).
2.3. Bacterial mRNA preparation/purification
Eukaryotic, mitochondrial and bacterial rRNA were depleted by using the Ribo-Zero™ Gold rRNA Removal Kit (Epidemiology) (Illumina, CA, USA), according to manufacturer’s instructions. Subsequently, the Dynabeads® mRNA Purification Kit for mRNA Purification from Total RNA preps (LifeTechnologies) was used, with and adapted protocol, to pull the eukaryotic mRNA out of the samples, leaving only the bacterial mRNA. The obtained bacterial mRNA was concentrated in a final volume of 13 μl, by using the RNeasy® MinElute™ Cleanup Kit (Qiagen, CA, USA), according to manufacturer’s instructions. Bacterial mRNA quality and concentration were determined by the Bioanalyzer (Agilent) equipment, where the absence of rRNA readings reflects the purity of the mRNA.
2.4. RNA-seq
Bacterial mRNA-enriched samples were subjected to library construction (TruSeq Stranded mRNA sample preparation kit, Illumina) and sequencing on an Illumina MiSeq sequencer using a paired-end (2 × 75 bp) strategy (at least 15 M reads were dedicated per sample), according to manufacturer’s instructions. FastQC analysis was used to assess reads quality and Bowtie2 was applied for mapping reads of each strain to the respective chromosome and plasmid DNA sequences obtained in a previous study (Borges et al., 2015). Cufflinks (version 2.1.1; http://cufflinks.cbcb.umd.edu/) tools were further applied to quantify the amount of transcripts of each chromosome and plasmid CDS normalized as fragments per kilobase of CDS per million mapped reads (FPKM), as previously described (Borges et al., 2015). Calculations of transcript amounts for each time point (before and after 10 and 30 min of rifampicin treatment) were based on two biological replicates. The use of a high throughput technology, such as the Illumina technology, is essential for the quantification of decaying mRNAs’ expression levels (Kristoffersen et al., 2012) as it potentiates the capture of low expressed genes and extremely labile transcripts. In this study, transcript counts were obtained for the three experimental conditions (T0, T10 and T30) for >99% of all annotated CDSs in both genomes.
2.5. cDNA generation and qPCR assays
In order to validate the obtained RNA-seq data, RNA decay kinetics was assessed in parallel by qPCR assays using cDNA samples generated at T0, T10 and T30, from both replicates of both strains, as previously described (Borges et al., 2010; Ferreira et al., 2013). Briefly, TaqMan® RT Reagents (Applied Biosystems, Branchburg, USA) were used as follows: 2.5 μM of random hexamers, 5.5 mM MgCl2, 500 μM of each dNTP, 1 × RT Buffer, 0.8 U/μl RNase inhibitor and 1.25 U/μl MultiScribe RT, in a final reaction volume of 50 μl. Cycling conditions were: 10 min at 25 °C, 15 min at 42 °C and 5 min at 99 °C. cDNA was stored at −20 °C until use.
DNA from a 48 h (pi) chlamydial culture was extracted (as described above) and subjected to eight two-fold serial dilutions in DNase-free water, in order to produce the DNA standard curves for the expression quantification. The use of DNA standard curves allows the cross-comparison of the expression levels among genes at each time-point that could not be achieved by using cDNA standard curves, given that DNA represents equal amounts of each single copy gene. Primers for the 16SrRNA and 21 other genes selected for quantification purposes were design using the Primer Express software (Applied Biosystems). The qPCR assays were performed using the LightCycler® 480 equipment, SYBR Green chemistry and optical plates and caps (Roche). The qPCR mixture consisted of 1 × LightCycler® 480 SYBR Green I Master (Roche), 400 nM of each primer and 5 μl of each DNA (standard curves) or cDNA (samples), in a final volume of 25 μl. Plates included one standard curve for each gene, cDNA duplicates, and “no RT” controls.
Gene expression was normalized by dividing the mean value of raw qPCR duplicates by the respective mean value of the 16SrRNA, for each condition (T0, T10 and T30), of both replicates of both strains. This strategy was used because 16SrRNA was previously shown to be a stable gene, being a valuable resource for expression normalization in C. trachomatis (Borges et al., 2010).
2.6. mRNA half-life time (t1/2) analysis
Each mRNA half-life time (t1/2) was calculated by using an adaptation of the “two-fold” decay step method (Selinger et al., 2003) based on the fit of an exponential decay between values obtained at the first time-point (before rifampicin addition; T0) and the values calculated t minutes (10 min and/or 30 min) after the transcriptional arrest (T1), using the formula: t1/2 = -ln2/k; where the rate of decay rate (k) was estimated as follows: k = ln(T1/T0)/t. Therefore, it was also required to establish the appropriate time interval to correctly apply the formula, which was determined by performing an overall screening of the “decay profile” of each transcript, by considering its mean value of FPKM for the T0 (no treatment), T10 (10 min of antibiotic treatment) and T30 (30 min of antibiotic treatment). In that regard, the criteria applied were: i) if expression at T0 > T10 > T30, we used the interval with the highest slope (usually was the T0-T10); ii) if T0 > T10 < T30, we used the T0-T10 interval, whether T0> T30 or T0 < T30, because we assumed that at 30 min after rifampicin treatment the blockage effect of this antibiotic may not be as absolute as after 10 min; iii) if T0 < T10> T30, we used T0-T30, but only if T0 > T30, otherwise we considered that there was no overall mRNA decay; and iv) if T0 < T10 < T30, we were not able to calculated mRNA decay at any interval, and therefore those transcripts were removed from the analysis.
Throughout the text, results were analyzed either in a gene-by-gene manner (assuming the gene designation of the D/UW3-CX genome annotation; NC_000117) or by functional categories (according to Heizer et al., 2006; with some exceptions; Supplementary Table).
3. Results and discussion
The main goal of the present study was to evaluate the whole transcriptome dynamics of C. trachomatis strains from the two biovars by using the RNA-seq technology. In particular, we aimed to: i) compare gene expression levels between four different serovar strains (D/CS637-11, E/CS1025-11, Ia/CS190-96 and L2b/CS19-08); ii) evaluate the decay rate of all mRNAs of two different biovar strains (E/CS1025-11 and L2b/CS19-08); and iii) correlate expression levels and half-life time (t1/2).
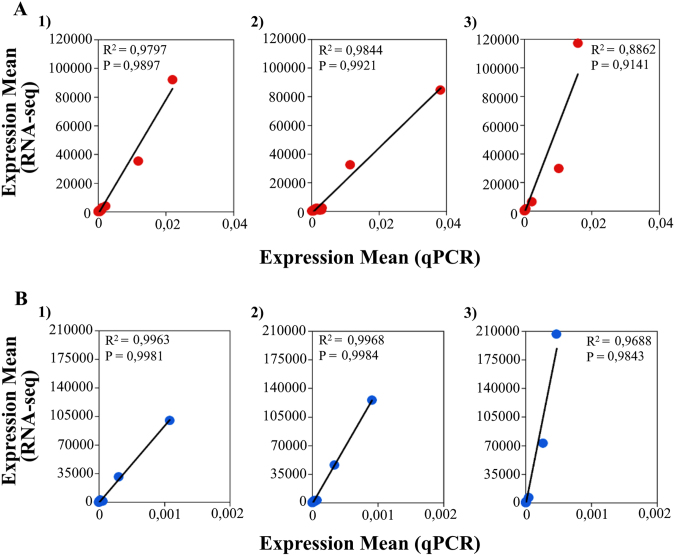
In a preliminary stage, we checked the reliability of the RNA-seq data through qPCR analysis of 22 transcripts to ensure that not only the initial mRNA levels of both strains were accurately determined by RNA-seq (T0), but also that the decay pattern of each transcript (T0 to T10 to T30) was also correctly determined. We obtained high reproducibility of the gene expression quantification results generated with the two distinct quantitative methods (R2 > 0.88 for all comparisons) (Fig. 1), which corroborates the suitability of the normalization strategy applied to the raw RNA-seq data.
Fig. 1.
Relation of the expression levels acquired by qPCR (horizontal axis) and RNA-seq (vertical axis), for two C. trachomatis strains: L2b/CS19/08 (A) and E/CS1025/11 (B). Each dot represents the relation of the expression values obtained by the two methods, for the same transcript, of that particular strain. For each strain is shown that relation regarding three different sets of transcripts: 1) All transcripts quantified for each strain; 2) transcripts with a calculated half-life time ≤ 30 min; and 3) transcripts with a calculated half-life time ≤ 10 min. Both the linear correlation coefficient (R2) and the Pearson correlation coefficient (P) are shown in each graph, and indicating strong correlation.
3.1. Expression analysis between the two C. trachomatis biovars
In this analysis, we compared the expression level of transcripts of C. trachomatis E/CS1025-11 and L2b/CS19-08 strains, and also of two additional strains (D/CS637-11 and Ia/CS190-96), for which RNA-seq data had been previously obtained (Borges et al., 2015). All four expression data sets were acquired at the mid-phase stage of the developmental cycle (when bacterial replication occurs) to ensure that the expression of the majority of the C. trachomatis genes is assessed (Belland et al., 2003; Humphrys et al., 2013).
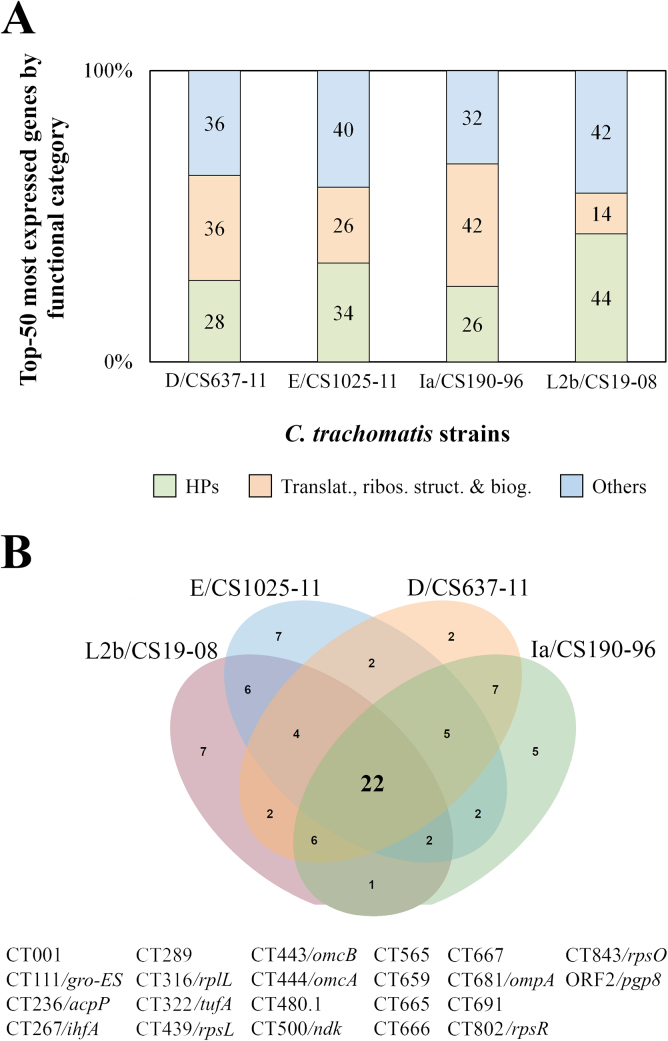
Firstly, we looked at the genes presenting the highest level of expression at the mid-phase for each strain, and found a considerable parallelism between strains (all expression data is available at Supplementary Table). In fact, seven genes were found among the 10 most expressed, regardless of the strain. This set includes three genes (CT443/omcB, CT444/omcA and CT681/ompA) encoding major constituents of the outer membrane of this species, whose high expression at replication stage is not surprising, and also three poorly characterized genes (CT001, CT267, CT500), whose transcription performance warrants further investigation. Finally, the ORF2/pgp8 genome region displayed high abundance of transcripts but this reflects the expression of its anti-sense sRNA (Albrecht et al., 2010; Ferreira et al., 2013). By extending the investigation to the top-50 most expressed genes, we observed that about 60–70% of those top ranked genes encode “Hypothetical proteins” (HPs) and proteins from the “Translation, ribosomal structure and biogenesis” functional category (Fig. 2A). The proportion of genes from each of the remainder functional categories within the top-50 ranking never exceeded 6%.
Fig. 2.
Distribution of the top-50 most expressed genes of each C. trachomatis strain. (A) Functional categories (colour coded) of the top-50 most expressed genes of the four strains. The proportion (in percentage) of each functional category is displayed in the vertical bars. (B) Venn’s diagram depicting the overlap scenario, among strains, of their top-50 most expressed genes. The 22 genes found in all four top-50 ranks are displayed underneath the diagram and are listed by ORF number.
Furthermore, 22 genes were found in all four top-50 ranks (Fig. 2B), where again the “HPs” were highly represented genes (7/22), followed by the “Translation, ribosomal structure and biogenesis” genes (5/22). All these 22 genes had already been pointed out as being highly expressed at the mid-phase stage of the developmental cycle of a trachoma biovar serovar E strain in a previous RNA-seq study (Humphrys et al., 2013). Considering our results on different serovar (and biovar) strains, these “core” genes certainly play a role in very conserved and essential mechanisms in C. trachomatis, at the mid-phase stage of the developmental cycle. Whereas for the members of the “Translation, ribosomal structure and biogenesis” functional category, this result may not be surprising, as at this stage there must be a tremendous demand of proteins playing a role in translation and metabolism, the high representation of HPs coding genes among the highly expressed genes is intriguing. In fact, C. trachomatis genes with unknown function are overrepresented in chlamydial genomes and are mostly genus- or species-specific (Griffiths et al., 2006; Stephens et al., 1998), which make them certainly important for chlamydial biology (Subtil et al., 2005; Valdivia, 2008), as corroborated by the present study.
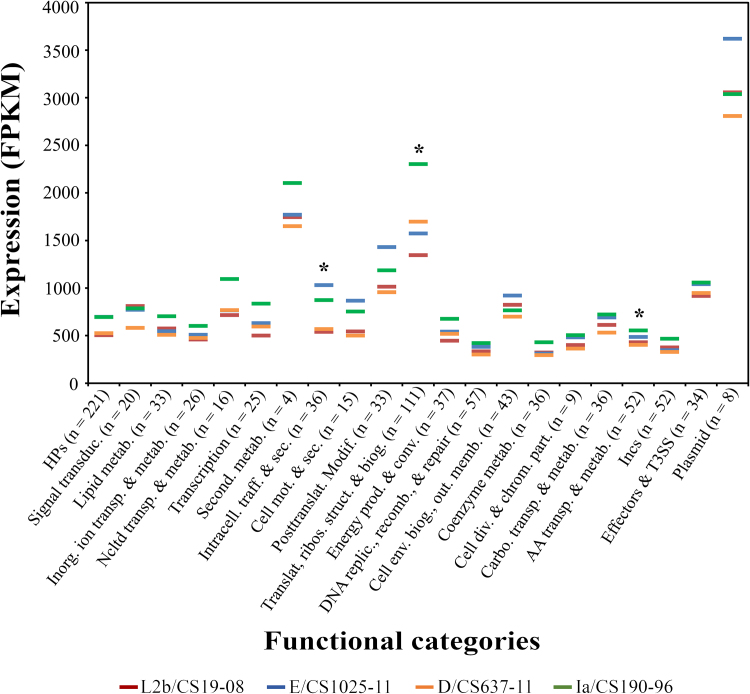
Subsequently, we evaluated the global gene expression differences between strains by calculating the median of the gene-by-gene expression differences for each strain pair. We verified that the Ia/CS190/96 had the highest median value of expression difference, being 36% and 32% more expressed then the D/CS637-11 and the L2b/CS19-08 strains, respectively. On the other hand, these two latter strains presented only ∼1% variation in the median of expression differences. We then assessed the existence of functional categories for which the observed global trend (Ia/CS190–96 > E/CS1025–11 > L2b/CS19–08 > D/CS637-11) is not applied. In this regard, the median of expression values was determined for each functional category, for each strain (Fig. 3).
Fig. 3.
Expression values by functional categories. The graph shows the medians of gene expression for 21 functional categories, for the four C. trachomatis strains (colour coded). Group sizes (displayed in the x axis) are similar among strains within each functional category. Asterisks (*) indicate the functional categories for which statistically significant differences (P < 0.05, one-way ANOVA test) were found between at least two strains. The expression median values displayed in the “Plasmid” functional category are a direct consequence of the tremendous transcript abundance of the sRNAs encoded in the antisense strand of two plasmid ORFs (Albrecht et al., 2010; Ferreira et al., 2013).
In agreement with the previous observation, both the L2b/CS19-08 and the D/CS637-11 strains presented the lowest median expression values across most functional categories, whereas Ia/CS190-96 was top-ranked for most of them (15/21). Nevertheless, there was a noteworthy exception as the E/CS1025-11 strain exhibited the highest expression medians for five functional categories, in particular in the “Plasmid genes” and “Cell envelope biogenesis, outer membrane” categories. When comparing the median values across functional categories, we found that four functional categories (“Secondary metabolites biosynthesis, transport, and catabolism”, “Posttranslational modification, protein turnover, chaperones”, “Translation, ribosomal structure and biogenesis”, and “Plasmid genes”) present higher medians than the remainder. As the plasmid encodes two highly expressed antisense sRNAs (Albrecht et al., 2010; Ferreira et al., 2013), the elevated expression values obtained for the “Plasmid genes” category are the result of the combined expression of both ORFs and the antisense sRNAs, and hence, the medians determined for this category are overestimated. There are five categories for which the medians were the lowest regardless the strain, namely, “DNA replication, recombination, and repair”, “Coenzyme metabolism”, “Cell division and chromosome partitioning”, “Amino acid transport and metabolism”, and “Incs or putative Incs”. Of note, the expression differences observed between functional categories, within each strain, should be eyed with caution, as they may be a consequence of the time point of the developmental cycle at which the gene expression was evaluated and the growth conditions applied, which most certainly impact the biological needs of the bacterium at a given moment. For instance, the observation of incs among the less expressed genes at the mid-stage of the developmental cycle likely reflects the fact that the majority of incs shows an early-cycle profile of expression (Almeida et al., 2012). On the other hand, expression differences between strains, within the same functional category, may suggest that dissimilar transcriptional regulation of functional-related genes is probably a determinant factor in the biological dissimilarities of the strains.
3.2. Half-life time analysis between different biovar strains
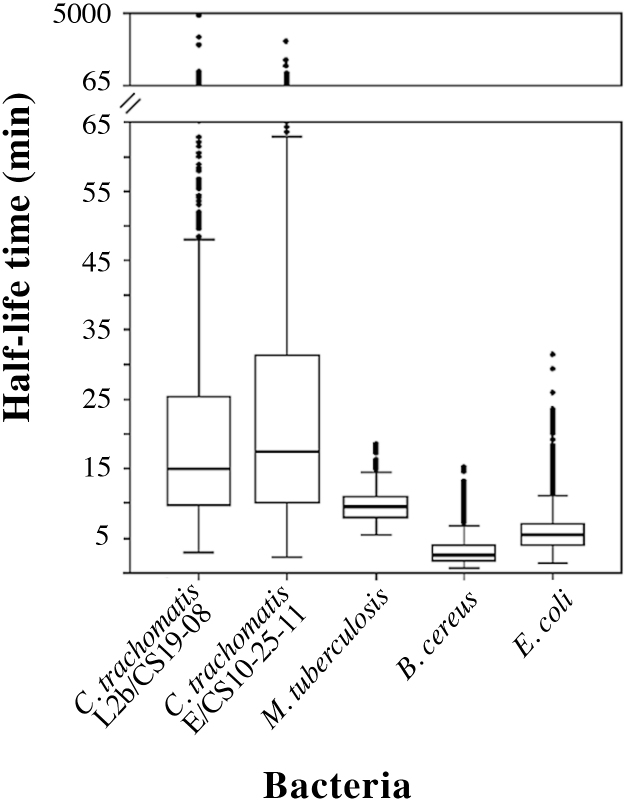
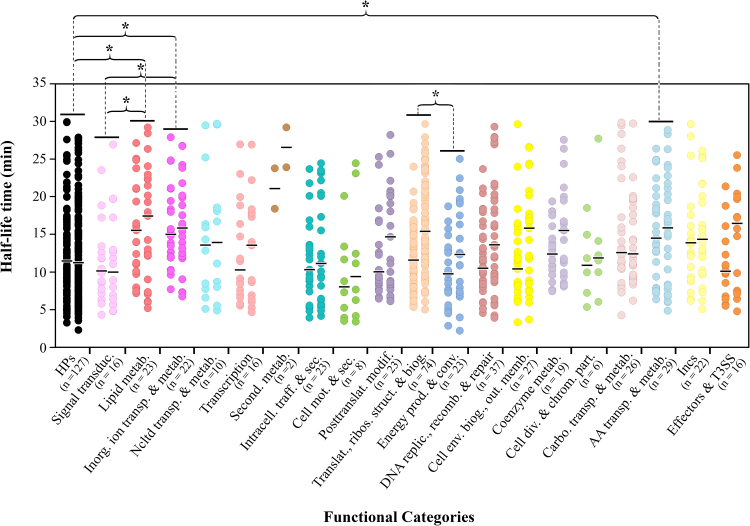
Transcripts’ degradation has an important role in the regulation of gene expression, where the coordinated action between the transcription initiation rate and the decay rate of each RNA, results in its steady-state abundance at a particular moment, under certain environmental conditions. Therefore, assessing transcripts stability may ultimately allow the inference of their role in the biology of bacteria. To date, several studies have already conducted global surveys of mRNA half-life times in several microorganisms (Wang et al., 2002; Bernstein et al., 2002; Hambraeus et al., 2003; Andersson et al., 2006; Barnett et al., 2007; Kristoffersen et al., 2012; Rustad et al., 2013; Liu et al., 2014; Chen et al., 2015), but none of them included an obligate intracellular pathogen. As such, we determined the t1/2 of the transcripts of two different biovar C. trachomatis strains (L2b/CS19-08 and E/CS1025-11) by measuring the transcript levels at 10 and 30 min post-treatment with the RNA polymerase blocking agent rifampicin. After defining the time interval that best fits the decay dynamics of each mRNA (see Section 2 Materials and methods), we observed that the global mRNA half-life time between L2b/CS19-08 and E/CS1025-11 was very similar (no statistically significant difference was found, one-way ANOVA test), with median t1/2 values of 15 min and 17 min, respectively (Fig. 4). Values of this magnitude had previously been reported for 5/10 genes with reliable T1/2 determination for the same-genus species, C. pneumoniae (Engström et al., 2010). In a previous study enrolling Mycobacterium (Rustad et al., 2013), it was speculated that, for different species of this genus, the differences of the mean t1/2 values observed could be linked to the length of the developmental cycle, i.e., slower growths would correlate with higher mRNA stability. A previous study (Rustad et al., 2013) argued that, the extended developmental cycle is a consequence of the strain’s inability to quickly regulate mRNAs abundance. Further analyses with multiple strains with dissimilar growth rates is needed to verify if this assumption applies to C. trachomatis species since, although LGV strains typically grow faster than the trachoma biovar strains, we observed no significant differences in the growth rate of these strains during our experiments.
Fig. 4.
Boxplots showing the distribution of mRNA t1/2 determined for different bacterial species: Escherichia coli (Bernstein et al., 2002), Bacillus cereus (Kristoffersen et al., 2012), Mycobacterium tuberculosis (Rustad et al., 2013), and C. trachomatis, E/CS1025-11 and L2b/CS19-08 strains (this study). Regarding the global half-life time of these bacteria, statistically significant differences (one-way ANOVA test) were found between the L2b/CS19-08 strain and E. coli, B. cereus, and M. tuberculosis (P < 0.01), whereas the E/CS1025-11 strain only differ from E. coli (P < 0.05), probably due to the wider range of t1/2 of this C. trachomatis strain.
The range of t1/2 values obtained for the obligate intracellular C. trachomatis strains was strikingly wider (especially for E/CS1025-11) than those previously obtained for other bacteria (Bernstein et al., 2002; Kristoffersen et al., 2012; Rustad et al., 2013), for which the t1/2 were also much lower, with medians of 9.3 min, 5.4 min and 2.6 min for Mycobacterium tuberculosis (facultative intracellular), Escherichia coli and Bacillus cereus (both extracellular), respectively (Fig. 4). Previous studies regarding the Gram-negative E. coli and the Gram-positive B. cereus attributed the t1/2 differences to key ribonucleases that each bacteria possesses, a feature believed to distinguish the two Gram groups (Commichau et al., 2009; Kristoffersen et al., 2012). However, when including our data from an obligate intracellular bacterium, strikingly larger differences between t1/2 are observed (Fig. 4). In fact, the two studied free-living bacteria possess very low transcripts t1/2 medians (≤ 5.4 min), the facultative intracellular bacterium M. tuberculosis presents an intermediate transcripts’ t1/2 median of ∼9.3 min, and the obligate intracellular C. trachomatis possesses much higher transcripts t1/2 medians (≥ 15 min). In support of this trend, the t1/2 median of M. tuberculosis contrasts with the one of its closely related free-living saprophyte M. smegmatis (5.2 min) (Rustad et al., 2013). Altogether, although the presence/absence of particular ribonucleases can ultimately determine differences in mRNA decay dynamics, our observations suggest that a major factor underlying the t1/2 differences among bacteria is their life-style. For instance, the fact that obligate intracellular organisms may face less drastic environmental changes than free-living bacteria could underlie the slower turn-over rate of the transcripts abundance observed for C. trachomatis. However, data from future studies using more bacterial species, both Gram-positive and −negative, with distinct life-styles, will certainly help to elucidate if this observed trend is a sampling bias or if it mirrors the actual association between bacterial life-style and mRNA stability.
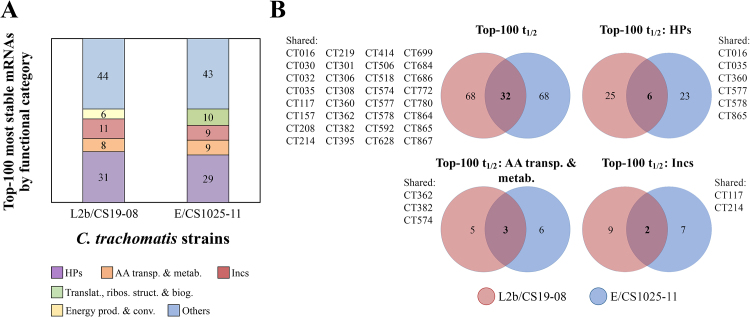
From the detailed analysis of t1/2 values detected for the two different biovar C. trachomatis strains, it was also striking to observe that both strains present a large number of highly stable genes (Fig. 4). To investigate the nature of those genes, we looked in detail for the 100 genes with the highest t1/2 values for each strain. The top-100 most stable transcripts presented t1/2 values above 45 min for both strains, and most of them (> 55%) belong to only four out of the 21 previously defined functional categories (Fig. 5A). In particular, three of this highly represented categories (HPs, “Incs or putative Incs”, and “Amino acid transport and metabolism”) are shared between strains (Fig. 5).
Fig. 5.
Top-100 most stable mRNAs of two different biovar C. trachomatis strains. (A) Functional category (colour coded) of the top-100 most stable mRNAs of the two different biovar C. trachomatis strains, L2b/CS19-08 and E/CS1025-11, where the four most represented functional categories are discriminated. The number of genes from each functional category present in the top-100 is displayed. (B) Venn’s diagram depicting the overlap between the L2b/CS19-08 (light red) and E/CS1025-11 (blue) strains of their top-100 most stable mRNAs, and of the mRNAs of the three functional categories (“HPs”, “AA transp. & metab.”, and “Incs”) most represented in both top-100 rankings. Each number represents the absolute quantity of genes shared by the indicated strains in each comparison.
Although we found this parallelism at the functional category level (Fig. 5A), the overlap was not so obvious at gene level as only 30 genes were shared by the two different biovar strains within this top-100 ranking. Of note, it must be stated that, despite the overrepresentation (30%) of “HPs” among the 100 most stable genes in both strains, this result must be interpreted with caution, as the “HPs” category is mostly composed by the unique C. trachomatis genes for which the biological function is yet to be established. For that reason, it is certainly highly heterologous in terms of gene function, encompassing representatives of several other categories. As such, any association between the huge mRNA stability and the scarce data available for some of them would be likely speculative. Besides all t1/2 determinations are performed through mathematical inferences, the t1/2 results beyond 30 min rely on extrapolation rather than on empirical observation of a two-fold decay during the studied time interval (0–30 min post-treatment). Thus, we further focused our analysis on transcripts presenting t1/2 values lower than 30 min for both strains (n = 525). Nonetheless, the pairwise comparison of the transcripts’ t1/2 revealed lack of correlation between the two strains (Pearson correlation coefficient, P = 0.546; R2 = 0.2978). As these t1/2 discrepancies can eventually be a result of the excessive sensitivity of the mathematical formula used to model the mRNA decay curve, we looked at the mRNA stability trends at functional category level, as this effect may be buffered. In fact, this approach is corroborated by previous genome-wide studies, which suggested some degree of correlation between gene function and mRNAs’ t1/2 in several organisms (Wang et al., 2002; Yang et al., 2003; Andersson et al., 2006). We observed statistically significant differences of t1/2 values between strains for three functional categories (Fig. 6): “Translation, ribosomal structure and biogenesis” (P < 0.001), “Energy production and conversion” (P < 0.05), and “DNA replication, recombination, and repair” (P < 0.05). Interestingly, for both strains, the “DNA replication, recombination, and repair” was also found to be one of the categories with the lowest medians of gene expression, whereas the “Translation, ribosomal structure and biogenesis” category was found to be one with the highest medians of gene expression, (see section “3.1 Expression analysis between the two C. trachomatis biovars”). However, both categories present statistically significant t1/2 differences between C. trachomatis strains from the two biovars, meaning that, regardless the mRNA abundance, both groups of housekeeping genes are differently regulated by these strains at the mid-stage of the developmental cycle. Taking into account the properties of these functional categories, the observed differences may account for dissimilar metabolic performance between two biovars’ strains potentially impacting the well-known development and pathogenicity dissimilarities. We also checked for significant differences in the t1/2 values between functional categories within each strain, and found that, regardless of the biovar, there were indeed some statistically significant differences (P < 0,05) (Fig. 6). This could suggest that transcripts acting in specific networks need to be maintained for longer periods, likely in order to guarantee that the production of specific proteins is not interrupted. This feature seems to be contingent on the species rather than on the biovar.
Fig. 6.
Half-life time by functional categories. The chart displays the half-life time values of the 525 transcripts yielding t1/2 < 30 min for both strains, according to their functional category (in different colours). For each functional category, t1/2 values for L2b/CS19-08 are represented on the left, whereas t1/2 values for E/CS1025-11 are represented on the right. The horizontal small dashes represent the median of the t1/2 values. Statistically significant differences of t1/2 values between functional categories (within the same strain) were found, and those shared by both strains are indicated with asterisks (P < 0.05; one-way ANOVA test).
3.3. Comparison between expression level and t1/2
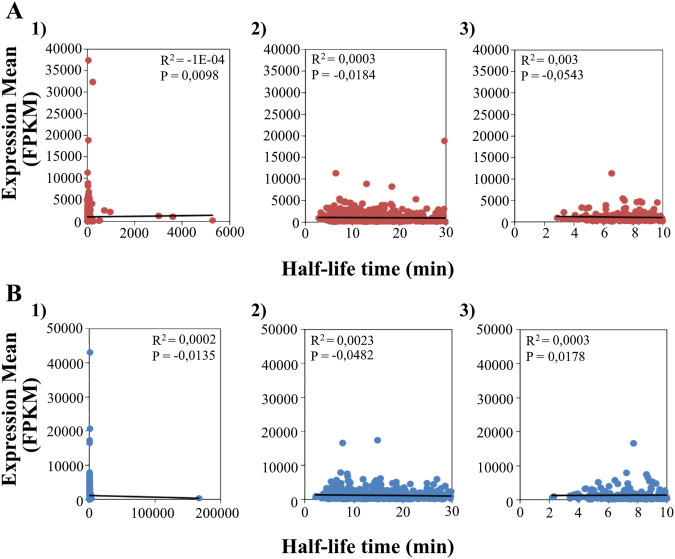
Finally, we evaluated whether the expression level at the mid-stage of the developmental cycle (T0) of a given transcript correlates with its t1/2 value. This analysis was performed for two different biovar strains (L2b/CS19-08 and E/CS1025-11). We found a lack of correlation between the stability of a transcript and its level of expression, as corroborated by the Pearson correlation coefficients calculated for the same pairs of comparisons (Fig. 7). Even when exclusively taking into account genes with the lowest t1/2 (< 10 min) (Fig. 7A.3 and B.3), i.e., those less prone to be biased by both the sensitivity of the formula applied for the t1/2 calculations and the period of time used, there was still no correlation between the level of expression and t1/2 values. Previous studies with different bacteria had also found no correlation or found a negative correlation between these two parameters (steady-state transcript abundance and t1/2) (Bernstein et al., 2002; Kristoffersen et al., 2012; Rustad et al., 2013), where the latter was suggested to be a consequence of the bacterial need for a quick turnover of highly abundant transcripts. We raise the hypothesis that, for an organism with a unique biphasic and multi-stage life-cycle as C. trachomatis, the overall lack of correlation may be just a consequence of the tight transcriptional control that is continuously operating during such a complex development. Another fact contributing to this trend may be the intricate host-pathogen interactions that take place for an obligate intracellular pathogen, which implies its continuous and dynamic adaptation. Nevertheless, it is noteworthy that we found sporadic cases in which a negative correlation was observed, although only four genes (CT157, CT219, CT421.2 and CT559) were common to both different biovar strains (analysis strictly focused on the first 100 upper and bottom ranked genes). Whereas CT421.2 and CT559 presented high expression levels and low mRNA stability, the opposite scenario was observed for CT219 and CT157. On the other hand, one study (Bernstein et al., 2002) also found a group of highly abundant transcripts that were highly stable, which does not fit the mentioned “negative correlation”. Curiously, although we found some transcripts following this pattern, none of them was shared by the two biovars (data not shown). As such, a highly abundant mRNA may or may not be as susceptible to degradation as a scarce mRNA, so transcript abundance per si does not seem to be a trigger for a quick-degradation.
Fig. 7.
Pairwise relation between the genes’ expression level, determined at the mid-stage of the developmental cycle (T0), and their t1/2. The three graphs, both in panel A and panel B, represent the relation obtained for three different groups of genes of L2b/CS19-08 and E/CS1025-11 strains, respectively. Those three groups of genes were defined according to the t1/2 they exhibited: 1) all transcripts with calculated t1/2; 2) transcripts with t1/2 ≤ 30 min; and 3) transcripts with t1/2 ≤ 10 min. Both the linear correlation coefficient (R2) and the Pearson correlation coefficient (P) are shown in each graph.
Overall, this study constitutes a first attempt to integrate and understand the global mRNA production and decay dynamics in C. trachomatis. While this exploratory work emphasizes that it would be valuable to conduct similar studies for other bacterial pathogens, the preliminary data generated here for C. trachomatis highlights important research questions that should be followed in this subject. For instance, it would be interesting to investigate a putative correlation between the growth rate and transcripts’ stability, and to get insights into the potential differential decay rates of transcripts belonging to polycistronic operons. Still, our data suggests that the specific interplay between transcripts’ production and degradation is likely driven by complex transcriptional networks that still need to be disclosed, and where specific RNA-binding proteins and small RNAs may play key roles, a notion that is becoming more consolidated (reviewed in Waters and Storz, 2009 and Oliva et al., 2015).
Declarations
Author contributions statement
Rita Ferreira, Vítor Borges: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Maria José Borrego: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
João Paulo Gomes: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper
Funding statement
Rita Ferreira was supported by a Ph.D. fellowship (SFRH/BD/68532/2010) from the Fundação para a Ciência e Tecnologia, Portugal.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at http://dx.doi.org/10.1016/j.heliyon.2017.e00364.
No additional information is available for this paper.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Abdelsamed H., Peters J., Byrne G.I. Genetic variation in Chlamydia trachomatis and their hosts: impact on disease severity and tissue tropism. Future Microbiol. 2013;8:1129–1146. doi: 10.2217/fmb.13.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht M., Sharma C.M., Reinhardt R., Vogel J., Rudel T. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res. 2010;38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F., Borges V., Ferreira R., Borrego M.J., Gomes J.P., Mota L.J. Polymorphisms in Inc Proteins and Differential Expression of inc Genes among Chlamydia trachomatis Strains Correlate with Invasiveness and Tropism of Lymphogranuloma Venereum Isolates. J. Bacteriol. 2012;194:6574–6585. doi: 10.1128/JB.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A., Lundgren M., Eriksson S., Rosenlund M., Bernander R., Nilsson P. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 2006;7:R99. doi: 10.1186/gb-2006-7-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett T., Bugrysheva J., Scott J. Role of mRNA stability in growth phase regulation of gene expression in the group A Streptococcus. J. Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belland R., Zhong G., Crane D., Hogan D., Sturdevant D., Sharma J., Beatty W., Caldwell H. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. 2003;100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein J., Khodursky A., Lin P., Lin-Chao S., Cohen S. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. 2002;99:9697–9702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V., Ferreira R., Nunes A., Nogueira P., Borrego M.J., Gomes J.P. Normalization strategies for real-time expression data in Chlamydia trachomatis. J. Microbiol. Methods. 2010;82:256–264. doi: 10.1016/j.mimet.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Borges V., Nunes A., Ferreira R., Borrego M.J., Gomes J.P. Directional Evolution of Chlamydia trachomatis towards Niche-Specific Adaptation. J. Bacteriol. 2012;194:6143–6153. doi: 10.1128/JB.01291-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V., Pinheiro M., Antelo M., Sampaio D., Vieira L., Ferreira R., Nunes A., Almeida F., Mota L., Borrego M. Chlamydia trachomatis In Vivo to In Vitro Transition Reveals Mechanisms of Phase Variation and Down-Regulation of Virulence Factors. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton M.J. Trachoma: an overview. Br. Med. Bull. 2007;84:99–116. doi: 10.1093/bmb/ldm034. [DOI] [PubMed] [Google Scholar]
- Chen H., Shiroguchi K., Ge H., Xie X. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 2015;11:781. doi: 10.15252/msb.20145794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commichau F., Rothe F., Herzberg C., Wagner E., Hellwig D., Lehnik-Habrink M., Hammer E., Volker U., Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creecy J., Conway T. Quantitative bacterial transcriptomics with RNA-seq. Curr. Opin. Microbiol. 2015;23:133–140. doi: 10.1016/j.mib.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N., Thomson N. Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 2010;13:619–624. doi: 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha M., Milho C., Almeida F., Pais S.V., Borges V., Mauricio R., Borrego M.J., Gomes J.P., Mota L.J. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol. 2014;14 doi: 10.1186/1471-2180-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehoux P., Flores R., Dauga C., Zhong G., Subtil A. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics. 2011:12. doi: 10.1186/1471-2164-12-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P., Bailey L., Onskog T., Bergström S., Johansson J. A comparative study of RNA and DNA as internal gene expression controls early in the developmental cycle of Chlamydia pneumoniae. FEMS Immunol. Med. Microbiol. 2010;58:244–253. doi: 10.1111/j.1574-695X.2009.00631.x. [DOI] [PubMed] [Google Scholar]
- Ferreira R., Borges V., Nunes A., Borrego M.J., Gomes J.P. Assessment of the load and transcriptional dynamics of Chlamydia trachomatis plasmid according to strains' tissue tropism. Microbiol. Res. 2013;168:333–339. doi: 10.1016/j.micres.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Ferreira R., Antelo M., Nunes A., Damião V., Borrego M.J., Gomes J.P. In silico scrutiny of genes revealing phylogenetic congruence with clinical prevalence or tropism properties of Chlamydia trachomatis strains. G3 (Bethesda) 2015;5:9–19. doi: 10.1534/g3.114.015354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georg J., Hess W. Cis-antisense RNA another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 2011;75:286–300. doi: 10.1128/MMBR.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes J.P., Nunes A., Bruno W.J., Borrego M.J., Florindo C., Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: Evidence for serovar Da recombination and correlation with tissue tropism. J. Bacteriol. 2006;188:275–286. doi: 10.1128/JB.188.1.275-286.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin S., McPherson J., McCombie W. Coming of age: ten years of next-generation sequencing technologies. Nature Rev. Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E., Ventresca M.S., Gupta R.S. BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species. BMC Genomics. 2006;7:14. doi: 10.1186/1471-2164-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- Hambraeus G., von Wachenfeldt C., Hederstedt L. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics. 2003;269:706–714. doi: 10.1007/s00438-003-0883-6. [DOI] [PubMed] [Google Scholar]
- Harris S.R., Clarke I.N., Seth-Smith H.M.B., Solomon A.W., Cutcliffe L.T., Marsh P., Skilton R.J., Holland M.J., Mabey D., Peeling R.W. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nature Genet. 2012;44:413–419. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann G., Honikel K., Knüsel F., Nüesch J. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim. Biophys. Acta. 1967;145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- Heizer E.J., Raiford D., Raymer M., Doom T., Miller R., Krane D. Amino acid cost and codon-usage biases in 6 prokaryotic genomes: a whole-genome analysis. Mol. Biol. Evol. 2006;23:1670–1680. doi: 10.1093/molbev/msl029. [DOI] [PubMed] [Google Scholar]
- Humphrys M., Creasy T., Sun Y., Shetty A., Chibucos M., Drabek E., Fraser C., Farooq U., Sengamalay N., Ott S. Simultaneous transcriptional profiling of bacteria and their host cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. The global dynamics of RNA stability orchestrates responses to cellular activation. BMC Biol. 2010;8:95. doi: 10.1186/1741-7007-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoffersen S., Haase C., Weil M., Passalacqua K., Niazi F., Hutchison S., Desany B., Kolstø A.B., Tourasse N.J., Read T.D. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol. 2012;13:R30. doi: 10.1186/gb-2012-13-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I., Toledo-Arana A., Gingeras T. An effort to make sense of antisense transcription in bacteria. RNA Biol. 2012;9:1039–1044. doi: 10.4161/rna.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Hatfull G. Mycobacterium smegmatis RNA polymerase: DNA supercoiling, action of rifampicin and mechanism of rifampicin resistance. Mol. Microbiol. 1993;8:277–285. doi: 10.1111/j.1365-2958.1993.tb01572.x. [DOI] [PubMed] [Google Scholar]
- Liu B., Deikus G., Bree A., Durand S., Kearns D., Bechhofer D. Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains. Mol. Microbiol. 2014;94:41–55. doi: 10.1111/mmi.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter E.I., Martens C., Hackstadt T. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp. Funct. Genomics. 2012;2012 doi: 10.1155/2012/362104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyairi I., Mahdi O., Ouellette S., Belland R., Byrne G. Different growth rates of Chlamydia trachomatis biovars reflect pathotype. J. Infect. Dis. 2006;194:350–357. doi: 10.1086/505432. [DOI] [PubMed] [Google Scholar]
- Nunes A., Borrego M.J., Nunes B., Florindo C., Gomes J.P. Evolutionary Dynamics of ompA, the Gene Encoding the Chlamydia trachomatis Key Antigen. J. Bacteriol. 2009;191:7182–7192. doi: 10.1128/JB.00895-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes A., Borrego M.J., Gomes J.P. Genomic features beyond Chlamydia trachomatis phenotypes: What do we think we know? Infect. Genet. Evol. 2013;16:392–400. doi: 10.1016/j.meegid.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Oliva G., Sahr T., Buchrieser C. Small RNAs, 5' UTR elements and RNA-binding proteins in intracellular bacteria: impact on metabolism and virulence. FEMS Microbiol. Rev. 2015;39:331–349. doi: 10.1093/femsre/fuv022. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Milos P. RNA sequencing: advances, challenges and opportunities. Nature Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peipert J.F. Clinical practice. Genital chlamydial infections. N. Engl. J. Med. 2003;349:2424–2430. doi: 10.1056/NEJMcp030542. [DOI] [PubMed] [Google Scholar]
- Pérez-Ortín J., Alepuz P., Moreno J. Genomics and gene transcription kinetics in yeast. Trends Genet. 2007;23:250–257. doi: 10.1016/j.tig.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Rustad T., Minch K., Brabant W., Winkler J., Reiss D., Baliga N., Sherman D. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic Acids Res. 2013;41:509–517. doi: 10.1093/nar/gks1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter J. Chlamydial infections (third of three parts) N. Engl. J. Med. 1978;298:540–549. doi: 10.1056/NEJM197803092981005. [DOI] [PubMed] [Google Scholar]
- Selinger D., Saxena R., Cheung K., Church G., Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C., Hoffmann S., Darfeuille F., Reignier J., Findeiss S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Sharma C., Vogel J. Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014;19:97–105. doi: 10.1016/j.mib.2014.06.010. [DOI] [PubMed] [Google Scholar]
- Song L., Carlson J.H., Whitmire W.M., Kari L., Virtaneva K., Sturdevant D.E., Watkins H., Zhou B., Sturdevant G.L., Porcella S.F. Chlamydia trachomatis Plasmid-Encoded Pgp4 Is a Transcriptional Regulator of Virulence-Associated Genes. Infect. Immun. 2013;81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010;11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- Steege D. Emerging features of mRNA decay in bacteria. RNA. 2000;6:1079–1090. doi: 10.1017/s1355838200001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R.S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R.L., Zhao Q.X. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Subtil A., Delevoye C., Balañá M.E., Tastevin L., Perrinet S., Dautry-Varsat A. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 2005;56:1636–1647. doi: 10.1111/j.1365-2958.2005.04647.x. [DOI] [PubMed] [Google Scholar]
- Thomason M., Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 2010;44:167–188. doi: 10.1146/annurev-genet-102209-163523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson N.R., Holden M.T.G., Carder C., Lennard N., Lockey S.J., Marsh P., Skipp P., O'Connor C.D., Goodhead I., Norbertzcak H. Chlamydia trachomatis: Genome sequence analysis of lymphogranuloma venereum isolates. Genome Res. 2008;18:161–171. doi: 10.1101/gr.7020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R.H. Chlamydia effector proteins and new insights into chlamydial cellular microbiology. Curr. Opin. Microbiol. 2008;11:53–59. doi: 10.1016/j.mib.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu C., Storey J., Tibshirani R., Herschlag D., Brown P. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L., Storz G. Regulatory RNAs in Bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright H.R., Turner A., Taylor H.R. Trachoma. Lancet. 2008;371:1945–1954. doi: 10.1016/S0140-6736(08)60836-3. [DOI] [PubMed] [Google Scholar]
- Yang E., van Nimwegen E., Zavolan M., Rajewsky N., Schroeder M., Magnasco M., Darnell J.E. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 2003;13:1863–1872. doi: 10.1101/gr.1272403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.