Abstract
A major mechanism of eukaryotic mRNA degradation initiates with deadenylation followed by decapping and 5′ to 3′ degradation. We demonstrate that the yeast EDC1 mRNA, which encodes a protein that enhances decapping, has unique properties and is both protected from deadenylation and undergoes deadenylation-independent decapping. The 3′ UTR of the EDC1 mRNA is sufficient for both protection from deadenylation and deadenylation-independent decapping and an extended poly(U) tract within the 3′ UTR is required. These observations highlight the diverse forms of decapping regulation and identify a feedback loop that can compensate for decreases in activity of the decapping enzyme. Surprisingly, the decapping of the EDC1 mRNA is slowed by the loss of Not2p, Not4p, and Not5p, which interact with the Ccr4p/Pop2p deadenylase complex. This indicates that the Not proteins can affect decapping, which suggests a possible link between the mRNA deadenylation and decapping machinery.
Keywords: deadenylation, decapping, mRNA decay, Not proteins
Introduction
The process of mRNA turnover is an important control point in gene expression. There are two general pathways by which polyadenylated mRNAs can be degraded in eukaryotic cells (reviewed in Parker and Song, 2004). In both cases, the degradation of the transcript begins with the shortening of the poly(A) tail at the 3′ end of the mRNA. Shortening of the poly(A) tail primarily leads to removal of the 5′ cap structure (decapping), which exposes the mRNA to 5′ to 3′ exonucleolytic degradation. Alternatively, following deadenylation, mRNAs can be degraded in a 3′ to 5′ direction by the cytoplasmic exosome.
Decapping is a critical control point in the 5′ to 3′ decay pathway since it both precedes and permits the degradation of the mRNA body. The decapping reaction requires two interacting proteins, Dcp1p and Dcp2p, which are thought to form the core of a decapping holoenzyme (reviewed in Coller and Parker, 2004). The general activity of this holoenzyme is regulated in both positive and negative ways. For example, the efficiency of the decapping reaction is enhanced by the activity of the Pat1p/Lsm1–7p complex and Dhh1p, a member of the DEAD box family of RNA helicases (Bonnerot et al, 2000; Bouveret et al, 2000; Tharun et al, 2000; Coller et al, 2001; Fischer and Weis, 2002). Although not required for decapping, these factors stimulate the decapping of both stable and unstable mRNAs suggesting they are general activators of the decapping reaction. In contrast, translation initiation factors can be inhibitors of decapping (Schwartz and Parker, 1999, 2000; Vilela et al, 2000; Ramirez et al, 2002).
Decapping is a site of control of mRNA turnover, and mRNAs differ in their mechanism of decapping. For example, Puf3p binds to the 3′ UTR of the COX17 mRNA and specifically enhances the rates of both deadenylation and decapping of this transcript (Olivas and Parker, 2000). Similarly, the rapid decapping of mRNAs containing aberrant translation termination codons, referred to as nonsense-mediated decay (NMD), requires Upf1p, Upf2p, and Upf3p, and is independent of the general activators of decapping, Pat1p, Dhh1p, and Lsm1p (reviewed in Coller and Parker, 2004; Maquat, 2004). In addition, NMD triggers deadenylation-independent decapping, where the need for deadenylation prior to decapping is bypassed (Muhlrad and Parker, 1994). The yeast Rps28b mRNA also undergoes deadenylation-independent decapping in a manner modulated by Edc3p (Badis et al, 2004), which can also enhance decapping of other mRNAs when decapping is limited (Kshirsagar and Parker, 2004). These observations suggest that there may be substantial variation in the mechanisms of decapping both with regard to the requirement for deadenylation and the role of trans-acting factors.
Two related proteins, Edc1p and Edc2p, were identified as proteins that enhance decapping. This was based on their identification as high-copy suppressors of decapping defects in strains containing temperature-sensitive alleles of DCP1 or DCP2 (Dunckley et al, 2001). Moreover, in sensitized strains where an edc1Δ or edc2Δ was combined with partially defective alleles of DCP1 or DCP2, mRNAs were further stabilized. This result indicated that these proteins play a role in compensating for defects in the function of the decapping enzyme. The Edc1 and Edc2 proteins are RNA binding proteins and can directly stimulate the activity of the decapping enzyme (Schwartz et al, 2003; Steiger et al, 2003). This suggests that Edc1p and Edc2p function to modulate the activity of decapping, either in response to general changes in decapping activity and/or on specific mRNAs.
In this work, we examined the mechanism of degradation of the EDC1 mRNA. We demonstrate that this mRNA undergoes deadenylation-independent decapping, specified by a poly(U) tract in its 3′ UTR. The decapping of the EDC1 mRNA is slowed by the loss of Not2p, Not4p, and Not5p, which interact with the Ccr4p/Pop2p deadenylase complex. This indicates that the Not proteins can affect decapping, which suggests a possible link between mRNA deadenylation and decapping. These observations also highlight the diverse forms of decapping regulation and identify a feedback loop that can compensate for decreases in activity of the decapping enzyme.
Results
The EDC1 mRNA has a relatively homogeneous poly(A) tail at steady state
During our initial analysis of the EDC1 gene, we examined the EDC1 mRNA on a high-resolution polyacrylamide Northern gel. In this experiment, the mRNA was first cleaved with RNaseH and an internal oligonucleotide (Figure 1A, o3′, position shown in cartoon) to shorten the mRNA length sufficiently to allow resolution of the average poly(A) tail length on the mRNA. We observed that at steady state, the EDC1 mRNA was a relatively discrete size (Figure 1B, lane 1), which was shortened by ∼45–55 nucleotides by a sequential and additional RNaseH-mediated cleavage of the mRNA (see below) with oligo(dT) to an 85–90-nucleotide mRNA fragment (Figure 1B, lane 3). These observations indicate that under steady-state conditions, the EDC1 mRNA possesses a relatively homogeneous poly(A) tail of ∼45–55 nucleotides. This poly(A) length is in contrast to typical yeast mRNAs, which show a distribution of poly(A) tail lengths of 10–75, as exemplified in Figure 1C for the MFA2 mRNA, with the specific distribution of poly(A) tail lengths being a function of the relative rates of deadenylation and decapping (Cao and Parker, 2001).
Figure 1.
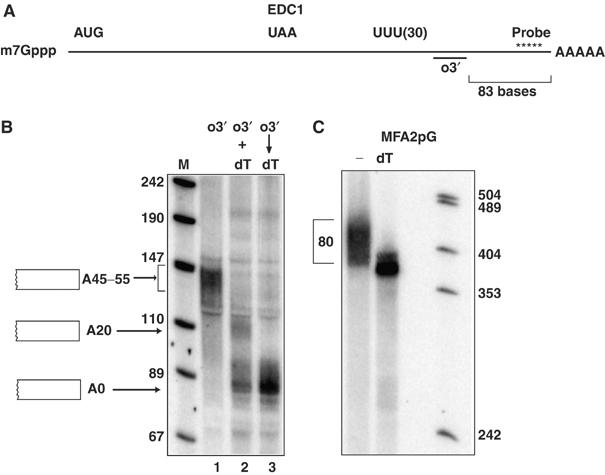
The EDC1 mRNA poly (A) tail has a uniform distribution at steady state. (A) Cartoon of the EDC1 mRNA with pertinent sequences and oligonucleotide locations (probe and o3′) marked. The Northern is probed with oRP1211 (Table III). Distances from the 3′ end of the oligonucleotides used for RNaseH reactions to the 3′ end of the mRNA are bracketed. (B) Polyacrylamide gel of the EDC1 mRNA in wild-type strain yRP683 cut with several combinations of oligonucleotides. Above each lane is the oligonucleotides with which the mRNAs in each lane were cut as shown in part A. Oligo 3′ (o3′)=oRP1209 (Table III). Simultaneous RNaseH cleavage with o3′ and dT and labeled (+) and serial cleavage are designated with an arrow. The markers and their sizes are shown to the left of the left-most lane. Cartooned to each side of the panel are the species of mRNA present in each band. Asterisks denote background bands. Lanes are labeled below for reference. (C) The MFA2pG mRNA from a wild-type strain yRP840 without (−) or with oligo(dT) cleavage. Size markers are shown on the right with their sizes labeled. The bracket designates the distribution of the poly(A) tail.
A significant technical issue is that the EDC1 mRNA contains an extended poly(U) tract that makes RNaseH-mediated cleavage by oligo(dT) inefficient. Specifically, if the mRNA is cleaved with both oligo(dT) and o3′ at the same time, we only observed partial removal of the poly(A) tail and detected both the species of 85–90 nucleotides, where the poly(A) tail is completely removed, and an ∼110-nucleotide EDC1 mRNA fragment, with approximately a 20-nucleotide poly(A) tail (Figure 1B, lane 2). However, our sequential reactions, where the mRNA is first cleaved with o3′, to separate the poly(U) tract from the poly(A) tail, followed by a denaturation step, and subsequent oligo(dT) cleavage led to efficient removal of the poly(A) tail and only detection of the fully deadenylated 85–90-nucleotide mRNA fragment (Figure 1B, lane 3). We interpret these results to indicate that the poly(U) tract in the EDC1 3′ UTR competes with oligo(dT) for binding to the poly(A) tail, and thereby inhibits RNaseH-mediated oligo(dT) cleavage. Thus, mRNAs with poly(U) tracts can be resistant to complete removal of their poly(A) tails by RNaseH and oligo(dT) by standard methods.
Because of its unique nature, we examined the size and distribution of the poly(A) tail on the EDC1 mRNA in strains defective in specific mRNA deadenylases or 3′ to 5′ exonucleases (Figure 2). These experiments revealed the following points. First, we observed that the EDC1 mRNA poly(A) tail did not change in strains lacking Pan2p compared to wild-type strains (Figure 2A, compare lanes 1 and 9). This is surprising since Pan2p is required for an initial, and rapid, deadenylation step that trims the poly(A) tail to an mRNA specific length, which then becomes a substrate for further deadenylation (Brown and Sachs, 1998). Thus, the EDC1 mRNA has its initial poly(A) tail length determined by a different mechanism. Second, we observed that the poly(A) tail length of the EDC1 mRNA did not change in strains lacking Pop2p, Ccr4p, or a ccr4Δ pan2Δ double mutant (Figure 2A, lanes 3, 5, and 7). This was also surprising since lesions in Ccr4 or Pop2 inactivate the major cytoplasmic deadenylase and typically result in mRNAs with a longer poly(A) tail, and strains lacking both Ccr4p and Pan2p show no deadenylation and accumulate fully adenylated mRNAs (Figure 2B; Tucker et al, 2001). Third, the poly(A) tail length of the EDC1 mRNA did not change in strains lacking Ski2p or Ski2p, Ski3p, Ski7p, and Ski8p, which are required for cytoplasmic exosome function (Figure 2A, lane 11 and data not shown). These results suggest that the metabolism of the EDC1 mRNA poly(A) tail is unique and raise the possibility that the EDC1 mRNA is not a substrate for deadenylation, and/or is degraded independently of poly(A) shortening.
Figure 2.
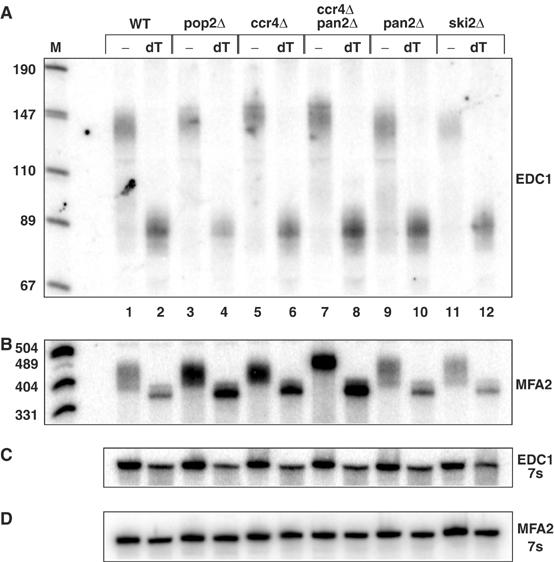
The poly(A) tail of the EDC1 mRNA does not change in strains deficient in mRNA deadenylation. (A) A polyacrylamide Northern gel probed with oRP1211 is shown after cleavage of the EDC1 mRNA with oligo o3′ (oRP1209) without (−) or with (dT) subsequent oligo(dT) cleavage. Deletion strains are marked across the top above each bracket set. WT=yRP840; pop2Δ=yRP1920; ccr4Δ=yRP1616; ccr4Δ pan2Δ=yRP1620; pan2Δ=yRP1619; ski2Δ=yRP1195. M represents the marker lane the band sizes of which are shown on the left. Lanes are numbered across the bottom for reference. (B) The MFA2 mRNA probed with oRP140 in the same deletion strains as panel A. Marker sizes are given on the left. (C, D) The 7s standard probed with oRP100 for Northerns in parts A and B, respectively.
The EDC1 mRNA undergoes deadenylation-independent decapping
The ∼45–55-nucleotide EDC1 poly(A) tail length seen at steady state can be explained in two different manners. First, the mRNA could exist as an adenylated species, but once it enters deadenylation it undergoes rapid poly(A) shortening and degradation such that intermediates in this process are not observed. Such behavior has been described for the yeast COX17 mRNA (Olivas and Parker, 2000). Alternatively, the EDC1 mRNA might be subjected to decapping prior to deadenylation. An experiment to determine if the decay of an mRNA requires deadenylation is to trap intermediates in the decay process and determine if they possess long poly(A) tails (Muhlrad and Parker, 1994). mRNA decay intermediates can be trapped by inserting a poly(G) tract in the 3′ UTR, which blocks the progression of the 5′ to 3′ exonuclease Xrn1p thereby trapping mRNA fragments in the process of degradation. If decay intermediates possess poly(A) tails similar to the full-length mRNA, then decay must have initiated without poly(A) shortening.
In order to trap decay intermediates of the EDC1 mRNA, we inserted a poly(G) tract into the 3′ UTR six nucleotides downstream of the translation termination codon, referred to as EDC1pG. The insertion of the poly(G) tract did not alter the behavior of the EDC1 mRNA either in terms of its poly(A) tail distribution or decay rates (data not shown). In addition, to allow manipulation of transcription, this construct was put under the regulation of the GAL1 UAS.
Examination of the EDC1pG mRNA on a polyacrylamide Northern gel at steady state revealed two important observations. First, we observed that the EDC1pG mRNA produced a full-length mRNA and accumulated an mRNA fragment that was the correct size to be the mRNA degraded 5′ to 3′ from the 5′ end to the 5′ side of the poly(G) tract and including a 45–55-nucleotide poly(A) tail (Figure 3, lane 1). The majority of the EDC1pG transcripts accumulated as the mRNA fragment, which is consistent with the rapid decay of the full-length EDC1pG mRNA and the stability of its decay fragment (see below). In addition to the adenylated fragment, a small amount of an mRNA species of the proper size to be a deadenylated form of the mRNA decay intermediate was seen (Figure 3, bottom arrow, left side). This is consistent with some deadenylation occurring on the mRNA fragment, albeit inefficiently (see below). A second important observation was that this mRNA fragment was shortened by treatment with oligo(dT) and RNaseH (compare lanes 1 and 2), which confirms that the decay fragment was indeed adenylated. The predominant species corresponds to the A20 form of the mRNA consistent with only partial removal of the poly(A) tail by RNaseH- and oligo(dT)-mediated cleavage. These results indicate that the mRNA fragment contains a poly(A) tail and argues that 5′ to 3′ degradation of this mRNA initiates prior to deadenylation. The production of two predominant species following oligo(dT) cleavage with RNaseH is consistent with our earlier results indicating that efficient oligo(dT) cleavage requires removal of the poly(U) tract from the same molecule undergoing oligo(dT)-mediated cleavage (Figure 1B).
Figure 3.
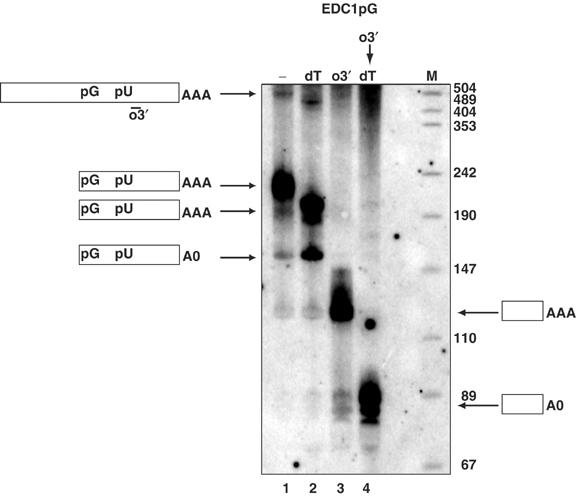
Decay products from the EDC1pG mRNA possess long poly(A) tails. This panel shows a Northern blot of the EDC1pG mRNA (pRP1173) in a wild-type strain yRP683 following different RNaseH- and oligo-directed cleavages specified as follows: no oligo (−), lane 1; oligo(dT) (dT), lane 2; EDC1 oligo (o3′) oPR1209, lane 3; serial EDC1 oRP1209 and oligo(dT) (o3′ → dT), lane 4. M designates the marker lane with the sizes given on the right next to the lane. Simple cartoons to each side show the species of mRNA present in the band designated with an arrow. pG represents the poly(G) structure and pU indicates the 3′ UTR poly(U) sequence.
To determine more precisely the poly(A) tail lengths on the mRNA and the trapped decay intermediate, we used RNaseH to sequentially cleave the mRNA, and the intermediate, with an oligonucleotide o3′, located 3′ of the poly(U) tract, which then allows complete removal of the poly(A) tail in a subsequent oligo(dT) cleavage step. It should be noted that because this oligonucleotide is 3′ of the inserted poly(G) tract, the full-length mRNA and the decay intermediate produce the same product following RNaseH-mediated cleavage. Following o3′ cleavage with RNaseH, we observed a single mRNA on the Northern blot of the size expected for a fully adenylated species, which was shortened by ∼45–55 nucleotides by sequential RNaseH cleavage with oligo(dT) (Figure 3, lanes 3 and 4). Because the mRNA fragment represents the bulk of the EDC1pG transcripts present, this observation indicates that the mRNA fragment contains a poly(A) tail. In addition, the ∼45–55-nucleotide-long poly(A) tail observed on the mRNA fragment is the same length as observed on the full-length mRNA (Figure 1). These observations demonstrate that the degradation of the EDC1 mRNA initiates independently of deadenylation.
The EDC1 mRNA poly(A) tail is protected from deadenylation
A surprising aspect of the EDC1 mRNA decay intermediate is that it was adenylated even at steady state. Other mRNAs that undergo deadenylation-independent decapping produce adenylated fragments that undergo deadenylation, thereby leading to deadenylated fragments at steady state (Cao and Parker, 2003). To determine the fate of the poly(A) tail on the EDC1 mRNA and the decay intermediate, we examined the poly(A) tail length on these species following transcriptional repression in a wild-type cell. In this experiment, we cleaved the mRNA with an oligonucleotide 5′ of the poly(G) tract. Although this type of cleavage does not allow for removal of the full poly(A) tail with oligo(dT) cleavage, it has two advantages. First, after cleavage, the full-length mRNA and mRNA fragments are different sizes, so whether they contain poly(A) tails can be assessed for each species. Second, we are still able to get partial removal of the poly(A) by oligo(dT)- and RNaseH-mediated cleavage so that we can determine if an RNA species contains a poly(A) tail. Thus the lanes with oligo(dT) treatment contain mRNAs where the poly(A) tail has only been partially removed. We observed that in wild-type cells, the full-length mRNA declined quickly (t1/2=4 min), but the mRNA fragment persisted in a fully adenylated form (Figure 4, left panel). This indicated that the poly(A) tail on the fragment is resistant to deadenylation.
Figure 4.
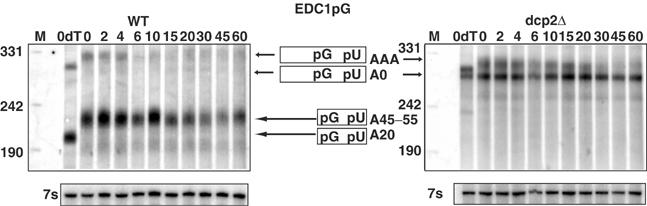
Analysis of decay and deadenylation of the EDC1pG mRNA (pRP1173) in wild-type strain yRP683 (WT) (left panel) and dcp2Δ strain yRP1358 (right panel). Strains were grown in galactose media and transcription was repressed at zero time by the addition of glucose. Samples were collected at various times after repression marked in minutes above each panel. To allow analysis on a polyacrylamide Northern of the poly(A) tail lengths on both full-length mRNA and the decay fragment, the mRNA was cleaved with an oligo 5′ of the poly(U) sequence, oRP1208, and RNaseH prior to analysis on the Northern gel. The zero minute time point in each panel was also cleaved with oligo(dT) (0dT) and is shown in the left-most lanes. M stands for the markers and their sizes are next to the lanes at the left of each panel. Arrows point to the corresponding mRNA species present in each band, which are cartooned between the Northern panels. pG represents the poly(G) structure and pU indicates the 3′ UTR poly(U) sequence. Below each panel is shown the 7s standard for the respective Northern.
To examine the fate of the poly(A) tail on the full-length mRNA, we analyzed the EDC1pG mRNA in a dcp2Δ strain, which will inhibit decapping and allow the full-length mRNA to persist. In dcp2Δ strains, the adenylated full-length mRNA (Figure 4, upper band, right panel) showed increased stability (t1/2=15 min) and no mRNA decay product was produced (Figure 4, right panel), indicating that the Dcp2p is required for decapping and degradation of this mRNA. In addition, two species of the full-length mRNA were observed, one possessed a 45–55-nucleotide poly(A) tail and the other corresponded to a completely deadenylated species. Note that the deadenylated species is extremely stable (Table I). The presence of deadenylated EDC1pG transcripts in a dcp2Δ strain indicated that the EDC1pG mRNA can be subject to deadenylation at some rate. However, the fully adenylated EDC1pG mRNA (the upper band in the dcp2Δ strain) persisted as a fully adenylated species, which only showed very little and slow deadenylation over the time course. These results suggest that deadenylation on the full-length mRNA is inefficient. These results in conjunction with Figure 2 indicate that the poly(A) tail of the EDC1 mRNA is resistant to deadenylation on both the full-length mRNA and the decay fragment.
Table 1.
mRNA characteristics in deletion strains
| Strain | Steady-state mRNA level | Detection of full-length deadenylated mRNA | Half-life (min) |
|---|---|---|---|
| WT | 1 | − | 4 |
| dcp1Δ | 11 × | +++ | >60 |
| dcp2Δ | 14 × | +++ | >60 |
| xrn1Δ | 17 × | ++ | >60 |
| dhh1Δ | 5 × | ++ | >60 |
| pat1Δ | 2.7 × | + | >60 |
| lsm1Δ | 2.5 × | + | >60 |
| upf1Δ | 1 | − | 4 |
| upf2Δ | 1 | − | 4 |
| upf3Δ | 1 | − | 4 |
| edc2Δ | 1 | − | ND |
| edc3Δ | 1 | − | ND |
| ski2Δ | 1.2 × | − | 4 |
| ccr4Δ | 1.1 × | − | ND |
| not2Δ | 2.9 × | + | ND |
| not3Δ | 1.3 × | − | ND |
| not4Δ | 4.5 × | ++ | 90 |
| not5Δ | 3 × | + | ND |
| ND=not determined. | |||
The EDC1 3′ UTR dictates deadenylation-independent decapping and protection from deadenylation
The experiments above revealed that the EDC1 mRNA undergoes deadenylation-independent decapping and also has a poly(A) tail that is resistant to deadenylation. To determine what portion of the EDC1 mRNA dictated these properties, we created and analyzed a series of chimeric genes. Replacement of the EDC1 promoter region with the GAL promoter still gave rise to deadenylation-independent decapping and little deadenylation (Figure 4). Similarly, replacement of the EDC1 coding region with HIS3 yielded a chimeric mRNA that still had a discrete poly(A) tail length of 45–55 residues (data not shown). These results suggested that the EDC1 3′ UTR dictated the novel properties of the EDC1 transcript. To test if the EDC1 3′ UTR was sufficient for the unique aspects of the EDC1 mRNA metabolism, we replaced the 3′ UTR of the normal MFA2 mRNA (under GAL control) with the EDC1 3′ UTR and examined the mRNAs produced. This reporter construct, MFA2pGE, also has a poly(G) tract inserted into its 3′ UTR upstream of the EDC1 mRNA 3′ UTR.
Several observations indicate that the MFA2pGE mRNA behaves like the EDC1 mRNA. In the first experiment, we cleaved the mRNA with an oligonucleotide 5′ of the poly(G) tract to allow us to separately assess the poly(A) status of the full-length mRNA and the decay intermediate. We observed that both the full-length MFA2pGE and its mRNA fragment were discrete species that were shortened by ∼25–30 nucleotides by oligo(dT) cleavage (Figure 5B, lanes 1 and 2). Because the oligo(dT) cleavage under these conditions leaves a residual poly(A) tail of approximately 20 residues, this result indicated they both possessed poly(A) tails of 45–55 nucleotides, similar to what is seen on the EDC1 mRNA. In contrast, the MFA2pG mRNA, which undergoes deadenylation-dependent decapping, shows heterogeneous poly(A) tails on the full-length mRNA ranging from A(10) to A(70) (Figure 1C) and an oligo(A) tail on the mRNA fragment of 0–12 residues (Decker and Parker, 1993; Muhlrad et al, 1994).
Figure 5.
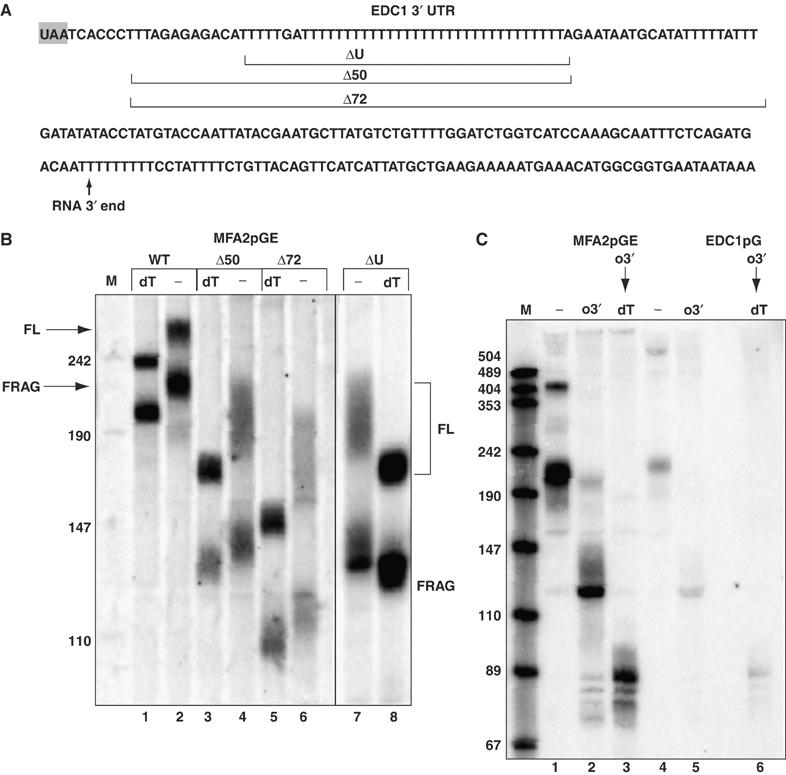
The EDC1 3′ UTR dictates its novel decay properties. (A) The sequence of the region of the 3′ UTR of the EDC1 mRNA. The stop codon is shaded gray. The approximate 3′ end of the mRNA as mapped is shown with an arrow. Areas deleted for analysis are marked within brackets and labeled with the number of bases deleted or ΔU for removal of the poly(U) sequence. (B) Polyacrylamide gel of the parental MFA2pGE transcript (pRP1169) and deletion constructs (pRP1170-Δ50, pRP1171-Δ72, pRP1172-ΔU) with oligo(dT) cleavage (dT) and without (−) in a wild-type strain yRP683. To allow analysis of the poly(A) tail lengths on both full-length mRNA and the decay fragment, the mRNA was cleaved with oligo oRP59 5′ of the poly(U) sequence and RNaseH prior to running the Northern gel. The species present in each band in the two outside lanes are labeled next to these lanes. FL is the full-length message and FRAG is the poly(G) containing decay intermediate. Marker sizes are shown on the left of the marker lane M. Constructs are given above each lane set and correspond to the sequences in part A. Lane numbers are given below the panel for reference. (C) Cleavage of mRNAs MFA2pGE (pRP1168) and EDC1pG (pRP1173) with oligo o3′ (oRP1209), which cleaves 3′ of the poly(U) sequence. Lanes are designated as follows: (−) no oligo cleavage; (o3′) oligo o3′ cleavage; (o3′ → dT) for serial cleavage. Markers (M) are shown on the left with the sizes given and lane numbers are given below the panel for reference.
We also examined the MFA2pGE mRNA without any cleavage or with cleavages 3′ of the poly(U) tract (Figure 5C). Without any cleavage, we detect both the full-length mRNA and the mRNA fragment, both of which are of the appropriate size to contain a 45–55-nucleotide poly(A) tail (Figure 5C, lane 1). Cleavage of the mRNAs with an oligonucleotide 3′ of the poly(U) tract and RNaseH produces a single predominant species (Figure 5C, lane 2), which is shortened by sequential cleavage with oligo(dT) (Figure 5C, lane 3). Additional evidence that the EDC1 3′ UTR is sufficient to protect an RNA from deadenylation is that following transcriptional repression, the mRNA fragment produced from the MFA2pGE transcript persists for over an hour with a 45–55-nucleotide poly(A) tail (data not shown). These observations indicate that the EDC1 3′ UTR dictates both deadenylation-independent decapping and resistance of the poly(A) tail to deadenylation.
A poly(U) tract within the 3′ UTR is required for deadenylation-independent decapping and protection from deadenylation
To determine what region of the EDC1 3′ UTR modulated nascent poly(A) tail lengths, we created deletions within the 3′ UTR of the MFA2pGE construct starting at the translation termination codon and extending 3′ (cartooned in Figure 5A). In addition, we precisely deleted a 38-nucleotide region of the EDC1 3′ UTR that contained 36 uridines. We examined the transcripts produced from the deletion constructs to determine whether the full-length mRNA (labeled FL) and the mRNA fragment (labeled FRAG) possessed long poly(A) tails, which is a marker for deadenylation-independent decapping. Moreover, because decay fragments that are not protected from deadenylation undergo poly(A) tail shortening, a poly(A) tail on the mRNA fragment is also indicative of protection from deadenylation.
Analysis of these deletions showed that all three deletions converted the mRNA into a more typical mRNA with a heterogeneous poly(A) tail on the full-length mRNA ranging approximately from 12 to 60 nucleotides, and an mRNA fragment with an oligo(A) tail of roughly 12 nucleotides (Figure 5B, lanes 3–8). These are features of deadenylation-dependent decapping (Decker and Parker, 1993; Cao and Parker, 2001). Deletions that went further 3′ into the 3′ UTR failed to produce any mRNA and presumably are deleting sequences critical for efficient 3′ end generation (data not shown). These results define the poly(U)-rich region of the EDC1 3′ UTR as being required for deadenylation-independent decapping and protection from deadenylation.
The EDC1 mRNA is affected by lesions affecting mRNA decapping
Given the unique properties of the EDC1 mRNA, we examined how its steady-state level and decay rate were affected in a range of mutations that affect mRNA decapping. In this experiment, we cleaved the endogenous EDC1 mRNA with RNaseH and an oligonucleotide 5′ of the poly(U) tract, thus allowing us to determine the poly(A) distribution on the full-length mRNA. We observed that strains lacking either subunit of the decapping enzyme (dcp1Δ or dcp2Δ), or the 5′ to 3′ exonuclease (xrn1Δ) showed a 4–10-fold increased level of the EDC1 mRNA accumulating in these strains (Figure 6). Half-life measurements also show an increased stability in these strains (Table I). Similar to other mRNAs, the EDC1 mRNA is a substrate for decapping by Dcp1p and Dcp2p and 5′ to 3′ decay by Xrn1p.
Figure 6.
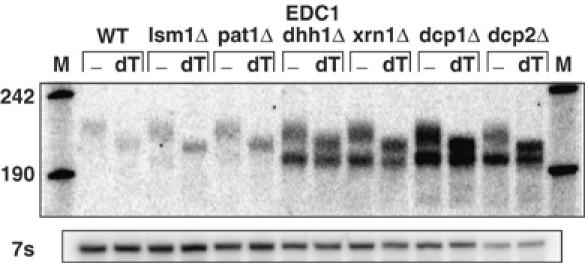
Mutations in decapping factors affect the EDC1 mRNA. Wild-type (WT) and strains deleted for specific mRNA decapping factors were analyzed on polyacrylamide gels showing the levels and adenylation status of the EDC1 mRNA. Deletion strains are marked above each set of bracketed lanes. Strains are as follows: WT=yRP840; lsm1Δ=yRP1410; pat1Δ=yRP1372; dhh1Δ=yRP1560; xrn1Δ=yRP1359; dcp1Δ=yRP1211; dcp2Δ=yRP1358. All lanes are cleaved with an oligo 5′ of the poly(U) oligo RP1208 (−). Oligo(dT) cleavage in addition to oligo oRP1208 lanes is marked above as (dT). Markers are shown in lane M and the sizes are given on the left. The lower panel is the same Northern probed with oRP100, the 7s loading standard.
The decay rate, steady-state level, and poly(A) tail distribution were not affected by the loss of the Upf1, Upf2, or Upf3 proteins, which are required for NMD (Table I). This demonstrates that the deadenylation-independent decapping seen with the EDC1 mRNA is distinct from that seen in NMD. We also observed that the steady-state level of the EDC1 mRNA was not affected by loss of two enhancers of decapping, Edc2p and Edc3p (Table I).
We also examined the role of the general activators of decapping, Lsm1p, Pat1p, and Dhh1p, on the decay of the EDC1 mRNA. These proteins are not required for decapping per se, but strains lacking these proteins show a decrease in decapping rate for a range of mRNAs—with pat1Δ strains having the most severe phenotype, lsm1Δ strains having a strong phenotype, and dhh1Δ strains showing a modest effect on mRNA decapping rates of the MFA2 mRNA (Beelman et al, 1996; Coller et al, 2001). These same factors also affect the EDC1 mRNA but in a slightly different manner. We observed that the steady-state level of the EDC1 mRNA was strongly affected by dhh1Δ (5–6 ×), whereas lsm1Δ and pat1Δ showed a more modest ∼2–3 × increase in steady-state mRNA levels (Figure 6 and Table I). Consistent with the change in steady-state levels, we observed that the EDC1 mRNA was more stable in the dhh1Δ, lsm1Δ, and pat1Δ strains (Table I). Interestingly, the lsm1Δ, pat1Δ, and dhh1Δ strains all show a strong decrease in EDC1 mRNA decay rate, but only the dhh1Δ strain shows a corresponding large increase in steady-state levels. This discrepancy between decay rates and steady-state levels is similar to what has been seen for some mRNAs in dcp1Δ strains (Muhlrad and Parker, 1999). We do not currently understand the basis for the differences in steady-state levels, but this clearly indicates that there must be additional factors that modulate mRNA steady-state levels. There are two simple possibilities. First, there might be a feedback mechanism such that EDC1 mRNA transcription is reduced in the lsm1Δ and pat1Δ strains. Alternatively, transcription may be proceeding at a normal pace but due to the specific defects in these strains, the nascent EDC1 mRNAs are degraded by an alternative mechanism, perhaps within the nucleus at a rate too fast to be transiently observed.
All of the above decapping lesions led to the accumulation of a deadenylated species of mRNA (Figure 6, bottom bands). The blocks to decapping seen in dcp1Δ, dcp2Δ, xrn1Δ, and dhh1Δ strains resulted in the production of a significant amount of this deadenylated form of the EDC1 mRNA (Figures 4 and 6), whereas the lsm1Δ and pat1Δ deletions yielded only small amounts (Figure 6). These results indicate that the EDC1 mRNA is able to undergo some form of extremely slow deadenylation, but only when decapping is blocked can these deadenylated mRNAs be produced sufficiently to be detected.
Decapping of the EDC1 mRNA is affected by lesions in Not2p, Not4p, and Not5p
In our survey of potential proteins involved in the decay of the EDC1 mRNA, we also looked at how its metabolism was affected in strains lacking Not2p, Not3p, Not4p, or Not5p. These Not proteins and the essential Not1p are found in an mRNA deadenylation complex (reviewed in Denis and Chen, 2003; Collart and Timmers, 2004). In addition, defects in Not proteins can have a small effect on deadenylation of the MFA2pG mRNA, suggesting that these proteins can be involved in mRNA degradation (Tucker et al, 2002). We observed that strains lacking Not2p, Not4p, and Not5p showed increases in the levels of EDC1 mRNA (Figure 7A), whereas not3Δ strains showed no significant differences in EDC1 mRNA levels. We also observed an accumulation of the deadenylated species of the EDC1 mRNA in not2Δ, not4Δ, and not5Δ strains (Table I). These phenotypes suggest that the absence of the Not proteins slows the decapping rate of the EDC1 mRNA. Consistent with that interpretation, the EDC1 mRNA showed a slower decay rate in a not4Δ strain following transcriptional repression using a temperature-sensitive allele of RNA polymerase II (Figure 7B). Because the EDC1 mRNA is only degraded by a decapping step, and these Not proteins can affect the decay of the EDC1 mRNA, we interpret these results to indicate that the Not proteins can affect decapping of at least some mRNAs.
Figure 7.
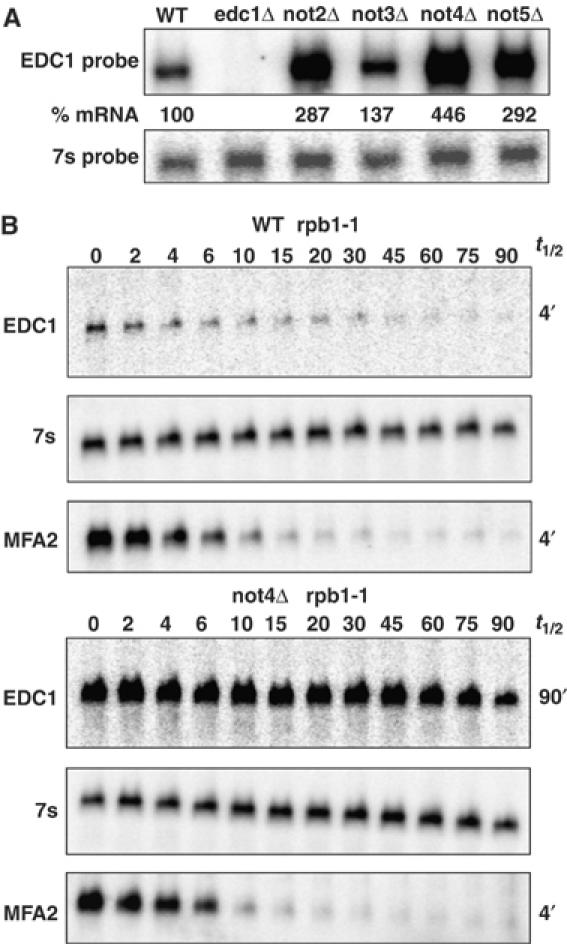
The not2Δ, not4Δ, and not5Δ strains show alterations in the decay of the EDC1 mRNA. (A) Steady-state levels of the EDC1 mRNA in wild-type (WT) (yRP840), not2Δ (yRP1670), not3Δ (yRP1671), not4Δ (yRP1672), and not5Δ (yRP1673) strains. Strains are given across the top of the agarose Northern. The upper panel is probed for the EDC1 mRNA, and the lower panel shows the standardizing 7s probe. Values determined for the %mRNA based on multiple experiments are given between the two panels. (B) Half-life measurements in temperature-sensitive RNA polymerase II (rpb1-1) mutant strains. Wild-type (WT) yRP582 or not4Δ yRP1806 strains containing a temperature-sensitive rpb1-1 mutation were shifted to 37°C and RNA was analyzed at various times in minutes following the shift, which are labeled above each panel set. The top panel shows the half-life of the EDC1 mRNA. The middle panel gives the 7s standardizing mRNA. The bottom panel shows the endogenous MFA2 mRNA to demonstrate the effectiveness of the rpb1-1 shut off. Half-life values for each set of experiments are given on the right of the respective panels.
Not proteins accumulate in P-bodies in a dcp1Δ strain
The experiments above argue that the Not2, Not4, and Not5 proteins can affect the decapping of the EDC1 mRNA. Recent experiments have identified specific cytoplasmic foci, termed P-bodies, that contain the mRNA decapping machinery and are sites of mRNA decapping and degradation (Ingelfinger et al, 2002; Lykke-Andersen, 2002; van Dijk et al, 2002; Sheth and Parker, 2003; Cougot et al, 2004). If Not proteins directly affect the decapping of mRNAs, then we predict they would, at least transiently, be associated with P-bodies. To address this issue, a C-terminal GFP fusion of each of the Not1–4 proteins was constructed and cellular localization of the chimeras determined in wild-type cells. These fusion proteins were judged to be functional because strains with the Not-GFP fusions behaved like wild-type strains and failed to show phenotypes seen in Not mutant (data not shown). Consistent with prior work (Tucker et al, 2002; Hu et al, 2003), the Not1–4 proteins were distributed in the cytoplasm (Figure 8). This observation can be explained by two possibilities. First, Not proteins do not enter P-bodies, and their effect on decapping is indirect. In this model, Not proteins would never concentrate in P-bodies. Second, Not proteins do enter P-bodies with mRNAs targeted for degradation, but the Not proteins leave P-bodies rapidly after the degradation of the mRNAs. In this latter model, the exit of Not proteins is sufficiently rapid that at steady state they appear to be distributed in the cytoplasm. This latter model predicts that Not proteins would be present in P-bodies if degradation of the mRNA within P-bodies was blocked.
Figure 8.
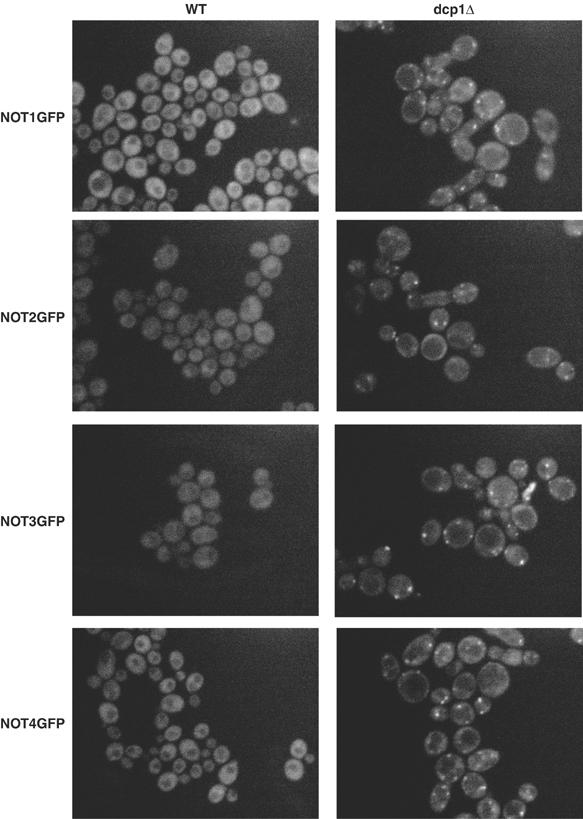
The NOT proteins localize to P-bodies in dcp1Δ strains. The respective NOT1–4-GFP fusion proteins are designated on the left of each row of pictures. Wild-type (WT) strains are shown in the left column for each fusion protein: not1=yRP1817; not2=yRP1818; not3=yRP1819; not4=yRP1820. The dcp1Δ strains are in the right column of pictures: not1=yRp1915; not2=yRP1916; not3=yRP1917; not4=yRP1918. All pictures have identical gain and normalization settings.
To distinguish these models, we localized the Not proteins in dcp1Δ strains, which are blocked at the enzymatic stage of decapping. Strikingly, we observed that Not1–4p are now found in discrete cytoplasmic foci, which colocalize with Dcp2p-RFP (Figure 8 and Supplementary Figure 1). The Not5-GFP fusion construct appeared to be nonfunctional in any strain background. However, given the interaction between the Not1–5 proteins and the effect of not5Δ on the EDC1 mRNA, we anticipate that Not5p also colocalizes within P-bodies in a dcp1Δ strain. This shows that Not proteins can localize to P-bodies when decapping is blocked and suggests that they play a direct role in the decapping reaction, at least for some mRNAs.
Discussion
The EDC1 mRNA undergoes deadenylation-independent decapping: a feedback mechanism for decapping homeostasis
Several observations indicate that the EDC1 mRNA undergoes deadenylation-independent decapping. The critical observation is that mRNA decay intermediates trapped by the insertion of a poly(G) tract contain poly(A) tails of the same length and distribution as the full-length mRNA (Figure 3). This demonstrates that the decay of the EDC1 mRNA initiates prior to deadenylation. In addition, the full-length mRNA possesses a relatively homogeneous poly(A) tail (Figure 1), and mutations in known cytoplasmic deadenylases have no effect on the EDC1 mRNA (Figure 2).
Several features of the deadenylation-independent decapping of the EDC1 mRNA create the potential for a regulatory feedback loop that could compensate for defects in decapping. First, the decay of the EDC1 mRNA is predominated by decapping rate, both by bypassing deadenylation and by being protected from deadenylation and 3′ to 5′ decay. A consequence of this circuitry is that this mRNA will respond linearly to changes in overall decapping rates. In contrast, an mRNA that also requires deadenylation for degradation, and can undergo 3′ to 5′ decay, would show a more modest response to changes in decapping rate (Cao and Parker, 2001). Second, Edc1p is a positive activator of decapping and can directly affect the activity of the decapping enzyme (Schwartz et al, 2003, Steiger et al, 2003). Thus, decreases in decapping rate will preferentially stabilize the EDC1 mRNA and produce more Edc1p, which will enhance the rate of ongoing decapping. This feedback model is also supported by the observations that the EDC1 gene is a high-copy suppressor of a Dcp1p conditional allele dcp1-2, and that the dcp1-2 edc1Δ strain shows a strong defect in decapping at the permissive temperature for dcp1-2 (Dunckley et al, 2001).
Deadenylation-independent decapping has also been reported for mRNAs that undergo NMD (Cao and Parker, 2003; Muhlrad and Parker, 1994) and for the Rps28b mRNA (Badis et al, 2004). Several observations indicate that the decapping of the EDC1 mRNA is distinct from that seen with substrates for NMD. First, the decapping of the EDC1 mRNA is independent of the Upf proteins (Table I), which are required for NMD (reviewed in Maquat, 2004). Second, the decapping of the EDC1 transcript is stimulated by Lsm1p, Pat1p, and Dhh1p (Figure 6 and Table I), which are not required for NMD. These results demonstrate that deadenylation-independent decapping can occur on some mRNAs independent of NMD. This implies that the EDC1 mRNA contains information that prevents the poly(A) tail from inhibiting decapping. Because the poly(A) binding protein (Pab1p) is required for inhibition of decapping by poly(A) tails (Caponigro and Parker, 1995), features of the EDC1 mRNA could either prevent Pab1p binding or block its ability to inhibit decapping once bound.
The EDC1 mRNA is protected from deadenylation
Three observations demonstrate that the poly(A) tail of the EDC1 mRNA is resistant to deadenylation and undergoes slow poly(A) shortening. First, no known deadenylases affect its poly(A) tail length (Figure 2). Second, in dcp2Δ strains where decapping is blocked, the fully adenylated EDC1 mRNA persists for over 20 min with only very slow shortening of the poly(A) tail (Figure 4). Second, mRNA decay intermediates trapped by the insertion of a poly(G) tract contain poly(A) tails that persist over periods of >30 min without any observable deadenylation (Figure 4). These observations suggest that specific information within the EDC1 mRNA not only prevents the poly(A) tail from inhibiting decapping but protects the poly(A) tail from deadenylation. A reasonable hypothesis is that the poly(A) tail is involved in novel molecular interactions that preclude its interaction with both mRNA deadenylases and the poly(A) binding protein.
A poly(U) tract in the EDC1 mRNA's 3′ UTR modulates its function
Two observations demonstrate that a poly(U) tract in the EDC1 3′ UTR is required for both deadenylation-independent decapping and inhibition of deadenylation. First, the EDC1 3′ UTR was sufficient to transfer the novel properties of the poly(A) tail to the MFA2 mRNA (Figure 5). Second, deletion analysis identified a poly(U)-rich region as being required for the function of the EDC1 3′ UTR in promoting deadenylation-independent decapping and inhibiting deadenylation (Figure 5). The poly(U) tract might interact with specific proteins that affect poly(A) tail metabolism, and/or the poly(U) tract might directly base-pair with the poly(A) tail. In the latter case, the poly(A) tail would then be sequestered and essentially nonfunctional.
A Role of Not proteins in affecting decapping: connecting decapping and deadenylation
Our results suggest that the Not proteins can enhance the rate of EDC1 mRNA decapping. First, strains lacking Not2p, Not4p, and Not5p show an increase in the steady-state levels of the EDC1 mRNA (Figure 7A). Second, measurement of decay rates showed that in a not4Δ strain, the increase in EDC1 mRNA levels can be attributed to a slower rate of mRNA degradation (Figure 7B). Because the EDC1 mRNA is not a substrate for deadenylation, these observations argue that the Not proteins affect decapping of the EDC1 transcript. Moreover, when decapping is blocked, Not proteins are concentrated in P-bodies, which are cytoplasmic sites of decapping (Figure 8 and Supplementary Figure 1). This suggests a model wherein Not proteins interact with mRNAs destined for degradation, localize to P-bodies, and then are released from the P-bodies following destruction of the mRNA. A function for the Not proteins in decapping suggests a possible link between deadenylation and decapping. The Not proteins are found in a complex with Ccr4p and Pop2p, which comprise the major cytoplasmic deadenylase (Tucker et al, 2001). Thus, the Not proteins provide a potential connection between the deadenylation and decapping machineries. Consistent with that connection, Dhh1p, which is a stimulator of decapping, also physically interacts with the Ccr4/Pop2/Not complex (Hata et al, 1998; Coller et al, 2001; Maillet and Collart, 2002). Because Not3p does not affect the EDC1 mRNA levels, and individual Not proteins show different effects on the deadenylation of the MFA2 mRNA (Tucker et al, 2002), an intriguing speculation is that individual Not proteins might play a role in modulating mRNA degradation and may differentially affect different mRNAs.
Diverse modes of controlling decapping
An emerging theme in mRNA decapping is the diversity of manners and protein complexes that recruit the decapping machinery to different mRNAs. For example, our results with the EDC1 mRNA suggest that this mRNA undergoes decapping in a manner stimulated by the Not proteins and including the normal regulators of decapping, Lsm1p, Pat1p, and Dhh1p. This is different from the majority of yeast mRNAs, which assemble a decapping complex consisting of the Lsm1 to Lsm7 proteins, Pat1p, Dhh1p, and Dcp1p/Dcp2p (reviewed in Coller and Parker, 2004). It is not yet clear whether the Not proteins are limited to being in the decapping complex on a small subset of mRNAs, or are part of the decapping complexes on many mRNAs, but only affect the decapping rate of some mRNAs because of different rate-limiting steps for different mRNAs in the complex process of decapping and decay. Additional modes of stimulating decapping also exist. For example, physical interactions between Upf1p and Dcp2p suggest that the process of NMD involves the assembly of a decapping complex wherein the decapping enzyme is recruited to the mRNA through interactions with Upf1p (He and Jacobson, 1995; Lykke-Andersen, 2002). Another specialized decapping complex appears to occur on the Rps28b mRNA, where, in an autoregulatory process, the Rps28b protein binds a stem–loop in the 3′ UTR and then interacts with Edc3p, which interacts with Dcp1p/Dcp2p and enhances their function (Badis et al, 2004). This diversity of regulation is only likely to grow and provides numerous opportunities for the differential regulation of decapping rates on individual mRNAs.
Materials and methods
Yeast strains were grown to mid-log in standard media. Half-life measurements, RNA extractions, and RNaseH reactions were performed as previously described (Muhlrad et al, 1994). Serial RNaseH reactions were performed by taking the completed RNaseH reaction with the first oligo and adding an appropriate amount of second oligo, denaturing again for 10 min at 80°C, annealing for 15 min at room temperature, and then adding new RNaseH and incubating at 30°C for 1 h before stoppage of the final reaction. Northerns were run using 25 μg of total RNA on 6%/8 M urea–polyacrylamide gels. Northern blots were probed for EDC1 with oRP1211, oRP140 for MFA2, or oRP100 for the scR1 ribosomal RNA and then viewed and quantitated on a Phosphorimager 9410.
C-terminal NOT1-5GFP fusions were made using PCR and their respective oligo pairs (shown in Table III) on a GFP template (Longtine et al, 1998) and transformed into yeast by standard methods. Correct fusion integrants were screened by PCR and Westerns.
Table 3.
Oligonucleotides used in this study
| oRP no. | Sequence |
|---|---|
| 100 | GTCTAGCCGCGAGGAAGG |
| 140 | ATATTGATTAGATCAGGAATTCC |
| 1208 | TGGCACTGCCATTACAACCCATTGG |
| 1209 | AAATAAAAATATGCATTATTCT |
| 1210 | GCATTCGTATAATTGGTACATAGGTATATATC |
| 1211 | AATTGCTTTGGATGACCAGATCC |
| 1212 | GGATTCAGACTTTAGAAGTGGCGCGCC |
| 1213 | GATTCTCTAGATGCCGATTCGCCATG |
| 1214 | GAGCTAGTACCGCTCCTTAAGCTAG |
| 1215 | GCATTCAGATCTACTCAACAGTGGG |
| 1216 | CCTTATGACGCACTAGGGAATGCTG |
| 1217 | CATGTGGGCATTGAAGCTGGAATAG |
| 1218 | GTGAAAGTAGTGACAAGTGTTGGCCATGGAACAGG |
| 1219 | CACCATCAATAGAAGGCAAACCCCTCTACAATCCA ACGCACGGATCCCCGGGTTAATTAA |
| 1220 | ATACAAGCTGTTCTATAGAACTTCAAGTTCGATTA CTCAAGAATTCGAGCTCGTTTAAAC |
| 1221 | GTGCCAAAGAGCTTTCTTATTGTTTTATAATGCTA TTATGCGGATCCCCGGGTTAATTAA |
| 1222 | AGATGTGCACACTATTTTTCTCAATTAATCATTAA AAAAAGAATTCGAGCTCGTTTAAAC |
| 1223 | GCAGCTCTTGAAACAATTGAAACAGGGAAAAATTA GTGTACGGATCCCCGGGTTAATTAA |
| 1224 | TCGCTATTCAACATGTTTGGTGGTTCGCTAAAGTG TCCCTGAATTCGAGCTCGTTTAAAC |
| 1225 | AAATCAACTAATCAACGGAAGGAAAATTATCGCCG GTAATCGGATCCCCGGGTTAATTAA |
| 1226 | GATGTAGACTGAGTCATTCGGATAAGAATTCCAAC CAACTGAATTCGAGCTCGTTTAAAC |
| 1227 | TAACGACTTCGTATATAATGAAGAAGATTTCGAAA AACTGCGGATCCCCGGGTTAATTAA |
| 1228 | GCTAGCAAGTAATGATGGTGCGGTCTACATCGAGT AAAAAGAATTCGAGCTCGTTTAAAC |
| 1229 | GGAATTCTAGATCACCCTTTAGAGAGACATT |
| 1230 | GGAATTCTAGAAGAATAATGCATATTTTTTATTT |
| 1231 | GGAATTCTAGAGATATATACCTATGTACCAA |
| 1232 | CCAAGTTTTTTATAATCACCCGGGAGAGAGACATT TTTGATTTT |
| 1233 | GCCAGCAACACGTAATAAATGCCCGGGTAGATATT GATTAGATGAG |
| 1234 | GGAATTCAAGCTTGAGAATCGCGAAACAATGG |
| 1235 | CAAAAATGTCTCTCTAAAGGGTGA |
Yeast strains were constructed by standard methods and are listed in Table II.
Table 2.
Yeast strains used in this study
| Strain no. yRP… | Genotype |
|---|---|
| 582 | MAT a, ura3-52, leu2, rpb1-1 |
| 840 | MAT a, leu2, his4, trp1, ura3, cup1∷LEU2PM |
| 841 | MAT alpha, leu2, lys2, trp1 ura3 cup1∷LEU2PM |
| 683 | MAT a, leu2, lys2, his4, ura3, trp1 |
| 684 | MAT alpha, leu2, lys2, his4, ura3, trp1 |
| 926 | MAT alpha, ura3, rpb1-1, leu2, xrn1Δ∷LEU2 |
| 1108 | MAT a, lue2, ura3, trp1, lys2, rpb1-1, dcp1Δ∷URA3 |
| 1195 | MAT a, his4, leu2, trp1, ura3, cup1∷LEU2PM, ski2∷LEU2 |
| 1198 | MAT a, his4, leu2, trp1, ura3, rpb1-1, ski2Δ∷LEU2 |
| 1211 | MAT a, lys2, leu2, trp1, his4, ura3, cup1∷URA3, dcp1Δ∷URA3 |
| 1358 | MAT a, his4, leu2, lys2, trp1, ura3, dcp2Δ∷TRP1 |
| 1359 | MAT alpha, his4, leu2, lys2, ura3, xrn1Δ∷URA3, cup1∷LEU2PM |
| 1372 | MAT a, trp1, ura3, leu2, his4, cup1∷LEU2PM, pat1Δ∷LEU2 |
| 1410 | MAT a, trp1, his4, ura3, leu2, cup1∷LEU2PM, lsm1Δ∷TRP1 |
| 1503 | MAT a, his3, ade2, leu2, lys2, trp1, ura3, edc1Δ∷HIS3, cup1∷LEU2PM |
| 1504 | MAT alpha, his4, leu2, trp1, ura3, edc2Δ∷NEO, cup1∷LEU2PM |
| 1560 | MAT a, his4, leu2, ura3, trp1, lys2, cup1∷LEU2PM, dhh1Δ∷URA3 |
| 1616 | MAT a, trp1, ura3, leu2, his4, cup1∷LEU2PM, ccr4Δ∷NEO |
| 1619 | MAT a, ura3, leu2, his4, cup1∷LEU2PM, pan2Δ∷URA3 |
| 1620 | MAT a, trp1, ura3, leu2, his4, cup1∷LEU2PM, ccr4Δ∷NEO, pan2Δ∷URA3 |
| 1670 | MAT alpha, leu2, lys2, trp1, ura3, cup1∷LEU2PM, not2Δ∷NEO |
| 1671 | MAT alpha, leu2, lys2, trp1, ura3, cup1∷LEU2PM, not3Δ∷NEO |
| 1672 | MAT alpha, leu2 trp1, ura3, lys2, cup1∷LEU2PM, not4Δ∷NEO |
| 1673 | MAT alpha, trp1, leu2, lys2, ura3, cup1∷LEU2PM, not5Δ∷NEO |
| 1798 | MAT a, rpb1-1, his4, ura3, leu2, pat1∷LEU2 |
| 1800 | MAT alpha, rpb1-1, his4, ura3, leu2, cup1∷LEU2PM, ccr4Δ∷NEO |
| 1802 | MAT a, rpb1-1, his4, trp1, ura3, leu2, dhh1Δ∷URA3 |
| 1804 | MAT a, rpb1-1, ura3, leu2, lys2, lsm1Δ∷TRP1, trp1 |
| 1805 | MAT alpha, his4, rpb1-1, lys2, ura3, leu2, dcp2Δ∷TRP1 |
| 1806 | MAT a, trp1, rpb1-1, ura3, not4Δ∷NEO |
| 1807 | MAT a, rpb1-1, his4, ura3, leu2,edc1Δ∷NEO |
| 1808 | MAT alpha, trp1, ura3, leu2, lys2, not4Δ∷NEO, edc1Δ∷NEO |
| 1817 | MAT a, trp1, leu2, ura3, his4, cup1∷LEU2PM, not1-GFPNEO |
| 1818 | MAT alpha, lys2, trp1, ura3, leu2, cup1∷ LEU2PM, not2-GFPNEO |
| 1819 | MAT a, trp1, leu2, ura3, his4, cup1∷LEU2PM, not3-GFPNEO |
| 1820 | MAT alpha, lys2, leu2, trp1, ura3, cup1∷ LEU2PM, not4-GFP-NEO |
| 1821 | MAT a, trp1, leu2, ura3, is4, cup1∷LEU2PM, not5-GFPNEO |
| 1831 | MAT a, rpb1-1, leu2, his4, trp1, ura3, lys2, upf1Δ∷URA3 |
| 1832 | MAT a, rpb1-1, leu2, lys2, his4, ura3, cup1∷LEU2PM, upf2Δ∷NEO |
| 1833 | MAT a, rpb1-1, leu2, ura3, cup1∷LEU2PM, upf3Δ∷NEO |
| 1915 | MAT a, leu2, cup1∷LEU2PM, lys2, trp1, ura3, dcp1∷URA3, not1-GFPNEO |
| 1916 | MAT alpha, leu2, cup1∷LEU2PM, trp1, his4, ura3, dcp1Δ∷URA3, not2-GFPNEO |
| 1917 | MAT a, lys2, trp1, ura3, dcp1∷URA3, leu2, cup1∷LEU2PM, not3-GFPNEO |
| 1918 | MAT a, lys2, leu2, cup1∷LEU2PM, trp1, ura3, dcp1∷URA3, not4-GFPNEO |
| 1919 | MAT alpha, trp1, lue2, cup1∷LEU2PM, ura3, dcp1∷URA3, not5-GFPNEO |
| 1920 | MAT alpha, leu2, ura3, trp1, lys2, pop2∷NEO, cup1∷LEU2PM |
Oligonucleotides used in this study are listed in Table III.
Plasmids were constructed by standard methods and consist of the following: pRP1189: expressing theEDC1 mRNA under GAL control on a CEN, TRP1, URA3 backbone.
pRP1173: expressing the EDC1pG mRNA under GAL control. The EDC1 gene was placed behind the GAL1 UAS and contains the complete coding sequence of EDC1. A poly(G) tract was added six bases after the stop codon and the included sequence extends through the 3′ end of the EDC1 mRNA. The entire construct is on a CEN URA3 plasmid backbone.
pRP1168: MFA2pGE—fusion of the EDC1 3′ UTR six bases after the stop codon at an artificially created SmaI site in EDC1 to an EcoRI filled site immediately after the poly(G) sequence in the 3′ UTR of the MFA2pG plasmid (pRP485). The entire construct is on a CEN URA3 plasmid.
pRP1169–1172: MFA2pGE and deletion constructs. The 3′ UTR sequence of EDC1 was amplified by PCR with oligos 1229, 1230, 1231, and 1232 to give either the full-length (WT), Δ50, Δ72, or ΔU plasmids 1169, 1170, 1171, or 1172, respectively. PCR products were cloned into XbaI and HindIII sites created from an XbaI insertion in pRP1168. Thus these plasmids contain the entire MFA2 coding region and part of its 3′ UTR, a poly(G) tract, and then the EDC1 mRNA 3′ end with or without the deletion sequences. All plasmids are on URA3 CEN backbones.
pRP1166: DCP2RFPT-TRP—this TRP1 CEN plasmid contains the promoter region through amino acid 300 of DCP2 from plasmid pRP1152 (a URA3 CEN plasmid), fused in-frame to the RFP gene and then to the PGK1 terminator and 3′ end of plasmid pRP1084 (Sheth and Parker, 2003). The DCP2RFP plasmid complements the RNA phenotype of a dcp2Δ strain.
Supplementary Material
Supplementary Figure 1
Acknowledgments
We thank Sam Ward's lab for use of their microscope and members of the Parker lab for intellectual contributions. This work was funded by the Howard Hughes Medical Institute.
References
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A (2004) Targeted mRNA degradation by deadenylation-independent decapping. Mol Cell 15: 5–15 [DOI] [PubMed] [Google Scholar]
- Beelman CA, Stevens A, Caponigro G, LaGrandeur TE, Hatfield L, Fortner DM, Parker R (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646 [DOI] [PubMed] [Google Scholar]
- Bonnerot C, Boeck R, Lapeyre B (2000) The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol Cell Biol 20: 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B (2000) A Sm-like protein complex that participates in mRNA degradation. EMBO J 19: 1661–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Sachs AB (1998) Poly (A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol 18: 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R (2001) Computational modeling of eukaryotic mRNA turnover. RNA 7: 1192–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Parker R (2003) Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113: 533–545 [DOI] [PubMed] [Google Scholar]
- Caponigro G, Parker R (1995) Multiple functions of the Poly (A) binding protein in mRNA decapping and deadenylation. Genes Dev 9: 2421–2432 [DOI] [PubMed] [Google Scholar]
- Collart MA, Timmers HT (2004) The eukaryotic Ccr4–not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 77: 289–322 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R (2004) Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R (2001) The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B (2004) Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Denis CL, Chen J (2003) The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Dunckley T, Tucker M, Parker R (2001) Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Weis K (2002) The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J 21: 2788–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Jacobson A (1995) Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 9: 437–454 [DOI] [PubMed] [Google Scholar]
- Hu WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 671–672 [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T (2002) The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar M, Parker R (2004) Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J (2002) Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Collart MA (2002) Interaction between Not1p, a component of the Ccr4–not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem 277: 2835–2842 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker C, Parker R (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ to 3′ degradation of the transcript. Genes Dev 8: 855–866 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R (1994) Premature translational termination triggers mRNA decapping. Nature 370: 578–581 [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R (1999) Recognition of yeast mRNAs as ‘nonsense containing' leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol Biol Cell 10: 3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas W, Parker R (2000) The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J 19: 6602–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Ramirez CV, Vilela C, Berthelot K, McCarthy JE (2002) Modulation of eukaryotic mRNA stability via the cap-binding tranlation complex eIF4F. J Mol Biol 318: 951–962 [DOI] [PubMed] [Google Scholar]
- Schwartz D, Decker CJ, Parker R (2003) The Enhancer of Decapping Proteins, Edc1p and Edc2p, bind RNA and stimulate activity of the decapping enzyme. RNA 9: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Parker R (1999) Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of yeast mRNAs. Mol Cell Biol 19: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Parker R (2000) mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R (2003) Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518 [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R (2002) Ccr4p is the catalytic sub-unit of a Ccr4/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C, Velasco C, Ptushkina M, McCarthy JE (2000) The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J 19: 4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


