Abstract
Fungi of the Metarhizium genus are a very versatile model for understanding pathogenicity in insects and their symbiotic relationship with plants. To establish a co-transformation system for the transformation of multiple M. robertsii genes using Agrobacterium tumefaciens, we evaluated whether the antibiotic nourseothricin has the same marker selection efficiency as phosphinothricin using separate vectors. Subsequently, in the two vectors containing the nourseothricin and phosphinothricin resistance cassettes were inserted eGFP and mCherry expression cassettes, respectively. These new vectors were then introduced independently into A. tumefaciens and used to transform M. robertsii either in independent events or in one single co-transformation event using an equimolar mixture of A. tumefaciens cultures. The number of transformants obtained by co-transformation was similar to that obtained by the individual transformation events. This method provides an additional strategy for the simultaneous insertion of multiple genes into M. robertsii.
Keywords: Agrobacterium, Co-transformation, Metarhizium, Nourseothricin
Fungi of the Metarhizium genus are a versatile study model based on their biotechnological applications, which have been implemented for over 100 years [1]. Aside from insects, these fungi have been demonstrated to be safely compatible with other organisms (plants, animals, and humans) due to their entomopathogenic status. This specificity has allowed organisms from the Metarhizium genus to be approved by different environmental agencies, including the United States Environmental Protection Agency [2] and the Canada Pest Management Regulatory Agency [3], for marketing and extensive field use.
Metarhizium fungi are the most widely studied entomopathogens at both the molecular and biochemical level [1]. These organisms are globally distributed, and they colonize a wide range of environments, including forests, savannas, marshes, coastal areas and deserts [1]. M. robertsii has become an excellent model organism for exploring several aspects of ecology and evolution, such as host specificity, changes, and specialization mechanisms [1,4,5]. In addition, Metarhizium-focused research is currently at the forefront of efforts to develop alternatives to chemical insecticides, such as biological control agents, for use in commercial agricultural products and disease vector control programs, and many of these agents are now on the market or in development [1,6,7].
Recently, the ecological impact of members of the Metarhizium genus and their potential as biological control agents has been reinforced by reports that Metarhizium fungi perform more environmental functions than previously believed. For example, Metarhizium fungi can interact with plants by colonizing their roots in a mycorrhizal and endophytic manner, establishing a symbiotic relationship with the plants, which stimulates their root growth [8], activates their defense system and increases their resistance to abiotic factors, such as salinity [9]. Metarhizium fungi also transfer nutrients, such as nitrogen, that are obtained from insect fungal parasites from the soil to the plant roots [10]. In turn, plants transfer carbon compounds to the fungi [11], indicating that Metarhizium fungi and plants have a mutualistic symbiotic relationship [11].
The numerous, diverse interactions between Metarhizium fungi and the environment has supported genomic sequencing of the species M. robertsii, M. acridum, M. rileyi, M. majus, M. brunneum, M. guizhouense, M. anisopliae, and M. album [12,13,14,15,16]. Molecular tools have been adapted to further elucidate the characteristics and differences of each of these Metarhizium species. For example, the use of exogenous selection markers have been successfully identified, such as benomyl [17], phosphinothricin [18], geneticin [19], chlorimuron ethyl [20] and recently, nourseothricin [21]. Thus far, Agrobacterium-mediated transformation is the most efficient and successfully applied technique in Metarhizium for exogenous DNA insertion and gene inactivation [22]. In this work, we evaluate the efficiency of the antibiotic nourseothricin as a selection marker, finding that its performance is equal to that of the herbicide phosphinothricin, which is the most selection marker used in Metarhizium fungi. Both nourseothricin and phosphinothricin were used to perform co-transformation mediated by Agrobacterium tumefaciens, which has not been reported in fungi, showing high efficiency in M. robertsii by simultaneously inserting multiple genes in separate vectors in a single event.
MATERIALS AND METHODS
Fungal and bacterial strains
The M. robertsii strain ARSEF 2575 was obtained from the United States Department of Agriculture Collection of Entomopathogenic Fungal Cultures (Ithaca, NY, USA). Cultures were grown on potato dextrose agar at 27℃ for 10 days for conidia harvest. Escherichia coli XL-Blue competent cells were used for vector construction using standard molecular techniques [23]. A. tumefaciens AGL1 was used as the carrier for the individually constructed vectors for fungal transformation as previously described [24].
Binary vector construction
The pPK2-BAR vector was constructed by inserting a PtrpC-BAR XbaI/EcoRI fragment from pBARKS1-SpeI previously cut and self-ligating at the SpeI sites from pBARKS1 [25] into the XbaI/EcoRI sites in binary pPK2 vector [26]. The pPK2-BAR-mCherry vector was constructed by amplifying the TtrpC fragment with the primers Ttrpc-XbaI-Up (5′-TCTCTAGAAAAGAAGGATTACCTCTAAACA-3′) and Ttrpc-SpeI-BamHI-Down (5′-GAGGATCCAGCAGACTAGTCACTTAACGTTACTGAAATCA-3′) from the pBARGEM7-2 vector [25] and then inserting the amplified fragment into the SpeI/BamHI sites in pPK2-BAR. The PgpdA-mCherry fragment was subsequently released from the pGG464 vector [27] and cloned into the EcoRI/BamHI sites. The pPK2-NTC vector was constructed by amplifying the nourseothricin-resistance cassette with the primers PNAT-XbaI (5′-TATCTAGAGCGGAGAGACGGA-3′) and 5NAT-BglII (5′-ATAGATCTTTAGGGGCAGGGCATG-3′) using the pA-NTC-OSCAR vector as a template [28] and then inserting the amplified fragment into the pPK2-BAR binary vector using the XbaI/BglII sites. The pPK2-NTC-eGFP vector was constructed by amplifying the GFP cassette with the primers 5CST-EGFP (5′-ATGAATTCGGGTAGCAAACGGTGGTCAAAG-3′) and 3CST-EGFP (5′-ATAGATCTCTGTCTGGTCTTCTACACGAAGGAA-3′) from the pFBENGFP vector [24] and then inserting the amplified fragments into the EcoRI/BglII sites in the pPK2-NTC plasmid. The T-DNA vector sequences of pPK2-BAR, pPK2-NTC, pPK2-NTC-eGFP, and pPK2-BAR-mCherry were deposited into the NCBI GenBank database under the accession numbers MF169981, MF169982, MF169983, and MF169984, respectively.
Fungal transformation
The four vectors generated were introduced individually into A. tumefaciens strain AGL1 cells. For the Metarhizium agro-transformation, we followed the protocol previously described by Fang and collaborators [24] with some modifications. Briefly, 1 × 106 conidia suspended in 0.01% Triton X-100 were placed in a 1.5-mL Eppendorf tube and centrifuged for 5min at 4,000 ×g. The Triton X-100 solution was removed to preserve the conidia buttons, and the buttons were then mixed with 1mL of A. tumefaciens cells, induced (OD660 = 0.7–0.8) and vortexed for 30 sec. Next, 200 µL of the mixture was spread on black filter paper to continue the protocol described by Fang et al. [24]. Nourseothricin (Gold Biotechnology, St. Louis, MO, USA) and phosphinothricin (herbicide Finale; Bayer Crop Science AG, Leverkusen, Germany) were used at concentrations of 350 µg/mL and 250 µg/mL, respectively, for individual selection on M-100 agar medium [24].
For co-transformation of M. robertsii mediated by A. tumefaciens, separate A. tumefaciens cultures containing the pPK2-NTC-eGFP and pPK2-BAR-mCherry vectors were induced with acetosyringone as described [24]. After the cells were induced (OD660 = 0.7–0.8), they were mixed equimolarly in a 1 : 1 ratio with M. robertsii conidia to continue the protocol described above. For double selection of the transformants, 350 µg/mL nourseothricin and 250 µg/mL phosphinothricin were simultaneously added on M-100 agar medium.
Transformant verification
The obtained transformants were verified by three passes on their respective plates containing the antibiotics. DNA extraction was then performed by grinding frozen mycelia and adding phenolchloroform [23]. Next, 100 ng of the extracted DNA was used as a template to verify the transformants using PCR to detect the presence of the corresponding resistance and fluorescence cassettes. The nourseothricin-resistance cassette was amplified using the oligonucleotides PNAT-XbaI and 5NAT-BglII, the eGFP cassette was amplified using the oligonucleotides 5CST-EGFP and 3CST-EGFP, the phosphinothricin-resistance cassette was amplified using the oligonucleotides PtrpC-XbaI (5′-ATTCTAGAATCGACAGAAGATGATATTGAAGGA-3′) and Bar-down (5′-TCAGATCTCGGTGACGGGC-3′), and the mCherry cassette was amplified using the oligonucleotides Pgpdashort (5′-GCGGAGAGACGGACGGAC-3′) and cherry-down (5′-TTACTTGTACAGCTCGTCCATGCC-3′). Protein expression of the fluorescent reporter protein was confirmed by visual inspection using a Nikon OPTIPHOT-2 microscope (Nikon, Tokyo, Japan) equipped with epifluorescent illumination.
RESULTS AND DISCUSSION
Co-transformation is defined as the simultaneous introduction of multiples genes in a cell, and the genes can be present on the same plasmid or on separate plasmids [29]. The main advantage of co-transformation for the transfer of multiples genes is that a single transformation event can result in the integration of multiples transgenes as opposed to sequential transformation, which requires multiple time-consuming transformation events [29]. Co-transformation can be adapted to a variety of transformation methods, such as electroporated, biolistic, protoplasts and Agrobacterium-mediated transformation [29,30,31]. Several different strategies have been employed in Agrobacterium-mediated co-transformation for the independent delivery of T-DNA in plants [31], but Agrobacterium-mediated co-transformation in fungi has not yet been reported. T-DNA can be held in mixed bacterial strains in which two T-DNA molecules are contained in separate binary vectors in different Agrobacterium cells or in a single bacterial strain, whereby two T-DNA molecules could either be held in distinct regions on the same binary plasmid or on separate binary plasmids [31].
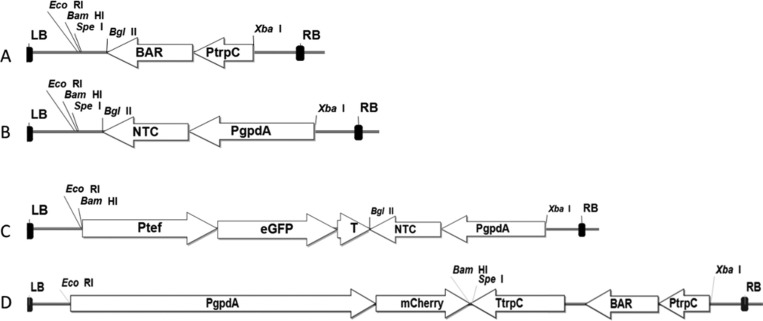
In this study, we used a mixed approach involving Agrobacterium containing two different T-DNA molecules on separate binary vectors to evaluate co-transformation mediated by Agrobacterium in M. robertsii. To identify a selection marker with the same efficiency as phosphinothricin for use in co-transformation, we evaluated the efficiency of the antibiotic nourseothricin, which has been used to transform M. robertsii, but its efficiency in this fungus has not been analyzed [21]. This antibiotic inhibits protein synthesis by inducing miscoding [32], and resistance is conferred by the nourseothricin acetyltransferase 1 (nat1) gene from Streptomyces noursei [32]. We determined that the concentration of nourseothricin required to inhibit M. robertsii growth on agar medium M-100 at 28℃ is 350 µg/mL. The plates were incubated for 20 days without observing spontaneous mutant resistance. Once the efficiency of nourseothricin in M. robertsii on M-100 agar medium was established, the binary vectors pPK2-NTC (Fig. 1A), pPK2-NTC-eGFP (Fig. 1B), pPK2-BAR (Fig. 1C), and pPK2-BAR-mCherry (Fig. 1D) were constructed as described in the materials and methods section.
Fig. 1. T-DNA vector construction. A, T-DNA in the pPK2-NTC vector for nourseothricin selection; B, T-DNA in the pPK2-BAR vector for phosphinothricin selection; C, T-DNA in the pPK2-NTC-eGFP vector for nourseothricin resistance using green fluorescence for visual selection; D, T-DNA in the pPK2-BAR-mCherry vector with phosphinothricin resistance using cherry fluorescence for visual selection. PgpdA, promoter of glyceraldehyde-3-phosphate dehydrogenase gene from Aspergillus nidulans; NTC, nourseothricin resistance nat1 gene from Filobasidiella neoformans; Ptef, promoter of translation elongation factor gene from Aureobasidium pullulans; eGFP, enhanced green fluorescent protein gene; T, termination region of the glucoamylase gene from Aspergillus awamori; PtrpC, promoter of tryptophan C gene from Aspergillus nidulans; BAR, phosphinothricin resistance gene from Streptomyces hygroscopicus; TtrpC, terminator of tryptophan C gene from Aspergillus nidulans; mCherry, cherry fluorescent protein gene; RB and LB are the right and left border of T-DNA, respectively.
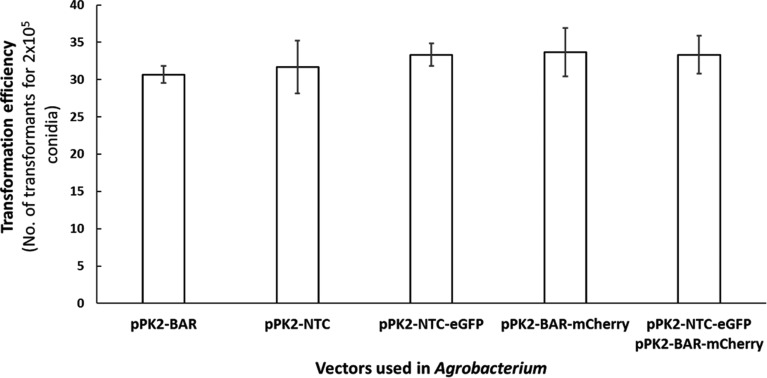
There was no difference in the number of transformants obtained using either nourseothricin or phosphinothricin as the selection agent in the Agrobacterium-mediated transformation experiments (Fig. 2), demonstrating that nourseothricin as well as phosphinothricin can be used as a dominant selection marker in Metarhizium.
Fig. 2. The transformation and co-transformation efficiency in Metarhizium robertsii using the vectors generated in this work. Each transformation was performed three times, and the transformants were counted. The bars represent the standard error.
These results lead us to perform an Agrobacterium-mediated co-transformation using both nourseothricin and phosphinothricin simultaneously to avoid pleiotropic effects. We co-transformed Metarhizium using the vectors pPK2-NTC-eGFP and pPK2-BAR-mCherry, and the number of resulting transformants was similar to that resulting from the individual transformation experiments in which the individual vectors were used. The high transformation efficiency using Agrobacterium-mediated co-transformation in M. robertsii suggests a synergistic effect. This transformation efficiency is consistent with co-transformation in plants, as a larger number of genes involved in the co-transformation reportedly increases the efficiency of the transformants obtained when Agrobacterium is used [31]. Transgene integration was confirmed by PCR (Fig. 3) and fluorescence microscopy (Fig. 4).
Fig. 3. Transformant confirmation analysis by PCR. The amplicons of two transformants (A, B) obtained by Agrobacterium-mediated co-transformation with the vectors pPK2-NTC-eGFP and pPK2-BAR-mCherry are shown. Lane M, DNA molecular size marker; lane A1–B2, NTC cassette, 1,435 bp; lane A3–B4, BAR cassette, 938 bp; lane A5–B6, GFP cassette, 1,736 bp; lane A7–B8, mCherry cassette, 1,561 bp.
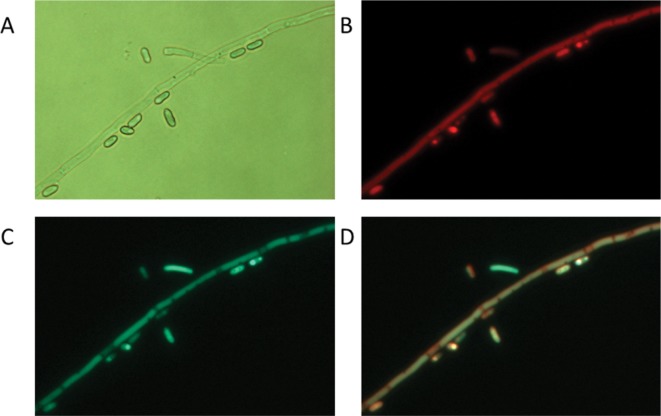
Fig. 4. Verification of Agrobacterium-mediated co-transformation using visual markers. The strain co-transformed with pPK2-NTC-eGFP and pPK2-BAR-mCherry vectors shows fluorescence in hyphae and conidia. A, Bright field image; B, Cherry fluorescence image; C, Green fluorescence image; D, Merge of the cherry (B) and green (C) images (A–D, ×400).
In this study, for the first time in fungi, we reported the use of Agrobacterium-mediated co-transformation to integrate multiple genes into the M. robertsii genome using mixed Agrobacterium cells containing two different T-DNA molecules on separate binary vectors, thus improving the genetic strategies for studying this fungus. The advantages of this co-transformation method can facilitate the study of Metarhizium by reducing valuable time spent on constructing complex or long T-DNA molecules in binary vectors and sequential transformations.
ACKNOWLEDGEMENTS
We gratefully received financial support from the following institutions: Universidad de Guanajuato (CIFOREA 89/2016), and Secretaría de Educación Pública (PROMEP UGTOPTC-327).
References
- 1.Roberts DW, St Leger RJ. Metarhizium spp., cosmopolitan insect-pathogenic fungi: mycological aspects. Adv Appl Microbiol. 2004;54:1–70. doi: 10.1016/S0065-2164(04)54001-7. [DOI] [PubMed] [Google Scholar]
- 2.Fang W, Vega-Rodríguez J, Ghosh AK, Jacobs-Lorena M, Kang A, St Leger RJ. Development of transgenic fungi that kill human malaria parasites in mosquitoes. Science. 2011;331:1074–1077. doi: 10.1126/science.1199115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Health Canada's Consumer Product Safety. Proposed Registration Decision for Metarhizium anisopliae strain F52 [Internet] Hamilton (ON): Health Canada's Consumer Product Safety; 2011. [cited 2011 Oct 27]. Available from: http://www.hc-sc.gc.ca/cps-spc/pest/part/consultations/_prd2011-13/prd2011-13-eng.php#a1. [Google Scholar]
- 4.Wang S, Leclerque A, Pava-Ripoll M, Fang W, St Leger RJ. Comparative genomics using microarrays reveals divergence and loss of virulence-associated genes in host-specific strains of the insect pathogen Metarhizium anisopliae. Eukaryot Cell. 2009;8:888–898. doi: 10.1128/EC.00058-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Fang W, Wang C, St Leger RJ. Insertion of an esterase gene into a specific locust pathogen (Metarhizium acridum) enables it to infect caterpillars. PLoS Pathog. 2011;7:e1002097. doi: 10.1371/journal.ppat.1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lomer CJ, Bateman RP, Johnson DL, Langewald J, Thomas M. Biological control of locusts and grasshoppers. Annu Rev Entomol. 2001;46:667–702. doi: 10.1146/annurev.ento.46.1.667. [DOI] [PubMed] [Google Scholar]
- 7.St Leger RJ, Wang C. Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl Microbiol Biotechnol. 2010;85:901–907. doi: 10.1007/s00253-009-2306-z. [DOI] [PubMed] [Google Scholar]
- 8.Sasan RK, Bidochka MJ. The insect-pathogenic fungus Metarhizium robertsii (Clavicipitaceae) is also an endophyte that stimulates plant root development. Am J Bot. 2012;99:101–107. doi: 10.3732/ajb.1100136. [DOI] [PubMed] [Google Scholar]
- 9.Khan AL, Hamayun M, Khan SA, Kang SM, Shinwari ZK, Kamran M, Ur Rehman S, Kim JG, Lee IJ. Pure culture of Metarhizium anisopliae LHL07 reprograms soybean to higher growth and mitigates salt stress. World J Microbiol Biotechnol. 2012;28:1483–1494. doi: 10.1007/s11274-011-0950-9. [DOI] [PubMed] [Google Scholar]
- 10.Behie SW, Zelisko PM, Bidochka MJ. Endophytic insect-parasitic fungi translocate nitrogen directly from insects to plants. Science. 2012;336:1576–1577. doi: 10.1126/science.1222289. [DOI] [PubMed] [Google Scholar]
- 11.Behie SW, Moreira CC, Sementchoukova I, Barelli L, Zelisko PM, Bidochka MJ. Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat Commun. 2017;8:14245. doi: 10.1038/ncomms14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G, et al. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 2011;7:e1001264. doi: 10.1371/journal.pgen.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Xiao G, Zheng P, Shang Y, Su Y, Zhang X, Liu X, Zhan S, St Leger RJ, Wang C. Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc Natl Acad Sci U S A. 2014;111:16796–16801. doi: 10.1073/pnas.1412662111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattemore JA, Hane JK, Williams AH, Wilson BA, Stodart BJ, Ash GJ. The genome sequence of the biocontrol fungus Metarhizium anisopliae and comparative genomics of Metarhizium species. BMC Genomics. 2014;15:660. doi: 10.1186/1471-2164-15-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staats CC, Junges A, Guedes RL, Thompson CE, de Morais GL, Boldo JT, de Almeida LG, Andreis FC, Gerber AL, Sbaraini N, et al. Comparative genome analysis of entomopathogenic fungi reveals a complex set of secreted proteins. BMC Genomics. 2014;15:822. doi: 10.1186/1471-2164-15-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang Y, Xiao G, Zheng P, Cen K, Zhan S, Wang C. Divergent and convergent evolution of fungal pathogenicity. Genome Biol Evol. 2016;8:1374–1387. doi: 10.1093/gbe/evw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernier L, Cooper RM, Charnley AK, Clarkson JM. Transformation of the entomopathogenic fungus Metarhizium anisopliae to benomyl resistance. FEMS Microbiol Lett. 1989;60:261–265. [Google Scholar]
- 18.Fang W, Pei Y, Bidochka MJ. A regulator of a G protein signalling (RGS) gene, cag8, from the insect-pathogenic fungus Metarhizium anisopliae is involved in conidiation, virulence and hydrophobin synthesis. Microbiology. 2007;153(Pt 4):1017–1025. doi: 10.1099/mic.0.2006/002105-0. [DOI] [PubMed] [Google Scholar]
- 19.Sevim A, Donzelli BG, Wu D, Demirbag Z, Gibson DM, Turgeon BG. Hydrophobin genes of the entomopathogenic fungus, Metarhizium brunneum, are differentially expressed and corresponding mutants are decreased in virulence. Curr Genet. 2012;58:79–92. doi: 10.1007/s00294-012-0366-6. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Wang F, Wei D. Chlorimuron ethyl as a new selectable marker for disrupting genes in the insect-pathogenic fungus Metarhizium robertsii. J Microbiol Methods. 2011;87:241–243. doi: 10.1016/j.mimet.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Cook D, Donzelli BG, Creamer R, Baucom DL, Gardner DR, Pan J, Moore N, Krasnoff SB, Jaromczyk JW, Schardl CL. Swainsonine biosynthesis genes in diverse symbiotic and pathogenic fungi. G3 (Bethesda) 2017;7:1791–1797. doi: 10.1534/g3.117.041384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu C, Zhang X, Qian Y, Chen X, Liu R, Zeng G, Zhao H, Fang W. A high-throughput gene disruption methodology for the entomopathogenic fungus Metarhizium robertsii. PLoS One. 2014;9:e107657. doi: 10.1371/journal.pone.0107657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 24.Fang W, Pei Y, Bidochka MJ. Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can J Microbiol. 2006;52:623–626. doi: 10.1139/w06-014. [DOI] [PubMed] [Google Scholar]
- 25.Pall ML, Brunelli JP. A series of six compact fungal transformation vectors containing polylinkers with multiple unique restriction sites. Fungal Genet Rep. 1993;40:59–62. [Google Scholar]
- 26.Covert SF, Kapoor P, Lee MH, Briley A, Nairn CJ. Agrobacterium tumefaciens-mediated transformation of Fusarium circinatum. Mycol Res. 2001;105:259–264. [Google Scholar]
- 27.Padilla-Guerrero IE, Barelli L, González-Hernández GA, Torres-Guzmán JC, Bidochka MJ. Flexible metabolism in Metarhizium anisopliae and Beauveria bassiana: role of the glyoxylate cycle during insect pathogenesis. Microbiology. 2011;157:199–208. doi: 10.1099/mic.0.042697-0. [DOI] [PubMed] [Google Scholar]
- 28.Paz Z, García-Pedrajas MD, Andrews DL, Klosterman SJ, Baeza-Montañez L, Gold SE. One step construction of Agrobacterium-recombination-ready-plasmids (OSCAR), an efficient and robust tool for ATMT based gene deletion construction in fungi. Fungal Genet Biol. 2011;48:677–684. doi: 10.1016/j.fgb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y. Gene transfer and genetically modified plants. In: Xu Y, editor. Molecular plant breeding. Bodmin: MPG Books Group; 2010. pp. 479–500. [Google Scholar]
- 30.St. Leger RJ, Shimizu S, Joshi L, Bidochka MJ, Roberts DW. Co-transformation of Metarhizium anisopliae by electroporation or using the gene gun to produce stable GUS transformants. FEMS Microbiol Lett. 1995;131:289–294. [Google Scholar]
- 31.Li FF, Wu SJ, Chen TZ, Zhang J, Wang HH, Guo WZ, Zhang TZ. Agrobacterium-mediated co-transformation of multiple genes in upland cotton. Plant Cell Tissue Organ Cult. 2009;97:225–235. [Google Scholar]
- 32.McDade HC, Cox GM. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39:151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]