ABSTRACT
A novel β-(1,3)-glucanase gene designated lamC, cloned from Corallococcus sp. strain EGB, contains a fascin-like module and a glycoside hydrolase family 16 (GH16) catalytic module. LamC displays broad hydrolytic activity toward various polysaccharides. Analysis of the hydrolytic products revealed that LamC is an exo-acting enzyme on β-(1,3)(1,3)- and β-(1,6)-linked glucan substrates and an endo-acting enzyme on β-(1,4)-linked glucan and xylan substrates. Site-directed mutagenesis of conserved catalytic Glu residues (E304A and E309A) demonstrated that these activities were derived from the same active site. Excision of the fascin-like module resulted in decreased activity toward β-(1,3)(1,3)-linked glucans. The carbohydrate-binding assay showed that the fascin-like module was a novel β-(1,3)-linked glucan-binding module. The functional characterization of the fascin-like module and catalytic module will help us better understand these enzymes and modules.
IMPORTANCE In this report of a bacterial β-(1,3)(1,3)-glucanase containing a fascin-like module, we reveal the β-(1,3)(1,3)-glucan-binding function of the fascin-like module present in the N terminus of LamC. LamC displays exo-β-(1,3)/(1,6)-glucanase and endo-β-(1,4)-glucanase/xylanase activities with a single catalytic domain. Thus, LamC was identified as a novel member of the GH16 family.
KEYWORDS: β-(1,3)-glucanase; Corallococcus sp. EGB; fascin-like module; GH16; broad substrate linkage specificity
INTRODUCTION
The enzymes known as β-(1,3)-glucanases, which are classified as endo-(1,3)-β-glucanases (EC 3.2.1.39) and exo-(1,3)-β-glucanases (EC 3.2.1.58), are widely distributed among higher plants, fungi, and bacteria. β-(1,3)-Glucanases catalyze the hydrolysis of β-(1,3)-glycosidic bonds in β-(1,3)-glucan, which is the main cell wall component in yeast and filamentous fungi and a structural polysaccharide (e.g., callose) in plants and is also found in exopolysaccharides produced by some bacteria (1). Based on their amino acid sequence similarity and secondary structure, β-(1,3)-glucanases are classified mainly into glycoside hydrolase family 16 (GH16) and GH17. However, these two families have the same hydrolytic mechanism with anomeric retention (2).
Numerous genes encoding β-(1,3)-glucanase have been cloned and characterized from different sources, including varieties of plants (3–5), bacteria, and archaea, such as Bacillus circulans (6), Paenibacillus (7, 8), Thermotoga neapolitana (1), Rhodothermus marinus (9), and Pyrococcus furiosus (10). Few glucanases exhibit broad substrate linkage specificity, although Lafond et al. cloned a gene from Podospora anserina that encodes a broad-specificity β-glucanase acting on β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans (11).
Many polysaccharide-degrading enzymes display a modular structure, in which a catalytic module is attached to one or more noncatalytic modules (8, 12, 13). The impact of the noncatalytic modules on the enzymatic properties of β-(1,3)-glucanase has been studied. Cheng et al. reported that the carbohydrate-binding module (CBM) repeats and Fa5/8C analogue could enhance the LamA hydrolytic activity of the catalytic module (8). Hong et al. also reported that the C-terminal CBM6 of a β-(1,3)-glucanase (Curd1) from Streptomyces sioyaensis enhanced the hydrolytic activity of the catalytic module against insoluble substrates (14). Members with a fascin-like module, including actin-bundling/cross-linking fascin proteins, have been identified in rice (Oryza sativa L.), the Hawaiian sea urchin (Tripneustes gratilla), the fruit fly (Drosophila), the South African clawed frog (Xenopus), mice, and humans (15–20). In eukaryotes, fascin is a 55-kDa actin-bundling protein with four homologous β-trefoil domains. The ability of fascin to bind and bundle filamentous actin plays a central role in the regulation of cell adhesion, migration, and invasion (21–23). The fascin-like module found in the glycoside hydrolases is not common in bacteria and has not been functionally characterized to date.
In the present study, we cloned a putative GH16 β-(1,3)-glucanase containing a fascin-like module near its N terminus from Corallococcus sp. strain EGB. Analysis of the sequence and the enzyme properties and kinetics revealed that the β-(1,3)-glucanase (LamC) is a novel GH16 member with broad substrate linkage specificity toward β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans and xylan. We also verified the function of the fascin-like module found in LamC.
RESULTS
Cloning of the β-(1,3)-glucanase gene and sequence analysis.
This specific β-(1,3)-glucanase (designated LamC, a laminarinase from Corallococcus sp. EGB) contains 438 amino acid residues and has a calculated pI of 5.26 and molecular mass of 47,040 Da. LamC contains several putative modules, including a predicted N-terminal signal peptide (residues 1 to 26), a fascin-like module (residues 56 to 182), and a β-(1,3)-glucanase catalytic module (residues 196 to 438) (Fig. 1a).
FIG 1.
Schematic overview of LamC and its derivatives. (a) Organization of the functional units of LamC and the module composition of the derivative proteins expressed in this study. SP, signal peptide; rLamC, a mature protein with a deletion in the signal peptide; rLamC-ΔN, a truncated protein with a deletion in the N-terminal fascin-like domain; rLamC-ΔC, a truncated derivative with a deletion of the C-terminal GH16 catalytic module. Every derivative protein has a Trx tag and a His6 tag fused to the N terminus. The ruler on the top represents the amino acid residue numbering. (b) The purity of the derivative LamC proteins is shown on a 12% SDS-PAGE gel. Approximately 1.5 μg protein was loaded in each lane. Lanes: M, protein molecular mass markers; 1, purified Trx-tag protein as a control; 2, purified rLamC-ΔC; 3, purified rLamC-ΔN; 4, purified rLamC.
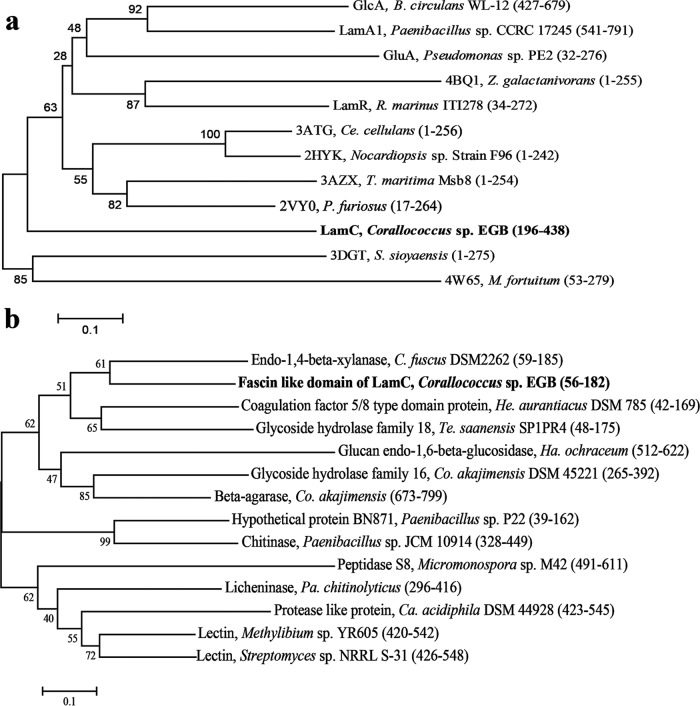
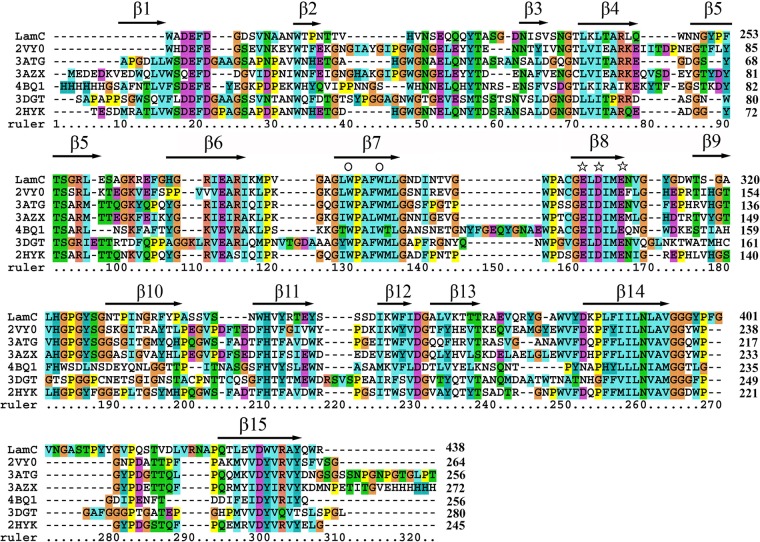
The BLASTP analysis showed that LamC shares the highest identity (92%) with the β-(1,3)-glucanase A1 in the genome of Corallococcus coralloides DSM 2259 (24), followed by the endo-β-(1,4)-xylanase from Cystobacter fuscus (62%) and the glycoside hydrolase family 16 protein from Herpetosiphon aurantiacus DSM785 (52%) (25). However, none of these proteins have been characterized. Among proteins with experimentally determined three-dimensional (3D) structures, the GH16 catalytic module of LamC (residues 196 to 438) showed the highest identity (38%) with the corresponding domain of the well-characterized endo-β-(1,3)-glucanase pfLamA (PDB 2VY0) (residues 17 to 264) from the hyperthermophilic archaeon P. furiosus (10), followed by 30.9% identity with the laminarinase TmLamCD (PDB 3AZX) (residues 1 to 254) from T. maritima MSB8 (26) and 26.2% identity with the endo-β-(1,3)-glucanase (PDB 3DGT) (residues 1 to 275) from S. sioyaensis (27) (Fig. 2a). Multiple alignments of the deduced GH16 domain amino acid sequences of LamC and other β-glucanases showed many conserved amino acids (Fig. 3). The amino acid sequence analysis indicated that LamC contained a single catalytic domain with the two catalytic residues (Glu304 and Glu309) that are highly conserved among GH16 members (10, 28).
FIG 2.
Sequence analysis of LamC. Phylogenetic analysis of the GH16 (a) and fascin-like modules (b) from different source proteins. The starting and ending amino acid positions are shown in brackets. The phylogenetic tree was constructed by the neighbor-joining algorithm based on the amino acid sequence alignment in MEGA7. The GH16 domain sequence of LamC was aligned with the following proteins: β-(1,3)-glucanase A1 (GlcA) from B. circulans (accession no. P23903), LamA from Paenibacillus sp. CCRC 17245 (accession no. ABJ15796), GluA from Pseudomonas sp. PE2 (accession no. BAC16331), beta-glucanase ZgLamA from Zobellia galactanivorans (PDB no. 4BQ1), laminarinase LamR from R. marinus (GenBank accession no. AAC69707), endo-β-(1,3)-glucanase from Cellulosimicrobium cellulans (PDB no. 3ATG), endo-β-(1,3)-glucanase from the alkaliphilic Nocardiopsis sp. strain F96 (PDB no. 2HYK), laminarinase from T. maritima Msb8 (PDB no. 3AZX), pfLamA from the hyperthermophilic archaeon P. furiosus (PDB no. 2VY0), endo-β-(1,3)-glucanase from S. sioyaensis (PDB no. 3DGT), and the glycoside hydrolase family protein from Mycobacterium fortuitum (PDB no. 4W65). The fascin-like module sequence of LamC was aligned with the following organisms and GenBank accession numbers: C. fuscus DSM 2262, EPX62475.1; H. aurantiacus DSM 785, ABX04162.1; Terriglobus saanensis SP1PR4, ADV82022.1; Paenibacillus sp. P22, CDN41972.1; Coraliomargarita akajimensis DSM 45221, ADE54011.1 and ADE54017.1; Paenibacillus chitinolyticus, WP_042225814.1; Catenulispora acidiphila DSM 44928, ACU70919.1; Haliangium ochraceum DSM 14365, ACY18721.1; Paenibacillus sp. JCM 10914, GAE07991.1; Micromonospora sp. M42, EWM66821.1; Methylibium sp. YR605, WP_047594263.1; and Streptomyces sp. NRRL S-31, WP_030753699.1.
FIG 3.
Alignment of the GH16 domain sequence of the GH16 family β-glucanases. The proteins (and PDB numbers) were as follows: LamC from Corallococcus sp. EGB; PDB no. 3ATG, endo-β-(1,3)-glucanase from Cellulosimicrobium cellulans; PDB no. 2HYK, endo-β-(1,3)-glucanase of from Nocardiopsis sp. strain F96; PDB no. 4BQ1, ZgLamA from Z. galactanivorans; PDB no. 3AZX, laminarinase from T. maritima Msb8; PDB no. 2VY0, pfLamA from P. furiosus; and PDB no. 3DGT, endo-β-(1,3)-glucanase from S. sioyaensis. The protein sequence alignments were generated using the MUSCLE alignment in MEGA 7.0 (38). Secondary structures are labeled based on their appearance in 2HYK following a previous annotation (28). The stars identify the catalytic amino acids, including Glu-304, Asp-306, and Glu-309 (LamC numbering), and the conserved Trp residues are marked by the symbol ○ above the column. The numbers on the right are the amino acid residue positions in the whole sequence. The colors highlight other identities between sequences.
BLAST searches in GenBank revealed that fascin-like modules are found in some bacterial genomes, but the functions of their corresponding proteins remain uncharacterized (Fig. 2b). Phylogenetic analysis based on sequence alignment of the fascin-like module indicated relationships between LamC and other proteins containing a fascin-like module. The deduced amino acid sequence of the LamC fascin-like module (residues 56 to 182) shared the highest identity (62%) with the homologous domain of endo-β-(1,4)-xylanase A precursor (residues 59 to 185) from C. fuscus DSM 2262, 41% identity with the lectin (residues 426 to 548) from Streptomyces sp. NRRL S-31, and 30% identity with the peptidase S8A (residues 491 to 611) from Micromonospora sp. M42.
Expression and purification of LamC derivatives.
Initial attempts to express LamC derivatives in Escherichia coli were hampered by the formation of inclusion bodies. This problem was overcome by addition of a Trx tag to the LamC derivatives. To characterize the function of the catalytic and noncatalytic modules, all LamC derivatives were Trx and His6 tagged on the N terminus and successfully expressed in redox-deficient E. coli Origami B(DE3) (Fig. 1a). The calculated molecular masses of the three expressed fusion proteins (rLamC-ΔC, rLamC-ΔN, and rLamC) are approximately 32.5, 47.0, and 67.8 kDa, respectively. Purification of the LamC derivatives was achieved by immobilized-metal affinity chromatography (IMAC) and analyzed by SDS-PAGE (Fig. 1b).
Effects of temperature and pH on enzymatic activity and stability.
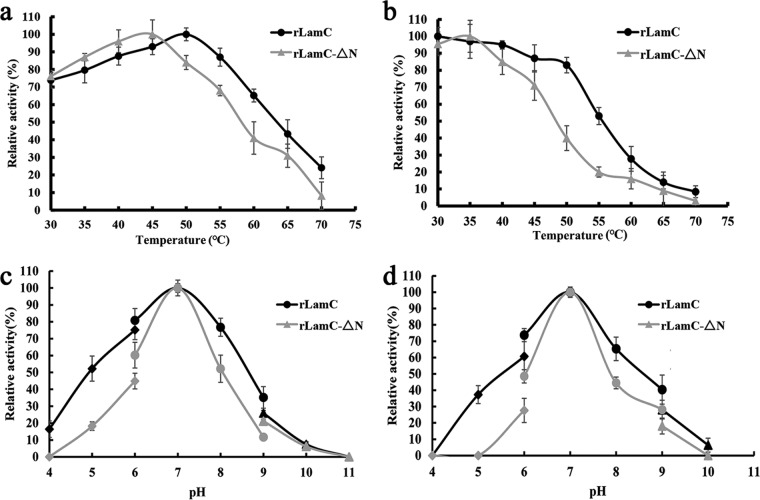
The enzymatic properties of rLamC and rLamC-ΔN were compared. The general properties of the LamC derivatives, including the optimal pH and temperature, were investigated against laminarin as the substrate. The optimal activity of rLamC was observed to be at 50°C and that of rLamC-ΔN at 45°C (Fig. 4a). Comparison of the thermal stability of rLamC and rLamC-ΔN showed that the rLamC containing the fascin-like module was more stable. rLamC retained more than 80% of its original activity after 30 min of incubation at 50°C, whereas rLamC-ΔN retained <40% of its original activity under the same conditions (Fig. 4b).
FIG 4.
Effects of temperature and pH on the activity and stability of rLamC and rLamC-ΔN. (a) To determine the optimal temperature, the recombinant enzymes were incubated in phosphate buffer (20 mM; pH 7.0) with 10 mg/ml laminarin substrate for 20 min at various temperatures (30 to 70°C). (b) Thermostability of rLamC and rLamC-ΔN. The residual activity was measured under optimal conditions after incubation of the enzyme at the indicated temperatures (30 to 70°C) for 30 min. (c) To determine the optimal pH, the recombinant enzymes were incubated with 10 mg/ml laminarin substrate for 20 min at 50°C in buffers of various pH values (4.0 to 10.0). (d) Stability of rLamC and rLamC-ΔN at different pH values. The residual enzyme activity was measured under optimal conditions after incubation of the purified enzyme with buffers with various pH values at 4°C for 24 h.
Deletion of the fascin-like module did not markedly affect the optimal pH. The optimal working pH toward laminarin was approximately 7.0 for both rLamC and rLamC-ΔN proteins (Fig. 4c). rLamC retained higher activity in the pH spectrum ranging from 4.0 to 10, whereas rLamC-ΔN retained <50% of its original activity at pH values outside its optimal pH (Fig. 4d). This finding is consistent with the result of the thermostability analysis, suggesting that the fascin-like module enhanced the enzymatic stability of LamC.
Kinetic properties of LamC.
Using polysaccharide substrates with various linkages, we characterized the enzymatic activities of rLamC and rLamC-ΔN (Table 1). Purified rLamC and rLamC-ΔN shared similar substrate spectra. They released soluble sugars from a wide range of β-linked polysaccharide substrates. No hydrolysis of amylose or α-dextran was observed. These data confirmed that LamC prefers β-(1,3)-linked glucans, and laminarin was the most favorable substrate for rLamC. The fascin-like module enhanced the hydrolytic activity toward β-(1,3)-glucan substrates. Notably, the enhancement was most pronounced for laminarin and pachyman.
TABLE 1.
Substrate specificity of rLamC and rLamC-ΔN
| Substratea | Solubility | Main linkage | Sp actb (U/nmol of protein) of: |
|
|---|---|---|---|---|
| rLamC | rLamC-ΔN | |||
| Laminarin | Soluble | β-1,3, β-1,6 (Glc) | 0.76 ± 0.04 | 0.11 ± 0.03 |
| Pachyman | Insoluble | β-1,3 (Glc) | 0.71 ± 0.08 | 0.057 ± 0.01 |
| Zymosan A | Insoluble | β-1,3, β-1,6 (Glc) | 0.59 ± 0.02 | 0.13 ± 0.01 |
| Salecan | Soluble | β-1,3, α-1,3 (Glc) | 0.22 ± 0.02 | 0.12 ± 0.01 |
| Curdlan | Insoluble | β-1,3 (Glc) | 0.14 ± 0.01 | 0.09 ± 0.01 |
| CMC | Soluble | β-1,4 (Glc) | 0.30 ± 0.04 | 0.22 ± 0.03 |
| Xylan | Insoluble | β-1,4 (Xyl) | 0.42 ± 0.02 | 0.27 ± 0.04 |
| Pustulan | Insoluble | β-1,6 (Glc) | 0.13 ± 0.01 | 0.08 ± 0.01 |
| α-Dextran | Soluble | α-1,6 (Glc) | NDc | ND |
| Amylose | Insoluble | α-1,4 (Glc) | ND | ND |
All substrates were used at a final concentration of 10 mg/ml, except salecan (5 mg/ml).
Activity ± standard error for triplicate samples.
ND, no activity detected.
The kinetic parameters of rLamC and rLamC-ΔN for laminarin hydrolysis were determined against laminarin. The Km values of rLamC and rLamC-ΔN were 1.4 and 2.6 mg/ml, respectively (Table 2), and rLamC showed higher Vmax values than rLamC-ΔN (Table 2), suggesting that deletion of the fascin-like module resulted in weaker binding to laminarin.
TABLE 2.
Kinetic constantsa of rLamC and rLamC-ΔN toward laminarin
| Enzyme | Km (mg/ml) | Vmax (U/nmol of protein) |
|---|---|---|
| rLamC | 1.4 ± 0.3 | 0.9 ± 0.1 |
| rLamC-ΔN | 2.6 ± 0.5 | 0.4 ± 0.1 |
Values are means ± standard errors.
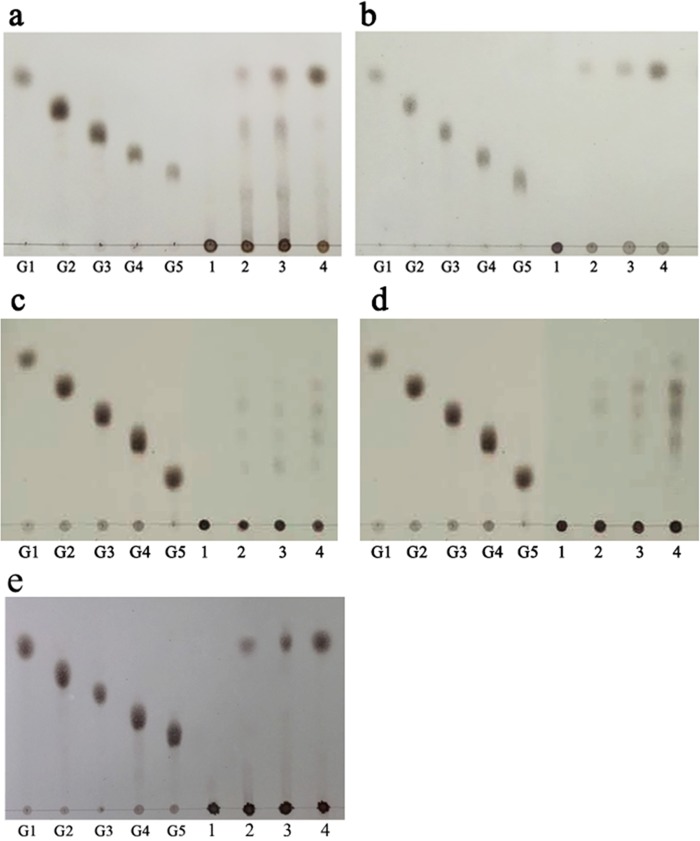
To evaluate the mode of action of LamC, hydrolytic products of polysaccharides (laminarin, pustulan, xylan, and carboxymethyl cellulose-sodium salt [CMC]) with different bond linkages were analyzed by thin-layer chromatography (TLC). Hydrolysis of β-(1,3)-glucan (laminarin and pachyman) or β-(1,6)-glucan (pustulan) yielded glucose as the major end product (Fig. 5a, b, and e). However, small amounts of cellotriose, cellotetraose, and cellopentaose were produced after 30 min of incubation with CMC substrate. When β-(1,4)-xylan was used as a substrate, oligosaccharides with degree of polymerization (DP) values of 2 and 3 were produced at the same time. The final products of CMC and xylan were a mixture of oligosaccharides with DP values of 1 to 4 at the end of the reaction (Fig. 5c and d). These results suggested that LamC contains exo-β-(1,3)/(1,6)-glucanase and endo-β-(1,4)-glucanase/β-(1,4)-xylanase activities. Similar modes of action were observed for a β-glucanase isolated from P. anserina (11).
FIG 5.
TLC analysis of the digestion products of rLamC with various different types of polysaccharide linkages for various time intervals. G1 to G5 show standard maltooligosaccharides. rLamC was incubated with 10 mg/ml laminarin (a), pustulan (b), CMC (c), xylan (d), and pachyman (e) at 30°C for 0 min, 30 min, 2 h, and 12 h (lanes 1 to 4, respectively). G1, glucose; G2, maltose; G3, maltotriose; G4, maltotetraose; G5, maltopentaose.
Mutation of the conserved residues of LamC.
Two catalytic glutamic acid residues (E304 and E309) conserved in the GH16 β-(1,3)-glucanase were simultaneously replaced with alanine in rLamC-Mutant. The recombinant rLamC and rLamC-Mutant were purified from E. coli by Ni2+-nitrilotriacetic acid (NTA) affinity chromatography. No β-xylanase or β-(1,3)-, β-(1,4)-, or β-(1,6)-glucanase activity was detected after 20 min of incubation at 50°C by the dinitrosalicylic acid (DNS) method (data not shown) even after an extended incubation time, indicating that the activities of LamC likely involve the same active site.
Function of the fascin-like module.
rLamC showed greater catalytic activities, particularly for the β-(1,3)-linked glucans laminarin, pachyman, and zymosan A, than did rLamC-ΔN (Table 1). To better understand the function of the noncatalytic fascin-like module of LamC, two approaches were taken to explore its potential polysaccharide-binding function. In the pulldown assay, the purified rLamC-ΔC and rLamC were incubated with the insoluble β-(1,3)-linked glucans pachyman, zymosan A, and α-1,4-linked amylose at 4°C for 1 h. The amounts of protein remaining in the supernatant (S) and coprecipitating with the substrate (P) were examined by SDS-PAGE (Fig. 6). The amount of protein in the pellet is an index of the binding affinity of the protein to the insoluble polysaccharides. As shown in Fig. 6, both rLamC-ΔC and rLamC had the ability to bind pachyman and zymosan A to various extents compared with bovine serum albumin (BSA), but they could not bind to amylose.
FIG 6.
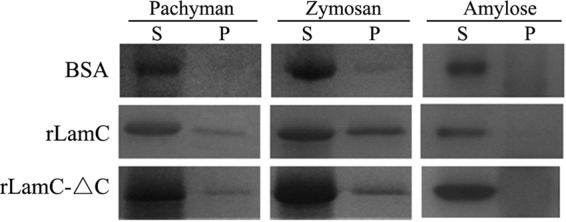
Binding assay of proteins toward insoluble polysaccharides (pachyman, zymosan A, and amylose). The purified rLamC-ΔC or rLamC (10 μg) and the indicated substrate (2 mg) were thoroughly mixed and incubated at 4°C for 1 h. The amounts of protein remaining in the supernatant (S) and coprecipitating with the substrate (P) were examined by SDS-PAGE. BSA and amylose were used as controls.
In the second approach, the ability to bind soluble polysaccharides was assessed by gel affinity electrophoresis. Under the assay conditions, laminarin slowed the migration rates of both rLamC-ΔC and rLamC (Fig. 7a and b). In contrast, CMC only retarded the mobility of rLamC and had no effect on rLamC-ΔC (Fig. 7a and c). The presence of soluble polysaccharides did not affect the mobility of BSA and Trx tag protein, excluding the nonspecific binding of soluble polysaccharides to the proteins. These results indicated that the fascin-like module could bind to laminarin but not to CMC. The retardation of the mobility of rLamC by CMC is caused by the interaction of the catalytic domain with CMC.
FIG 7.
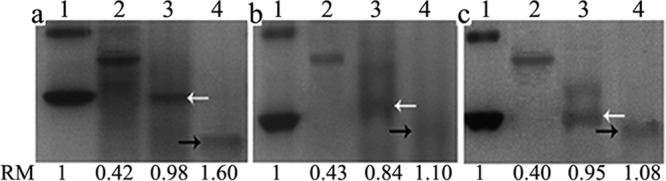
Retardation of protein mobility in native polyacrylamide gels with embedded soluble polysaccharides. The indicated proteins were separated by native 12% PAGE without polysaccharide (a), native PAGE including 0.2% (wt/vol) laminarin (b), and native PAGE including 0.2% (wt/vol) CMC (c) in the separation gel. The relative mobility (RM) of each protein compared with BSA and Trx tag protein under the given conditions is indicated. Lanes: 1, samples (5 μg) of BSA; 2, purified Trx tag protein, used as a control; 3, rLamC-ΔC; 4, purified rLamC. White and black arrows indicate the migration positions of rLamC-ΔC and rLamC, respectively.
The actin binding activity of the fascin-like module from LamC (rLamC-ΔC) was assessed at high-speed (224,000 × g) cosedimentation with F-actin as described previously by Jansen et al. (29). After centrifugation for 30 min, the amounts of protein remaining in the supernatant and pellet were analyzed using 12% SDS-PAGE (see Fig. S1 in the supplemental material). rLamC-ΔC was still present in the supernatant after centrifugation (lane 2), whereas filamentous actin (F-actin) was almost entirely in the pellet (lane 6). Note that there was no rLamC-ΔC in the pellet either when run with F-actin (lane 9) or in the absence of F-actin (lane 3). Those results indicated that the fascin-like module of LamC does not bind to F-actin.
DISCUSSION
This study identified a novel β-(1,3)-glucanase (LamC) comprising 438 amino acid residues from Corallococcus sp. EGB. In addition to a GH16 catalytic module, this β-(1,3)-glucanase also contained a fascin-like module (amino acids 56 to 182) located at its N terminus. The domain organization of LamC is similar to that of GH5BG (a β-glucosidase cloned from rice [Oryza sativa L.] seedlings), which is composed of a 19-amino-acid prepeptide, a fascin-like module, and a β-(1,3)-glucosidase module (20). Successful expression of three LamC derivatives in E. coli allowed biochemical functional characterization of this novel protein.
Modules of GH16 are present in bacterial β-(1,3)- and β-(1,3-1,4)-glucanases. However, LamC showed unusually broad specificity. The catalytic module of LamC catalyzed the hydrolysis of β-(1,3)- and β-(1,6)-linked glucans in an exo-mode action and that of β-(1,4)-linked glucan and xylan by endo-mode action. Few glycoside hydrolases with similar activity have been reported. Lafond et al. reported a broad-specificity GH131 β-glucanase, PaGluc131A, that shared similar action modes on substrates [i.e., exo-mode on β-(1,3)/(1,6)-glucan and endo-mode on β-(1,4)-glucan] (11). However, PaGluc131A could not hydrolyze xylan, although it showed weak activity on pNP-β-d-xylopyranoside (0.03%). Shi et al. characterized a bifunctional enzyme (XynBE18) that had xylanase-glucanase activity with only one catalytic domain (30). Its catalytic mechanism was explained as XynBE18 having a larger substrate-binding cleft that allowed the binding of larger substrates, such as barley β-glucan and lichenin. Further, they stated that for endo-acting glycoside hydrolases with a triosephosphate isomerase (TIM)-like barrel structure, such as endoglucanases, xylanases, and β-(1,3-1,4)-glucanases, a larger cleft or groove allows the binding of several sugar units and permits the enzyme to hydrolyze a variety of substrates. This model may also explain the broad specificity of LamC, but this possibility requires further structural analysis of the LamC-substrate complex.
A fascin-like module has been identified near the N terminus of a GH5 β-glucosidase (GH5BG) from rice (Oryza sativa L.), but the function of the fascin-like module has not been described (20). Little is known about the role of the fascin-like module in glycoside hydrolases. Therefore, we sought to determine whether the fascin-like module could assist LamC in binding hydrolyzable polysaccharides. The binding assays suggested that the fascin-like module can bind laminarin and insoluble pachyman and zymosan A (Fig. 6 and 7) and that its presence enhanced the hydrolytic activity toward those substrates (Table 1) and the enzymatic stability of the catalytic module (Fig. 4).
Fascin is the main actin filament bundling protein in protrusive cellular structures such as filopodia and dendrites, and it contains four β-trefoil domains (23). Jansen et al. concluded that fascin contains two major actin-binding sites for bundling actin filaments, coinciding with regions of high sequence conservation in β-trefoil domains 1 and 3, based on a high-resolution crystal structure (29). Molecules of glycerol and polyethylene glycol could also be bound in pockets and clefts located within the two major actin-binding sites, while the macroketone binding site is located in β-trefoil domain 4 (29). Fascin was recently found to interact with β-catenin (31) and Rab35 (32), but the binding sites on fascin remain unknown. These results suggested that each module of fascin has a different role in binding different molecules, including proteins and other organic compounds. Liu et al. also reported that each gelsolin-like domain of Caenorhabditis elegans gelsolin, another actin binding protein, plays distinct roles in actin filament binding, severing, and capping, although amino acid sequences of the four gelsolin-like domains are highly homologous (33). The lack of cosedimentation of rLamC-ΔC and F-actin indicated that the fascin-like module of LamC could not bind to F-actin (Fig. S1 in the supplemental material). We presumed that the fascin-like module of LamC is more similar to β-trefoil domain 2 or β-trefoil domain 4 than to the two actin-binding sites (β-trefoil domains 1 and 3). Thus, it is reasonable that the fascin-like module of LamC binds polysaccharides rather than F-actin.
In conclusion, we identified and characterized a novel β-(1,3)-glucanase (LamC) from Corallococcus sp. EGB; this enzyme shows exo-mode activity toward β-(1,3)- and β-(1,6)-linked glucans and endo-mode activity on a β-(1,4)-linked glucan and xylan within a single catalytic domain. The fascin-like module of LamC is identified as a novel β-(1,3)-linked glucan-binding module.
MATERIALS AND METHODS
Strains, media, plasmids, and chemicals.
Corallococcus sp. EGB (CCTCC no. M2012528) was cultivated in CTT medium (pH 7.6), consisting of 1% (wt/vol) Casitone, 8 mM MgSO4, 10 mM Tris-HCl, and 1 mM potassium phosphate. Escherichia coli DH5α (Invitrogen, Carlsbad, CA, USA) and the plasmid pMD19-T (TaKaRa, Otsu, Japan) were used for gene cloning. The host strain E. coli Origami B(DE3) and pET32a(+) (Novagen, Damstad, Germany) were used for expression of LamC derivatives. E. coli Origami B(DE3) was cultivated in Luria-Bertani (LB) broth or on agar plates containing 15 μg/ml kanamycin, 12.5 μg/ml tetracycline, and 50 μg/ml ampicillin for protein expression. The His6-tagged protein was purified by immobilized-metal affinity chromatography (IMAC) using an Ni2+-nitrilotriacetic acid (NTA) resin (Qiagen, Hilden, Germany). The DNA purification kit, isopropyl-β-d-thiogalactopyranoside (IPTG), restriction endonucleases, T4 DNA ligase, PrimeSTAR HS DNA polymerase, and LA Taq DNA polymerase with GC buffer and deoxynucleoside triphosphates (dNTPs) were purchased from TaKaRa.
Laminarin (from Laminaria digitata), barley β-glucan, zymosan A, birchwood xylan, CMC, and α-dextran were purchased from Sigma-Aldrich (St. Louis, MO, USA). Pachyman was purified from commercial fruiting bodies of the basidiomycete Poria cocos (34). Salecan, which is a novel soluble β-(1,3)-glucan, was prepared from Agrobacterium sp. ZX09 as described by Xiu et al. (35). Filamentous actin (F-actin; Cytoskeleton, Denver, CO, USA) was used for analyzing the binding activity of the fascin-like module with F-actin.
Molecular cloning of the β-(1,3)-glucanase-encoding gene.
Genomic DNA was extracted from Corallococcus sp. EGB cells using the method described by Kaiser et al. (36). In accordance with a putative β-(1,3)-glucanase A1 (GenBank accession no. AFE08907) sequence from C. coralloides DSM 2259 (24), the full-length β-(1,3)-glucanase gene was PCR amplified from the chromosomal DNA of Corallococcus sp. EGB with the F1 and R1 primers (Table 3). The PCR amplification was performed with 32 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 80 s, followed by a 10-min extension at 72°C. The amplified PCR products were purified, ligated into the pMD19-T simple vector, and sequenced by Invitrogen Corporation.
TABLE 3.
Oligonucleotide primers used in the PCR to amplify the desired DNA fragments
| Amplified fragment | Primer sequence (5′→3′)a |
|---|---|
| LamC | F1, ATGGCGACGAGAGCCGTGAAGG |
| R1, CTAGCGCCACTGGTAGGCG | |
| rLamC-ΔC | F2, CCGGAATTCCTGAAGGCGTGCGCAACGCAG |
| R2, CCGCTCGAGCTAGAAGGAGAACGCCTCCCAGC | |
| rLamC-ΔN | F3, CCGGAATTCTGGGCCGACGAGTTCGAC |
| R3, CCGCTCGAGCTAGCGCCACTGGTAGGCG | |
| rLamC | F4, CCGGAATTCGCTTCGCGGGACGCGGCGC |
| R4, CCGCTCGAGCTAGCGCCACTGGTAGGCG | |
| rLamC-Mutantb | F5, GCGGGGCGCTCGACATCATGGCGAACGTCG |
| R5, CGACGTTCGCCATGATGTCGAGCGCCCCGC |
Underlined sequences within the primers are the EcoRI and XhoI restriction sites.
The two mutated nucleotides are underlined and in boldface.
Bioinformatic analysis.
The nucleotide sequences were assembled by the DNAMAN software package (version 5.2.2; Lynnon BioSoft, San Ramon, CA, USA). The signal peptide was predicted in the SignalP 4.0 server (http://www.cbs.dtu.dk/services/SignalP). The DNA and protein sequence alignments were performed via the National Center for Biotechnology Information (NCBI) with the programs BLASTN and BLASTP (http://www.ncbi.nlm.nih.gov/BLAST), respectively. The conserved domains and the GH family classification were identified via the website http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml (37). The protein sequence alignments were generated with the MUSCLE alignment in MEGA 7.0 software (38). The distance matrix for nucleotides was calculated by Kimura's two-parameter model (39). The phylogenetic tree was constructed using the neighbor-joining algorithm in MEGA 7.0 and assessed using 1,000 bootstrap replications (40). The molecular mass and the isoelectric point (pI) were calculated via the ExPASy Proteomics server (41).
Construction of the expression vectors.
Three LamC derivatives (designated rLamC-ΔC, rLamC-ΔN, and rLamC) containing one or two modules were designed based on analysis of the conserved domains (Fig. 1a). The corresponding coding regions were PCR amplified from the 1.3-kb cloned DNA with the indicated primer pairs (Table 3). The PCR amplifications with the PrimeSTAR HS DNA Polymerase were performed in 32 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 to 2 min, followed by a 10-min extension at 72°C. The PCR-amplified fragments introduced an EcoRI site at the 5′ end and an XhoI site at the 3′ end. The PCR products were digested with EcoRI and XhoI and then inserted into the pET32a(+) expression plasmid with an N-terminal Trx-tag and a His6 tag to generate pET-rLamΔC, pET-rLamC-ΔN, and pET-rLamC. After transformation into E. coli DH5α, the positive transformants were screened and verified by DNA sequencing. The expression constructs were used to transform E. coli Origami B(DE3) competent cells for protein expression.
Protein expression and purification.
For expression of LamC derivatives, selected clones were cultured in liquid LB broth at 37°C until the optical density at 600 nm (OD600) reached 0.5 to 0.6. Bacterial culture was induced with a final concentration of 0.2 mM IPTG at 18°C for 16 h. The cells were harvested, washed, resuspended in an equilibration buffer (20 mM sodium phosphate buffer, pH 7.0, and 1 mM phenylmethylsulfonyl fluoride [PMSF]) at 4°C, and lysed by ultrasonication (Insonator M201; Kubota, Japan). The lysate was centrifuged at 12,580 × g for 20 min, and the supernatant was used as a crude enzyme solution. Expressed proteins were purified with Ni2+-NTA resin (42) according to the manufacturer's instructions. All protein samples were analyzed by SDS-PAGE.
Initial enzyme activity assay and determination of protein concentration.
Activity screening against the various substrates was performed by DNS assays (43). Unless otherwise indicated, the enzyme reaction mixture containing suitably diluted enzyme and different polysaccharide substrates at a final concentration of 10 mg/ml in 20 mM sodium phosphate buffer (pH 7.0) was incubated at 50°C for 20 min. The amount of reducing sugar was determined spectrophotometrically at 540 nm. One unit of enzyme activity was defined as the amount of enzyme required to release reducing sugars equivalent to 1 μmol of glucose per min under the test conditions. Hydrolytic activities against insoluble substrates were evaluated after gently mixing the reaction mixtures during incubation. Then, the mixture was centrifuged at 12,580 × g at 4°C for 10 min. The reducing sugars in the supernatant were measured by the method described by Li et al. (44). The protein concentration was photodensitometrically determined by the Bradford method using BSA as the standard (45).
Enzymatic characterization.
Against laminarin as the substrate, the optimal temperature for the β-(1,3)-glucanase activity was determined over the range of 30 to 70°C in 20 mM phosphate buffer (pH 7.0). Enzyme thermostability was determined by measuring the residual activity after preincubation of the enzyme in 20 mM phosphate buffer (pH 7.0) at 30 to 70°C without substrate for 30 min. The optimal pH for β-(1,3)-glucanase activity was assessed in several buffers at 50°C. The following buffers were used: 20 mM citrate buffer, pH 4.0 to 6.0; 20 mM phosphate buffer, pH 6.0 to 9.0; and 20 mM glycine–NaOH buffer, pH 9.0 to 10.0. To measure the pH stability, the enzyme was incubated at 4°C for 24 h in different buffers, and the residual activity was determined against laminarin at 50°C for 20 min.
The substrate specificity was determined using the DNS method with various carbohydrates (10 mg/ml, laminarin, curdlan, pachyman, zymosan A, pustulan, xylan, CMC, α-dextran, and 5 mg/ml salecan) under the optimal conditions for each enzyme.
To determine the apparent kinetic parameters against laminarin under the optimal conditions for each enzyme, the initial velocities were measured for laminarin concentrations ranging from 0.5 to 3 mg/ml in accordance with the reaction rate of LamC and varied linearly with the substrate concentration. The Km and Vmax values were obtained from Lineweaver-Burk plots (46).
Site-directed mutagenesis.
To investigate whether the β-(1,3)-, β-(1,4)-, and β-(1,6)-glucanase and xylanase activities of LamC were derived from the same active center, site-directed mutagenesis of the catalytic residues was performed by overlap extension PCR (47). The forward and reverse primers (flanking primers) were F4 and R4, respectively. The double mutations E304A and E309A were generated in rLamC-Mutant with the internal primers F5 and R5, listed in Table 3. The resulting mutant plasmids were confirmed by DNA sequencing and transformed into E. coli Origami B(DE3) cells. The expression, purification, and enzyme activity assay of the mutant β-(1,3)-glucanase (rLamC-Mutant) followed the same procedure as the one described for wild-type LamC.
Detection of hydrolytic products.
Purified rLamC and various substrates (laminarin, pustulan, xylan, and CMC) at a final concentration of 10 mg/ml were incubated in 1 ml of 20 mM phosphate buffer (pH 7.0) at 30°C for various time intervals. The hydrolytic products were examined by TLC on silica gel 60 plates (Merck, Germany) using n-butanol-methanol-H2O (8:4:3, vol/vol/vol) as the solvent system (48). The reaction products were visualized by spraying a sulfuric acid-methanol (1:1, vol/vol) solution onto the plate, followed by baking at 95°C for 10 min.
Binding activity assays.
The binding of the noncatalytic fascin-like module (rLamC-ΔC) to insoluble polysaccharides was determined by using a pulldown assay (49). The purified rLamC-ΔC or rLamC (10 μg) and the indicated substrate (2 mg) were thoroughly mixed in 200 μl of 20 mM phosphate buffer (pH 7.0) at 4°C with gentle shaking. After centrifugation at 1,000 × g for 5 min, the pellet was washed once and resuspended in 200 μl of the same buffer. The proteins in the supernatant and pellet were analyzed by 12% SDS-PAGE. BSA was used as a negative control.
The binding of rLamC-ΔC to soluble polysaccharides was assayed by affinity electrophoresis with 2 mg/ml laminarin and CMC incorporated into a native 12% polyacrylamide gel (PAGE) (50). Electrophoresis was performed at 80 V at 4°C. Retardation of protein migration on the gel would depend on the binding potency of the protein to laminarin and CMC.
Fascin is the main actin filament bundling protein in filopodia. In order to examine whether the fascin-like module from LamC has binding activity toward F-actin, a cosedimentation using rLamC-ΔC and F-actin was performed (29). rLamC-ΔC was first centrifuged at 224,000 × g for 30 min to remove potential aggregates. F-actin (15 μM) was incubated with 10 μM rLamC-ΔC for approximately 8 h at room temperature in G buffer (2 mM Tris-HCl [pH 7.4], 0.2 mM CaCl2, 0.2 mM ATP, 1 mM dithiothreitol [DTT], 1 mM NaN3). Samples were centrifuged for 30 min at 224,000 × g for binding experiments. The amounts of protein remaining in the supernatant and pellet were analyzed by 12% SDS-PAGE.
Accession number(s).
The sequence for the novel β-(1,3)-glucanase gene (lamC) cloned from Corallococcus sp. EGB was deposited into the GenBank database under the accession number KX583630.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Science Foundation of China (no. 31400056 and 31560031), the Natural Science Foundation of Jiangsu Province (no. BK 20140687), and the Postdoctoral Science Foundation of China (no. 2016M591859).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01016-17.
REFERENCES
- 1.Zverlov VV, Volkov IY, Velikodvorskaya TV, Schwarz WH. 1997. Highly thermostable endo-1,3-beta-glucanase (laminarinase) Lam A from Thermotoga neapolitana: nucleotide sequence of the gene and characterization of the recombinant gene product. Microbiology 143:1701–1708. doi: 10.1099/00221287-143-5-1701. [DOI] [PubMed] [Google Scholar]
- 2.Henrissat B, Bairoch A. 1993. New families in the classification of glycoside hydrolases based on amino acid sequence similarities. Biochem J 293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucciaglia PA, Smith AG. 1994. Cloning and characterization of Tag1, a tobacco anther β-1,3-glucanase expressed during tetrad dissolution. Plant Mol Biol 24:903–914. doi: 10.1007/BF00014444. [DOI] [PubMed] [Google Scholar]
- 4.Chye M-L, Cheung K-Y. 1995. Beta-1,3-glucanase is highly-expressed in laticifers of Hevea brasiliensis. Plant Mol Biol 29:397–402. doi: 10.1007/BF00043663. [DOI] [PubMed] [Google Scholar]
- 5.Oh HY, Yang MS. 1995. Nucleotide sequence of genomic DNA encoding the potato beta-1,3-glucanase. Plant Physiol 107:1453. doi: 10.1104/pp.107.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yahata N, Watanabe T, Nakamura Y, Yamamoto Y, Kamimiya S, Tanaka H. 1990. Structure of the data encoding β-1,3-glucanase A1 of Bacillus circulans WL-12. Gene 86:113–117. doi: 10.1016/0378-1119(90)90122-8. [DOI] [PubMed] [Google Scholar]
- 7.Hong T-Y, Meng M. 2003. Biochemical characterization and antifungal activity of an endo-1,3-β-glucanase of Paenibacillus sp. isolated from garden soil. Appl Microbiol Biotechnol 61:472–478. doi: 10.1007/s00253-003-1249-z. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Y-M, Hong T-Y, Liu C-C, Meng M. 2009. Cloning and functional characterization of a complex endo-β-1,3-glucanase from Paenibacillus sp. Appl Microbiol Biotechnol 81:1051–1061. doi: 10.1007/s00253-008-1617-9. [DOI] [PubMed] [Google Scholar]
- 9.Spilliaert R, Hreggvidsson GO, Kristjansson JK, Eggertsson G, Palsdottir A. 1994. Cloning and sequencing of a Rhodothermus marinus gene, bglA, coding for a thermostable β-glucanase and its expression in Escherichia coli. Eur J Biochem 224:923–930. doi: 10.1111/j.1432-1033.1994.00923.x. [DOI] [PubMed] [Google Scholar]
- 10.Ilari A, Fiorillo A, Angelaccio S, Florio R, Chiaraluce R, van der Oost J, Consalvi V. 2009. Crystal structure of a family 16 endoglucanase from the hyperthermophile Pyrococcus furiosus—structural basis of substrate recognition. FEBS J 276:1048–1058. doi: 10.1111/j.1742-4658.2008.06848.x. [DOI] [PubMed] [Google Scholar]
- 11.Lafond M, Navarro D, Haon M, Couturier M, Berrin JG. 2012. Characterization of a broad-specificity beta-glucanase acting on beta-(1,3)-, beta-(1,4)-, and beta-(1,6)-glucans that defines a new glycoside hydrolase family. Appl Environ Microbiol 78:8540–8546. doi: 10.1128/AEM.02572-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charnock SJ, Bolam DN, Turkenburg JP, Gilbert HJ, Ferreira LM, Davies GJ, Fontes CM. 2000. The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry 39:5013–5021. doi: 10.1021/bi992821q. [DOI] [PubMed] [Google Scholar]
- 13.Henrissat B, Davies GJ. 2000. Glycoside hydrolases and glycosyl transferases. Families, modules, and implications for genomics. Plant Physiol 124:1515–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong T-Y, Cheng C-W, Huang J-W, Meng M. 2002. Isolation and biochemical characterization of an endo-1,3-β-glucanase from Streptomyces sioyaensis containing a C-terminal family 6 carbohydrate-binding module that binds to 1,3-β-glucan. Microbiology 148:1151–1159. doi: 10.1099/00221287-148-4-1151. [DOI] [PubMed] [Google Scholar]
- 15.Kane R. 1976. Actin polymerization and interaction with other proteins in temperature-induced gelation of sea urchin egg extracts. J Biol Chem 71:704–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson J, O'Hare K. 1991. Structure and transcription of the singed locus of Drosophila melanogaster. Genetics 129:1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holthuis JC, Schoonderwoert VT, Martens GJ. 1994. A vertebrate homolog of the actin-bundling protein fascin. Biochim Biophys Acta 1219:184–188. doi: 10.1016/0167-4781(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 18.Edwards RA, Herrera-Sosa H, Otto J, Bryan J. 1995. Cloning and expression of a murine fascin homolog from mouse brain. J Biol Chem 270:10764–10770. doi: 10.1074/jbc.270.18.10764. [DOI] [PubMed] [Google Scholar]
- 19.Ono S, Yamakita Y, Yamashiro S, Matsudaira PT, Gnarra JR, Obinata T, Matsumura F. 1997. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J Biol Chem 272:2527–2533. doi: 10.1074/jbc.272.4.2527. [DOI] [PubMed] [Google Scholar]
- 20.Opassiri R, Pomthong B, Akiyama T, Nakphaichit M, Onkoksoong T, Cairns MK, Cairns JRK. 2007. A stress-induced rice (Oryza sativa L.) β-glucosidase represents a new subfamily of glycoside hydrolase family 5 containing a fascin-like domain. Biochem J 408:241–249. doi: 10.1042/BJ20070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams JC. 2004. Roles of fascin in cell adhesion and motility. Curr Opin Cell Biol 16:590–596. doi: 10.1016/j.ceb.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Edwards RA, Bryan J. 1995. Fascins, a family of actin bundling proteins. Cell Motil Cytoskeleton 32:1–9. doi: 10.1002/cm.970320102. [DOI] [PubMed] [Google Scholar]
- 23.Jayo A, Parsons M. 2010. Fascin: a key regulator of cytoskeletal dynamics. Int J Biochem Cell Biol 42:1614–1617. doi: 10.1016/j.biocel.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 24.Huntley S, Zhang Y, Treuner-Lange A, Kneip S, Sensen CW, Søgaard-Andersen L. 2012. Complete genome sequence of the fruiting myxobacterium Corallococcus coralloides DSM. 2259. J Bacteriol 194:3012–3013. doi: 10.1128/JB.00397-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss H, Nett M, Domin N, Martin K, Maresca JA, Copeland A, Lapidus A, Lucas S, Berry KW, Glavina Del Rio T, Dalin E, Tice H, Pitluck S, Richardson P, Bruce D, Goodwin L, Han C, Detter JC, Schmutz J, Brettin T, Land M, Hauser L, Kyrpides NC, Ivanova N, Göker M, Woyke T, Klenk HP, Bryant DA. 2011. Complete genome sequence of the filamentous gliding predatory bacterium Herpetosiphon aurantiacus type strain (114-95T). Stand Genomic Sci 5:356–370. doi: 10.4056/sigs.2194987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeng W-Y, Wang N-C, Lin C-T, Shyur L-F, Wang AH-J. 2011. Crystal structures of the laminarinase catalytic domain from Thermotoga maritima MSB8 in complex with inhibitors: essential residues for β-1,3- and β-1,4-glucan selection. J Biol Chem 286:45030–45040. doi: 10.1074/jbc.M111.271213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong T-Y, Hsiao Y-Y, Meng M, Li TT. 2008. The 1.5 Å structure of endo-1,3-β-glucanase from Streptomyces sioyaensis: evolution of the active-site structure for 1,3-β-glucan-binding specificity and hydrolysis. Acta Crystallogr D Biol Crystallogr 64:964–970. doi: 10.1107/S0907444908021550. [DOI] [PubMed] [Google Scholar]
- 28.Fibriansah G, Masuda S, Koizumi N, Nakamura S, Kumasaka T. 2007. The 1.3 Å crystal structure of a novel endo-β-1,3-glucanase of glycoside hydrolase family 16 from alkaliphilic Nocardiopsis sp. strain F96. Proteins 69:683–690. doi: 10.1002/prot.21589. [DOI] [PubMed] [Google Scholar]
- 29.Jansen S, Collins A, Yang C, Rebowski G, Svitkina T, Dominguez R. 2011. Mechanism of actin filament bundling by fascin. J Biol Chem 286:30087–30096. doi: 10.1074/jbc.M111.251439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi P, Tian J, Yuan T, Liu X, Huang H, Bai Y, Yang P, Chen X, Wu N, Yao B. 2010. Paenibacillus sp. strain E18 bifunctional xylanase-glucanase with a single catalytic domain. Appl Environ Microbiol 76:3620–3624. doi: 10.1128/AEM.00345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao YS, Edwards RA, Tubb B, Wang S, Bryan J, McCrea PD. 1996. Beta-catenin associates with the actin-bundling protein fascin in a noncadherin complex. J Cell Biol 134:1271–1281. doi: 10.1083/jcb.134.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang J, Fonovic M, Suyama K, Bogyo M, Scott MP. 2009. Rab35 controls actin bundling by recruiting fascin as an effector protein. Science 325:1250–1254. doi: 10.1126/science.1174921. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Klaavuniemi T, Ono S. 2010. Distinct roles of four gelsolin-like domains of Caenorhabditis elegans gelsolin-like protein-1 in actin filament severing, barbed end capping, and phosphoinositide binding. Biochemistry 49:4349–4360. doi: 10.1021/bi100215b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito H, Misaki A, Harada T. 1968. A comparison of the structure of curdlan and pachyman. Agric Biol Chem 32:1261–1269. [Google Scholar]
- 35.Xiu A, Kong Y, Zhou M, Zhu B, Wang S, Zhang J. 2010. The chemical and digestive properties of a soluble glucan from Agrobacterium sp. ZX09. Carbohydr Polym 82:623–628. doi: 10.1016/j.carbpol.2010.05.027. [DOI] [Google Scholar]
- 36.Kaiser D, Manoil C, Dworkin M. 1979. Myxobacteria: cell interactions, genetics, and development. Annu Rev Microbiol 33:595–639. doi: 10.1146/annurev.mi.33.100179.003115. [DOI] [PubMed] [Google Scholar]
- 37.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607. In Walker JM. (ed), The proteomics protocol handbook. Humana Press, Totowa, NJ. [Google Scholar]
- 42.Janknecht R, De Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. 1991. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc Natl Acad Sci U S A 88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 44.Li Z, Wu J, Zhang B, Wang F, Ye X, Huang Y, Huang Q, Cui Z. 2015. AmyM, a novel maltohexaose-forming α-amylase from Corallococcus sp. strain EGB. Appl Environ Microbiol 81:1977–1987. doi: 10.1128/AEM.03934-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Chem 72:248–254.35. [DOI] [PubMed] [Google Scholar]
- 46.Dowd JE, Riggs DS. 1965. A comparison of estimates of Michaelis-Menten kinetic constants from various linear transformations. J Biol Chem 240:863–869. [PubMed] [Google Scholar]
- 47.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 48.Hansen SA. 1975. Thin-layer chromatographic method for the identification of mono-, di- and trisaccharides. J Chromatogr A 107:224–226. doi: 10.1016/S0021-9673(00)82770-3. [DOI] [Google Scholar]
- 49.Cheng Y-M, Hsieh F-C, Meng M. 2009. Functional analysis of conserved aromatic amino acids in the discoidin domain of Paenibacillus β-1,3-glucanase. Microb Cell Fact 8:62. doi: 10.1186/1475-2859-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomme P, Creagh AL, Kilburn DG, Haynes CA. 1996. Interaction of polysaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC. 1. Binding specificity and calorimetric analysis. Biochemistry 35:13885–13894. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.