Abstract
We identified two genes related to fungicide resistance in Fusarium fujikuroi through random mutagenesis. Targeted gene deletions showed that survival factor 1 deletion resulted in higher sensitivity to fungicides, while deletion of the gene encoding F-box/WD-repeat protein increased resistance, suggesting that the genes affect fungicide resistance in different ways.
Keywords: Bakanae disease, F-box, Fungicide resistance, Fusarium fujikuroi, Survival factor
The ascomycete fungus Fusarium fujikuroi Nirenberg (sexual stage Gibberella fujikuroi) causes bakanae or foolish seedling disease in rice. The defining symptoms of the disease are the formation of elongated rice stems in response to gibberellins produced by the fungus, and the poor grain ripening in infected rice [1,2]. Infected seeds act as the primary inoculum, sometimes leading to seedling death, but usually resulting in the aforementioned symptoms [3].
Bakanae disease has been successfully controlled in Korea by seed disinfection using several fungicides such as prochloraz, tebuconazole, and benomyl. However, recently the disease has been reemerging, resulting in severe yield losses [1,4,5]. This may be in part due to higher temperatures during the rice nursery period than in the past, but the occurrence of fungicide-resistant strains in Korean rice fields has had even greater impact. Strains resistant to fungicides have been frequently isolated, and the rate of resistance is greater than 30% [5,6,7]. Despite the importance of F. fujikuroi fungicide resistance in agriculture, studies examining resistance mechanisms have been limited.
To understand the mechanism of its fungicide resistance, we generated mutants of the fungicide-susceptible F. fujikuroi field strain B14 [1] through random mutagenesis. The fungal strain was maintained on potato dextrose agar (PDA) and complete medium (CM) [2]. Conidia were produced in carboxymethyl cellulose (CMC) medium [8]. To generate the construct for random mutagenesis, a fragment including a hygromycin resistance cassette and the open reading frame of green fluorescent protein lacking a promoter was amplified from pIGPAPA with primers hyg1F and gfp1F (Table 1) [9]. The amplicon was added to fungal protoplasts generated from germinating conidia for transformation. Mycelial blocks of the wild-type strain B14 were inoculated in CMC liquid medium and cultivated at 25℃ and 200 rpm for 5 days. Filtrated fungal conidia produced in CMC were inoculated into 50mL of YPG (10 g peptone, 3 g yeast extract, 20 g glucose per 1 L) liquid medium and grown at 25℃ and 200 rpm for 12 hr. Mycelia harvested by filtration were incubated in 35mL of 1M NH4Cl containing Driselase (15mg/mL) to generate protoplasts. Further transformation steps were performed as previously described [10]. Transformants were selected on regeneration medium (1 g casein, 1 g yeast extract, 342 g sucrose, and 15 g agar per 1 L) containing hygromycin (75 µg/mL). Hygromycin-resistant transformants were moved to PDA supplemented with the minimum inhibitory concentration (MIC) level of prochloraz (23% EC; Kyungnong, Seoul, Korea) for the complete inhibition of mycelial growth of the wild-type fungal strain, and incubated at 25℃ for 7 days in the dark. The MIC level for prochloraz was 0.5 µg/mL, and each experiment was repeated twice with 3 replicates each.
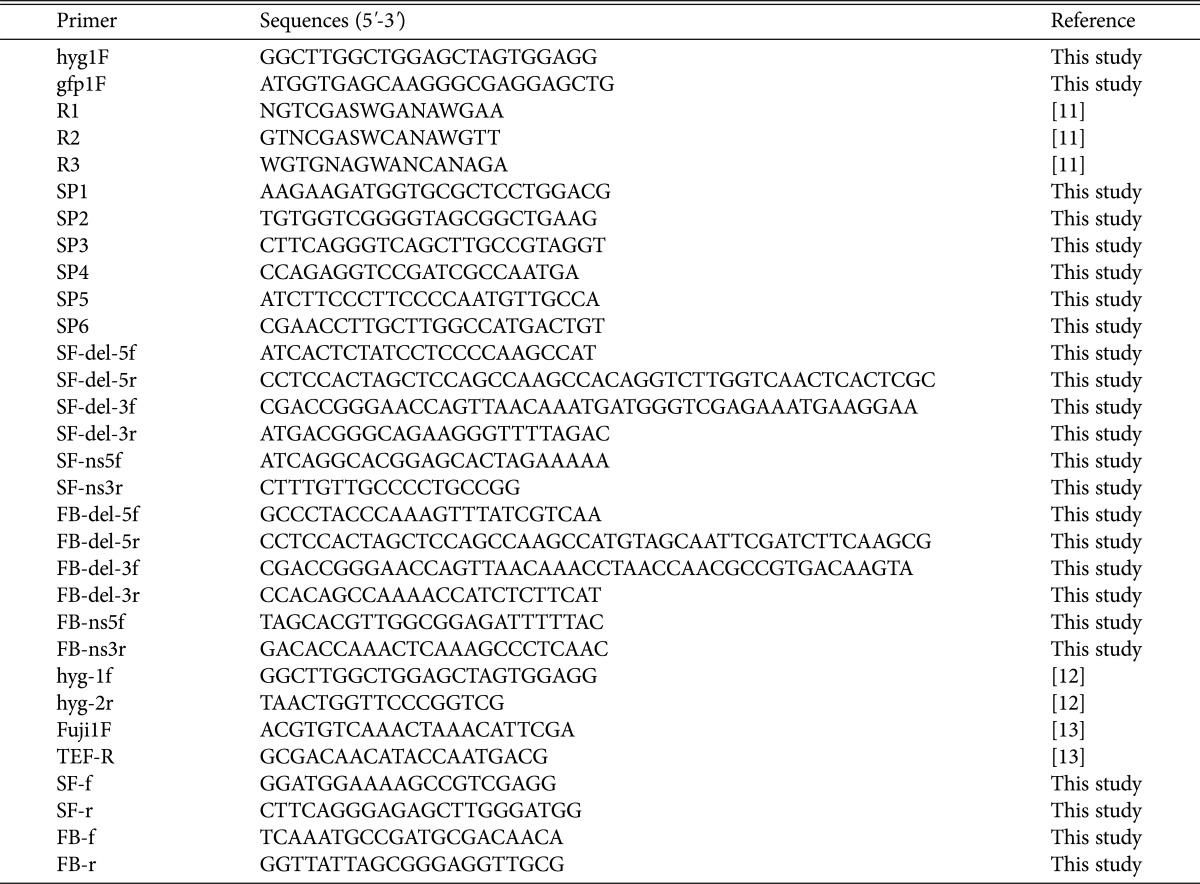
Table 1. Primers used in this study.
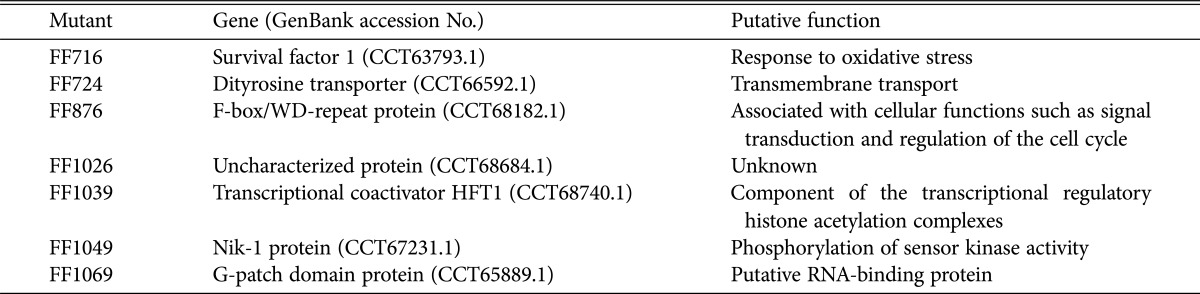
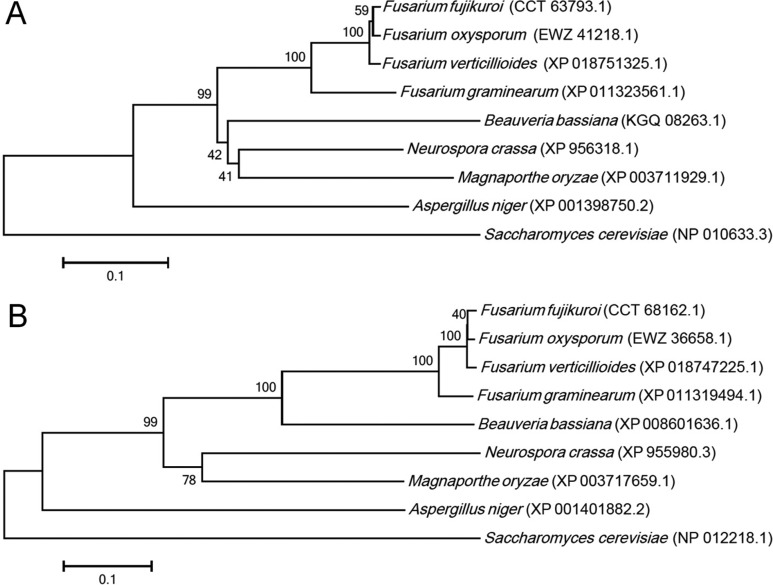
We generated 3,000 transformants that carried hygromycin resistance, and of these, 7 mutant strains showed resistance against prochloraz. The vector insertion sites in these mutants were identified through thermal asymmetric interlaced PCR [11]. The genomic DNA of fungicide-resistant mutant strains was isolated from mycelia grown in CM for 3 days at 25℃. PCR was performed with three arbitrary degenerate primers (R1, R2, and R3) and three specific primers (SP1-6) designed from vector sequences introduced into the mutant strains (Table 1). After the third PCR, the amplicons were cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) for sequencing and the sequences analyzed using the Basic Local Alignment Search Tool (BLAST; National Center for Biotechnology Information, Bethesda, MD, USA) using the blastn algorithm (Table 2). Fungicide resistance in 5 of the 7 mutants was mitotically instable, as consecutive regeneration resulted in the loss of fungicide resistance. Two mutant strains, FF716 and FF876, showed concrete resistance after several rounds of mitotic regeneration, suggesting that the gain-of-function phenotype displayed in these mutants were retained through asexual reproduction. In the FF716 mutant strain, the vector was integrated into the promoter region of putative survival factor 1, which encodes a 377 amino acid protein, and the vector in the FF876 mutant strain was integrated into the coding region of an F-box/WD-repeat protein with 958 amino acids. Both genes were highly conserved among filamentous fungi (Fig. 1).
Table 2. Genes rescued from mutants that were resistant to the fungicide prochloraz.
Fig. 1. Phylogenetic analyses of survival factor 1 (A) and F-box/WD-repeat protein (B) in Fusarium fujikuroi and other known fungal species. The amino acid sequences were aligned with ClustalW, and MEGA software ver. 6.0 was used to perform 1,000 bootstrap phylogenetic analyses using the neighbor-joining method.
To confirm whether these two genes are responsible for fungicide resistance, we performed targeted gene deletion. Constructs were prepared through double joint PCR [14]. In brief, 5′ and 3′ flanking regions of each target gene were amplified by PCR using primer pairs, del-5f/del-5r and del-3f/del-3r, from the wild-type strain, and a hygromycin-resistance cassette, used as a selective marker for transformation, was amplified from pIGPAPA with primers hyg-1f and hyg-2r. These 3 amplicons were mixed in a 1 : 1 : 2 molar ratio and fused in the second PCR step. In the third PCR step, the mixture was amplified using the nested primers ns5f and ns3r. The final amplicon was added to fungal protoplasts for transformation. Information on the primers used in this study is provided in Table 1.
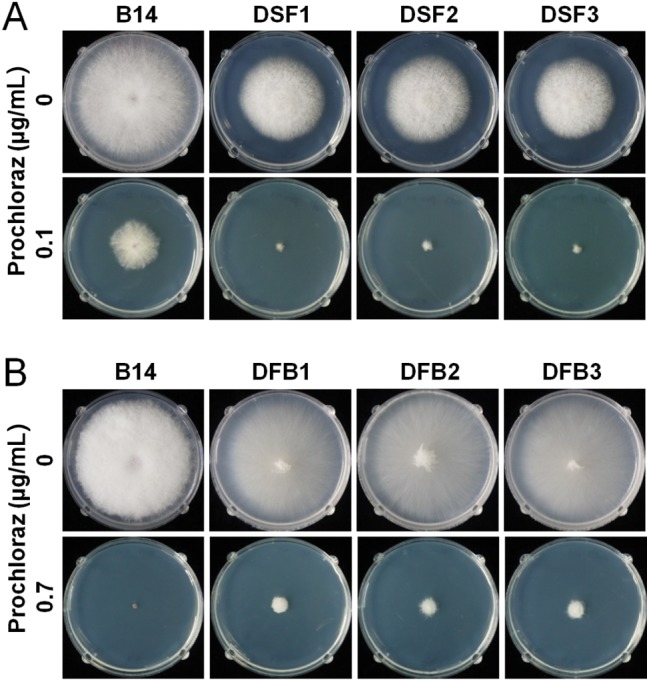
More than 20 mutant strains with deletion of either gene were selected, and 3 independent mutants for each gene were further analyzed for biological characterization. The survival factor 1 deletion mutants (DSF1-3) showed reduced mycelial growth on PDA, and the mutant strains were more sensitive to prochloraz than the wild-type strain (Fig. 2A). The F-box/WD-repeat protein deletion mutants (DFB1-3) had a similar mycelial growth rate but reduced aerial mycelia compared to the wild-type strain. Prochloraz resistance was increased compared to the wild-type strain, and the mutants survived up to 0.7 µg/mL of prochloraz (Fig. 2B).
Fig. 2. Prochloraz sensitivity of Fusarium fujikuroi mutant strains with deletion of survival factor 1 (A) and F-box/WD-repeat protein (B). Each strain was inoculated on potato dextrose agar supplemented without or with prochloraz and incubated at 25℃ for 3 days. B14, wild-type F. fujikuroi strain; DSF1–3, independent mutants with deletion of survival factor 1; DFB1–3, independent mutants with deletion of F-box/WD-repeat protein.
Morphological and physiological characteristics are commonly changed by mutations, which can destroy gene function or cause changes in gene expression. In this study, random mutagenesis resulted in identification of 2 genes responsible for prochloraz resistance in F. fujikuroi. In the FF876 mutant, the mutagenesis vector was integrated in the gene encoding an F-box/WD-repeat protein, and deletion of the gene caused increased prochloraz resistance. In the FF716 mutant, the vector was integrated in the promoter region of survival factor 1, and its deletion resulted in decreased prochloraz resistance. These results indicate that survival factor 1 and F-box/WD-repeat protein may positively and negatively regulate prochloraz resistance in F. fujikuroi. Further studies will examine the biological functions and resistance mechanisms of these genes.
ACKNOWLEDGEMENTS
This study was supported by grants from the Rural Development Administration (PJ0098912015) and from the Strategic Initiative for Microbiomes in Agriculture and Food, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea.
References
- 1.Hwang IS, Kang WR, Hwang DJ, Bae SC, Yun SH, Ahn IP. Evaluation of bakanae disease progression caused by Fusarium fujikuroi in Oryza sativa L. J Microbiol. 2013;51:858–865. doi: 10.1007/s12275-013-3472-3. [DOI] [PubMed] [Google Scholar]
- 2.Leslie JF, Summerell BA. The Fusarium laboratory manual. Ames (IW): Blackwell Publishing; 2006. [Google Scholar]
- 3.Sunder S. Satyavir. Survival of Fusarium moniliforme in soil, grains and stubbles of paddy. Indian Phytopathol. 1998;51:47–50. [Google Scholar]
- 4.Jeon YA, Yu SH, Lee YY, Park HJ, Lee S, Sung JS, Kim YG, Lee HS. Incidence, molecular characteristics and pathogenicity of Gibberella fujikuroi species complex associated with rice seeds from Asian countries. Mycobiology. 2013;41:225–233. doi: 10.5941/MYCO.2013.41.4.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SH, Park MR, Kim YC, Lee SW, Choi BR, Lee SW, Kim IS. Degradation of prochloraz by rice bakanae disease pathogen Fusarium fujikuroi with differing sensitivity: a possible explanation for resistance mechanism. J Korean Soc Appl Biol Chem. 2010;53:433–439. [Google Scholar]
- 6.Shin MU, Kang HJ, Lee YH, Kim HT. Detection for the resistance of Fusarium spp. Isolated from rice seeds to prochloraz and cross-resistance to other fungicides inhibiting sterol biosynthesis. Korean J Pestic Sci. 2008;12:277–282. [Google Scholar]
- 7.Youn K, Choi HW, Shin DB, Jung B, Lee J. Resistance of Fusarium fujikuroi isolates to hydrogen peroxide and its application for fungal isolation. Res Plant Dis. 2015;21:227–230. [Google Scholar]
- 8.Cappellini RA, Peterson JL. Macroconidium formation in submerged cultures by a non-sporulating strain of Gibberella zeae. Mycologia. 1965;57:962–966. [Google Scholar]
- 9.Horwitz BA, Sharon A, Lu SW, Ritter V, Sandrock TM, Yoder OC, Turgeon BG. A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet Biol. 1999;26:19–32. doi: 10.1006/fgbi.1998.1094. [DOI] [PubMed] [Google Scholar]
- 10.Kim JE, Jin J, Kim H, Kim JC, Yun SH, Lee YW. GIP2, a putative transcription factor that regulates the aurofusarin biosynthetic gene cluster in Gibberella zeae. Appl Environ Microbiol. 2006;72:1645–1652. doi: 10.1128/AEM.72.2.1645-1652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- 12.Lee J, Leslie JF, Bowden RL. Expression and function of sex pheromones and receptors in the homothallic ascomycete Gibberella zeae. Eukaryot Cell. 2008;7:1211–1221. doi: 10.1128/EC.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amatulli MT, Spadaro D, Gullino ML, Garibaldi A. Molecular identification of Fusarium spp. associated with bakanae disease of rice in Italy and assessment of their pathogenicity. Plant Pathol. 2010;59:839–844. [Google Scholar]
- 14.Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]