ABSTRACT
Lactococcus lactis is one of the most commonly used lactic acid bacteria in the dairy industry. Activation of competence for natural DNA transformation in this species would greatly improve the selection of novel strains with desired genetic traits. Here, we investigated the activation of natural transformation in L. lactis subsp. cremoris KW2, a strain of plant origin whose genome encodes the master competence regulator ComX and the complete set of proteins usually required for natural transformation. In the absence of knowledge about competence regulation in this species, we constitutively overproduced ComX in a reporter strain of late competence phase activation and showed, by transcriptomic analyses, a ComX-dependent induction of all key competence genes. We further demonstrated that natural DNA transformation is functional in this strain and requires the competence DNA uptake machinery. Since constitutive ComX overproduction is unstable, we alternatively expressed comX under the control of an endogenous xylose-inducible promoter. This regulated system was used to successfully inactivate the adaptor protein MecA and subunits of the Clp proteolytic complex, which were previously shown to be involved in ComX degradation in streptococci. In the presence of a small amount of ComX, the deletion of mecA, clpC, or clpP genes markedly increased the activation of the late competence phase and transformability. Altogether, our results report the functionality of natural DNA transformation in L. lactis and pave the way for the identification of signaling mechanisms that trigger the competence state in this species.
IMPORTANCE Lactococcus lactis is a lactic acid bacterium of major importance, which is used as a starter species for milk fermentation, a host for heterologous protein production, and a delivery platform for therapeutic molecules. Here, we report the functionality of natural transformation in L. lactis subsp. cremoris KW2 by the overproduction of the master competence regulator ComX. The developed procedure enables a flexible approach to modify the chromosome with single point mutation, sequence insertion, or sequence replacement. These results represent an important step for the genetic engineering of L. lactis that will facilitate the design of strains optimized for industrial applications. This will also help to discover natural regulatory mechanisms controlling competence in the genus Lactococcus.
KEYWORDS: competence, sigma factor, ComX, Clp protease, lactic acid bacteria, Lactococcus, natural DNA transformation
INTRODUCTION
Lactococcus lactis is a lactic acid bacterium of great industrial interest, which is used as the main dairy starter species in various cheese preparations (e.g., cheddar, gouda, and brie) and fermented milk products (e.g., quark, buttermilk, and sour cream) (1). It is also exploited as a host for heterologous protein production and as a delivery platform for therapeutic molecules (2, 3). Genome editing in this species is currently carried out via electroporation, with a highly variable strain-to-strain efficiency (4). Chromosomal modifications are usually performed by simple or double crossover homologous recombination using either suicide or thermosensitive plasmids (for a review, see reference 5). More recently, sophisticated tools, such as recombineering, were also developed for chromosomal editing in this species (6). However, all these approaches are generally time-consuming as they require either plasmid construction or extensive screening procedures for mutant selection. The activation of competence for natural DNA transformation in L. lactis would significantly enhance the efficiency of genome modification for the design of improved strains. For instance, as recently shown for the related species Streptococcus thermophilus, PCR-assembled linear DNA fragments can be added as donor DNA to a transformable strain in order to achieve any kind of chromosomal modification (i.e., point mutation, insertion, and deletion) (7–10).
DNA acquisition by natural transformation is widespread among prokaryotes and has been reported in more than 80 species (11). For instance, it has been well characterized in many Gram-positive bacteria such as Bacillus subtilis (12) and several streptococci (e.g., Streptococcus pneumoniae, Streptococcus mutans, and Streptococcus thermophilus) (for a recent review, see reference 13). Commonly, competence for natural transformation is recognized to fulfill various nonexclusive functions: genome plasticity, DNA repair, and/or nutrition (14, 15).
In streptococci, competence for DNA transformation is induced in response to secreted signaling peptides referred to as competence pheromones, or alarmones (13). The production of this class of cell-to-cell communication molecules is initiated in response to specific environmental stresses or conditions that trigger the coordination of physiological functions (e.g., competence, predation, and biofilm formation) (13, 14). At a threshold concentration, competence pheromones activate the master regulator ComX (alternative sigma factor σX), which ultimately leads to a major transcriptional reprogramming of cells (commonly known as the late competence phase), including the induction of genes strictly required for DNA transformation (16, 17). ComX binds a specific DNA sequence, named Com-box or Cin-box, which is typically located in promoters of late competence (com) genes and/or operons responsible for DNA uptake (i.e., comG, comF, and comE operons), DNA protection (ssb), and DNA recombination (e.g., recA, dprA, and coiA) (18, 19).
While the late competence steps are well conserved among Gram-positive bacteria, the early steps leading to competence activation (early competence phase) differ from species to species (11, 20). In streptococci, two major peptide-based signaling pathways—i.e., ComCDE and ComRS—have been identified so far (21). Within the mitis and anginosus groups of streptococci (S. pneumoniae as paradigm), the competence signaling peptide (CSP, or mature ComC) triggers a phosphorylation cascade between the histidine kinase ComD and the response regulator ComE, both belonging to a classical two-component system (22). Then, the phosphorylated form of ComE activates comX transcription (22). Within the salivarius, mutans, pyogenes, bovis, and suis groups of streptococci, another regulation mechanism is operational (S. thermophilus as paradigm). This system involves the ComX induction peptide (XIP, or mature ComS) which is internalized by the oligopeptide transporter Opp, binds to and activates the regulator ComR, which in turn induces comX transcription (8, 21, 23–27).
Even though streptococci and lactococci belong to the same family, namely, the Streptococcaceae, little is known about competence development in L. lactis. Indeed, there is no experimental evidence showing that natural DNA transformation is functional in the genus Lactococcus. However, homologues of comX and of all late com genes essential for natural transformation were identified in the genome of L. lactis (28–30). All these late com genes/operons feature an upstream and conserved Com-box, suggesting that they are under ComX control (30). Their transcriptional activation upon comX overexpression was validated in strain IL1403 of the subspecies lactis (30). In addition, specific growth conditions have been reported to activate com genes, such as carbon starvation in strain IL1403 (31) and the plant-borne strain KF147 (subspecies lactis) (32), or during cheese-making conditions in strain MG5267 (subspecies cremoris) (33). However, attempts to validate the functionality of natural transformation in KF147 were unsuccessful (32). Finally, although all late com genes essential for natural transformation were identified in L. lactis, some are present as putative pseudogenes in different strains (28–30). For instance, mutations resulting in a potentially inactive dprA gene were observed in many L. lactis strains, especially in subspecies lactis (30). Since DprA inactivation was shown to strongly decrease competence efficiency in S. pneumoniae (34, 35), natural DNA transformation could be impaired in many L. lactis strains.
In the present study, we investigated the occurrence of late com genes essential for natural transformation in five complete genomes of subspecies cremoris strains and identified strain KW2 as containing a complete set of com genes required for natural DNA transformation. In the absence of signaling systems orthologous to ComCDE or ComRS, activation of the late competence phase was fostered by either constitutive or inducible expression of comX, which resulted in the activation of all major late com genes essential for DNA transformation. Importantly, we demonstrated that natural transformation is functional in strain KW2 and requires the DNA uptake machinery.
RESULTS AND DISCUSSION
L. lactis subsp. cremoris KW2 contains all late com genes essential for DNA transformation.
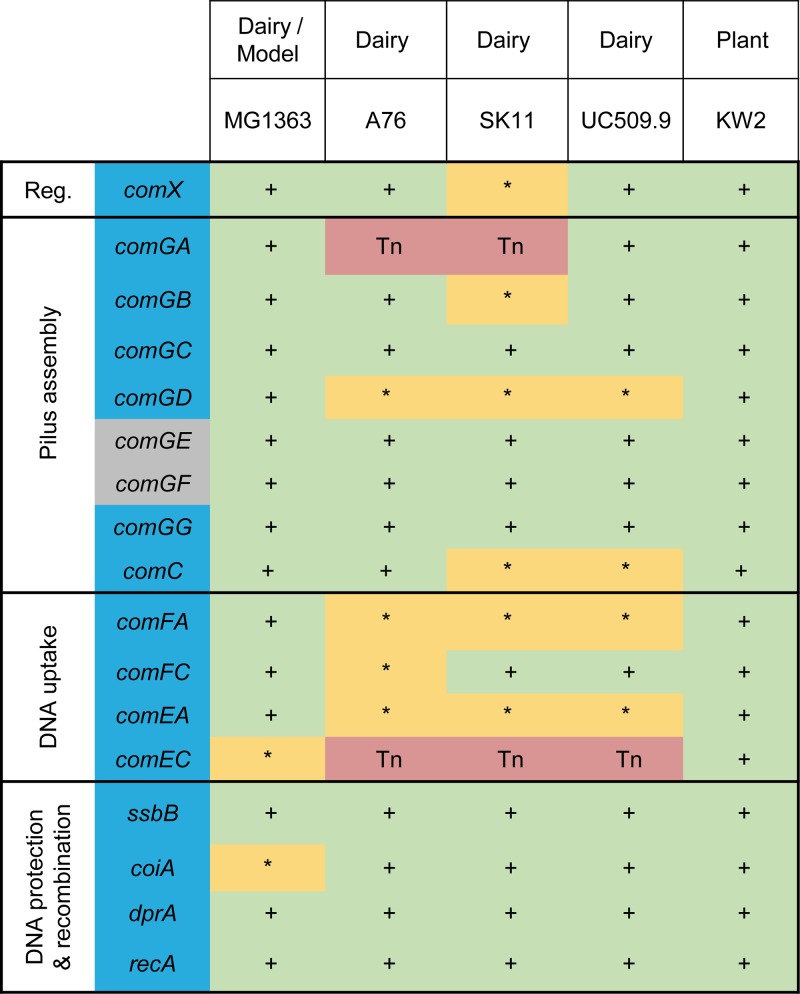
Among L. lactis strains, genomic variability was previously investigated for comX and dprA alleles (30). Although all strains (31/31) displayed an intact copy of comX, the dprA gene integrity was variable between the two subspecies: 50% of the lactis strains (10/20) contained nonsense mutations in dprA, while all cremoris strains (11/11) harbored an intact and potentially functional dprA gene (30). Since dprA is required for high transformation levels in streptococci, its integrity in cremoris strains prompted us to further analyze the genomic content in com genes in this subspecies. The presence of the minimal set of late com genes (nearly) essential for DNA transformation (17 candidate genes including comX) (Fig. 1; see Table S1 in the supplemental material) was examined in five fully sequenced genomes of cremoris strains (i.e., strains MG1363, A76, SK11, UC509.9, and KW2). This in silico analysis revealed that the genomes of strains A76, SK11, and UC509.9 (cheese or dairy origin) contain a high number of pseudogenes among key com genes (between five and eight incomplete late com genes) due to transposon insertion or frameshift events (nucleotide insertion or deletion). In particular, the presence of transposable elements in comGA (pilus biogenesis) and/or comEC (DNA translocation) genes strongly suggests that natural transformation is no longer functional in those strains, as described in streptococci (20, 36). Although the set of full-length com genes in the laboratory strain MG1363 is larger, mutations in comEC (nucleotide insertion) and coiA (nonsense mutation) probably impair its ability to be competent (29). These mutations were also found in the genome of its isogenic derivative NZ9000, which strongly suggests that they do not result from DNA sequencing errors. In contrast to other strains, strain KW2 of plant origin (corn fermentation) (37) contains the whole set of known essential late genes required to fulfill natural DNA transformation (Fig. 1), making it a good candidate to further study the functionality of competence in the cremoris subspecies.
FIG 1.
Late com genes in the complete genomes of L. lactis subsp. cremoris strains MG1363, A76, SK11, UC509.9, and KW2. The strain origin is indicated above each name. The essential late genes for DNA transformation identified in S. pneumoniae (17) are highlighted in blue. Two genes of the comG operon, which are potentially essential for DNA transformation, are highlighted in gray. Gene-associated function in DNA transformation is indicated on the left. Reg., regulation. The complete and incomplete status of late genes is based on blastp and tblastn similarity searches using orthologues of S. pneumoniae TIGR4 and S. thermophilus LMD-9 and default parameters. Symbols and abbreviations: + (green), presence of a complete gene; * (yellow), incomplete gene due to nucleotide change, insertion, or deletion resulting in a premature stop codon; Tn (red), gene disrupted by the insertion of at least one transposon.
Induction of the comGA promoter by constitutive comX expression.
To test the ability of ComX to induce natural transformation in strain KW2, a plasmid-borne copy of comX under the control of the constitutive lactococcal P32 promoter (plasmid pGIBLD001) was introduced into a KW2-derivative reporter strain (BLD101). This strain chromosomally encodes a PcomGA-luxAB transcriptional fusion that we used here as a proxy for competence activation.
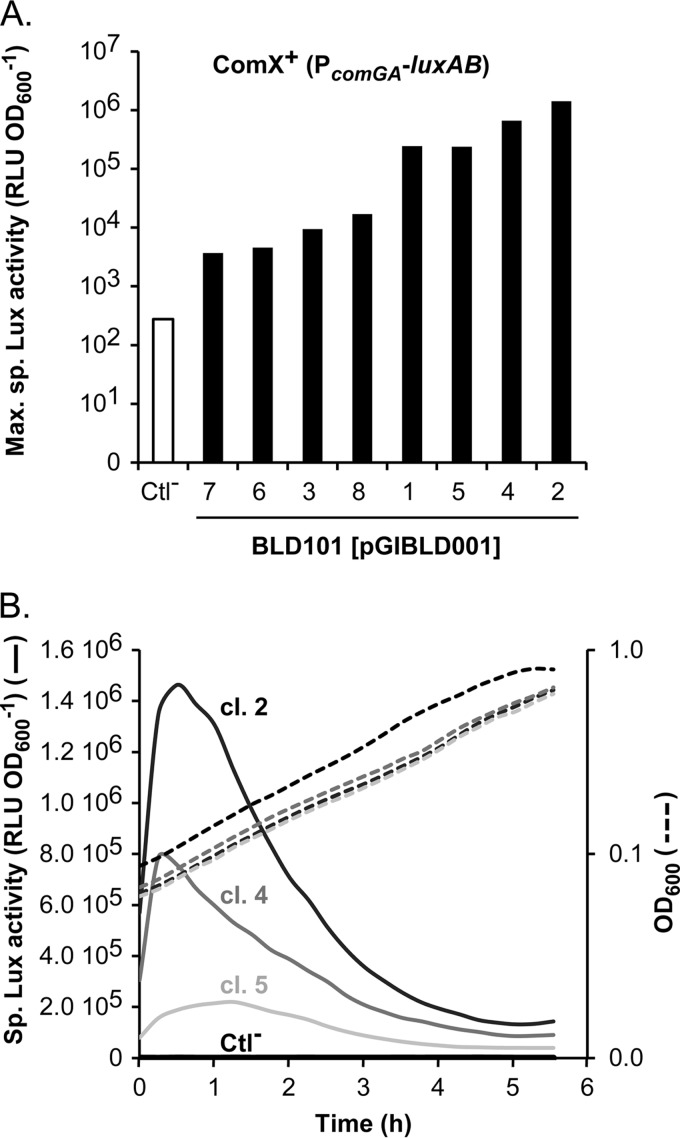
Eight clones (1 to 8) of the reporter strain carrying pGIBLD001 (P32-comX, henceforth named ComX+) were randomly selected and their specific luciferase (Lux) activity was monitored in liquid, chemically defined medium (CDM) supplemented with glucose (CDMG). This medium was previously shown to be permissive for competence development in various streptococcal species (8, 23, 25, 26, 38–40). To ensure reproducibility of the assay, exponentially growing cells in complex medium (M17G conditions) were washed and inoculated in fresh CDMG before starting the experiment. As expected, all tested ComX+ clones displayed between 101- and 104-fold-higher specific Lux activities than the control strain carrying the empty vector [BLD101(pG+host9), hereafter named Ctl−] (Fig. 2A). The heterogeneity in Lux activity that we observed between clones was previously described in similarly engineered ComX+ S. thermophilus cells and could reflect a physiological disturbance due to a toxic effect of the constitutive ComX production (7). Intriguingly, the specific Lux activity peaked approximately after 1 h of growth in CDMG (Fig. 2B), which is counterintuitive for a constitutive expression system. However, this discrepancy might be explained by different mechanisms such as ComX degradation and/or aggregation, or instability and/or inactivity of the Lux reporter protein. These results show that the competence sigma factor is functional for the activation of the comG operon from strain KW2 when it is constitutively produced.
FIG 2.
Activation of the late promoter PcomGA by constitutive comX overexpression. (A) Maximum specific luciferase (Lux) activity (RLU OD600−1) emitted by eight independent clones (clones 1 to 8) of the ComX+ strain [BLD101(pGIBLD001), PcomGA-luxAB] compared to the control strain (Ctl−) carrying the empty vector [BLD101(pG+host9)]. (B) Kinetics of specific Lux activity (solid lines) during growth (OD600, dotted lines) for the control strain (Ctl−, black lines) and three selected ComX+ clones [BLD101(pGIBLD001), clones [cl.] 2, 4, and 5; gray lines]. Cells were grown in CDMG. The results of one representative experiment of two independent replicates are shown.
Constitutive comX expression activates the late com regulon.
To further investigate the ComX regulon in L. lactis strain KW2, the transcriptomes of Ctl− and clone 2 of the ComX+ strain that exhibits the strongest Lux activity (Fig. 2A) were compared by RNA sequencing. Cells were grown as reported above and collected in early exponential growth phase.
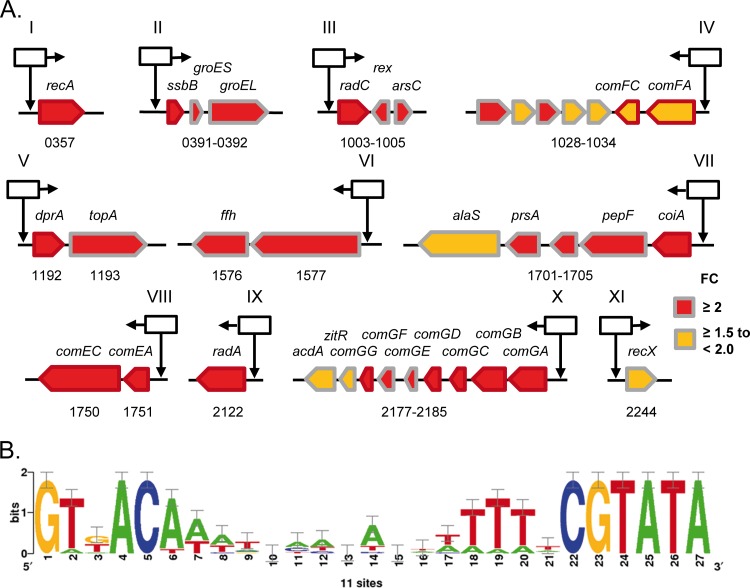
Besides comX (fold change [FC] = 19.8), 96 genes were considered upregulated (FC ≥ 2.0) in the ComX+ strain versus Ctl−. In addition, we took into account eight genes that are upregulated with a FC ≥ 1.5 and are located downstream of a predicted ComX-binding motif (see Table S2 in the supplemental material). In total, 11 putative Com-boxes were identified directly upstream of 11 gene clusters (I to XI) that gather 35 upregulated genes (Fig. 3A; see also Tables S2 and S3 in the supplemental material). These genes, except for cluster VI, are usually described as late com or competence-associated genes (Fig. 3A). Similarly to the Com-box consensus sequence previously reported in L. lactis subsp. lactis IL1403 (30), the Com-box of strain KW2 (total length, 27 bp) consists of two stretches of highly conserved nucleotides (Fig. 3B). With the exception of comC encoding the pre-pilin leader peptidase that is preceded by a predicted Com-box (see Table S3 in the supplemental material), all the late com genes previously identified in S. pneumoniae as essential for DNA transformation are activated upon constitutive comX expression (see Table S2 in the supplemental material) (17). Similarly, previous transcriptomic analyses failed to identify comC as upregulated by ComX in S. thermophilus (41). In addition, >400 genes were found to be downregulated (FC ≤ 0.5) by the constitutive comX expression. They are mainly involved in carbon catabolism, nitrogen metabolism, or cell envelope biogenesis (data not shown), suggesting that the physiology of the cell is significantly affected upon artificial induction of comX. It is important to note that the extent of the ComX regulon revealed here could partially result from physiological adaptations and/or compensatory mutations due to the strong constitutive production of ComX. Altogether, this transcriptomic analysis showed that comX is capable of controlling the expression of (nearly) all essential late com genes for DNA transformation in strain KW2, which suggests that natural DNA transformation might be functional in the ComX+ strain.
FIG 3.
Upregulated late com genes preceded by a predicted Com-box upon constitutive comX expression. (A) Genetic organization of 11 loci (I to IX), including all the essential late genes for DNA transformation which are upregulated in clone 2 of the ComX+ strain [BLD101(pGIBLD001)], based on transcriptomic analyses. Pentagons represent open reading frames with respect to their sizes and orientations. Red-outline pentagons indicate genes essential for genetic transformation. Red- and yellow-filled pentagons represent, respectively, genes with an FC ≥ 2 and an FC between ≥1.5 and <2 as reported in the RNA sequencing analysis (see Table S2 in the supplemental material). The gene symbols and locus tag number are indicated above and below the pentagons, respectively. Black arrows topped with black-outlined squares indicate the position of predicted Com-boxes; horizontal arrows indicate the orientation of the motif. (B) Consensus of the predicted Com-box motif (27 bp) of strain KW2 (see Table S3) based on the sites identified in the 11 loci reported in panel A.
Constitutive comX expression induces natural transformation.
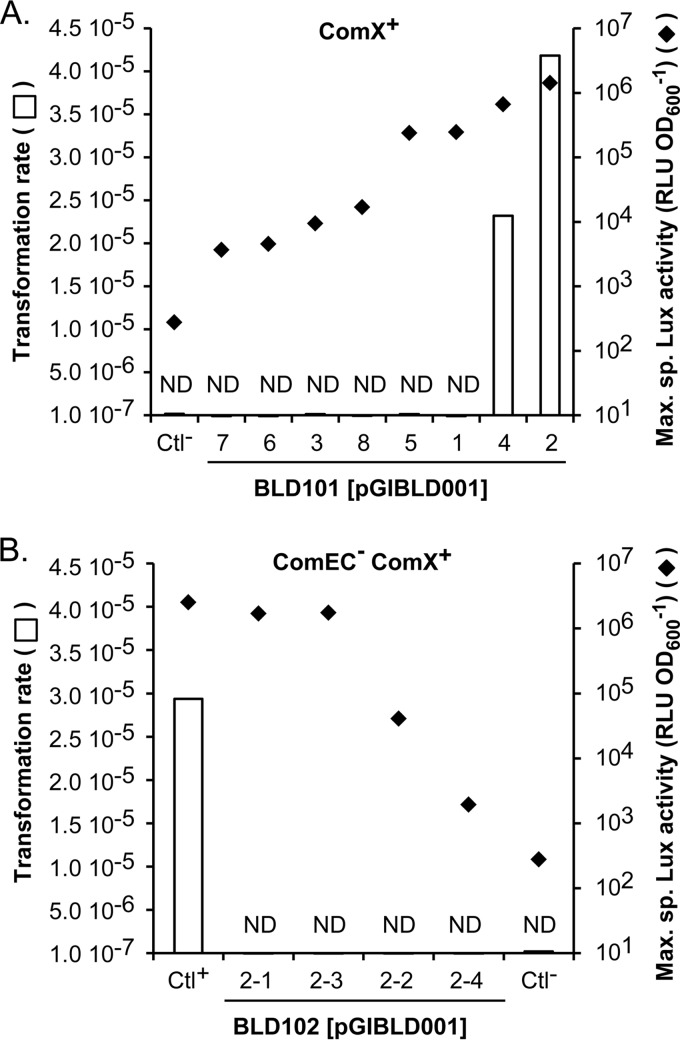
Because experimental evidence of natural transformation events was lacking in L. lactis, we first tested the transfer of single point mutations in the chromosome of the ComX+ strain. For this purpose, we used PCR-amplified fragments as transforming DNAs that encompass a mutated rpsL allele from a streptomycin-resistant (Strr) strain of L. lactis subsp. cremoris MG1363. Besides the strA1 mutation known to confer the Strr phenotype in B. subtilis (K56I substitution in ribosomal protein S12) (42), the MG1363 rpsL allele also contained two additional silent polymorphisms located at positions −11 (A→T) and −128 (G→T) relative to the strA1 mutation (see Fig. S1 in the supplemental material). To ensure efficient recombination, the strA1 allele was combined with upstream and downstream KW2 recombination arms of ∼1.85 kb. Transformation assays (see standard protocol in Materials and Methods) were performed with the eight previously selected clones of the ComX+ strain and Ctl− as a negative control. Remarkably, clones 2 and 4 of the ComX+ strain that displayed the highest PcomGA activation yielded mutation frequencies ∼15-fold higher than the background level. This background corresponds to the level of spontaneous mutation that was calculated in the absence of DNA for each individual transformation assay. After its subtraction, a transformation rate of up to 4 × 10−5 (∼104 transformants ml−1) was obtained for clone 2 (Fig. 4A). By sequencing the rpsL gene of 10 Strr derivatives of clone 2, we systematically observed the cotransfer of strA1 and the nearby −11 mutation. In two cases, the −128 mutation was also cotransferred. The chimeric nature of the rpsL gene (i.e., the presence of strA1 and −11 mutations without the −128 mutation) in some Strr ComX+ derivatives of KW2 ultimately demonstrates that a recombination process occurred between the exogenous and chromosomal DNA. In contrast, −11 and/or −128 mutations were never observed in the rpsL gene of spontaneous Strr mutants (20 sequenced clones) obtained in the negative-control experiments (i.e., assays performed in the absence of DNA or with the control strain carrying the empty vector in the presence of DNA). These results show that exogenous DNA can enter KW2 cells and be integrated in their chromosome by homologous recombination when a certain threshold of comX expression is reached.
FIG 4.
DNA transformation of the strA1 allele upon constitutive comX expression. (A) Transformation rate (white bars) and maximum specific luciferase (Lux) activity (black diamonds, RLU OD600−1, as reported in Fig. 2) of eight clones (clones 1 to 8) of the ComX+ strain [BLD101(pGIBLD001), PcomGA-luxAB] compared to the negative-control strain (Ctl−) carrying the empty vector [BLD101(pG+host9)]. (B) Transformation rate (white bars) and maximum specific luciferase (Lux) activity (black diamonds, RLU OD600−1) of four clones (clones 2-1 to 2-4) of the ComEC− ComX+ strain [BLD102(pGIBLD001), PcomGA-luxAB] compared to the positive [Ctl+, BLD101(pGIBLD001) clone 2] and negative [Ctl−, BLD101(pG+host9)] control strains. Transformability was assessed according to the standard protocol described in Materials and Methods using strA1-carrying PCR products as donor DNA. ND, transformation rate below the detection level of spontaneous Strr mutants (<10−7). The results of one representative experiment of two independent replicates are shown.
To test whether the observed horizontal DNA transfer in ComX+ cells was mediated by natural transformation and not by phage transduction or conjugation, we next investigated the implication of ComEC in this process. This protein was previously shown to be essential for DNA transformation in streptococci since it channels single-stranded DNA through the membrane (20, 36). To create the ComEC− ComX+ reporter strain [BLD102(pGIBLD001)], clone 2 of the ComX+ strain was transformed by PCR products encompassing the comEC gene disrupted by the insertion of a chloramphenicol resistance cassette (P32-cat; see Materials and Methods). Four clones (2-1 to 2-4) of the ComEC− ComX+ strain were validated by PCR for the correct insertion of P32-cat in the comEC gene. Transformation assays with the mutated rpsL allele showed that the frequencies of emergence of Strr clones in all tested ComEC− ComX+ derivatives were similar to the background level observed for spontaneous rpsL mutation frequencies (<10−7) (Fig. 4B). Although heterogeneity in PcomGA activation was observed between clones as reported above for the ComX+ strain, half of the ComEC− ComX+ derivative clones (i.e., clones 2-1 and 2-3) displayed a maximum specific Lux activity similar to the transformable ComX+ clones (>1.0 × 106 relative light units [RLU] per optical density at 600 nm [OD600]) (Fig. 4B). This shows that the transformation defect in these ComEC− ComX+ clones does not result from an insufficient ComX production. Taken together, these results demonstrate that natural DNA transformation can be artificially activated in strain KW2.
Inducible comX expression stabilizes activation of natural transformation.
Constitutive ComX production presents drawbacks since a high variability in PcomGA activation and transformability was observed between different clones of BLD101(pGIBLD001) (Fig. 2 and 4). This variability was assigned to a toxic effect resulting from the constitutive expression of comX. Consequently, a range of compensatory mutations can arise either in the expression plasmid (e.g., alteration of P32 promoter) or in the chromosome (e.g., suppressor mutations) in order to reduce ComX production or activity. To overcome this effect, the identification of a tightly controlled and inducible expression system for ComX production was needed. Our first attempts using the well-established nisin-controlled expression (NICE) system were unsuccessful due to an excessive basal activity of the PnisA promoter (data not shown). As an alternative, we chose the xylose-inducible promoter of the xylose permease gene (xylT). PxylT was previously exploited to develop a convenient inducible system for the expression of recombinant proteins in L. lactis subsp. lactis NCDO2118 (43). This promoter was shown in Gram-positive bacteria to be repressed by the CcpA-mediated catabolite repression in the presence of glucose and to be strongly induced by XylR in the presence of xylose (44, 45).
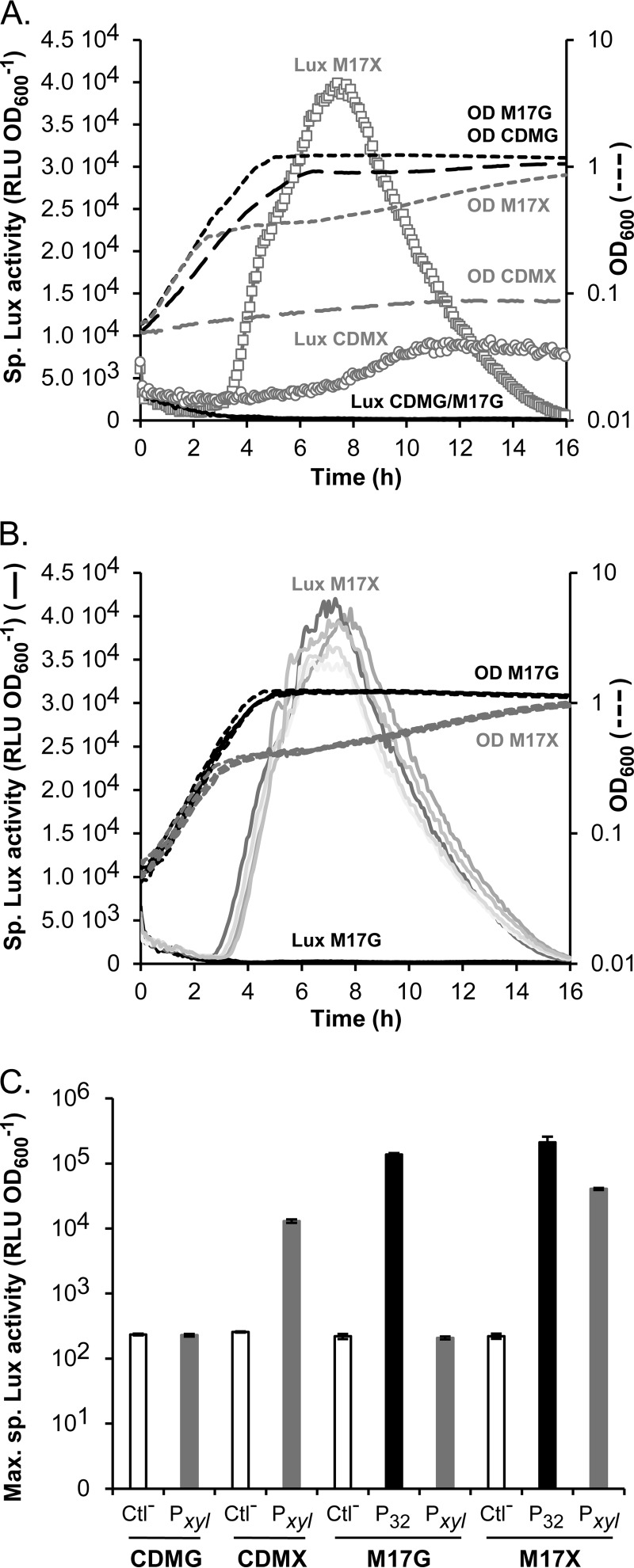
To test the ability of PxylT to control comX expression, we replaced the P32 promoter located on pGIBLD001 by the endogenous PxylT. The resulting plasmid pGIFPT001 was transformed by electroporation in the reporter strain BLD101. Due to a slow growth of strain KW2 and its derivatives in CDM with xylose as sole carbon source (CDMX), growth and PcomGA activity were also monitored in rich medium containing either glucose (M17G) or xylose (M17X) (Fig. 5A). In CDM conditions, growth of BLD101(pGIFPT001) on xylose (CDMX) led to an ∼60-fold increase in PcomGA activity compared to glucose (CDMG). The kinetics of PcomGA activation was slow and coherent with a lower growth rate in CDMX compared to CDMG (Fig. 5A). In M17 conditions, an ∼200-fold increase in PcomGA activity was observed in M17X compared to M17G. In contrast to the constitutive system, all the clones grown in M17X behaved similarly regarding PcomGA activation (Fig. 5B and see Fig. S2 in the supplemental material). The growth of BLD101(pGIFPT001) in M17X was diauxic compared to M17G (Fig. 5A and B). This diauxic shift may be due to the presence of additional sugars in the rich medium M17 that are preferentially consumed by the cells. In M17X, Lux activity was only detected after this shift, indicating that xylose is metabolized at this stage. Lux activity reached a maximum (4.1 × 104 ± 0.1 × 104 RLU OD600−1) after ∼7 h (Fig. 5A and B), which is only 5-fold lower than with the constitutive expression system in the same M17X conditions [BLD101(pGIBLD001), clone 2; 2.1 × 105 ± 0.4 × 105 RLU OD600−1] (Fig. 5C). Strain BLD101(pGIBLD001) (clone 2) showed a maximum specific Lux activity ∼10-fold lower when cultured in M17G or M17X compared to CDMG (compare Fig. 2A and 5C). Importantly, strain BLD101(pGIFPT001) grown with glucose (CDMG or M17G) is unable to activate PcomGA, showing that PxylT remains efficiently silent under these conditions (Fig. 5C).
FIG 5.
Activation of the late promoter PcomGA by inducible comX expression. (A) Kinetics of specific Lux activity during growth (OD600) of strain BLD101(pGIFPT001) grown in CDMG, CDMX, M17G, and M17X. The symbols for OD600 in CDMG (OD CDMG), CDMX (OD CDMX), M17G (OD M17G), and M17X (OD M17X) are long black dashes, long gray dashes, short black dashes, and short gray dashes, respectively. The symbols for Lux activity in CDMG (Lux CDMG) and M17G (Lux M17G) are solid black lines; the symbols in CDMX (Lux CDMX) and M17X (Lux M17X) are circles and squares, respectively. One representative experiment of three independent replicates. (B) Kinetics of specific Lux activity (solid lines) during growth (OD600, dashed lines) of five randomly selected clones of strain BLD101(pGIFPT001) grown in M17G and M17X. The symbols for OD600 in M17G (OD M17G) and M17X (OD M17X) are black and gray dashes, respectively. The symbols for Lux activity in M17G (Lux M17G) and M17X (Lux M17X) are gray and black solid lines, respectively. (C) Maximum specific luciferase (Lux) activity (RLU OD600−1) emitted by strains BLD101(pG+host9) (Ctl−, negative control, white bars), BLD101(pGIFPT001) (Pxyl, gray bars), and BLD101(pGIBLD001) (clone 2; P32, black bars) grown in CDMG, CDMX, M17G, and M17X. Mean values (n = 3) ± the standard deviations are indicated.
Finally, M17 and CDM growth conditions were compared for transformability. Transformation rates of strain BLD101(pGIFPT001) were calculated after 6 and 24 h of culture in M17G/M17X and CDMG/CDMX. The mutated rpsL allele was used as donor DNA. In contrast to M17G, CDMG, and CDMX, natural DNA transformation was shown to be functional in M17X with a transformation rate that correlates with the window of induction: 5.5 × 10−6 ± 1.9 × 10−6 after 6 h and 3.3 × 10−3 ± 0.5 × 10−3 after 24 h. These results support the previous observation that a threshold of PcomGA activation needs to be reached in order to activate natural DNA transformation.
Overall, these results show that the controlled expression of comX leads to a more robust activation of natural DNA transformation in strain KW2. In addition, DNA transformation is observed at a much lower level of PcomGA activation than with the constitutive system, which suggests that controlled expression better mimics natural conditions of competence activation.
The Clp machinery represses PcomGA activation and transformability.
In closely related streptococci such as S. pyogenes and S. thermophilus, the proteolytic Clp machinery was shown to degrade ComX, thereby preventing competence induction in various suboptimal or nonpermissive growth conditions (41, 46, 47). The adaptor protein MecA, the ATPase subunits ClpC and ClpE, and the serine protease ClpP have been shown to participate in this degradation process (e.g., MecA-ClpCP in S. thermophilus and S. mutans or ClpEP in S. pneumoniae; for a review, see reference 13). Interestingly, transcriptomic analysis revealed that mecA is upregulated in the ComX+ strain (see Table S2 in the supplemental material), and this might indicate that the MecA-Clp machinery still exerts a negative control on ComX levels.
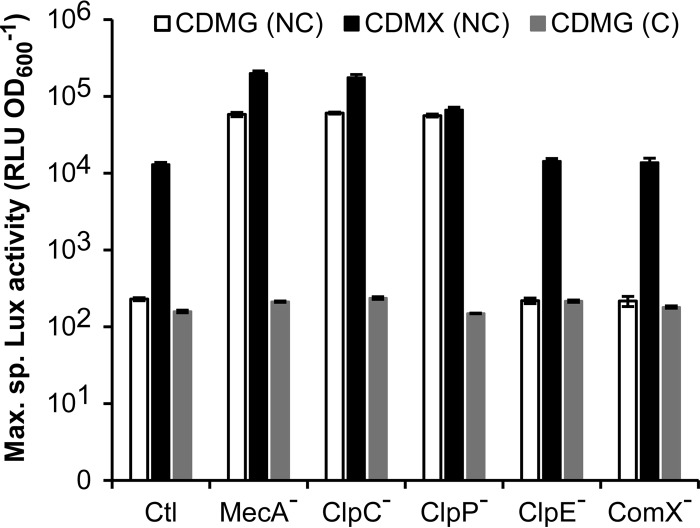
To investigate the potential contribution of MecA and Clp homologues in the control of competence, we deleted mecA, clpC, clpE, clpP, or comX genes by replacing their coding sequence with a chloramphenicol resistance cassette in strain BLD101(pGIFPT001) (see Materials and Methods). We used the comX mutant as a negative control. To monitor the impact of these gene deletions on PcomGA activity, three independent mutants per gene were cured of their thermosensitive plasmid pGIFPT001 and were compared to noncured clones (Fig. 6). Plasmid-free mutants harboring the different gene deletions did not show any significant increase in Lux activity (Fig. 6), suggesting that any known subunit of the Clp machinery involved in competence control is able to repress PcomGA in the tested conditions. However, inactivation of any partner of the MecA-ClpCP machinery (MecA−, ClpC−, or ClpP−) in strain BLD101(pGIFPT001) induced PcomGA under repression conditions (CDMG) and to a slightly higher extent in induction conditions (CDMX), compared to the negative-control strains (Fig. 6). This suggests that MecA, ClpC and ClpP are involved in the proteolytic degradation of ComX. Interestingly, these three mutant strains were able to be transformed by the mutated rpsL allele when grown in CDMG, while the control strain BLD101(pGIFPT001) could not. The observed difference between cured and noncured clones was allocated to a weak leakage of the PxylT promoter in the absence of xylose, allowing a net ComX accumulation when its posttranslational degradation system is deficient. However, when intact, the MecA-ClpCP machinery is sufficient to keep the ComX concentration below the activating threshold. Together, these results suggest that the MecA-ClpCP machinery plays a significant role in the negative control of competence in strain KW2.
FIG 6.
Impact of the inactivation of members of the MecA-Clp machinery on PcomGA activation. The maximum specific luciferase (Lux) activity (RLU OD600−1) of the reporter strain BLD101 (Ctl) and its isogenic mutant strains (MecA−, ClpC−, ClpP−, ClpE−, and ComX− [used as a negative control]), cured or not from plasmid pGIFPT001, were determined. Noncured (NC) strains grown in CMDG and CDMX are represented in white and black, respectively. Cured (C) strains obtained by temperature upshift and grown in CDMG are in gray. Mean values (n = 3) ± the standard deviations are indicated.
Concluding remarks.
Our work reports on functional natural DNA transformation in the species L. lactis. The developed tools based on the controlled expression of comX from a thermosensitive vector provide a potent and flexible method to edit L. lactis genomes with single point mutations, sequence insertions, or sequence replacements. The use of in vitro-assembled PCR products as donor DNA for transformation is also of great interest since it allows the generation of mutant strains in a 1-week time frame.
The activation of natural transformation by the mere activation of the presumptive master regulator(s) of competence has been attempted in many Gram-positive bacteria, including bacilli (ComK) (48–50), streptococci (ComX/SigX) (7, 9, 51, 52), lactobacilli (SigH) (53), staphylococci (SigH and/or ComK) (54), and Listeria spp. (SigH and/or ComK) (55, 56), but with a low to moderate success rate so far. The absence of activation of natural transformation was explained in many cases by a partial activation of the required set of late genes (e.g., Staphylococcus aureus, Listeria monocytogenes, and Lactobacillus sakei) (53–56), the implication of more than one master regulator (e.g., S. aureus) (54), the requirement of a coactivator (e.g., ComW in S. pneumoniae) (52), or a limited access of DNA to the uptake machinery by cell wall compounds (e.g., S. pyogenes) (47, 51). Among species of Streptococcaceae that were not shown to develop spontaneous competence, this strategy was only applied successfully to S. thermophilus (7, 9).
We fruitfully applied this experimental design to activate natural transformation in L. lactis subsp. cremoris KW2, a strain isolated from plant material. The in silico analysis of key late competence genes in the four other sequenced dairy isolates of cremoris strains suggests that competence is either affected or impaired. In agreement, our attempts to apply our transformation protocol to strain MG1363, which contains only two pseudogenes (point or insertion mutations) among essential late genes for DNA transformation, were unsuccessful (data not shown). Regarding strains belonging to the subspecies lactis (sequenced strains IL1403, CV56, KF147, and IO-1), two environmental isolates (KF147 and IO-1) contain the whole set of late genes, while the two others harbor one pseudogene (i.e., dprA in IL1403 of dairy origin and comGA in CV56 from vaginal flora) (data not shown). From this analysis, it is likely that all of the above-mentioned strains of dairy origin are unable to develop an efficient transformation process, potentially due to their domestication in the milk niche. Nevertheless, the inactivation of com genes could be restricted to only one pseudogene such as found in IL1403, which suggests that a repair strategy could be attempted for the more promising strains.
An important outstanding issue is the discovery of suitable conditions for the spontaneous development of competence in L. lactis and the identification of the molecular mechanism(s) controlling competence activation or repression. Given that no genuine orthologues for competence signaling proteins (i.e., ComDE and ComR streptococci) have been found yet in L. lactis, we decided to target the well-known Clp machinery involved in the negative control of competence induction in both bacilli and streptococci (12, 13). Interestingly, our results suggest that the MecA-ClpCP machinery might be involved in the degradation of ComX as reported in B. subtilis, S. thermophilus, and S. mutans (13), thereby contributing to the posttranslational control of competence development in L. lactis.
To conclude, this work reveals that natural DNA transformation is functional in the L. lactis species. This enables various chromosomal modifications that would favor the genome plasticity in this species, ruling out an exclusive nutritional role as previously proposed (30). This important step will not only facilitate the design of novel strains for industrial applications but will pave the way for the discovery of the regulatory mechanisms responsible for its control.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown with shaking at 37°C in lysogeny broth (57). Plasmids derived from pMG36e (58) and pG+host9 (59) were constructed in E. coli strains TG1 (57) and EC1000 (60), respectively. L. lactis was cultivated in M17 (Becton Dickinson) or CDM (61) at 30°C without agitation. M17 and CDM were supplemented with either 0.5% (wt/vol) glucose or 1% (wt/vol) xylose (named M17G, CDMG, and M17X, CDMX, respectively). Solid agar plates were prepared by adding 2% (wt/vol) agar to the medium. When required, 5 μg ml−1 of erythromycin, 1 mg ml−1 of streptomycin, or 10 μg ml−1 of chloramphenicol was added to the medium for L. lactis, and 250 μg ml−1 of erythromycin, 250 μg ml−1 of ampicillin, or 10 μg ml−1 of chloramphenicol was added to the medium for E. coli.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F'[traD36 proAB+ lacIq lacZΔM15] | 57 |
| EC1000 | Kmr; recA+; MC1000 containing a copy of the repA gene from pWV01 in its chromosome | 60 |
| L. lactis | ||
| MG1363 | Laboratory strain, dairy origin | 72 |
| KW2 | Wild-type isolate from corn fermentation | 37 |
| BLD101 | KW2 kw2_0563::PcomGA-luxAB | This study |
| BLD102 | BLD101 comEC::P32-cat | This study |
| FT103 | BLD101 comX::P32-cat | This study |
| FT107 | BLD101 mecA::P32-cat | This study |
| FT105 | BLD101 clpC::P32-cat | This study |
| FT106 | BLD101 clpE::P32-cat | This study |
| FT104 | BLD101 clpP::P32-cat | This study |
| Plasmids | ||
| pGEM-T Easy | Apr; cloning vector | Promega |
| pG+host9 | Emr Ts | 59 |
| pMG36eT | Emr; E. coli-L. lactis shuttle vector containing the P32 constitutive promoter from L. lactis | 64 |
| pJIM4900 | Emr Ts; pG+host9 derivative containing the luxAB genes of Photorhabdus luminescens | E. Guédon (laboratory collection) |
| pSEUDOPusp45GFP | Emr; suicide vector containing the llmg_pseudo_10(kw2_0563)::Pusp45-gfp+ insertion cassette | 65 |
| pUC18Cm | Apr Cmr; pUC18 derivative containing the P32-cat cassette | 73 |
| pUC18Ery | Apr Emr; pUC18 derivative containing an erythromycin resistance marker | 67 |
| pNZ5319 | Emr Cmr; pACYC184 derivative containing the P32-cat cassette surrounded by lox sites | 68 |
| pGIBLD101 | Emr Ts; pG+host9 derivative containing the llmg_pseudo_10 (kw2_0563)::PcomGA-luxAB insertion cassette | This study |
| pGIBLD001 | Emr Ts, pG+host9 derivative carrying comX under the control of the constitutive promoter P32 | This study |
| pGIFPT001 | Emr Ts, pG+host9 derivative carrying comX under the control of the inducible promoter PxylT | This study |
| pGIBLD201 | Apr; pGEM-T Easy derivative carrying the rpsL* gene | This study |
| pGIBLD102 | Apr Emr Cmr; pUC18Ery derivative allowing the insertion of P32-cat at the comEC locus | This study |
Emr, Apr, Cmr, and Kmr: erythromycin, ampicillin, chloramphenicol, and kanamycin resistance, respectively. Ts, thermosensitive RepA protein.
Detection of absorbance and luminescence.
Growth (OD600) and luciferase (Lux) activity were monitored at 10-min intervals in a Varioskan Flash multi-mode reader (Thermo Fisher) as previously described (8). The luciferase activity is expressed in RLU, and the specific luciferase activity is expressed in RLU OD600−1.
DNA techniques and electrotransformation.
General molecular biology techniques were performed according to the instructions provided by Sambrook et al. (57). Electrotransformation of E. coli (62) and L. lactis (4) was performed as previously described. The electrotransformed cells of L. lactis were immediately resuspended in 1 ml of M17G, followed by incubation for 1 h at 30°C. Chromosomal DNA of L. lactis was prepared as previously described (63). PCRs were performed with the Phusion DNA polymerase (NEB) in a GeneAmp PCR system 2400 (Applied Biosystems). The primers used in this study were purchased from Eurogentec and are listed in Table 2.
TABLE 2.
Primers used in this study
| Function and primer | Sequence (5′–3′) | Target or source |
|---|---|---|
| Construction of pGIBLD001 (P32-comX) | ||
| ComXSDLLCup | AAAAGAGCTCAATTATGAAAAAGAGG | comX |
| ComXSDLLCdown | AAAACTGC AGTTAATCATCATCTCG | comX |
| pMGP32UpMfeI | ATATCAATTGGTCCTCGGGATAT GATAAG | P32-comX cassette from pGIBLD011 |
| pMGTerDown | GACTTTGAACCTCAACTCC | P32-comX cassette from pGIBLD011 |
| Construction of pGIFPT001 (PxylT-comX) | ||
| pGhxylTcomXmaI | GTGGATCCCCCGGGCTGCAGGGTAGCGCAGAACGAGATTCACTTG | PxylT |
| pxylTcomXrec | GATAGTAACTCCTTAATTTTTATTTGC | PxylT |
| pGhxlTorfXMGrec | GCAAATAAAAATTAAGGAGTTACTATCATGGCAATCGTTTCAGCAGAAAAATTCG | pGIBLD001 |
| pGhxlTorfXMGXmaI | CCTGCAGCCCGGGGGATCCAC | pGIBLD001 |
| Construction of the PcomGA-luxAB reporter strain | ||
| LuxLLCf1 | ATAGTCTCGAGTTTAAGCAATTGAATCGCTAG | PcomGA promoter and pGIBLD012 |
| LuxLLCr1 | GCAAAAAGTTTCCAAATTTCATACTAGAATATACGCAATTTG | PcomGA promoter |
| LuxLLCf2 | CAAATTGCGTATATTCTAGTATGAAATTTGGAAACTTTTTGC | luxAB |
| LuxLLCr2 | GCGAAAGGATCCCTATTAGGTATATTCCATGTGG | luxAB |
| P3pseudoLLC | GCTCCCTCGAGGGCGGCTCTGTTGGATTAATATATGG | pGIBLD012 |
| Sequencing of rpsL | ||
| RpsLUnivUP | ATGCCTACAATTAACCAAT | rpsL |
| RpsLUnivDN | CACCGTATTTAGAACGG | rpsL |
| Amplification of rpsL | ||
| LLCdacARpsL | AGTAGTATCAGCACTGACAGC | rpsL |
| LLCfusARpsL | ACACCTTTGTTCTTGAAGG | rpsL |
| Construction of the comEC disruption mutant | ||
| ComECLLCUp | AAAGAGCTCAAAATAAAAATGAAATTATGG | comEC |
| ComECLLCDown | AAAGCTAGCGGGAAAAAATTGTGAATTAC | comEC |
| CatUpSpeI | AAAAACTAGTGCAGTTTAAATTCGGTCCTCGG | P32-cat cassette from pNZ5319 |
| CatDownSpeI | AAAAACTAGTGTACAGTCGGCATTATCTCAT | P32-cat cassette from pNZ5319 |
| Amplification of P32-cat cassette for construction of mecA, clpC, clpE, clpP, and comX deletion mutants | ||
| fgt02Fcat | TCCTCGGGATATGATAAGATTAATAG | P32-cat cassette from pUC18cm |
| fgt02RVcat | TCTCATATTATAAAAGCCAGTCATTAG | P32-cat cassette from pUC18cm |
| Construction and validation of the mecA deletion mutant | ||
| fgt01FmecArec | CTTTAATGATGGAATGATTG | mecA |
| fgt01RVmecArec | CTATTAATCTTATCATATCCCGAGGATCCATATAACTATATGAAACC | mecA |
| fgt03FmecArec | CTAATGACTGGCTTTTATAATATGAGACTTAGAAAAATCTAAATATGGTTG | mecA |
| fgt03RVmecArec | GAAGATTTTTAATTTCAAGTGTAG | mecA |
| MecAKOF | TCAGTACCGAAAAACGAATG | mecA |
| MecAKORV | ATTTACCAGTTCCGTTAGG | mecA |
| Construction and validation of the clpC deletion mutant | ||
| ClpCUPF | CTTTGGGTTCTAATTTATC | clpC |
| ClpCUPRVRec | CTATTAATCTTATCATATCCCGAGGACGTTGGTGTATATTTTAC | clpC |
| ClpCDownFRec | CTAATGACTGGCTTTTATAATATGAGATAGAAATAAAGGAAAGGAC | clpC |
| ClpCDownRV | TTGCTTTAAGGATAGTTTC | clpC |
| ClpCFdiag | AGAAGCCAATAATGACGATG | clpC |
| ClpCRVdiag | AGAATTCTGATGATGCACAGTC | clpC |
| Construction and validation of the clpE deletion mutant | ||
| ClpEUPF | CAGAGGACAGTAATATTTTT | clpE |
| CP_clpEUPRVRec | CTATTAATCTTATCATATCCCGAGGACTCCTCTTAATTCTCAGTAAT | clpE |
| ClpEDNFRec | CTAATGACTGGCTTTTATAATATGAGAGTCAGTAAAATAGTATTAGTGACA | clpE |
| ClpEDNR | GATGCTGGAACAATATTT | clpE |
| ClpEFdiag | CAAGGAACAGTGGAGCTTTTA | clpE |
| ClpERVdiag | GTAATTGATCCTGTTGGAGTTG | clpE |
| Construction and validation of the clpP deletion mutant | ||
| ClpPUPF | AAAGGTTGAATTTTCTG | clpP |
| ClpPUPRVRec | CTATTAATCTTATCATATCCCGAGGAAGATATGGACTTAATTTAGG | clpP |
| ClpPDNFRec | CTAATGACTGGCTTTTATAATATGAGAAATAAGCAATAAAGTCCTAG | clpP |
| ClpPDNR | TCCTTTACAGTTTTTAGATG | clpP |
| ClpPFdiag | AGAGGAGTTGTTCAAGAAGAAAG | clpP |
| ClpPRVdiag | AAATTACTTGGAGTTAAGGCC | clpP |
| Construction and validation of the comX deletion mutant | ||
| ComXUPF | GAAAAACGAAATTCAAAC | comX |
| ComXUPRVRec | CTATTAATCTTATCATATCCCGAGGAGGGAATTCTATTATAATGTTG | comX |
| ComXDNFRec | CTAATGACTGGCTTTTATAATATGAGAAGTTAAAACGGATACATAAG | comX |
| ComXDNR | TTAAGAGATTAGCTAAACATC | comX |
| ComXRVdiag | AACAGCTCAACGATTCTTTC | comX |
| ComXFdiag | ATTCCTTAGAAAGGAGGTGATC | comX |
Construction of plasmid pGIBLD001 for constitutive comX expression.
As a representative of the cremoris subspecies, the comX gene from the laboratory strain MG1363 was initially chosen. ComX proteins of this subspecies are highly conserved with at least 98% of identity. The comX gene was amplified by PCR using the primers ComXSDLLCup and ComXSDLLCdown and inserted into plasmid pMG36eT (64) under the control of the constitutive P32 promoter by SacI/PstI cloning, yielding the intermediate plasmid pGIBLD011. The P32-comX fusion from pGIBLD011 was amplified by PCR with the primers pMGP32UpMfeI and pMGTerDown, digested by MfeI and KpnI, and cloned into the EcoRI/KpnI-digested thermosensitive pG+host9 vector. The resulting plasmid was named pGIBLD001.
Construction of plasmid pGIFPT001 for inducible comX expression.
The xylT promoter region (PxylT) of KW2 was identified by homology with PxylT from L. lactis subsp. lactis NCDO2118 described by Miyoshi et al. (43). PxylT from KW2 was amplified by PCR using primers pGhxylTcomXmaI and pxylTcomXrec. In addition, pGIBLD001 plasmid was amplified by PCR using the primers pGhxlTorfXMGrec and pGhxlTorfXMGXmaI to remove the P32 promoter. These two PCR fragments were fused by overlapping PCR using the primers pGhxylTcomXmaI and pGhxlTorfXMGXmaI, digested by XmaI, and self-ligated. The resulting plasmid was named pGIFPT001.
Construction of PcomGA-luxAB reporter strain.
The PcomGA promoter was amplified by PCR from chromosomal DNA of L. lactis subsp. cremoris KW2 with primers LuxLLCf1 and LuxLLCr1 (PCR1 product). The luxAB genes were amplified by PCR from plasmid pJIM4900 with primers LuxLLCf2 and LuxLLCr2 (PCR2 product). The PcomGA-luxAB fusion was created by overlapping PCR using PCR1 and PCR2 products and primers LuxLLCf1 and LuxLLCr2. The resulting fusion was cloned into plasmid pSEUDOPusp45GFP (65) using the restriction enzymes XhoI and BamHI, yielding the intermediate plasmid pGIBLD012. In order to remove the Pusp45 promoter, the entire vector except the Pusp45 promoter was amplified by inverse PCR with the primers P3pseudoLLC and luxLLCf1 and self-ligated after XhoI digestion. The insertion cassette (llmg_pseudo_10::PcomGA-luxAB) was excised from the resulting plasmid and cloned into the pG+host9 thermosensitive vector using the restriction enzymes KpnI and EagI. The final plasmid pGIBLD101 was then electrotransformed in strain KW2 and used to integrate the PcomGA-luxAB cassette at locus kw2_0563 (llmg_pseudo_10 in MG1363) by double homologous recombination as previously described (66), resulting in the reporter strain BLD101 (KW2 PcomGA-luxAB).
Isolation of a rpsL mutant conferring resistance to streptomycin.
Spontaneous streptomycin-resistant MG1363 clones were isolated on plates containing streptomycin (1 mg ml−1). After sequencing of the rpsL gene with the primers RpsLUnivUP and RpsLUnivDN, one spontaneous mutant containing the K56I mutation into the ribosomal protein S12 was selected (see Fig. S1 in the supplemental material). A 3.7-kb fragment containing the rpsL mutated gene was amplified by PCR with the primers LLCdacARpsL and LLCfusARpsL and cloned into the pGEM-T Easy vector (Promega), yielding plasmid pGIBLD201. This plasmid was used as a template to amplify a 3.7-kb DNA fragment by PCR with the primers LLCdacARpsL and LLCfusARpsL. This DNA fragment was used as donor DNA in natural transformation assays.
Standard natural transformation assay.
The BLD101 reporter strain (KW2 PcomGA-luxAB) carrying the pGIBLD001 plasmid [BLD101(pGIBLD001)] was grown overnight in M17G containing erythromycin at 30°C. Then, 1.5 ml of the preculture was diluted in 8.5 ml of fresh M17G medium containing erythromycin to restart the culture. After 2 h of growth, the cells were washed twice in distilled water, and the OD600 was adjusted to 0.05 in CDM containing erythromycin and supplemented with either 5% (vol/vol) glycerol or 5% (wt/vol) mannitol used as potential osmostabilizers. Typically, 5 μg of DNA was added in 300 μl of inoculated medium, and the culture was further incubated for 6 h at 30°C. For the BLD101 reporter strain carrying the pGIFPT001 plasmid [BLD101(pGIFPT001)], the protocol was slightly modified. After the washing steps with distilled water, the cells were resuspended in M17X (without osmostabilizers) instead of CDM. In addition, the incubation in the presence of DNA was extended from 6 to 24 h. The cells were then spread onto M17G agar plates supplemented with appropriate antibiotics, and the CFU were counted after 48 h of incubation. The transformation frequency was calculated as the number of antibiotic-resistant CFU ml−1 divided by the total number of viable CFU ml−1. In the case of streptomycin-resistant transformants, the antibiotic-resistant CFU ml−1 corresponds to the number of transformants obtained in the presence of DNA minus the number of spontaneous transformants obtained under conditions where no DNA is added in the culture. The transfer of the mutation conferring streptomycin resistance was confirmed by DNA sequencing of the rpsL gene after its amplification by PCR using the primers RpsLUnivUP and RpsLUnivDN.
Disruption of comEC by natural transformation.
A comEC-containing DNA fragment of ∼3.2 kb was amplified by PCR with the primers ComECLLCUp and ComECLLCDown. Then, the PCR product was digested by SacI and NheI and cloned into the SacI/XbaI-digested suicide plasmid pUC18Ery (67), yielding the intermediate plasmid pGIBLD013. To generate a comEC disruption cassette that allows the selection of double crossing-over recombinants, the P32-cat fusion conferring resistance to chloramphenicol was cloned in the middle of the comEC gene. For this purpose, the P32-cat cassette was amplified by PCR from plasmid pNZ5319 (68) with primers CatUpSpeI and CatDownSpeI. The amplification product was digested by SpeI and cloned into the XbaI-digested pGIBLD013, yielding plasmid pGIBLD102. This suicide plasmid was used to generate high quantity of donor DNA by PCR amplification for comEC disruption. The resulting donor DNA was used to transform clone 2 of strain BLD101(pGIBLD001). The insertion of the P32-cat cassette in the comEC gene of transformants was validated by PCR (Table 2).
Deletion of mecA, clpC, clpE, clpP, and comX genes by natural transformation.
The mecA, clpC, clpE, clpP, and comX genes were similarly inactivated by the exchange of their CDSs by the P32-cat cassette using double crossing-over events. For this purpose, overlapping PCR products containing P32-cat flanked by two recombination arms of ∼1.5 kb (upstream and downstream homologous regions) were generated as previously reported (8). Briefly, upstream, downstream, and P32-cat fragments were separately amplified by PCR, purified, mixed in equimolar concentration, and assembled by overlapping PCR by using the most external primers (see the list of primers in Table 2). The obtained overlapping PCR product (5 μg) was used as donor DNA for natural transformation of strain BLD101(pGIFPT001). The correct insertion of P32-cat in each targeted locus of the transformants was validated by PCR (see the list of primers in Table 2). To evaluate the impact of the deleted gene in the absence of comX expression, the thermosensitive vector pGIFPT001 was cured by growing the strains overnight at 37°C without erythromycin. The cultures were subsequently diluted and plated on M17G agar without erythromycin at 30°C. The resulting colonies were streaked in parallel on M17G plates with or without erythromycin. The absence of plasmid pGIFPT001 in Erys clones was validated by PCR. To evaluate the impact of these different deletions in the presence of low comX expression, the noncured mutant strains were analyzed in the absence of the xylose inducer.
Transcriptome analyses.
BLD101(pG+host9) (control strain with empty vector, Ctl−) and BLD101(pGIBLD001) (clone 2, ComX+) strains were grown in CDMG supplemented with 5% mannitol. When cultures reached the mid-log phase (OD600 of ∼0.3), the cells were harvested and washed twice in ice-cold phosphate-buffered saline buffer. An OD600 of ∼0.3 was chosen in order to obtain a sufficient amount of cells for RNA extraction with a reasonable PcomGA induction (60% of the maximum level). Pellets were frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated by using an RNeasy minikit (Qiagen). RNA was treated with DNase, purified using the RNA cleanup protocol from the RNeasy minikit (Qiagen), and stored at −80°C. The integrity of the RNA was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Removal of rRNA was performed using the RiboMinus bacterial module (Invitrogen), and the transcriptome library for sequencing was constructed according to the NEXTflexRapid directional RNA-Seq library protocol (Bioo Scientific). The prepared library (insert size of 130 to 580 bp) was validated for quality by running an aliquot on an Agilent High Sensitivity DNA kit chip. Sequencing was performed on an Illumina NextSeq 500 instrument with a 75-nt paired-end protocol. The Illumina raw reads were quality checked using by FastQC v2.2 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). RNA extraction, library construction, and Illumina sequencing were performed by Genotypic Technology (Bangalore, India).
Treatment of the Illumina sequence data for abundance estimation and differential expression between control and ComX+ strains was performed with the DESeq2 (69), edgeR (70), and DEXUS (71) packages. Genes were considered upregulated in the ComX+ strain when the calculated FC was ≥2.0 with the three packages. When the upregulated genes were preceded by a predicted ComX-binding motif (Com-box), adjacent downstream genes with a calculated FC ≥ 1.5 with the three packages were also retained (see Tables S2 and S3 in the supplemental material).
Accession number(s).
The complete data set from this study is available in the GEO database (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE86476).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully thank J. W. Veening and W. J. Kelly for providing plasmid pSEUDOPusp45GFP and strain KW2, respectively. We warmly thank Johann Mignolet for critically reading the manuscript.
P.H. is Senior Research Associate at Fonds De La Recherche Scientifique (FNRS).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01074-17.
REFERENCES
- 1.Cavanagh D, Fitzgerald GF, McAuliffe O. 2015. From field to fermentation: the origins of Lactococcus lactis and its domestication to the dairy environment. Food Microbiol 47:45–61. doi: 10.1016/j.fm.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Bahey-El-Din M, Gahan CG, Griffin BT. 2010. Lactococcus lactis as a cell factory for delivery of therapeutic proteins. Curr Gene Ther 10:34–45. doi: 10.2174/156652310790945557. [DOI] [PubMed] [Google Scholar]
- 3.Morello E, Bermudez-Humaran LG, Llull D, Sole V, Miraglio N, Langella P, Poquet I. 2008. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J Mol Microbiol Biotechnol 14:48–58. doi: 10.1159/000106082. [DOI] [PubMed] [Google Scholar]
- 4.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills DA. 2001. Mutagenesis in the postgenomics era: tools for generating insertional mutations in the lactic acid bacteria. Curr Opin Biotechnol 12:503–509. doi: 10.1016/S0958-1669(00)00254-8. [DOI] [PubMed] [Google Scholar]
- 6.van Pijkeren JP, Britton RA. 2014. Precision genome engineering in lactic acid bacteria. Microb Cell Fact 13(Suppl 1):S10. doi: 10.1186/1475-2859-13-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomqvist T, Steinmoen H, Havarstein LS. 2006. Natural genetic transformation: a novel tool for efficient genetic engineering of the dairy bacterium Streptococcus thermophilus. Appl Environ Microbiol 72:6751–6756. doi: 10.1128/AEM.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol 192:1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontaine L, Dandoy D, Boutry C, Delplace B, de Frahan MH, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. Development of a versatile procedure based on natural transformation for marker-free targeted genetic modification in Streptococcus thermophilus. Appl Environ Microbiol 76:7870–7877. doi: 10.1128/AEM.01671-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dandoy D, Fremaux C, de Frahan MH, Horvath P, Boyaval P, Hols P, Fontaine L. 2011. The fast milk acidifying phenotype of Streptococcus thermophilus can be acquired by natural transformation of the genomic island encoding the cell envelope proteinase PrtS. Microb Cell Fact 10(Suppl 1):S21. doi: 10.1186/1475-2859-10-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. 2014. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12:181–196. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 12.Hamoen LW, Venema G, Kuipers OP. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 13.Fontaine L, Wahl A, Flechard M, Mignolet J, Hols P. 2015. Regulation of competence for natural transformation in streptococci. Infect Genet Evol 33:343–360. doi: 10.1016/j.meegid.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu Rev Microbiol 60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 15.Palchevskiy V, Finkel SE. 2006. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol 188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol 181:5004–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peterson SN, Sung CK, Cline R, Desai BV, Snesrud EC, Luo P, Walling J, Li H, Mintz M, Tsegaye G, Burr PC, Do Y, Ahn S, Gilbert J, Fleischmann RD, Morrison DA. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol Microbiol 51:1051–1070. doi: 10.1046/j.1365-2958.2003.03907.x. [DOI] [PubMed] [Google Scholar]
- 18.Campbell EA, Choi SY, Masure HR. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol 27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 19.Luo P, Morrison DA. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J Bacteriol 185:349–358. doi: 10.1128/JB.185.1.349-358.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Claverys JP, Martin B, Polard P. 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol Rev 33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 21.Fontaine L, Goffin P, Dubout H, Delplace B, Baulard A, Lecat-Guillet N, Chambellon E, Gardan R, Hols P. 2013. Mechanism of competence activation by the ComRS signalling system in streptococci. Mol Microbiol 87:1113–1132. doi: 10.1111/mmi.12157. [DOI] [PubMed] [Google Scholar]
- 22.Martin B, Soulet AL, Mirouze N, Prudhomme M, Mortier-Barriere I, Granadel C, Noirot-Gros MF, Noirot P, Polard P, Claverys JP. 2013. ComE/ComE∼P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol Microbiol 87:394–411. doi: 10.1111/mmi.12104. [DOI] [PubMed] [Google Scholar]
- 23.Gardan R, Besset C, Gitton C, Guillot A, Fontaine L, Hols P, Monnet V. 2013. Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J Bacteriol 195:1845–1855. doi: 10.1128/JB.02196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks LR, Mashburn-Warren L, Federle MJ, Hakansson AP. 2014. Streptococcus pyogenes biofilm growth in vitro and in vivo and its role in colonization, virulence, and genetic exchange. J Infect Dis 210:25–34. doi: 10.1093/infdis/jiu058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol 78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morrison DA, Guedon E, Renault P. 2013. Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus. J Bacteriol 195:2612–2620. doi: 10.1128/JB.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zaccaria E, van Baarlen P, de Greef A, Morrison DA, Smith H, Wells JM. 2014. Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS One 9:e99394. doi: 10.1371/journal.pone.0099394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolotin A, Wincker P, Mauger S, Jaillon O, Malarme K, Weissenbach J, Ehrlich SD, Sorokin A. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis subsp. lactis IL1403. Genome Res 11:731–753. doi: 10.1101/gr.GR-1697R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wydau S, Dervyn R, Anba J, Dusko ES, Maguin E. 2006. Conservation of key elements of natural competence in Lactococcus lactis subsp. FEMS Microbiol Lett 257:32–42. doi: 10.1111/j.1574-6968.2006.00141.x. [DOI] [PubMed] [Google Scholar]
- 31.Redon E, Loubiere P, Cocaign-Bousquet M. 2005. Transcriptome analysis of the progressive adaptation of Lactococcus lactis to carbon starvation. J Bacteriol 187:3589–3592. doi: 10.1128/JB.187.10.3589-3592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ercan O, Wels M, Smid EJ, Kleerebezem M. 2015. Genome-wide transcriptional responses to carbon starvation in nongrowing Lactococcus lactis. Appl Environ Microbiol 81:2554–2561. doi: 10.1128/AEM.03748-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachmann H, de Wilt L, Kleerebezem M, van Hylckama Vlieg JE. 2010. Time-resolved genetic responses of Lactococcus lactis to a dairy environment. Environ Microbiol 12:1260–1270. doi: 10.1111/j.1462-2920.2010.02168.x. [DOI] [PubMed] [Google Scholar]
- 34.Berge M, Mortier-Barriere I, Martin B, Claverys JP. 2003. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol Microbiol 50:527–536. doi: 10.1046/j.1365-2958.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 35.Mirouze N, Berge MA, Soulet AL, Mortier-Barriere I, Quentin Y, Fichant G, Granadel C, Noirot-Gros MF, Noirot P, Polard P, Martin B, Claverys JP. 2013. Direct involvement of DprA, the transformation-dedicated RecA loader, in the shut-off of pneumococcal competence. Proc Natl Acad Sci U S A 110:E1035–E1044. doi: 10.1073/pnas.1219868110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laurenceau R, Pehau-Arnaudet G, Baconnais S, Gault J, Malosse C, Dujeancourt A, Campo N, Chamot-Rooke J, Le CE, Claverys JP, Fronzes R. 2013. A type IV pilus mediates DNA binding during natural transformation in Streptococcus pneumoniae. PLoS Pathog 9:e1003473. doi: 10.1371/journal.ppat.1003473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly WJ, Altermann E, Lambie SC, Leahy SC. 2013. Interaction between the genomes of Lactococcus lactis and phages of the P335 species. Front Microbiol 4:257. doi: 10.3389/fmicb.2013.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol 194:3774–3780. doi: 10.1128/JB.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardan R, Besset C, Guillot A, Gitton C, Monnet V. 2009. The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J Bacteriol 191:4647–4655. doi: 10.1128/JB.00257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmann H, Santos F, Kleerebezem M, van Hylckama Vlieg JE. 2007. Luciferase detection during stationary phase in Lactococcus lactis. Appl Environ Microbiol 73:4704–4706. doi: 10.1128/AEM.02807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutry C, Wahl A, Delplace B, Clippe A, Fontaine L, Hols P. 2012. Adaptor protein MecA is a negative regulator of the expression of late competence genes in Streptococcus thermophilus. J Bacteriol 194:1777–1788. doi: 10.1128/JB.06800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hosoya Y, Okamoto S, Muramatsu H, Ochi K. 1998. Acquisition of certain streptomycin-resistant (str) mutations enhances antibiotic production in bacteria. Antimicrob Agents Chemother 42:2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyoshi A, Jamet E, Commissaire J, Renault P, Langella P, Azevedo V. 2004. A xylose-inducible expression system for Lactococcus lactis. FEMS Microbiol Lett 239:205–212. doi: 10.1016/j.femsle.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 44.Lokman BC, Heerikhuisen M, Leer RJ, van den Broek A, Borsboom Y, Chaillou S, Postma PW, Pouwels PH. 1997. Regulation of expression of the Lactobacillus pentosus xylAB operon. J Bacteriol 179:5391–5397. doi: 10.1128/jb.179.17.5391-5397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miwa Y, Nakata A, Ogiwara A, Yamamoto M, Fujita Y. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res 28:1206–1210. doi: 10.1093/nar/28.5.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahl A, Servais F, Drucbert AS, Foulon C, Fontaine L, Hols P. 2014. Control of natural transformation in salivarius streptococci through specific degradation of σX by the MecA-ClpCP protease complex. J Bacteriol 196:2807–2816. doi: 10.1128/JB.01758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mashburn-Warren L, Morrison DA, Federle MJ. 2012. The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone. J Bacteriol 194:4589–4600. doi: 10.1128/JB.00830-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kovacs AT, Smits WK, Mironczuk AM, Kuipers OP. 2009. Ubiquitous late competence genes in Bacillus species indicate the presence of functional DNA uptake machineries. Environ Microbiol 11:1911–1922. doi: 10.1111/j.1462-2920.2009.01937.x. [DOI] [PubMed] [Google Scholar]
- 49.Kovacs AT, Eckhardt TH, van Hartskamp M, van Kranenburg R, Kuipers OP. 2013. Functional analysis of the ComK protein of Bacillus coagulans. PLoS One 8:e53471. doi: 10.1371/journal.pone.0053471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mironczuk AM, Kovacs AT, Kuipers OP. 2008. Induction of natural competence in Bacillus cereus ATCC 14579. Microb Biotechnol 1:226–235. doi: 10.1111/j.1751-7915.2008.00023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Opdyke JA, Scott JR, Moran CP Jr. 2003. Expression of the secondary sigma factor sigmaX in Streptococcus pyogenes is restricted at two levels. J Bacteriol 185:4291–4297. doi: 10.1128/JB.185.15.4291-4297.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo P, Li H, Morrison DA. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol Microbiol 54:172–183. doi: 10.1111/j.1365-2958.2004.04254.x. [DOI] [PubMed] [Google Scholar]
- 53.Schmid S, Bevilacqua C, Crutz-Le Coq AM. 2012. Alternative sigma factor sigmaH activates competence gene expression in Lactobacillus sakei. BMC Microbiol 12:32. doi: 10.1186/1471-2180-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fagerlund A, Granum PE, Havarstein LS. 2014. Staphylococcus aureus competence genes: mapping of the SigH, ComK1, and ComK2 regulons by transcriptome sequencing. Mol Microbiol 94:557–579. doi: 10.1111/mmi.12767. [DOI] [PubMed] [Google Scholar]
- 55.Medrano R V, Morikawa K. 2016. Listeria monocytogenes σH contributes to expression of competence genes and intracellular growth. J Bacteriol 198:1207–1217. doi: 10.1128/JB.00718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, Orsi RH, Boor KJ, Wiedmann M, Guariglia-Oropeza V. 2016. An advanced bioinformatics approach for analyzing RNA-seq data reveals σH-dependent regulation of competence genes in Listeria monocytogenes. BMC Genomics 17:115. doi: 10.1186/s12864-016-2432-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 58.van de Guchte M, van der Vossen JM, Kok J, Venema G. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 55:224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sissler M, Delorme C, Bond J, Ehrlich SD, Renault P, Francklyn C. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc Natl Acad Sci U S A 96:8985–8990. doi: 10.1073/pnas.96.16.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dower WJ, Miller JF, Ragsdale CW. 1988. High-efficiency transformation of Escherichia coli by high-voltage electroporation. Nucleic Acids Res 16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferain T, Hobbs JN Jr, Richardson J, Bernard N, Garmyn D, Hols P, Allen NE, Delcour J. 1996. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J Bacteriol 178:5431–5437. doi: 10.1128/jb.178.18.5431-5437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fontaine L, Hols P. 2008. The inhibitory spectrum of thermophilin 9 from Streptococcus thermophilus LMD-9 depends on the production of multiple peptides and the activity of BlpG(St), a thiol-disulfide oxidase. Appl Environ Microbiol 74:1102–1110. doi: 10.1128/AEM.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Overkamp W, Beilharz K, Detert Oude WR, Solopova A, Karsens H, Kovacs A, Kok J, Kuipers OP, Veening JW. 2013. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl Environ Microbiol 79:6481–6490. doi: 10.1128/AEM.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biswas I, Gruss A, Ehrlich SD, Maguin E. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol 175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Kranenburg R, Marugg JD, van Swam II, Willem NJ, de Vos WM. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol 24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 68.Lambert JM, Bongers RS, Kleerebezem M. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl Environ Microbiol 73:1126–1135. doi: 10.1128/AEM.01473-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klambauer G, Unterthiner T, Hochreiter S. 2013. DEXUS: identifying differential expression in RNA-Seq studies with unknown conditions. Nucleic Acids Res 41:e198. doi: 10.1093/nar/gkt834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goffin P, Lorquet F, Kleerebezem M, Hols P. 2004. Major role of NAD-dependent lactate dehydrogenases in aerobic lactate utilization in Lactobacillus plantarum during early stationary phase. J Bacteriol 186:6661–6666. doi: 10.1128/JB.186.19.6661-6666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.