Abstract
In an ongoing survey of Korean indigenous fungi, three fungal strains belonging to the Sordariomycetes were isolated from soil samples. These strains were designated KNU16-001, KNU16-002, and KNU16-009, and identified as Ambrosiella grosmanniae, Acremonium sclerotigenum, and Trichocladium asperum, respectively, based on morphological characterization and phylogenetic analysis using internal transcribed spacer region sequences of ribosomal DNA. This is the first report of these species in Korea.
Keywords: Acremonium, Ambrosiella, Soil fungi, Sordariomycetes, Trichocladium
Fungi are one of the most important groups of soil microbes that play diverse and often critical roles in the ecosystem [1,2]. Fungi perform a range of important ecological functions associated with the cycling of all major elements (e.g., C, N, P, S), and this cycling affects the structure and functions of soil ecosystems [1,2,3]. In Korea, the composition of soil fungal communities at the phylum level (Ascomycota and Basidiomycota) is probably similar to that reported worldwide, but variations at the species level reflect local environmental conditions, plant and animal communities, and land use. Detailed knowledge of the structure of fungal communities has important implications in various fields such as agriculture, environment, and human health [4,5,6].
It is well known that the Sordariomycetes is the second largest class of the phylum Ascomycota [7]. Based on literature until end of 2015, the Sordariomycetes included 6 subclasses, 32 orders, 105 families, 1,331 genera, and over 10,000 species [8]. The Sordariomycetes have a cosmopolitan distribution and include mostly terrestrial taxa, although several members can be found in aquatic habitats [7,9]. Members of this class of fungi are known as pathogens and endophytes of plants, disease-causing agents in arthropods and mammals, as well as mycoparasites and saprobes involved in decomposition and nutrient cycling. Some species of the Sordariomycetes are economically important biocontrol agents [10], producers of bioactive compounds with medicinal applications [11], and extremely useful organisms for biotechnological industries [12].
During screening for unrecorded fungal species in soil samples collected from various fields around Korea, we isolated few dozen morphologically distinct strains. Among them, three strains belonging to the Sordariomycetes, namely, KNU16-001, KNU16-002, and KNU16-009, were identified as Ambrosiella grosmanniae, Acremonium sclerotigenum, and Trichocladium asperum, respectively. In this report, we describe the isolation and identification of these strains and discuss potential directions for further study. To the best of our knowledge, these fungi have not been previously reported in Korea.
MATERIALS AND METHODS
Soil sampling and isolation of fungi
Soil samples were collected in 2016 from fields at various locations in Daegu (35°53′45.23″ N, 128°35′10.94″ E), Nonsan (36°21′43.38″ N, 127°22′02.61″ E), and Jeonju (35°79′32.46″ N, 127°10′86.82″ E) in Korea. The samples were taken from a depth of 10–15 cm, air-dried, and stored in plastic bags at 4℃ until use. Fungi were isolated using a conventional dilution plating technique. For this purpose, 1 g of each soil sample was suspended in 10 mL of sterile distilled water, and the prepared suspension was vortexed, serially diluted, and spread onto potato dextrose agar (PDA; Difco Laboratories, Detroit, MI, USA) plates. The plates were incubated for 3–7 days at 25℃, until fungal colony growth was observed. Single colonies on these plates were purified by transferring them onto new plates, followed by incubation on PDA at 25℃. The pure cultures were maintained on PDA slants at 4℃ for future use.
Morphological characterization
Morphological characteristics of isolates KNU16-001, KNU16-002, and KNU16-009 were observed on PDA (MB Cell, Los Angeles, CA, USA) agar. The strains were inoculated on 9-cm petri dishes and incubated at 25℃ in the dark for 14 days. After incubation, colony characteristics such as color, shape, and size were recorded. Conventional slide culture technique was applied to observe the conidia of the isolated fungi [13]. A PDA block, 5 mm × 5 mm, was cut from a clean PDA plate and placed inside the lid. The sides of the block were inoculated with the culture, and a sterilized cover glass was placed over the block. Then, the lid was placed back on the PDA plate, and the plate was incubated at 25℃ in the dark for 20 days. When the fungus had grown onto the cover glass, it was removed from the PDA block, placed on an objective slide, and fixed with 85% lactic acid. The morphology of the isolate was examined under an Olympus BX50 light microscope (Olympus, Tokyo, Japan).
DNA extraction, PCR, sequencing, and phylogenetic analysis
Total genomic DNA was extracted from isolates KNU16-001, KNU16-002, and KNU16-009 by using the HiGene Genomic DNA Prep Kit (BIOFACT, Daejeon, Korea), per manufacturer's instructions. The internal transcribed spacer (ITS) regions, including the 5.8S rRNA region, were amplified with primers ITS1 (5′-TCCGTAGGTGAACCTGCG-3′) and ITS4 (5′-CCTCCGCTTATTGATATGC-3′) [14] under the following conditions: 95℃ for 2 min, followed by 35 cycles consisting of 95℃ for 30 sec, 55℃ for 45 sec, and 72℃ for 1 min, and one final step of extension at 72℃ for 7 min. The amplified PCR products were sequenced using an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The sequences were compared with reference ITS sequences from the GenBank database at the National Center for Biotechnology Information (NCBI), using the basic local alignment search tool (BLAST). Evolutionary distance matrices based on the neighbor-joining algorithm were calculated using Kimura's two-parameter model [15]. Tree topology was inferred by the neighbor-joining method in the program MEGA7 [16], with bootstrap values based on 1,000 replications.
RESULTS AND DISCUSSION
In a survey of the diversity of microfungi in soils collected from various regions in Korea, several fungal strains were isolated and identified based on phylogenetic analysis and morphological characterization of the isolates. Currently, sequence-based identification is considered the new “gold standard” for fungal species delimitation and identification, and the nuclear ribosomal DNA operon (nrDNA) has been used extensively for this purpose [18]. ITS1 was found between the 18S rRNA and the 5.8S rRNA genes, and ITS2 between the 5.8S rRNA and 28S rRNA genes. The two spacers, ITS1 and ITS2, and the 5.8S rRNA gene are usually referred to as the ITS region, and this region is the most common target for species-level identification [19]. Three isolates, KNU16-001, KNU16-002, and KNU16-009, were subjected to molecular identification based on PCR amplification of ITS regions and their molecular identifications were confirmed based on morphological and culture characteristics of the isolates. The nucleotide sequences were deposited in GenBank, with accession numbers LC228051, LC228050, and LC228052 for KNU16-001, KNU16-002, and KNU16-009, respectively.
Ambrosiella grosmanniae
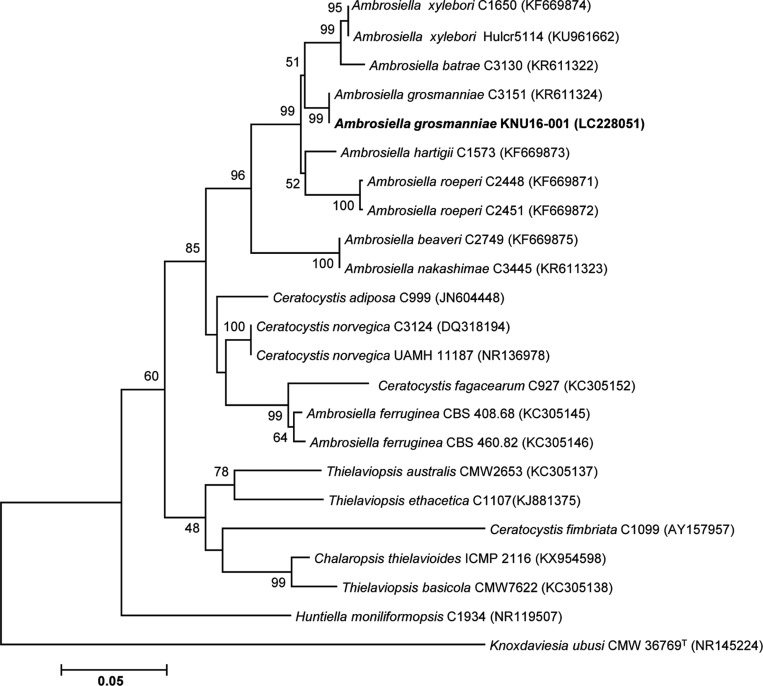
The first isolated strain, designed KNU16-001, was assigned to the genus Ambrosiella. A 585-bp ITS region sequence was obtained for the isolate. BLAST searches against the GenBank database showed that the ITS sequence of KNU16-001 had 100% identity with that of Ambrosiella grosmanniae strain C3151 (KR611324). In the phylogenetic tree based on the neighbor-joining algorithm (Fig. 1), strain KNU16-001 occupied a position within the genus Ambrosiella and clustered together with A. grosmanniae, indicating their closest relationship at the species level.
Fig. 1. Neighbor-joining phylogenetic tree, based on internal transcribed spacer region sequences, shows the phylogenetic position of Ambrosiella grosmanniae KNU16-001 among members of the genus Ambrosiella and other representatives of the Sordariomycetes. The strain isolated in this study is shown in boldface. Bootstrap values (based on 1,000 replications) greater than 50% are shown at the branch points. Accession numbers are shown in parentheses. Knoxdaviesia ubusi CMW 36769T was used as an outgroup. Bar, 0.05 substitutions per nucleotide position.
The genus Ambrosiella was first invalidly described by Brader [20] as A. xylebori, but no type was designated. Later, Von Arx and Hennebert [21] illustrated and designated a type for the genus and species based on Brader's isolate. The genus Ambrosiella accommodates species belonging to the Ceratocystidaceae within the class Sordariomycetes those are morphologically characterized as having conidiophores single to aggregated in sporodochia, hyaline, unbranched or sparingly branched, one-celled to septate, and producing terminal aleurioconidia or chains of conidia from phialides [22].
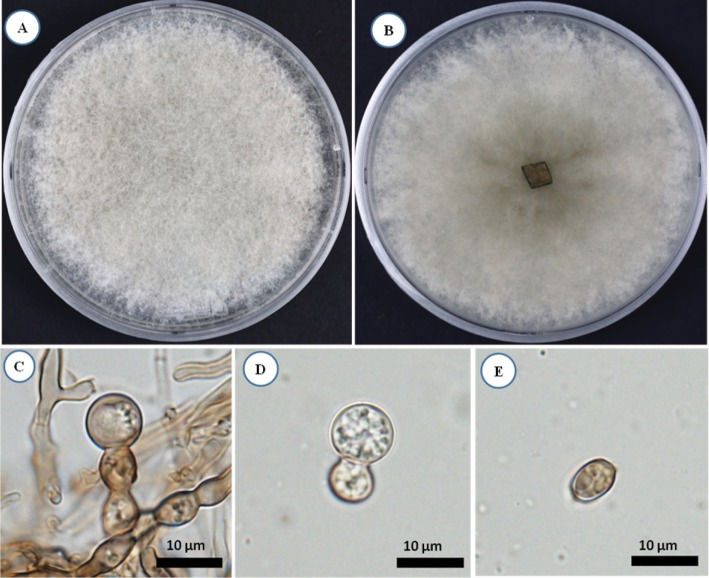
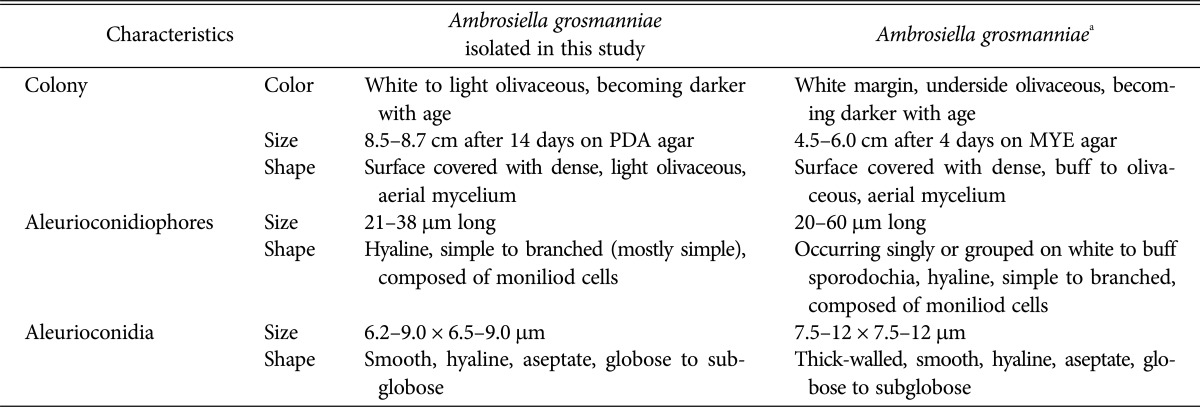
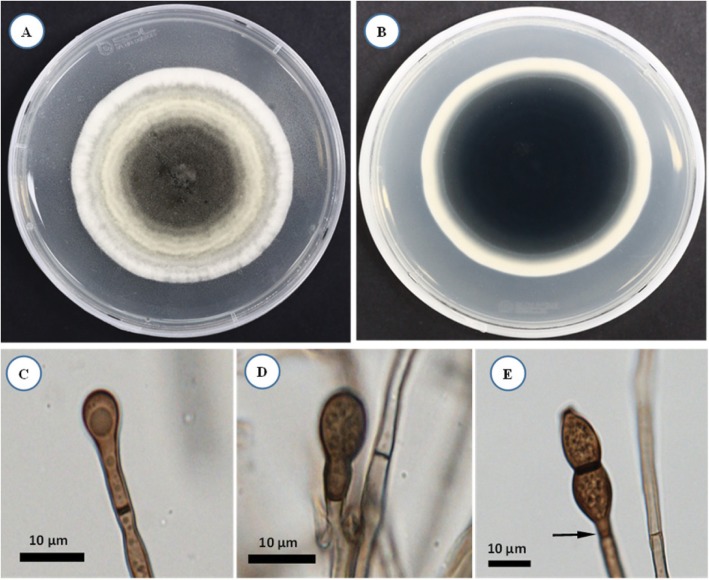
After 14 days of incubation on PDA agar, KNU16-001 colony was 8.5–8.7 cm in diameter (Fig. 2A and 2B). The isolate produced white to olivaceous aerial mycelium and light olivaceous pigment at the bottom of the medium; the color became darker with age. Aleurioconidiophores occurred singly or grouped, composed of moniliod cells (chain of cells), were hyaline and 21–38 µm in size (Fig. 2C). Aleurioconidia were smooth, hyaline, aseptate, globose to subglobose, and 6.2–9.0 × 6.5–9.0 µm in size (Fig. 2D and 2E). As shown in Table 1, these morphological characteristics of isolate KNU16-001 were completely consistent with those previously reported for Ambrosiella grosmanniae [17], thus strongly supporting the results of phylogenetic analysis of KNU16-001 based on the ITS sequence. Besides that, KNU16-001 can be distinguished morphologically from phylogenetically closely related A. xylebori by its light olivaceous pigmentation and by absence of a straight, hyphoid aleurioconidiophore with a single attached aleurioconidium that is unique feature of A. xylebori. In contrast to KNU16-001, other closely related species A. batrae was characterized by the presence of both aleurioconidia borne in chains and terminal aleurioconidia that tear away with conidiophore cells attached [17]. This is the first report of Ambrosiella grosmanniae isolated from soil in Korea.
Fig. 2. Morphology of Ambrosiella grosmanniae KNU16-001 observed using a light microscope. A, Colony in front; B, Colony in reverse; C, Microscopic image of aleurioconidiophores; D, E, Microscopic images of aleurioconidia (scale bars: C–E = 10 µm).
Table 1. Morphological characteristics of Ambrosiella grosmanniae isolated in this study.
PDA, potato dextrose agar; MYE, malt yeast extract.
aSource of description [17].
Acremonium sclerotigenum
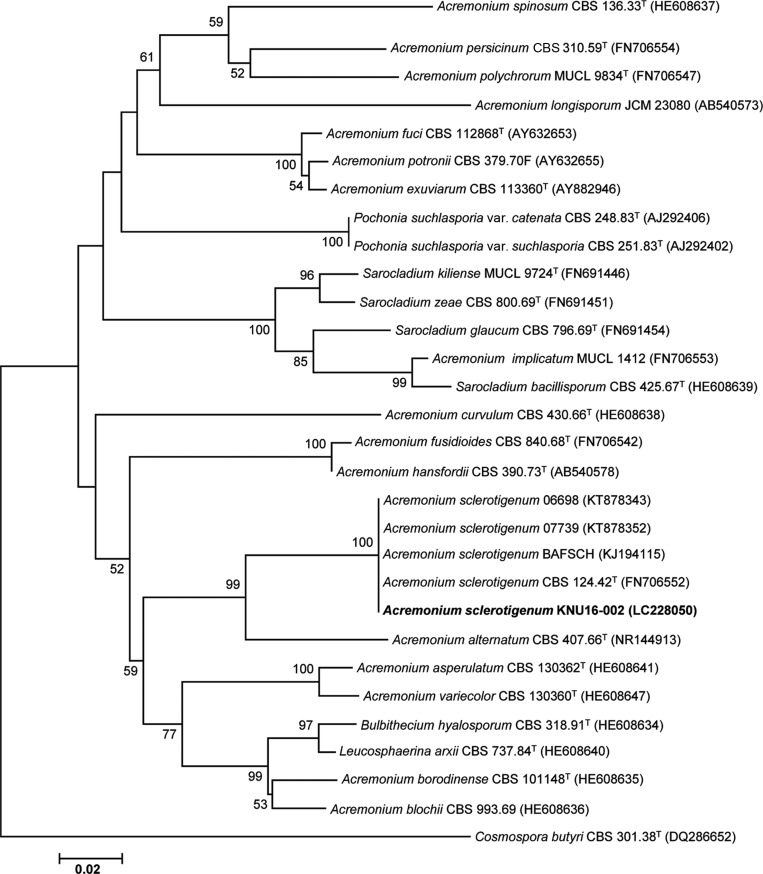
The second isolated strain, designated KNU16-002, was assigned to the genus Acremonium. An ITS region sequence of 585 bp was obtained for the isolate. BLAST searches against the GenBank database showed that the ITS sequence of KNU16-002 had 100% identity with several Acremonium sclerotigenum strains such as A. sclerotigenum CBS 124.42T (FN706552), A. sclerotigenum BAFSCH (KJ194115), A. sclerotigenum 06698 (KT878343), and A. sclerotigenum 07739 (KT878352). In the neighbor-joining phylogenetic tree, strain KNU16-002 occupied a position within the genus Acremonium and clustered together with A. sclerotigenum strains, indicating their closest relationship at the species level (Fig. 3).
Fig. 3. Neighbor-joining phylogenetic tree, based on internal transcribed spacer region sequences, shows the phylogenetic position of Acremonium sclerotigenum KNU16-002 among members of the genus Acremonium and other representatives of the Sordariomycetes. The strain isolated in this study is shown in boldface. Bootstrap values (based on 1,000 replications) greater than 50% are shown at the branch points. Accession numbers are shown in parentheses. Cosmospora butyri CBS 301.38T was used as an outgroup. Bar, 0.02 substitutions per nucleotide position.
The genus Acremonium was erected by Link, with A. alternatum Link as the type species, and currently contains more than 100 species, mainly belonging to the Hypocreales within the class Sordariomycetes [23]. Acremonium spp. are common soil and plant saprophytes [24], but some species are known as opportunistic human and animal pathogens [25] or as plant pathogens [26]. The species of this genus are morphologically characterized by colonies with slow or moderate growth; thin hyphae; simple or poorly basitonously branched, orthotropic phialides, usually gradually tapering toward the tip; and conidia that are usually small, unicellular, and aggregated in slimy heads, chains, or both [24,27].
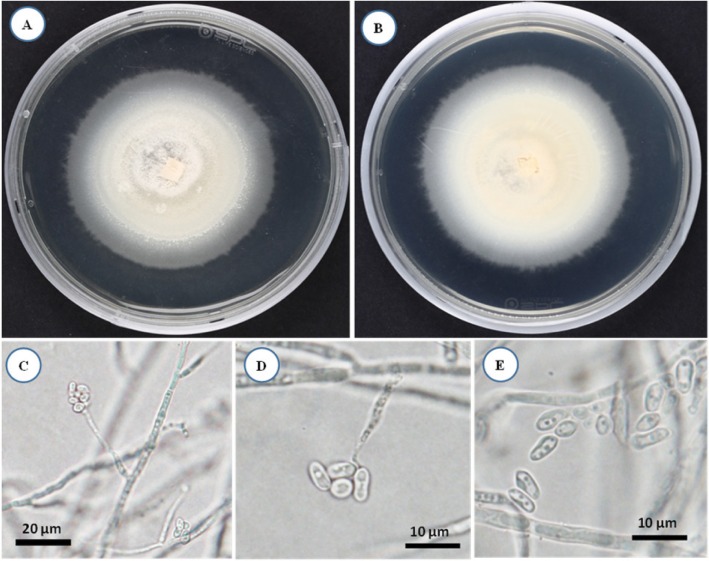
The colony formed by isolate KNU16-002 was whitish and floccose in appearance, and reached a diameter of 6.5 cm after 14 days incubation on PDA agar at 25℃ (Fig. 4A and 4B). Phialides were simple without branching, formed from substratum and fasciculated aerial hyphae, and were chromophilic near the base (Fig. 4C and 4D). Conidia were aggregated in heads, cylindrical, or tapering towards the tips and slightly fusiform, smooth-walled, one-celled, and hyaline (Fig. 4E). Morphological characteristics of isolate KNU16-009 were in good agreement with the abovementioned description of the genus Acremonium and most closely matched the characteristics of A. sclerotigenum (Table 2) [27,28]. Similar to other strains of the species [28], A. sclerotigenum KNU16-009 distinguished morphologically from other Acremonium species by its conidial arrangement with slimy heads. Thus, the results of phylogenetic analysis of KNU16-009 were well supported by its morphological characteristics. Although this is a common fungal species with a cosmopolitan distribution, this is the first report of A. sclerotigenum in Korea.
Fig. 4. Morphology of Acremonium sclerotigenum KNU16-002 observed using a light microscope. A, Colony in front; B, Colony in reverse; C, D, Microscopic images of phialides; E, Microscopic image of solitary conidia (scale bars: C = 20 µm, D, E = 10 µm).
Table 2. Morphological characteristics of Acremonium sclerotigenum isolated in this study.
Trichocladium asperum
On the basis of morphological characterization and phylogenetic analysis third isolated strain KNU16-009 was assigned to Trichocladium Harz. Isolate KNU16-009 was subjected to molecular identification based on sequence analysis of the ITS regions. A search of the GenBank database via BLAST revealed that the ITS region sequence of KNU16-009 exhibited high similarity (99–100%) with various T. asperum strains. This relationship was also evident from the phylogenetic tree constructed using ITS region sequences. The isolate KNU16-009 clustered together with T. asperum strains, thus confirming their closest relationship at the species level (Fig. 5).
Fig. 5. Neighbor-joining phylogenetic tree, based on internal transcribed spacer region sequences, shows the phylogenetic position of Trichocladium asperum KNU16-009 among members of the genus Trichocladium and other representatives of the Sordariomycetes. The strain isolated in this study is shown in boldface. Bootstrap values (based on 1,000 replications) greater than 50% are shown at the branch points. Accession numbers are shown in parentheses. Zygosaccharomyces bailii CBS 1097 was used as an outgroup. Bar, 0.05 substitutions per nucleotide position.
The genus Trichocladium was introduced by Harz, with T. asperum Harz as its type species [29]. Currently, MycoBank lists 41 described species for the genus. Members of the genus Trichocladium reproduce asexually and are dematiaceous (dark pigmented) hyphomycetes within the family Chaetomiaceae. Molecular studies supported the affinity of this genus with the order Sordariales within the class Sordariomycetes [30]. Following the principal of “one fungus, one name” and considering that T. asperum occurs in the same clade as Chaetomium [30], Reblova et al. [31] proposed that the name Chaetomium Kunze 1817 be used rather than Trichocladium Harz 1871. However, the authors emphasized that “eventually all generic names proposed for protection will be evaluated by the Nomenclature Committee for Fungi (NCF), and formally accepted or not at the Nomenclature Section meeting of the next International Botanical Congress in 2017.” This recommendation had not been formally accepted yet at the time of writing; therefore, we used the generic name Trichocladium in this paper.
The genus Trichocladium is characterized by effuse colonies, mycelium partly superficial partly immersed; stroma absent; conidiophores micronematous, semi-, macronematous, mononematous, straight or flexuous, pale brown; conidia solitary, dry, acrogenous or acropleurogenous, simple, clavate, cylindrical, rounded at the apex, obovoid or pyriform, smooth or verrucose, and thick walled with one to many dark brown transverse septa [29,32].
The mycelium of isolate KNU16-009 was approximately 6.5 cm in diameter after 14 days incubation on PDA at 25℃ (Fig. 6A and 6B). Aleurioconidia were oval, straight, dark brown, mostly two-celled, and approximately 26.5 × 9.5 µm in size (Fig. 6C-6E). Morphological characteristics of isolate KNU16-009 were in good agreement with the above-mentioned description of the genus Trichocladium and most closely matched the characteristics of T. asperum (Table 3) [29,32]. Moreover, rhexolytic secession is a specific characteristic of T. asperum conidia. Sometimes two closely located septa can develop, and secession can occur at this “separating cell” [30]. In Fig. 6E, the arrow indicates the location of the double septa of the eventual site of rhexolytic secession that was also found in the isolate. Morphological characteristics obtained here support the results of phylogenetic analysis of KNU16-009 as belonging to T. asperum. The isolate shared unique morphological features with T. asperum summarized by Goh and Hyde [32] that differentiated it from other Trichocladium species. T. asperum has not been previously reported from Korea, and this is the first detailed report of its isolation and identification in Korea.
Fig. 6. Morphology of Trichocladium asperum KNU16-009 observed using a light microscope. A, Colony in front; B, Colony in reverse; C–E, Microscopic images of aleurioconidia (scale bars: C–E = 10 µm).
Table 3. Morphological characteristics of Trichocladium asperum isolated in this study.
Among members of the genera Acremonium and Trichocladium there are producers of bioactive compounds with antifungal activity, such as pyrrocidines A and B [33], and antibacterial activity, such as furanone [34] and trichocladinols A–C, which showed modest cytotoxic effects against the human tumor cell lines HeLa and MCF-7 [35]. In contrast, data regarding similar activities of Ambrosiella spp. are practically nonexistent. Thus, further investigation of Ambrosiella grosmanniae KNU16-001, Acremonium sclerotigenum KNU16-002, and T. asperum KNU16-009 would be worthwhile.
ACKNOWLEDGEMENTS
This work was supported by a grant (No. NIBR 2015-01205) from the National Institute of Biological Resources, funded by the Ministry of Environment (MOE) of the Republic of Korea; and by the Brain Pool Program of 2016 (Grant No. 162S-4-3-1727) through the Korean Federation of Science and Technology Societies (KOFST), funded by the Ministry of Science, ICT and Future Planning, Republic of Korea. This work is also partially supported by Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ011631042017) Rural Development Administration, Republic of Korea.
References
- 1.Taylor DL, Sinsabaugh RL. Soil microbiology, ecology and biochemistry. Waltham (MA): Academic Press; 2015. The soil fungi: occurrence; pp. 77–109. [Google Scholar]
- 2.Anderson IC, Cairney JW. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol. 2004;6:769–779. doi: 10.1111/j.1462-2920.2004.00675.x. [DOI] [PubMed] [Google Scholar]
- 3.Gadd GM. Mycotransformation of organic and inorganic substrates. Mycologist. 2004;18:60–70. [Google Scholar]
- 4.Wall DH, Nielsen UN, Six J. Soil biodiversity and human health. Nature. 2015;528:69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 5.Verbruggen E, van der Heijden MG, Rillig MC, Kiers ET. Mycorrhizal fungal establishment in agricultural soils: factors determining inoculation success. New Phytol. 2013;197:1104–1109. doi: 10.1111/j.1469-8137.2012.04348.x. [DOI] [PubMed] [Google Scholar]
- 6.Gauthier GM, Keller NP. Crossover fungal pathogens: the biology and pathogenesis of fungi capable of crossing kingdoms to infect plants and humans. Fungal Genet Biol. 2013;61:146–157. doi: 10.1016/j.fgb.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia. 2006;98:1076–1087. doi: 10.3852/mycologia.98.6.1076. [DOI] [PubMed] [Google Scholar]
- 8.Maharachchikumbura SN, Hyde KD, Jones EB, McKenzie EH, Bhat JD, Dayarathne MC, Huang SK, Norphanphoun C, Senanayake IC, Perera RH, et al. Families of Sordariomycetes. Fungal Divers. 2016;79:1–317. [Google Scholar]
- 9.Jones EB, Suetrong S, Sakayaroj J, Bahkali AH, Abdel-Wahab MA, Boekhout T, Pang KL. Classification of marine Ascomycota, Basidiomycota, Blastocladiomycota and Chytridiomycota. Fungal Divers. 2015;73:1–72. [Google Scholar]
- 10.Kaewchai S, Soytong K, Hyde KD. Mycofungicides and fungal biofertilizers. Fungal Divers. 2009;38:25–50. [Google Scholar]
- 11.Martinez-Klimova E, Rodriguez-Pena K, Sanchez S. Endophytes as sources of antibiotics. Biochem Pharmacol. 2017;134:1–17. doi: 10.1016/j.bcp.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Adrio JL, Demain AL. Fungal biotechnology. Int Microbiol. 2003;6:191–199. doi: 10.1007/s10123-003-0133-0. [DOI] [PubMed] [Google Scholar]
- 13.de Meyer EM, de Beer ZW, Summerbell RC, Moharram AM, de Hoog GS, Vismer HF, Wingfield MJ. Taxonomy and phylogeny of new wood- and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia. 2008;100:647–661. doi: 10.3852/07-157r. [DOI] [PubMed] [Google Scholar]
- 14.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego (CA): Academic Press; 1990. pp. 315–322. [Google Scholar]
- 15.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayers CG, McNew DL, Harrington TC, Roeper RA, Fraedrich SW, Biedermann PH, Castrillo LA, Reed SE. Three genera in the Ceratocystidaceae are the respective symbionts of three independent lineages of ambrosia beetles with large, complex mycangia. Fungal Biol. 2015;119:1075–1092. doi: 10.1016/j.funbio.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 18.Summerbell RC, Levésque CA, Seifert KA, Bovers M, Fell JW, Diaz MR, Boekhout T, de Hoog GS, Stalpers J, Crous PW. Microcoding: the second step in DNA barcoding. Philos Trans R Soc Lond B Biol Sci. 2005;360:1897–1903. doi: 10.1098/rstb.2005.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brader L. Étude de la relation entre le scolyte des rameaux du caféir, Xyleborus compactus Eichh. (X. morstatti Hag.), et sa plante-hote. Mededelingen (Landbouwhogeschool Wageningen) 1964;64:1–109. [Google Scholar]
- 21.Von Arx JA, Hennebert GL. Deux champignons ambrosia. Mycopathol Mycol Appl. 1965;25:309–315. [Google Scholar]
- 22.Harrington TC, Aghayeva DN, Fraedrich SW. New combinations in Raffaelea, Ambrosiella, and Hyalorhinocladiella, and four new species from the redbay ambrosia beetle, Xyleborus glabratus. Mycotaxon. 2010;111:337–361. [Google Scholar]
- 23.Summerbell RC, Gueidan C, Schroers HJ, de Hoog GS, Starink M, Rosete YA, Guarro J, Scott JA. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud Mycol. 2011;68:139–162. doi: 10.3114/sim.2011.68.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. 2nd ed. Eching: IHW-Verlag; 2007. [Google Scholar]
- 25.Das S, Saha R, Dar SA, Ramachandran VG. Acremonium species: a review of the etiological agents of emerging hyalohyphomycosis. Mycopathologia. 2010;170:361–375. doi: 10.1007/s11046-010-9334-1. [DOI] [PubMed] [Google Scholar]
- 26.Oh SY, Nam KW, Yoon DH. Identification of Acremonium acutatum and Trichothecium roseum isolated from grape with white stain symptom in Korea. Mycobiology. 2014;42:269–273. doi: 10.5941/MYCO.2014.42.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo Piccolo S, Alfonzo A, Giambra S, Conigliaro G, Lopez-Llorca LV, Burruano S. Identification of Acremonium isolates from grapevines and evaluation of their antagonism towards Plasmopara viticola. Ann Microbiol. 2015;65:2393–2403. [Google Scholar]
- 28.Perdomo H, Sutton DA, García D, Fothergill AW, Cano J, Gené J, Summerbell RC, Rinaldi MG, Guarro J. Spectrum of clinically relevant Acremonium species in the United States. J Clin Microbiol. 2011;49:243–256. doi: 10.1128/JCM.00793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hughes SJ. Trichocladium Harz. Trans Br Mycol Soc. 1952;35:152–157. [Google Scholar]
- 30.Hambleton S, Nickerson NL, Seifert KA. Leohumicola, a new genus of heat-resistant hyphomycetes. Stud Mycol. 2005;53:29–52. [Google Scholar]
- 31.Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, et al. Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales) IMA Fungus. 2016;7:131–153. doi: 10.5598/imafungus.2016.07.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goh TK, Hyde KD. A synopsis of Trichocladium species, based on the literature. Fungal Divers. 1999;2:101–118. [Google Scholar]
- 33.Wicklow DT, Roth S, Deyrup ST, Gloer JB. A protective endophyte of maize: Acremonium zeae antibiotics inhibitory to Aspergillus flavus and Fusarium verticillioides. Mycol Res. 2005;109(Pt 5):610–618. [PubMed] [Google Scholar]
- 34.Hijikawa Y, Matsuzaki M, Suzuki S, Inaoka DK, Tatsumi R, Kido Y, Kita K. Re-identification of the ascofuranone-producing fungus Ascochyta viciae as Acremonium sclerotigenum. J Antibiot (Tokyo) 2017;70:304–307. doi: 10.1038/ja.2016.132. [DOI] [PubMed] [Google Scholar]
- 35.Guo H, Sun B, Gao H, Niu S, Liu X, Yao X, Che Y. Trichocladinols A-C, cytotoxic metabolites from a Cordyceps-colonizing ascomycete Trichocladium opacum. Eur J Org Chem. 2009;32:5525–5530. [Google Scholar]