ABSTRACT
New York City (NYC) is pioneering green infrastructure with the use of bioswales and other engineered soil-based habitats to provide stormwater infiltration and other ecosystem functions. In addition to avoiding the environmental and financial costs of expanding traditional built infrastructure, green infrastructure is thought to generate cobenefits in the form of diverse ecological processes performed by its plant and microbial communities. Yet, although plant communities in these habitats are closely managed, we lack basic knowledge about how engineered ecosystems impact the distribution and functioning of soil bacteria. We sequenced amplicons of the 16S ribosomal subunit, as well as seven genes associated with functional pathways, generated from both total (DNA-based) and expressed (RNA) soil communities in the Bronx, NYC, NY, in order to test whether bioswale soils host characteristic bacterial communities with evidence for enriched microbial functioning, compared to nonengineered soils in park lawns and tree pits. Bioswales had distinct, phylogenetically diverse bacterial communities, including taxa associated with nutrient cycling and metabolism of hydrocarbons and other pollutants. Bioswale soils also had a significantly greater diversity of genes involved in several functional pathways, including carbon fixation (cbbL-R [cbbL gene, red-like subunit] and apsA), nitrogen cycling (noxZ and amoA), and contaminant degradation (bphA); conversely, no functional genes were significantly more abundant in nonengineered soils. These results provide preliminary evidence that urban land management can shape the diversity and activity of soil communities, with positive consequences for genetic resources underlying valuable ecological functions, including biogeochemical cycling and degradation of common urban pollutants.
IMPORTANCE Management of urban soil biodiversity by favoring taxa associated with decontamination or other microbial metabolic processes is a powerful prospect, but it first requires an understanding of how engineered soil habitats shape patterns of microbial diversity. This research adds to our understanding of urban microbial biogeography by providing data on soil bacteria in bioswales, which had relatively diverse and compositionally distinct communities compared to park and tree pit soils. Bioswales also contained comparatively diverse pools of genes related to carbon sequestration, nitrogen cycling, and contaminant degradation, suggesting that engineered soils may serve as effective reservoirs of functional microbial biodiversity. We also examined both total (DNA-based) and expressed (RNA) communities, revealing that total bacterial communities (the exclusive targets in the vast majority of soil studies) were poor predictors of expressed community diversity, pointing to the value of quantifying RNA, especially when ecological functioning is considered.
KEYWORDS: 16S RNA, environmental microbiology, metagenomics, microbial ecology, soil microbiology, urban ecology
INTRODUCTION
Rapid urbanization has emerged as a primary driver of 21st century anthropogenic global change, threatening the biodiversity associated with critical ecological functions (1, 2). Among the biological communities most impacted by urban development are soil microbes, the bacteria, archaea, and fungi that collectively process the majority of nutrient and energy transformations in terrestrial ecosystems (3). Microbial lineages control plant growth by fixing and transforming atmospheric C and N (4, 5), regulating the fluxes of important greenhouse gases, including N2O, CO2, and methane (6, 7), and modulating human exposure to air-, water-, and soil-borne pollutants, like NOX, polychlorinated biphenyls (PCBs), heavy metals, and diverse hydrocarbons (8–11). Rarely considered for conservation, urban soil communities are highly fragmented, if not completely paved over, and subject to multiple acute stresses from compaction and salinization to diverse pollution inputs (12). Just as the degradation of urban soil biodiversity threatens to degrade ecosystem functioning, conservation and management of soil microbial communities hold the potential to enhance functions performed by bacteria and other soil microbiota.
Recognizing the economic and social value of ecosystem services, local governments are increasingly integrating biological communities into civic infrastructure (13). The decision in New York City (NYC), NY, to maintain tap water quality by preferentially investing in watershed ecosystems rather than filtration infrastructure is among the most well-known examples of green infrastructure (GI) approaches (14). NYC is investing extensively in urban soils as part of its GI plan for stormwater management. The city will ultimately install up to $1.5 billion in green roofs, rain gardens, and bioswales designed to divert or delay precipitation runoff from the sewer system to reduce combined sewer overflows, which presently discharge nearly 30 billion gallons of raw sewage and polluted runoff into New York waterways (15). These installations exemplify an emerging subgroup of urban microhabitats: engineered soils designed and maintained for ecofunctional performance to optimize stormwater infiltration and retention. Several elements of engineered GI soils are likely to modify microbial community composition and diversity compared to other nonengineered soil types, including urban parks, tree pits, and medians. Bioswales are protected from compaction and host-maintained plant communities, and they contain soils with specified particle size, pH, and nutrient content (16). These soils are also designed to absorb runoff containing trace metals, hydrocarbons, and other organic pollutants that may select for microbial taxa with the capacity to metabolize these inputs (17). Official city literature implicitly presumes GI installations will maintain functional microbial communities, asserting that these soils will accrue cobenefits, including the support of plant biodiversity and improved air, water, and soil quality ecosystem services that all depend in part on microbial metabolic processes (18).
Despite their importance, no published studies have quantified the microbial diversity of bioswale soils. However, it is clear from the varied effects of tillage, crop rotation, and crop succession on soil bacterial diversity (19) and functional gene differences (20) that land management practices can shape soil microbial diversity patterns. Although only a few studies have examined fine-scale patterns of urban microbial diversity, they suggest that soil communities are biogeographically structured across urban microhabitats. The microbial biodiversity of Central Park, for example, mirrors that found at global scales, with the community structure corresponding to variations in soil characteristics (21). Similarly, bacterial communities in medians (22) and fungal communities in green roofs (23) are distinct from those in city parks, indicating how urban land management can impact soil biodiversity. These findings provide urban, local-scale corroboration of studies describing biogeographically structured soil microbial communities across regional and global scales (24–26).
Here, we extend the scope of urban bacterial soil surveys based on the ribosomal RNA (rRNA) 16S gene to two unexamined types of engineered soil habitats, right-of-way and streetside bioswales, to test the hypothesis that engineered soils have distinct bacterial communities compared to nonengineered urban soils. To determine whether engineered soils have enhanced functional diversity, we used tagged amplicon sequencing to target seven genes underlying several functions related to nutrient cycling and contaminant degradation. We sampled both RNA and DNA to assess whether total diversity and composition predictably represented expressed communities. Each site was also sampled before and after precipitation events in order to account for any effect of runoff intake on community diversity.
RESULTS
Diversity patterns of total communities.
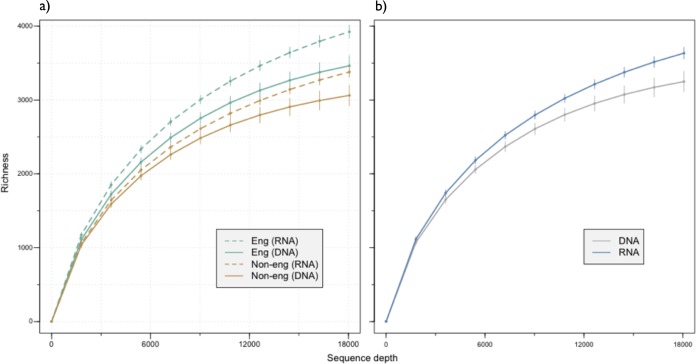
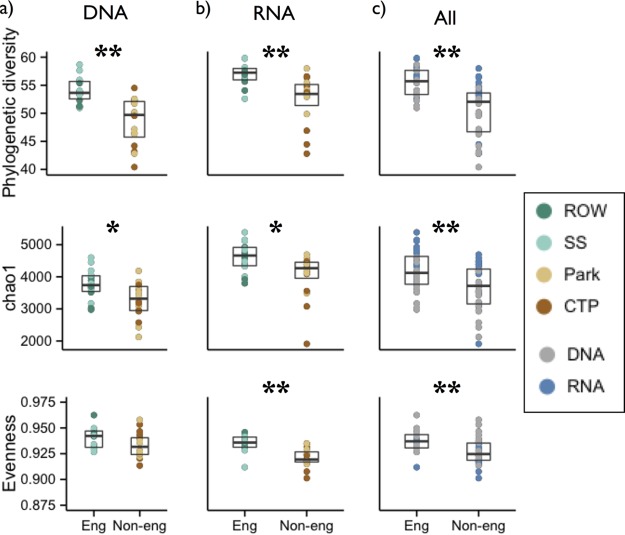
A total of 7,728 bacterial, 40 archaeal, and 202 unassigned 16S phylotypes were distributed across all samples, with an average of 2,620 (±630) phylotypes per site. Accumulation curves indicate that the majority of the resident phylotypes were sampled (Fig. 1). Based on 16S phylotypes derived from genomic DNA, total bacterial communities in engineered sites had greater phylogenetic diversity, Chao1 richness, and Shannon diversity than those in nonengineered sites (Fig. 2). Phylogenetic diversity did not vary when sampled from different time points, suggesting these results are robust to temporal variability. All samples were taken in the summer, however, so a potential seasonal effect cannot be ruled out.
FIG 1.
Accumulation curves indicating the number of phylotypes as a function of sequence coverage, with 95% confidence intervals. Phylotype accumulation given for each of the four soil types sampled (a) and for RNA versus DNA phylotypes across all samples (b). Eng, engineered; Non-eng, nonengineered.
FIG 2.
Alpha diversity results for engineered and nonengineered soil communities for DNA (a), RNA (b), and all (c) phylotypes. Three alpha diversity measures are shown: phylogenetic diversity (top row), Chao1 phylotype richness (middle row), and evenness (bottom row). Individual samples are colored by soil type (a and b) and template (c). Significance of differences was tested using the Wilcoxon test; **, P < 0.005; *, P < 0.05. Park, park lawns; CTP, conventional tree pits.
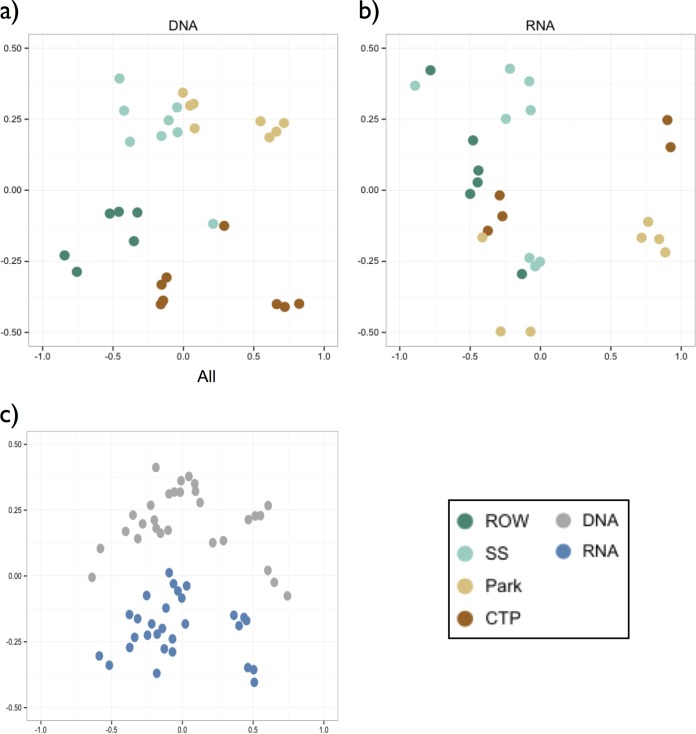
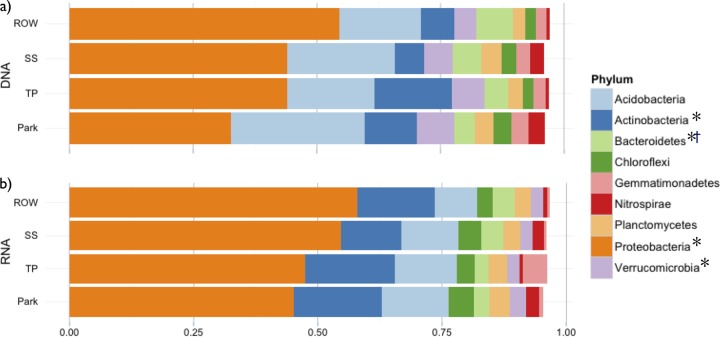
Bacterial communities in engineered soils were compositionally distinct from those in nonengineered sites (P < 0.001; Fig. 3a). Among taxa with greater than 1% relative abundance, Proteobacteria and Bacteroidetes were significantly more abundant in engineered sites, while nonengineered sites had greater abundances of Actinobacteria and Verrucomicrobia (Fig. 4a). In total, 29.4% of all phyla sampled in the study were differentially abundant (n = 34), as were 32% of all bacterial classes (n = 99). Most phyla, however, were relatively rare in all soils (75% of phyla had less than 1% average abundance). The most differentially abundant classes (fold change >4; P < 0.05) were Anaerolineae (Chloroflexi), Bacteroidia (Bacteroidetes), and Holophagae (Acidobacteria), all significantly more abundant in engineered sites, and Acidobacteria (Acidobacteria), which was more abundant in nonengineered sites. Among individual phylotypes, just over 1/10 were differentially abundant in engineered versus nonengineered sites (11.9%), with the slight majority (56%) being more abundant in nonengineered sites. Although many phylotypes were rare (36% of phylotypes had abundance of <1%), sites did not have significantly different evenness (P = 0.08), indicating that community differences were not primarily driven by abundance patterns among rare taxa. Despite the clear differences among bioswale and nonengineered soils, all sites were dominated by just five bacterial classes, which together encompassed greater than 47% of all phylotypes in each site. These included four Proteobacteria classes: Alphaproteobacteria (12.9% average abundance), Betaproteobacteria (12.5%), Deltaproteobacteria (10.7%), and Gammaproteobacteria (9.2%), as well as Acidobacteria-6 (9.2%) in the phylum Acidobacteria.
FIG 3.
Nonmetric multidimensional scaling (NMDS) ordination plots for DNA (a), RNA, colored by soil type (b), and all samples (c), colored by template. ANOSIM R values are 0.31 (a), 0.26 (b), and 0.39 (c), and clustering was significant in all cases with a P value of <0.001.
FIG 4.
Relative abundance of DNA (a) and RNA (b) phylotypes in all four soil types, for all phyla with greater than 1% overall abundances. Phyla with significantly different phylotype abundances in engineered versus nonengineered sites are indicated for DNA (*) and RNA (†) (significance adjusted for false-discovery rate of P < 0.05). TP, tree pits.
Diversity patterns of expressed communities.
Differences in community composition and diversity between engineered and nonengineered soils were evident for expressed (RNA) as well as total (DNA) communities. For both DNA and RNA, engineered sites had significantly greater phylogenetic diversity, richness (Chao1), and Shannon diversity (Fig. 2). Unlike with DNA, engineered sites had more even RNA phylotype abundances (DNA P = 0.06, RNA P < 0.001). Differences in RNA community structure were also concordant with DNA; in both cases, samples from each of the four soil types were clustered, and engineered sites had a distinct community structure (Fig. 3b).
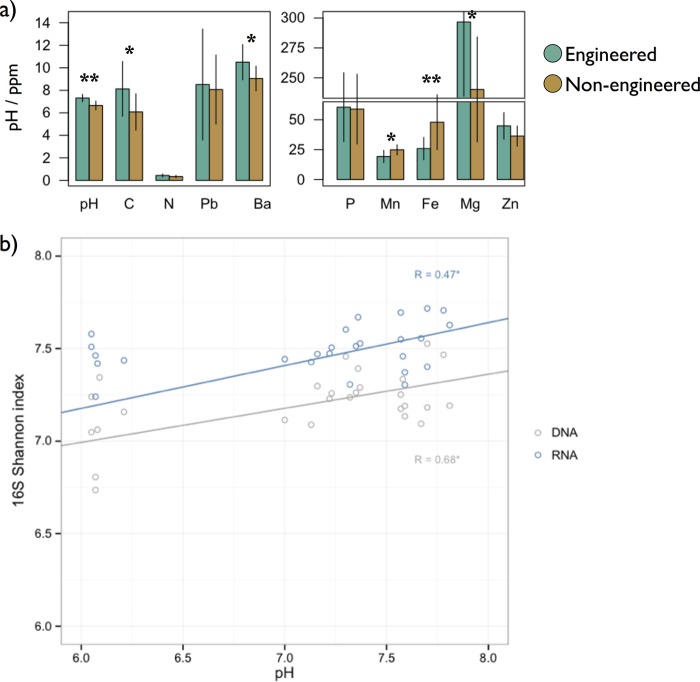
Although alpha and beta diversity analyses of DNA and RNA point to similar qualitative conclusions about how bacterial communities in engineered and nonengineered soils differ, RNA and DNA community composition patterns were nevertheless distinct (Fig. 3c). Compositional differences appeared to be driven in part by high alpha diversity of RNA phylotypes (Fig. 2c). RNA and DNA phylogenetic diversity values were highly correlated (R = 0.61, P < 0.001), as was richness, although to a lesser degree (Chao1, R = 0.378, P < 0.05). In contrast, RNA Shannon index values, which take into account richness as well as evenness, were not predictive of their corresponding DNA values (R = 0.19, P = 0.32). Evenness was also not correlated (R = −0.11, P = 0.56).
Active communities included relatively more rare taxa than total communities, as indicated by greater evenness of RNA phylotypes, and several highly abundant phyla were also differentially abundant in RNA and DNA. For example, Acidobacteria, the second most abundant phylum in total communities, was third to Actinobacteria in active communities in all four soil types (Fig. 4). In general, there was little evidence that taxa were consistently more or less abundant in RNA than in DNA, as differential abundance patterns were highly varied within and across taxonomic groups.
Soil environmental associations.
Bioswale sites had significantly higher pH than parks and conventional tree pits (Fig. 5a). Variation in pH was positively correlated with the phylogenetic diversity of DNA phylotypes (R = 0.71, P < 0.001), which was otherwise not predicted by geographic distance or any other nutrient, salt, or trace metal. RNA diversity was similarly associated with pH (R = 0.66, P < 0.001), and unlike for DNA, also with Zn and Ni content (R = 0.40, P < 0.05; and R = 0.39, P < 0.05, respectively), and negatively with Fe (R = −0.57, P < 0.001) (Fig. 5b). Associations were identical when using Shannon values, indicating that these patterns were robust to the alpha diversity metric used. Differences in community structure of DNA phylotypes were not predicted by pH, geographic distance, or edaphic characteristics (rM [Mantel r coefficient] < 0.1, P > 0.15). In contrast, differences in community composition of RNA phylotypes were associated with changes in N and C content (rM = 0.25, P < 0.01; and rM = 0.31, P < 0.01, respectively). No other variable significantly explained differences in community structure despite significant variation in nutrient and trace metal content across soil microhabitats: C content was higher in engineered soils, as were Mg and Ba levels, while nonengineered sites had greater Fe and Mn contents (Fig. 5a).
FIG 5.
Physical soil characteristics. (a) Differences in mean pH value and parts per million (ppm) of select nutrients and metals in engineered and nonengineered soils. **, two-tailed t test, P < 0.001; *, P < 0.05. (b) Significant Pearson correlations (P < 0.01) of pH with 16S Shannon diversity values based on DNA (gray) and RNA (blue).
Functional gene diversity.
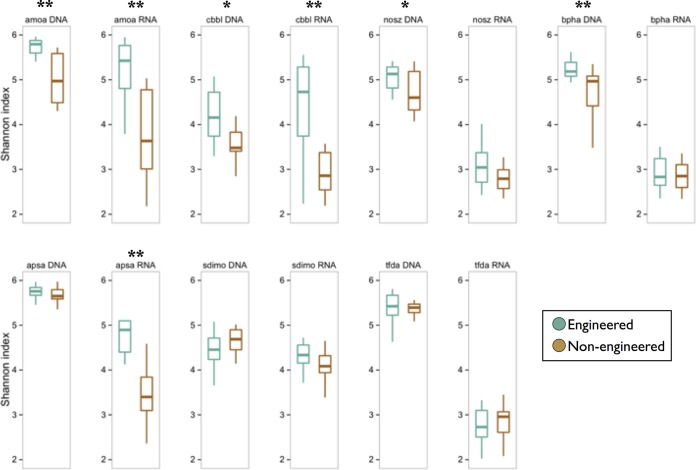
Nonengineered soils and engineered communities had significantly different compositions of functional gene phylotypes for all target genes, whether based on DNA or RNA. Alpha diversity patterns among soil types were more varied. Looking at DNA, diversity was higher in engineered sites for pollution degradation (bphA), carbon fixation (cbbL-R [cbbL gene, red-like subunit]), and N cycling (amoA and nosZ) genes (Fig. 6). The richness of soluble di-iron monooxygenase (SDIMO) genes (hydrocarbon degradation) was higher in nonengineered sites. These patterns held for expressed gene profiles (RNA), except for SDIMO genes and nosZ, which were indistinct among engineered and nonengineered sites. RNA profiles of the sulfur cycling gene apsA were distinct in engineered sites, but DNA profiles were not. Of the genes examined in this study, only the herbicide degradation gene tfdA had indistinct profiles across all sites. For all target genes, average DNA alpha diversity across all sites was greater than for RNA, although the difference was not significant for cbbL-R.
FIG 6.
Box plots of mean Shannon diversity values for engineered and nonengineered soil bacterial communities. Significance of differences was tested using the Wilcoxon test; **, P < 0.001; *, P < 0.05. Whiskers indicate standard errors.
Functional gene richness did not correspond to environmental variation, with one exception: bphA was significantly correlated with pH for DNA only (P < 0.01). Differences in N and C content helped explain the dissimilarity of functional gene profiles for one N cycling gene (amoA; N rM = 0.171, P < 0.01; C rM = 0.114, P < 0.05; Mantel-Spearman). Neither variation in soil pH nor spatial distance was associated with functional gene profile dissimilarity among sites. Communities with greater 16S diversity tended to have greater N cycling (nosZ and amoA), sulfur cycling (apsA), and herbicide degradation (tfdA) diversity values. 16S diversity was not correlated with diversity levels for other genes. Functional gene similarities were not correlated with 16S similarity for any gene based on DNA. For RNA, the only gene with dissimilarity patterns correlated to 16S was tfdA (rM = 0.180, P < 0.05).
DISCUSSION
Bioswales have distinct bacterial communities.
Efforts to integrate urban green spaces into civic infrastructure represent an implicit investment in the ecological functioning of soil communities, and our data demonstrate that engineered soils indeed maintain considerable microbial biodiversity and functional potential compared to other urban soils. Right-of-way and streetside bioswales had distinct bacterial communities that were more phylogenetically diverse than nonengineered soil types. The significantly higher diversity of bioswale bacterial communities is notable in comparison to previous surveys of NYC soils, which demonstrated that bacteria in medians (22) and fungi in green roofs (23) had similar and lower diversity, respectively, than those in city parks. The diverse, differentiated bacterial communities in engineered soils may be due to the intentional inflow of stormwater, which has a higher diversity of resources and stressors than precipitation, the primary water input for most nonengineered urban soils (17). Several other features also distinguish engineered and nonengineered soils, including designer soils, managed plant communities, and protection from compaction. Based predominantly on studies of macroorganisms, urbanization has been thought to have a homogenizing effect on biodiversity (27), but our results provide further evidence that fragmented urban soils can maintain heterogeneous and relatively diverse bacterial communities.
A key question regarding the distinct bacterial communities in engineered soils concerns the environmental drivers responsible for these differences, especially given that neither alpha nor beta diversity was correlated with spatial distance. For DNA phylotypes, pH was the only significant predictor of alpha diversity, corroborating findings from other bacterial surveys on both local and global scales (28). Neither pH nor any other variable in the study, however, explained differences in the composition of total communities, regardless of the distance metric used. The diversity of expressed communities, in contrast, was generally more associated with edaphic characteristics. Sites with similar N and C contents tended to have more similar expressed communities, and engineered sites tended to have higher N and C contents, although the difference was significant only for C. The phylogenetic diversity of expressed communities was predicted not only by pH but also by trace metals which are negatively associated with Fe and positively associated with Zn and Ni, perhaps as a result of selection for unique lineages tolerant of heavy metals. It is important to note that we cannot exclude an effect of plant diversity, which was generally greater in bioswale sites than in the grass monocultures and sparse vegetation in city park lawns and tree pits, respectively. Broader surveys that encompass a more complete representation of urban soil types will be required to more thoroughly parse the drivers of urban microbial biogeography patterns, which may include soil and plant characteristics not examined here.
Functional potential in engineered soils.
The identities of many of the highly abundant taxa suggest that bioswale communities metabolically respond to stormwater-associated resources. Four of the five most abundant bacterial phylotypes in engineered sites (each more than 20 times more abundant than in nonengineered soils) were closely related to taxa known to respond to pollution and nutrient loads associated with stormwater runoff. These include species known from soils contaminated with hydrocarbons and chlorinated solvents (PRR-10 [29]) and petroleum-based fuels (Uliginosibacterium [30]), as well as taxa with methanotrophic (Methylococcaceae [31]) and iron-oxidizing (Crenothrix [32, 33]) capabilities. Functional gene profiles also point to enriched metabolic capacity in bioswale soils, as five of the seven functional genes in the study had significantly greater alpha diversity in bioswales than in nonengineered soils for either DNA or RNA (Fig. 6). In no case did nonengineered sites have significantly greater functional gene diversity than engineered sites. For several genes, the diversity in engineered versus nonengineered sites was even greater in RNA communities than in DNA communities, suggesting these soils may be more metabolically active with respect to nitrification, carbon fixation, and sulfur cycling (involving genes amoA, cbbL, and apsA, respectively).
While our functional gene data suggest that engineered soils have greater diversity of genes underlying several ecological functions, evidence is conflicting as to whether changes in metatranscriptomic transcript levels directly indicate ecological functioning (34). In most community-level functional gene studies, including the present one, the translation rate, availability of oxygen and other essential substrates, and any dependence on competing and alternate pathways or environmental condition are unknown, obscuring the link between transcript diversity and function. Ammonia monooxygenase (amoA), for example, can oxidize ammonium, methane, or other compounds, depending on a range of factors (35). Nevertheless, there is reason to believe that communities with comparatively diverse functional gene profiles generally have enhanced functional performance. In a recent meta-analysis, of the 59 out of 419 functional gene studies that quantified process rates, there was a weak but significant positive association between gene abundances and corresponding functions (34). More specifically, each of our target genes has been previously linked to changes in function, through associations between gene diversity and either quantified processes or soil amended with target substrates (36–40). Overall, we found that engineered soils had more diverse bacterial communities with richer repositories of functional genes, and although there is reason to suspect that this is tied to functional performance, future work will be required to determine whether high functional gene diversity represents enhanced ecological functioning, greater functional resilience in response to dynamic environments or stresses, or simply greater redundancy in metabolic networks.
Engineered sites had greater phylogenetic diversity and tended to have greater functional diversity, but the phylogenic diversity of a site did not predict its functional diversity: 16S community composition was not correlated with the beta-diversity patterns of any functional genes. Alpha diversity patterns between 16S phylotypes and functional genes were more varied, with community phylogenetic diversity weakly correlated with functional alpha diversity for only 3 of 7 genes (amoA, nosZ, and bphA). This corresponds with a previous comparison of soils from different biomes, which showed that although phylogenetic diversity (PD) was correlated with functional gene diversity, the association was driven by the inclusion of desert soils and disappeared when they were excluded (25). Our findings highlight that although certain soil types may host diverse bacterial communities, e.g., bioswales at the local scale or tropical forests at the biome scale, this does not necessarily correspond to functional diversity, which must be assessed directly.
Total and active communities.
Quantifying both DNA and RNA diversity allowed us to directly assess differences in expressed, or active, communities compared to total communities, which may also include dormant or recently expired taxa (41, 42). The vast majority of soil surveys are based on genomic DNA; yet, in this study, diversity and community composition patterns based on genomic DNA were, at best, inconsistent predictors of differences among expressed communities. The degree of differentiation of expressed communities was not predictable from the total community data. These uncoupled community structure estimates were driven by clear differences in the relative abundances of major taxa based on whether RNA or DNA was examined. For example, Proteobacteria and Acidobacteria were clearly the most abundant phyla among DNA phylotypes in all four soil types, as has been found consistently in other NYC soils (21, 22). However, in RNA-based communities, Actinobacteria were more abundant than Acidobacteria, suggesting that rank abundances of microbial communities based on DNA may be positively misleading with respect to the composition of expressed communities. Supporting this view, DNA and RNA community evenness values were also uncorrelated, indicating that the relative abundances of active taxa are not predicted by their abundances in total communities. The only alpha diversity metrics that were correlated among RNA and DNA samples were phylogenetic diversity and, to a lesser extent, richness. So although shifts in DNA community structure were unrelated to shifts in the composition of RNA communities, more phylogenetically diverse total communities had more diverse expressed communities. Evenness and Shannon diversity indices, in contrast, were not correlated for RNA and DNA, suggesting that inferences about the diversity of expressed communities may be better reflected in DNA by phylogenetic diversity than other metrics. It is important to note that rRNA sampling is unlikely to recover a fully accurate picture of taxa that are active at the time of sampling, due to potential disconnects between rRNA and growth rates, taxonomic variability in the relationship between abundances of rRNA concentration, and the potential expression of rRNA in dormant cells (43). Even considering these caveats, RNA is probably more indicative than DNA of active communities, and the dissimilar diversity profiles of each underline that the 16S results from genomic DNA should be interpreted cautiously with respect to inferences about how communities actively respond to dynamically varying environments.
A surprising aspect of our results was that RNA bacterial communities were significantly more diverse than DNA communities for every alpha diversity metric except evenness. Although counter to the expectation that the expressed community at a given time represents a fraction of the diversity of the total potential community (44, 45), other studies have similarly found that RNA diversity exceeds DNA diversity for both bacteria (46) and fungi (47). Mikkonen et al. (46) attributed their results to the nature of the remediating soils they examined, hypothesizing that greater RNA diversity indicated a contemporaneously diversifying community relative to DNA, which reflected a previous low-diversity highly stressed state. The persistent presence of comparatively diverse RNA communities in other contexts, however, demands a more general explanation. Part of the answer may lie in community evenness patterns. Although RNA phylotype richness is higher, DNA communities have greater evenness. Thus, the greater richness and PD of RNA communities appear to be driven by the greater relative abundances of numerous rare taxa that are so low in total communities as to be undetectable. Indeed, it has previously been shown that rare bacterial taxa are disproportionately active compared to common taxa (48). We also cannot exclude an effect of posttranscription processes, like alternative splicing, that may cause expressed transcripts to appear more diverse than their genomic templates (49).
Conclusions.
Understanding how stresses associated with intensifying urbanization are likely to impact the biological diversity of soil microbial communities is critical to conserving the ecological functions these communities provide. As soils continue to be featured in civic infrastructure, restoration, and remediation projects, characterizing how specific urban land use practices shape microbial composition and function is an increasingly practical matter. We find that the management practices embodied in NYC's green infrastructure plan for stormwater management maintain distinct total and active microbial communities with rich repositories of ecologically functional genes. Combined with broader surveys of urban microhabitats as well as future research clarifying the links between total and active community diversity, functional gene diversity, and ecological functions, these results point to the prospect that monitoring and managing the metabolic potential of microbial communities could emerge as viable elements of sustainable urban planning.
MATERIALS AND METHODS
Soil sampling.
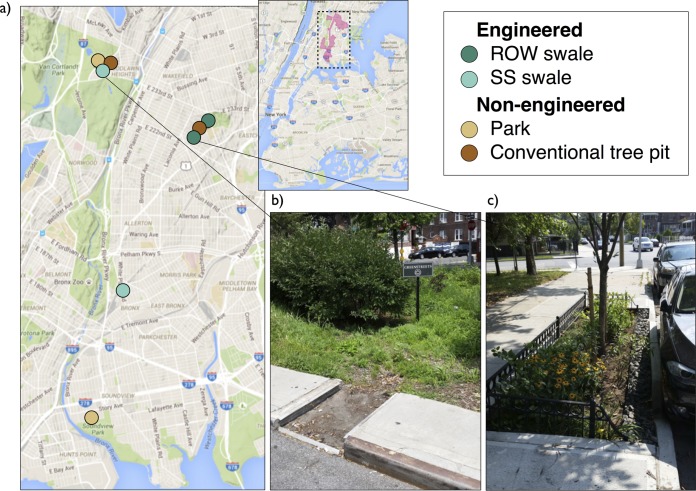
Sampling for this study was centered on the Bronx River sewershed in the Bronx, NY, a priority area for environmental improvement. All sites were between 40.81 and 40.9N, 73.84 and 73.88W, with average annual precipitation and mean temperature of 1,100 mm and 16.4°C, respectively. Within the sewershed, NYC has installed green infrastructure, including green roofs, right-of-way (ROW) bioswales, and streetside (SS) bioswales. The two bioswale types differ mainly in size: ROW swales are small installations with inlets and outlets integrated into sidewalks to resemble modified tree pits, while SS swales are relatively large rain gardens often occupying an entire corner of an intersection. Soil samples were taken from four engineered soil sites (two ROW swales and two SS swales) and four nonengineered sites, including two city park lawns and two sidewalk tree pits (Fig. 7). Sampling from each site was replicated at four different time points: twice preceding and twice following precipitation events, in July and August 2014. For each sample, four 10-cm-deep soil cores were taken using a 2.5-cm-diameter soil corer and homogenized on site using UV-sterilized 2-mm sieves. Soil for RNA extraction was collected into 25-ml cryogenic tubes, placed directly into an on-site portable liquid nitrogen Dewar flask, and stored at −80°C until extraction. The remainder of the soil was placed into Whirl-Pak bags and stored at −20°C to await DNA extractions. Soil pH and nutrient data were measured using methods described by McGuire et al. (23).
FIG 7.
Sample sites. (a) Engineered and nonengineered soil sites were sampled in the Bronx River sewershed, shaded pink in the inset map, in the Bronx, NY. Engineered sites included streetside (SS) swales (b) and right-of-way (ROW) swales (c). Nonengineered sites include park lawns and conventional tree pits. (The base map is from Google Maps.).
RNA and DNA sequencing.
Genomic DNA (gDNA) and RNA were extracted using PowerSoil extraction kits (Mo Bio, Carlsbad, CA), according to the manufacturer's protocols, with minor modifications (28). gDNA was removed from RNA extractions using DNase treatment (Mo Bio RTS DNase kit), and cDNA was synthesized using a high-capacity cDNA reverse transcriptase kit (Life Technologies, Carlsbad, CA). Using both gDNA and cDNA as templates, eight target loci were PCR amplified in duplicate using GoTaq polymerase (Promega, Madison, WI). The V4-V5 region of the 16S gene was amplified using the 515F and 806R primers (50). Seven functional loci were also amplified. These include four C cycling genes related to CO2 fixation (cbbL-R) and degradation of hydrocarbons (SDIMO genes), polychlorinated bisphenols (PCBs; bphA), and phenoxy herbicides (tfdA). Two N cycling genes related to nitrification (amoA) and denitrification (nosZ) and one gene involved in sulfur oxidation and carbon fixation (apsA) were also targeted. A single pair of previously published degenerate primers was chosen for each functional locus (Table 1). Amplicons were quantified using PicoGreen double-stranded DNA (dsDNA) fluorometry assays. For each sample, equimolar volumes of DNA and RNA amplicons for each of the eight target loci were pooled, ligated with multiplex barcoded Illumina adapters, and sequenced on a single Illumina MiSeq lane by the New York Medical Center Genomics Core Laboratory (Valhalla, NY).
TABLE 1.
Descriptions, primers, and references for each target functional gene
| Gene(s) | General function | Specific function | Primer name | Primer sequence | Reference |
|---|---|---|---|---|---|
| C cycling | |||||
| cbbL-R | CO2 fixation | Large subunit of form I red-like RubisCO | cbbLR1F | AAGGAYGACGAGAACATC | 60 |
| cbbLR1intR | TGCAGSATCATGTCRTT | ||||
| SDIMO genes | Hydrocarbon degradation | Alpha subunit of soluble di-iron monooxygenases; oxidizes diverse hydrocarbons, including common urban pollutants like propene and vinyl chloride | NVC66 | CCANCCNGGRTAYTTRTTYTCRAACCA | 8 |
| NVC57 | CAGTCNGAYGARKCSCGNCAYAT | ||||
| bphA | PCB degradation | Biphenyl dioxygenase; aerobic degradation of PCBS, a priority urban pollutant | bphAf668-3 | GTTCCGTGTAACTGGAARTWYGC | 61 |
| bphAr1153-2 | CCAGTTCTCGCCRTCRTCYTGHTC | ||||
| N cycling | |||||
| tfdA | Phenoxy herbicide degradation | Oxoglutarate dioxygenase; degrades phenolic acids found in many herbicides | TfdA421dF | ACSGARTTCKSIGACATGC | 62 |
| TfdA778vkR | AGCGGTTGTCCCACATCAC | ||||
| nosZ | Denitrification | Subunit of nitrous oxide reductase; catalyzes reduction of N2O to N2 | nosZ2F | CGCRACGGCAASAAGGTSMSSGT | 63 |
| nosZ2R | CAKRTGCAKSGCRTGGCAGAA | ||||
| amoA | Nitrification | Active site of ammonia monooxygenases; oxidizes ammonia | A189 | GNGACTGGGACTTCTGG | 64 |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC | ||||
| S cycling | |||||
| apsA | Sulfur oxidation and carbon fixation | Adenosine phosphosulfate; oxidizes sulfur to sulfate | apsAF | TGGCAGATMATGATYMACGG | 65 |
| apsAR | GGGCCGTAACCGTCCTTGAA | ||||
Sequence processing.
Low-quality reads were removed using a maximum E value of 1, resulting in sequences with an average of no more than one base error per sequence. 16S reads were identified using UBLAST (51) to match reads to the Greengenes database (52) at an E value of 1 × e−10 and query coverage of 0.75. Functional genes were identified using a similar procedure, except that gene-specific databases were downloaded from the Functional Gene Repository (53) if available, or from the NCBI database. After dereplication and removal of singletons, sequences were clustered at 97% similarity to identify phylotypes (operational taxonomic units [OTUs]) using UPARSE (54). Reads were then mapped to representative phylotype sequences to generate an OTU table. QIIME (55) was used to assign taxonomic names with the Ribosomal Database Project database and resolve phylogenetic relationships among 16S phylotypes using FastTree (56). Sequence abundances in each OTU table were normalized in two ways: rarefaction based on the minimum read count per sample, and cumulative sum scaling (CSS) to standardize counts by dividing up to a percentile determined empirically, using the phyloseq (57) and metagenomeSeq (58) R packages, respectively. Richness and community similarity values based on rarefied and CSS OTU tables were highly correlated, and normalized CSS tables were used for downstream analysis, since by retaining rare phylotypes, they provide more robust estimates of differential abundance by providing a better sampled null distribution (58).
Statistical analysis.
Alpha diversity was assessed using the Chao1 richness estimator, Faith's phylogenetic diversity (59), and the Shannon diversity index. Community dissimilarity, or beta diversity, was assessed using nonmetric multidimensional scaling of Bray-Curtis distance matrices. Unifrac distances were also calculated for 16S, but since the two are highly correlated (Spearman's r = 0.93, P < 0.001), only Bray distances are reported. Analysis of similarity (ANOSIM) was used to test whether community composition differed among categories. Differential abundance was determined based on normalized OTU tables and a statistical model accounting for undersampling, and differences were tested using multiple Kruskal-Wallis tests with false-discovery rate correction (58). Wilcoxon tests were used to test differences in mean alpha diversity values. Statistical associations between Bray-Curtis distances and environmental variation were determined using Mantel tests. All statistical analyses were conducted in the R environment (www.r-project.org) using the vegan, phyloseq, and metagenomeSeq packages.
Accession number(s).
These sequence data have been submitted to the NCBI Sequence Read Archive under accession numbers SRP082234 and PRJNA339153.
ACKNOWLEDGMENTS
We thank the NYC Parks Department and Department of Environmental Protection for permission to access sampling sites. We also thank Noah Fierer and Albert Barberan for helpful discussions.
This research was funded by the National Science Foundation (Coastal SEES 1325185 to K.L.M.).
We declare no conflicts of interest.
REFERENCES
- 1.Pickett ST, Cadenasso ML, Grove JM, Boone CG, Groffman PM, Irwin E, Kaushal SS, Marshall V, McGrath BP, Nilon CH, Pouyat RV, Szlavecz K, Troy A, Warren P. 2011. Urban ecological systems: Scientific foundations and a decade of progress. J Environ Manage 92:331–362. doi: 10.1016/j.jenvman.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Kaye JP, Groffman PM, Grimm NB, Baker LA, Pouyat RV. 2006. A distinct urban biogeochemistry? Trends Ecol Evol 21:192–199. doi: 10.1016/j.tree.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Madsen ELE. 2011. Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol 22:456–464. doi: 10.1016/j.copbio.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Canfield DE, Glazer AN, Falkowski PG. 2010. The evolution and future of Earth's nitrogen cycle. Science 330:192. doi: 10.1126/science.1186120. [DOI] [PubMed] [Google Scholar]
- 5.Trivedi P, Anderson IC, Singh BK. 2013. Microbial modulators of soil carbon storage: integrating genomic and metabolic knowledge for global prediction. Trends Microbiol 21:641–651. doi: 10.1016/j.tim.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Schimel JP, Schaeffer SM. 2012. Microbial control over carbon cycling in soil. Front Microbiol 3:348. doi: 10.3389/fmicb.2012.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pouyat R, Groffman P, Yesilonis I, Hernandez L. 2002. Soil carbon pools and fluxes in urban ecosystems. Environ Pollut 116:S107–S118. doi: 10.1016/S0269-7491(01)00263-9. [DOI] [PubMed] [Google Scholar]
- 8.Coleman NV, Bui NB, Holmes AJ. 2006. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol 8:1228–1239. doi: 10.1111/j.1462-2920.2006.01015.x. [DOI] [PubMed] [Google Scholar]
- 9.Wong CSC, Li X, Thornton I. 2006. Urban environmental geochemistry of trace metals. Environ Pollut 142:1–16. doi: 10.1016/j.envpol.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 10.Meijer SN, Ockenden WA, Sweetman A, Breivik K, Grimalt JO, Jones KC. 2003. Global distribution and budget of PCBs and HCB in background surface soils: implications for sources and environmental processes. Environ Sci Technol 37:667–672. doi: 10.1021/es025809l. [DOI] [PubMed] [Google Scholar]
- 11.Pulleman M, Creamer R, Hamer U, Helder J, Pelosi C, Pérès G, Rutgers M. 2012. Soil biodiversity, biological indicators and soil ecosystem services—an overview of European approaches. Curr Opin Environ Sustain 4:529–538. doi: 10.1016/j.cosust.2012.10.009. [DOI] [Google Scholar]
- 12.De Kimpe CR, Morel J-L. 2000. Urban soil management: a growing concern. Soil Sci 165:31. doi: 10.1097/00010694-200001000-00005. [DOI] [Google Scholar]
- 13.Tzoulas K, Korpela K, Venn S, Yli-Pelkonen V, Kazmierczak A, Niemela J, James P. 2007. Promoting ecosystem and human health in urban areas using green infrastructure: a literature review. Landsc Urban Plan 81:167–178. doi: 10.1016/j.landurbplan.2007.02.001. [DOI] [Google Scholar]
- 14.Pires M. 2004. Watershed protection for a world city: the case of New York. Land Use Policy 21:161–175. doi: 10.1016/j.landusepol.2003.08.001. [DOI] [Google Scholar]
- 15.The City of New York. 2010. NYC green infrastructure plan: a sustainable strategy for clean waterways. The City of New York, New York, NY: http://www.nyc.gov/html/dep/pdf/green_infrastructure/NYCGreenInfrastructurePlan_LowRes.pdf. [Google Scholar]
- 16.The City of New York. 2016. Standard designs and guidelines for green infrastructure practices. The City of New York, New York, NY: http://www.nyc.gov/html/dep/pdf/green_infrastructure/bioswales-standard-designs.pdf. [Google Scholar]
- 17.Gasperi J, Garnaud S, Rocher V, Moilleron R. 2008. Priority pollutants in wastewater and combined sewer overflow. Sci Total Environ 407:263–272. doi: 10.1016/j.scitotenv.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 18.The City of New York. 2017. NYC green infrastructure plan. The City of New York, New York, NY: http://www.nyc.gov/html/dep/html/stormwater/using_green_infra_to_manage_stormwater.shtml. [Google Scholar]
- 19.Souza RC, Cantão ME, Vasconcelos ATR, Nogueira MA, Hungria M. 2013. Soil metagenomics reveals differences under conventional and no-tillage with crop rotation or succession. Appl Soil Ecol 72:49–61. doi: 10.1016/j.apsoil.2013.05.021. [DOI] [Google Scholar]
- 20.Xue K, Wu L, Deng Y, He Z, Van Nostrand J, Robertson PG, Schmidt TM, Zhou J. 2013. Functional gene differences in soil microbial communities from conventional, low-input, and organic farmlands. Appl Environ Microbiol 79:1284–1292. doi: 10.1128/AEM.03393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramirez KS, Leff JW, Barberan A, Bates ST, Betley J, Crowther TW, Kelly EF, Oldfield EE, Shaw EA, Steenbock C, Bradford MA, Wall DH, Fierer N. 2014. Biogeographic patterns in below-ground diversity in New York City's Central Park are similar to those observed globally. Proc Biol Sci 281:20141988. doi: 10.1098/rspb.2014.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reese AT, Savage A, Youngsteadt E, McGuire KL, Koling A, Watkins O, Frank SD, Dunn RR. 2015. Urban stress is associated with variation in microbial species composition-but not richness-in Manhattan. ISME J 10:751–760. doi: 10.1038/ismej.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ, Gedallovich SM, Discenza J, Rangamannar R, Koshner JA, Massmann AL, Orazi G, Essene A, Leff JW, Fierer N. 2013. Digging the New York City Skyline: soil fungal communities in green roofs and city parks. PLoS One 8:e58020. doi: 10.1371/journal.pone.0058020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delmont TO, Simonet P, Vogel TM. 2012. Describing microbial communities and performing global comparisons in the omic era. ISME J 6:1625–1628. doi: 10.1038/ismej.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, Owens S, Gilbert JA, Wall DH, Caporaso JG. 2012. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci U S A 109:21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayer EJ, Wagner M, Oliver AE, Pywell RF, James P, Whiteley AS, Heard MS. 2013. Grassland management influences spatial patterns of soil microbial communities. Soil Biol Biochem 61:61–68. doi: 10.1016/j.soilbio.2013.02.012. [DOI] [Google Scholar]
- 27.McKinney ML. 2006. Urbanization as a major cause of biotic homogenization. Biol Conserv 127:247–260. doi: 10.1016/j.biocon.2005.09.005. [DOI] [Google Scholar]
- 28.Lauber CL, Hamady M, Knight R, Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schabereiter-Gurtner C, Saiz-Jimenez C, Piñar G, Lubitz W, Rölleke S. 2004. Phylogenetic diversity of bacteria associated with Paleolithic paintings and surrounding rock walls in two Spanish caves (Llonín and La Garma). FEMS Microbiol Ecol 47:235–247. doi: 10.1016/S0168-6496(03)00280-0. [DOI] [PubMed] [Google Scholar]
- 30.Udotong IR, Uko MP, Udotong J. 2015. Microbial diversity of a remote aviation fuel-contaminated sediment of a lentic ecosystem in Ibeno, Nigeria. J Environ Anal Toxicol 5:320. [Google Scholar]
- 31.Bowman J. 2006. The methanotrophs—the families Methylococcaceae and Methylocystaceae, p 266–289. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes: volume 5: Proteobacteria: alpha and beta subclasses. Springer New York, NY. [Google Scholar]
- 32.Stoecker K, Bendinger B, Schoning B, Nielsen PH, Nielsen JL, Baranyi C, Toenshoff ER, Daims H, Wagner M. 2006. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci U S A 103:2363–2367. doi: 10.1073/pnas.0506361103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emerson D, Fleming EJ, McBeth JM. 2010. Iron-oxidizing bacteria: an environmental and genomic perspective. Annu Rev Microbiol 64:561–583. doi: 10.1146/annurev.micro.112408.134208. [DOI] [PubMed] [Google Scholar]
- 34.Rocca JD, Hall EK, Lennon JT, Evans SE, Waldrop MP, Cotner JB, Nemergut DR, Graham EB, Wallenstein MD. 2015. Relationships between protein-encoding gene abundance and corresponding process are commonly assumed yet rarely observed. ISME J 9:1693–1699. doi: 10.1038/ismej.2014.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arp DJ, Stein LY. 2003. Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit Rev Biochem Mol Biol 38:471–495. doi: 10.1080/10409230390267446. [DOI] [PubMed] [Google Scholar]
- 36.Helbling DE, Ackermann M, Fenner K, Kohler H-PE, Johnson DR. 2011. The activity level of a microbial community function can be predicted from its metatranscriptome. ISME J 6:902–904. doi: 10.1038/ismej.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petersen DG, Blazewicz SJ, Firestone M, Herman DJ, Turetsky M, Waldrop M. 2012. Abundance of microbial genes associated with nitrogen cycling as indices of biogeochemical process rates across a vegetation gradient in Alaska. Environ Microbiol 14:993–1008. doi: 10.1111/j.1462-2920.2011.02679.x. [DOI] [PubMed] [Google Scholar]
- 38.Hügler M, Gärtner A, Imhoff JF. 2010. Functional genes as markers for sulfur cycling and CO2 fixation in microbial communities of hydrothermal vents of the Logatchev field. FEMS Microbiol Ecol 73:526–537. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, Zhao X, Liang Y, Li G, Zhou J. 2013. Microbial functional genes reveal selection of microbial community by PAHs in polluted soils. Environ Chem Lett 11:11–17. doi: 10.1007/s10311-012-0370-6. [DOI] [Google Scholar]
- 40.Keshri J, Mishra A, Jha B. 2013. Microbial population index and community structure in saline-alkaline soil using gene targeted metagenomics. Microbiol Res 168:165–173. doi: 10.1016/j.micres.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen KM, Johnsen PJ, Bensasson D, Daffonchio D. 2007. Release and persistence of extracellular DNA in the environment. Environ Biosafety Res 6:37–53. doi: 10.1051/ebr:2007031. [DOI] [PubMed] [Google Scholar]
- 42.Carini P, Marsden PJ, Leff JW, Morgan EE, Strickland MS, Fierer N. 2016. Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat Microbiol 2:16242. doi: 10.1038/nmicrobiol.2016.242. [DOI] [PubMed] [Google Scholar]
- 43.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. 2013. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanzén A, Jørgensen SL, Bengtsson MM, Jonassen I, Øvreås L, Urich T. 2011. Exploring the composition and diversity of microbial communities at the Jan Mayen hydrothermal vent field using RNA and DNA. FEMS Microbiol Ecol 77:577–589. doi: 10.1111/j.1574-6941.2011.01138.x. [DOI] [PubMed] [Google Scholar]
- 45.Myrold DD, Zeglin LH, Jansson JK. 2014. The potential of metagenomic approaches for understanding soil microbial processes. Soil Sci Soc Am J 78:3. doi: 10.2136/sssaj2013.07.0287dgs. [DOI] [Google Scholar]
- 46.Mikkonen A, Santalahti M, Lappi K, Pulkkinen A-M, Montonen L, Suominen L. 2014. Bacterial and archaeal communities in long-term contaminated surface and subsurface soil evaluated through coextracted RNA and DNA. FEMS Microbiol Ecol 90:103–114. doi: 10.1111/1574-6941.12376. [DOI] [PubMed] [Google Scholar]
- 47.Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Větrovský T, Žifčáková L, Šnajdr J, Rídl J, Vlček Č, Voříšková J. 2011. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6:248–258. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helm M. 2006. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR. 2013. FunGene: the functional gene pipeline and repository. Front Microbiol 4:291. doi: 10.3389/fmicb.2013.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 55.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paulson JN, Stine OC, Bravo HC, Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol Conserv 61:1–10. doi: 10.1016/0006-3207(92)91201-3. [DOI] [Google Scholar]
- 60.Selesi D, Pattis I, Schmid M, Kandeler E, Hartmann A. 2007. Quantification of bacterial RubisCO genes in soils by cbbL targeted real-time PCR. J Microbiol Methods 69:497–503. doi: 10.1016/j.mimet.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 61.Lehtinen T, Mikkonen A, Sigfusson B, Ólafsdóttir K, Ragnarsdóttir KV, Guicharnaud R. 2013. Bioremediation trial on aged PCB-polluted soils—a bench study in Iceland. Environ Sci Pollut Res 21:1759–1768. doi: 10.1007/s11356-013-2069-z. [DOI] [PubMed] [Google Scholar]
- 62.Zaprasis A, Liu YJ, Liu SJ, Drake HL, Horn MA. 2009. Abundance of novel and diverse tfdA-like genes, encoding putative phenoxyalkanoic acid herbicide-degrading dioxygenases, in soil. Appl Environ Microbiol 76:119–128. doi: 10.1128/AEM.01727-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Warneke S, Schipper LA, Matiasek MG, Scow KM, Cameron S, Bruesewitz DA, McDonald IR. 2011. Nitrate removal, communities of denitrifiers and adverse effects in different carbon substrates for use in denitrification beds. Water Res 45:5463–5475. doi: 10.1016/j.watres.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen J-P, Zhang L-M, Zhu Y-G, Zhang J-B, He J-Z. 2008. Abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea communities of an alkaline sandy loam. Environ Microbiol 10:1601–1611. doi: 10.1111/j.1462-2920.2008.01578.x. [DOI] [PubMed] [Google Scholar]
- 65.Yousuf B, Kumar R, Mishra A, Jha B. 2014. Unravelling the carbon and sulphur metabolism in coastal soil ecosystems using comparative cultivation-independent genome-level characterisation of microbial communities. PLoS One 9:e107025. doi: 10.1371/journal.pone.0107025. [DOI] [PMC free article] [PubMed] [Google Scholar]