ABSTRACT
Escherichia coli serotype O157:H7 is a zoonotic food- and waterborne bacterial pathogen that causes a high hospitalization rate and can cause life-threatening complications. Increasingly, E. coli O157:H7 infections appear to originate from fresh produce. Ruminants, such as cattle, are a prominent reservoir of E. coli O157:H7 in the United States. California is one of the most agriculturally productive regions in the world for fresh produce, beef, and milk. The close proximity of fresh produce and cattle presents food safety challenges on a uniquely large scale. We performed a survey of E. coli O157:H7 on 20 farms in California to observe the regional diversity and prevalence of E. coli O157:H7. Isolates were obtained from enrichment cultures of cow feces. Some farms were sampled on two dates. Genomes from isolates were sequenced to determine their relatedness and pathogenic potential. E. coli O157:H7 was isolated from approximately half of the farms. The point prevalence of E. coli O157:H7 on farms was highly variable, ranging from zero to nearly 90%. Within farms, generally one or a few lineages were found, even when the rate of isolation was high. On farms with high isolation rates, a single clonal lineage accounted for most of the isolates. Farms that were visited months after the first visit might have had the same lineages of E. coli O157:H7. Strains of E. coli O157:H7 may be persistent for months on farms.
IMPORTANCE This survey of 20 cow-calf operations from different regions of California provides an in depth look at resident Escherichia coli O157:H7 populations at the molecular level. E. coli O157:H7 is found to have a highly variable prevalence, and with whole-genome sequencing, high prevalences in herds were found to be due to a single lineage shed from multiple cows. Few repeat lineages were found between farms in this area; therefore, we predict that E. coli O157:H7 has significant diversity in this area beyond what is detected in this survey. All isolates from this study were found to have pathogenic potential based on the presence of key virulence gene sequences. This represents a novel insight into pathogen diversity within a single subtype and will inform future attempts to survey regional pathogen populations.
KEYWORDS: California, Escherichia coli, O157:H7, STEC, cattle, food-borne pathogens, genomics
INTRODUCTION
California's agricultural valleys and the surrounding foothill regions are some of the most agriculturally productive regions in the United States. California accounts for a large proportion of the produce that is consumed raw or minimally processed in the United States, including more than 200 specialty crops (1). Cow-calf beef operations are often located in close proximity to produce production fields at the transition between foothill grasslands and valley agricultural production. Given the potential juxtaposition of thousands of beef cattle with many of California's produce-growing regions and the concern of cattle functioning as an environmental source of E. coli O157:H7 (2), we determined the prevalence, associated risk factors, and identity of resident E. coli O157:H7 strains in a statewide cross-section of cow-calf operations.
Over nine million cases of foodborne disease occur annually in the United States from 31 major pathogens, which result in over one thousand deaths (3). Gram-negative bacteria cause many of these illnesses, with nontyphoidal Salmonella being the second most common cause of enteric infection at 11% and the leading cause of mortality. Shiga toxin-producing Escherichia coli (STEC) serotype O157:H7 infections are less common, but have a high hospitalization rate (46%, approximately 2,138 cases) and rank third in the number of estimated hospitalizations behind nontyphoidal Salmonella (19,336 cases) and Campylobacter spp. (8,463 cases) (3).
STEC describes E. coli that has at least one copy of the Shiga toxin gene (stx1 or stx2), which is carried on a lysogenic lambdoid bacteriophage (5). A particularly dangerous subset of these strains is enterohemorrhagic E. coli (EHEC), of which E. coli O157:H7 is the most common. These strains are associated with hemorrhagic colitis and hemolytic uremic syndrome (5). These complications are due to the action of Stx1 and/or Stx2 on the kidneys and are most likely to appear in young children, the elderly, and the immunocompromised (6). There are several different recognized subtypes of Stx1 and Stx2, some of which differ by only a few amino acids and have different toxicities (7).
Although the incidence of E. coli O157:H7 human infection has fallen since the mid-1990s, it remains a major public health risk, and the consumption of fresh produce is now recognized as a leading cause of foodborne illness and disease-related hospitalization in the United States (8, 9). Regarding bacterial foodborne illness, 27% of illnesses and 35% of hospitalizations are attributed to the consumption of contaminated produce (10). Moreover, a recent study found that the prevalence of STEC (O157 and other STEC serotypes) may have increased for preharvest leafy green vegetables in California, especially in the Central Coast region, where isolation rates from leafy green vegetables increased from less than 0.1% to 2.5% from 2007 to 2013, speculated to be the result of one or more complex mechanisms associated with agricultural landscape modification (11). E. coli O157:H7 contamination of produce is thought to occur through multiple mechanisms, such as contaminated irrigation water sources, fecal deposition from animals, and using improperly composted animal manure as a soil amendment, with leaf surface contamination the higher concern but uptake through plant roots a remote but possible mechanism following high levels of soil contamination (12, 13).
Ruminants are the major natural reservoir for STEC, with cattle being the one that is most significant for North America (2, 14). The observed point prevalences of key serotypes of STEC such as E. coli O157:H7 in cattle operations have varied widely (14, 15). This is partly due to differences in the ecology of E. coli O157:H7 in feedlot populations compared with its epidemiology in extensively grazed systems, such as rangeland cow-calf operations (16). A recent 3-year survey in Central Coastal California found that the point prevalence for E. coli O157:H7 in rangeland cattle fecal samples ranged from 0 to 4%, with a few herds experiencing an occasional temporary increase in fecal point prevalence of this pathogen (16). Individual cattle seem to clear strains of E. coli O157:H7, but these can persist for extended periods within cattle populations (17–19). Some of this temporality may be strain dependent, potentially influenced by virulence factors (4, 20, 21). In addition, some cattle may temporarily shed elevated E. coli O157:H7 levels in their feces, thereby increasing the risk of contamination (22–24).
This work identifies several herd management and environmental risk factors associated with the point prevalence of E. coli O157:H7 infection in cow-calf herds across California and characterizes the distribution of key genomic features of E. coli O157:H7 shared within and between these beef cattle herds.
RESULTS
Herd-level point prevalence.
Twenty cow-calf operations were enrolled in the project from 14 different California counties: Butte, Contra Costa, Humboldt, Kern, Lassen, Madera, Modoc, Mono, San Joaquin, San Luis Obispo, Solano, Stanislaus, Tulare, and Yuba (Fig. 1). This geographical region represents a wide range of climatic and environmental conditions, ranging from the mild interior coastal valleys and arid and hot annual grasslands in the southern Sierra Nevada foothills to the colder high elevation rangeland in the Northeast part of the state. One thousand four hundred twenty-four cattle were sampled during 39 separate herd visits between May 2012 and September 2013.
FIG 1.
Map of California showing the counties from which cow-calf herds were sampled. Counties which had a herd return at least one E. coli O157:H7 isolate are indicated in gray. Counties with no positive samples are filled with diagonal lines. Map created using ArcMap 10.5 (ESRI, Redlands, CA).
Nine (45%) of the 20 ranches enrolled in the project had one or more cattle shedding E. coli O157:H7 (Table 1). The point prevalence between adult cattle and calves was not significantly different (P > 0.10), with 4.7% (34/726) of adult cattle and 5.2% (34/654) of calves having detectable levels of E. coli O157:H7 in fecal samples at the time of herd sampling. Among calves, there appeared to be a higher point prevalence among weaned calves (11.9%) than among preweaned calves (3.3%) (Table 2), an observation similar to that from previous work (25, 26). Despite this low overall point prevalence at the population level, point prevalence per herd visit was highly variable within and across herds. Specifically, all cattle tested negative during 27 (69%) of the 39 herd visits at these 20 ranches. Limiting the sample size to ca. 40 animals per herd visit may have resulted in false-negative herd visits if the point prevalence was below detectable levels. Among the 12 herd visits with one or more positive cattle, the point prevalence ranged from 2.4% to 7.5% for 10 of these positive herds. By contrast, two different ranches had 37% (herd P) and 88% (herd A) of cattle test positive on a single occasion. Interestingly, both of these high point prevalence herds had a subsequent negative (herd P, 0/41) or low (herd A, 2/38 [5.3%]) point prevalence herd visit the following year. As discussed further below, herd A retested positive at a low point prevalence with a strain of E. coli O157:H7 that was highly related to the group of strains isolated earlier in the year.
TABLE 1.
Source and genomic characterization of E. coli O157:H7 strains isolated from cow-calf operations in California from 2012 to 2013
| County (herd IDa) | Sampling dates (mo/day/year) | Herd characteristics |
E. coli O157:H7 characteristics |
||||
|---|---|---|---|---|---|---|---|
| No. of animals (A:H:C)b | Stocking density | Prevalence (% [no./total]) | No. of isolates | Cladec | stx type(s) | ||
| Yuba (A) | 05/11/2012 | ND:ND:ND | ND | 87.5 (35/40) | 35 | 8 | 2a, 2c |
| 04/18/2013 | 25:0:25 | 2.27 | 5.2 (2/38) | 2 | 8 | 2a, 2c | |
| San Joaquin (B) | 05/14/2012 | 30:0:30 | 1.71 | 2.9 (1/35) | 1 | 8 | 2a, 2c |
| Stanislaus (C) | 02/19/2013 | 110:0:110 | 0.44 | 7.5 (3/40) | 3 | 3 | 1, 2a |
| Stanislaus (E) | 05/24/2012 | 243:0:243 | 32.40 | 5.0 (2/40) | 4 | 8 | 2a, 2c |
| 02/12/2013 | 140:0:130 | 0.68 | 7.5 (3/40) | 1 | 6 | 2c | |
| San Luis Obispo (N) | 02/14/2013 | 100:75:140 | 1.97 | 5.6 (2/36) | 1 | 6 | 2c |
| 1 | 8 | 2a, 2c | |||||
| Madera (P) | 11/10/2012 | 66:0:60 | 0.26 | 37.2 (16/43) | 4 | 2 | 1, 2a |
| 12 | 7 | 2c | |||||
| Tulare (Q) | 11/27/2012 | 800:0:675 | 0.12 | 2.5 (1/40) | 1 | 8 | 2a, 2c |
| 01/09/2013 | 800:0:675 | 0.12 | 5.0 (2/40) | 2 | 8 | 2a, 2c | |
| Tulare (R) | 12/08/2012 | 120:30:150 | 1.07 | 2.4 (1/42) | 1 | 2 | 1, 2a |
| Madera (S) | 06/05/2013 | 80:19:0 | 0.45 | 2.4 (1/41) | 1 | 8 | 2a, 2c |
TABLE 2.
Distribution of E. coli O157:H7 shedding in 20 California cow-calf operations between 2012 and 2013
| Characteristic | E. coli O157:H7 prevalence (% [no/total]) | Odds ratio | P valuea | 95% CIb |
|---|---|---|---|---|
| Age of cow (mo.) | ||||
| 0.1–3.0 | 5.4 (9/168) | 1.15 | 0.84 | 0.48–2.51 |
| 3.1–6.0 | 2.7 (6/219) | 0.57 | 0.29 | 0.19–1.41 |
| 6.1–12.0 | 7.1 (19/267) | 1.56 | 0.18 | 0.82–2.87 |
| Adultc | 4.7 (34/726) | 1.0 | —d | — |
| NAe | 2.3 (1/44) | — | — | — |
| Status | ||||
| Calf, not weaned | 3.3 (16/509) | 0.66 | 0.23 | 0.34–1.25 |
| Calf, weaned | 11.9 (18/145) | 2.88 | 0.002 | 1.48–5.43 |
| Adultc | 4.7 (34/726) | 1.0 | — | — |
| NA | 2.3 (1/44) | — | — | — |
| Season for herd visit | ||||
| Spring | 6.8 (40/585) | 2.11 | 0.04 | 1.05–4.63 |
| Summer | 0.5 (1/222) | 0.13 | 0.04 | 0.003–0.91 |
| Fall | 5.9 (17/289) | 1.80 | 0.19 | 0.78–4.33 |
| Winterc | 3.4 (11/328) | 1.0 | — | — |
| Length of calving season (days)f | ||||
| 20–50 | 4.8 (19/396) | 13.73 | <0.001 | 2.16–574 |
| 51–70 | 8.8 (44/499) | 26.34 | <0.001 | 4.43–1070 |
| 71–90 | 3.1 (5/162) | 8.65 | 0.06 | 0.96–413 |
| ≥91c | 0.4 (1/274) | 1.0 | — | — |
| NA | 0.0 (0/93) | — | — | — |
| No. of pasture or rangeland rotations during previous 8 weeks | ||||
| 1–2 | 2.9 (27/942) | 1.14 | 1.0 | 0.39–4.60 |
| 3–4 | 1.9 (3/157) | 0.76 | 1.0 | 0.11–4.60 |
| 5–6c | 2.5 (4/159) | 1.0 | — | — |
| NA | 21 (35/166) | — | — | — |
| Stocking densityg previous 8 weeks | ||||
| 0.003–1.0c | 3.4 (27/799) | 1.0 | — | — |
| >1.0–2.0 | 1.4 (3/221) | 0.39 | 0.16 | 0.08–1.30 |
| >2.0–10.0 | 1.7 (2/117) | 0.50 | 0.52 | 0.07–2.00 |
| >10.0 | 19.1 (37/194) | 6.7 | <0.001 | 3.90–11.80 |
| NA | 21 (35/40) | — | — | — |
| Cattle access to surface water source(s) previous 8 weeks | ||||
| Yes | 3.9 (25/639) | 2.91 | 0.007 | 1.30–7.14 |
| Noc | 1.4 (9/652) | 1.0 | — | — |
| NA | 26 (35/133) | — | — | — |
P values based on exact logistic regression.
CI, confidence interval.
Referent category.
—, not applicable.
NA, not available (data missing or unknown for herd).
Number of days for 75% of all calves to be born.
Total head/acreage.
Cattle management and environmental risk factors.
Based on bivariate or crude odds ratio analyses of individual risk factors, the factors associated with fecal shedding of E. coli O157:H7 included postweaned calves, the season of herd visit, the length of the calving season, the stocking density, and cattle access to surface drinking water sources (Table 2). Only the season of the herd visit, the length of the calving season, the stocking density, and cattle access to surface drinking water sources were significantly associated with fecal shedding of E. coli O157:H7 in the final logistic regression model (Table 3). To conserve degrees of freedom given the relative rarity of testing positive for E. coli O157:H7 for most sampling events, both the stocking density and the length of the calving season were treated as continuous variables in the final logistic regression model.
TABLE 3.
Multivariate logistic regression model for herd-level risk factors associated with fecal shedding of E. coli O157:H7 in 20 California cow-calf operations between 2012 and 2013
| Factor | Coefficient | Odds ratio | P valuea | 95% CI |
|---|---|---|---|---|
| Stocking density of herdb | 0.077 | 1.08 | 0.02 | 1.01–1.15 |
| Length of calving season (days)c | −0.037 | 0.96 | 0.01 | 0.94–0.99 |
| Season for herd visit | ||||
| Spring | −1.96 | 0.14 | 0.001 | 0.05–0.43 |
| Summer | −3.10 | 0.05 | 0.02 | 0.003–0.62 |
| Fall | −0.32 | 0.73 | 0.56 | 0.25–2.14 |
| Winterd | 0.0 | 1.0 | —e | — |
| Cattle access to surface water source(s) | ||||
| Nod | 0.0 | 1.0 | — | — |
| Yes | 1.44 | 4.2 | 0.01 | 1.38–13.04 |
| Constant | 0.18 | 0.03 | 0.04–0.88 |
P values adjusted for potential intraherd correlation of E. coli O157:H7 shedding status.
Adult cattle, heifers, and calves all counted as individual head of cattle.
Number of days for 75% of calves to be born.
Referent category.
—, not applicable.
The model's coefficients are interpreted as follows. For each additional head of cattle per acre, the odds of testing positive for an individual animal increased by approximately 8% (e0.077 × 1 = 1.08). A 10-fold increase in the stocking density would increase the odds 2.2-fold (or 220%). Similarly, a statistical analysis of two large epidemiological surveys of over 400 farms in Scotland found that for each 10-fold increase in herd size, the odds of the farm testing positive for fecal E. coli O157:H7 increased approximately 4-fold, or 400% (27). For each additional day needed to complete 75% of all calvings on a ranch, the odds of an individual animal testing positive decreased by approximately 4% (e−0.037 × 1 = 0.96). In contrast to numerous studies on cattle in dairy and feedlot production systems that found higher odds of cattle testing positive during summer than during winter, the odds of testing positive for fecal E. coli O157:H7 were 2 to 3 times lower for cattle from these cow-calf operations tested during spring (March to May) or summer (June to August) than for cattle tested during the winter months (December to February) (25, 28). The odds of testing positive for fecal E. coli O157:H7 were 4.2 times higher for beef cattle on ranches that used surface water sources for drinking than for cattle on ranches without surface water access (odds ratio [OR], 4.2). Interestingly, a recent history of movement between pastures or rangeland locations was not significantly associated with fecal shedding of E. coli O157:H7 (Table 2), which is in contrast to related findings in Scotland where recent movement of cattle onto the farm was an identified risk factor for this pathogen (26, 27, 29). It is important to interpret these findings from the final logistic regression model as statistical associations, given those data were collected in a cross-sectional manner. Moreover, missing or unknown data for several variables from a few herds reduced the effective sample size in the final logistic regression model to 1,258 cattle. The pseudo-R2 and the Hosmer-Lemeshow goodness-of-fit test were 17.5% and 0.051, respectively, indicating that the final model has limitations in predicting the positive status of infected cattle.
Whole-genome sequencing quality and isolate classification.
To describe the individual E. coli O157:H7 isolates from these herds and their population structure, a single draft E. coli O157:H7 genome was sequenced from each positive sample. We sequenced a single isolate from each positive sample to avoid introducing clonally related isolates through enrichment. Genome assemblies had an average coverage of 36.8 for contigs with length greater than 500 (see Table S1 in the supplemental material). An average of ca. 5.46 Mb of total sequence was found in ca. 348 contigs, which is typical of E. coli O157:H7 genomes (30–32).
An in silico multilocus sequence typing (MLST) analysis revealed that all E. coli isolates belonged to sequence-type ST11 complex, which includes mostly E. coli O157:H7 strains and a small proportion of O55:H7 strains (data not shown) (33). Some isolates were either additional isolates from a single enrichment culture or were determined to not be E. coli and are not included in the statistics presented above.
We classified our strains according to the single nucleotide polymorphism (SNP)-based clade designations defined in the report by Manning et al. (34) to aid in the interpretation and association with previous literature. We found strains from clades 2, 6, 7, and 8 in our survey (Table 1; see also Data Set S1). Clades 7 and 8 were the most heavily represented with 12 and 37 isolates, respectively. These results reflect large populations of individual lineages observed at each of two farms, the two combined accounting for more than 50% of the isolates in this study. Clades 2 and 8 have been linked to major outbreaks in the United States, while clades 6 and 7 have been implicated less, with clade 6 linked to clinical disease and 7 linked to asymptomatic carriage, though both have been associated with cases of hemolytic uremic syndrome (HUS) (34, 35).
Isolates from individual herds were closely related.
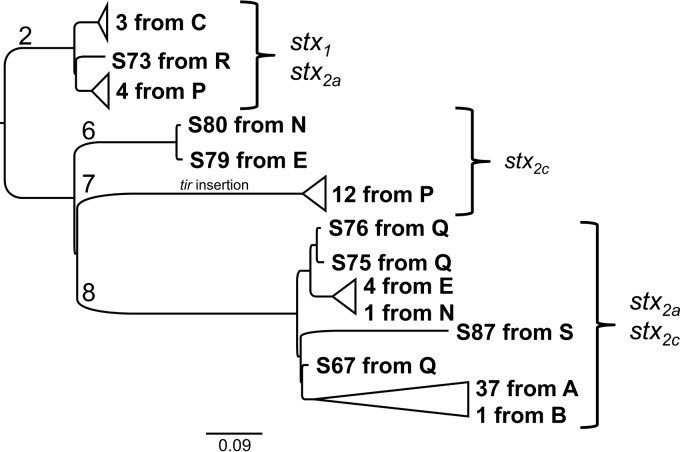
A phylogenetic analysis based on single nucleotide polymorphisms indicated that populations of E. coli O157:H7 within a herd tended to be dominated by a single sequence type. In the two herds with the largest number of positive samples, 37 of 39 (95%) isolates from herd A and 18 of 22 (82%) isolates from herd P belonged to a single phylogenetic lineage (Fig. 2; see also Fig. S1). From individual visits and from herds in general, isolates belonged to a maximum of two distinct genetic lineages. These data indicate that E. coli O157:H7 strain diversity within herds tends to be low within cow-calf herds in California. Our data set includes only limited temporal data, but isolates S85 and S86 from herd A were collected 11 months after the original sampling, and both group phylogenetically into the major clade observed previously (Fig. S1).
FIG 2.
SNP-based collapsed phylogenetic tree of the strains used in this study. The scale bar represents the number of nucleotide substitutions per site. Groups of strains have a triangle connector whose length represents the range of nucleotide substitution rates within the group. Brackets group strains based on stx gene alleles. The introduction of a short insertion into the tir gene is indicated with text on the corresponding branch. The tree without collapsed branches is presented as Fig. S1 in the supplemental material.
Herds with larger numbers of E. coli O157:H7 isolates did not exhibit correspondingly higher numbers of distinct genetic lineages; instead, these herds had multiple isolates from a single lineage. Most visits that returned a positive sample contained a single lineage except for two—two lineages were returned from herd N (n = 2) and herd P (n = 16) (Table 1; see also Fig. S1). This suggests that the point prevalence of E. coli O157:H7 infection within a cow-calf herd does not positively correlate with an increased diversity of strains. A possible explanation is that herds with a higher point prevalence of E. coli O157:H7 may have high levels of interanimal transmission of one or, alternatively, they may have a common external source that infects multiple cattle within an immunologically naive herd.
To ensure that our phylogenies were not overinfluenced by mobile genetic elements and fragmented genome sequences, a core genome was used to extract core E. coli genes from our genomes, and then the gene sequences that are highly prevalent (full-length sequences present in >95% of the new genomes) were used to create a phylogeny as described above (see Fig. S2) (36) (http://www.ecogene.org/old/topic.php?topic_id=358). The one key difference between this core-genome-derived phylogeny (Fig. S2) and the whole-genome-derived phylogeny (Fig. S1) is that a subclade within the strains from herd A is dissolved, and these strains are now indistinguishable from the rest of clade A. This strongly suggests that the appearance of this subclade was due to a mobile genetic element, but we have not discerned its identity. We interpret this to mean that the strains from clade A belong to a single lineage.
Genetic relatedness to outbreak strains of E. coli O157:H7.
Recently, many genome sequences from known outbreaks have been released, which allows newer isolates to be phylogenetically compared to strains from previous outbreaks. A set of genomes related to E. coli O157:H7 Sakai was downloaded from the NCBI genome database, as well as several additional genomes of serotype O55 and O145, to test whether our isolates might be similar or identical to previous outbreaks (Table S2). This set contains strains both isolated from and not isolated from outbreaks. We used an allelic approach with Ridom SeqSphere(+) (Ridom GmbH, Münster, Germany) where each gene from Sakai was screened in silico against the genomes from the cattle isolates, and then the identity of the alleles present in each genome was used as a measure of relatedness tied to genetic content.
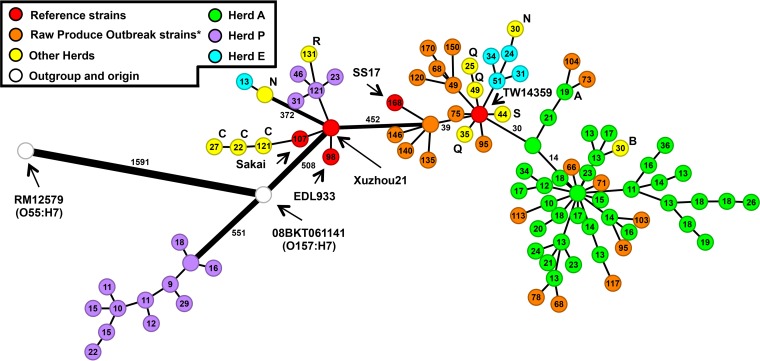
Of the E. coli O157:H7 isolates we sequenced, all were very closely related to isolates from previous outbreaks, with the exception of the major clade from herd P (Fig. 3; see also Fig. S3). For presentation, we limited our displayed data to only a few key strains in the E. coli O157:H7 literature, several notable intermediates, and an outbreak from lettuce in salad bars in a multistate outbreak (37). This representation is not meant to suggest we have found the strains implicated in any outbreak directly, but simply to ascertain whether the genetic content of our strains is or is not similar to isolates that have caused outbreaks, thus providing evidence for their pathogenicity. The topology of this representation matches well with the phylogenetic tree presented in Fig. 2, with most major branches preserved.
FIG 3.
Minimum-spanning tree (MST) of the strains used in this study plus selected strains from the NCBI genome database. The MST was created using the number of allelic differences between genome sequences, using E. coli O157:H7 Sakai as the reference and not counting gene absences. The identities of the strains are indicated by color, with the exception of yellow-filled circles, which have the letter of the corresponding herd adjacent. Connections are shown by black lines, with the allelic distance to the nearest interior node indicated within the circle, except for those along the main path relative to strain O8BKT061141, which are indicated along the line. Line thickness is increased for connections with many differences: >250, double; >500, triple; >1,000, quadruple. A detailed tree with all isolate names is presented in Fig. S3 in the supplemental material. *, the raw produce outbreak strains are from a previous report (39).
The major group of isolates from herd A and another group comprising isolates from herds E, N, Q, and S are intermixed with strains from a 2011 salad bar outbreak (Fig. 3) along with two well-documented strains, TW14359 and SS17 (38, 39). TW14359 belongs to clade 8 E. coli O157:H7, a hypervirulent variety found in a 2006 spinach outbreak (30). They have numbers of allelic differences that are typical of isolates from this study and are similar to each other, showing a close genetic relationship and suggesting that these bovine strains have the pathogenic potential to cause foodborne outbreaks.
Only the major group of strains associated with herd P did not closely resemble a known outbreak strain in the NCBI E. coli O157:H7 Sakai genome group. These isolates have only stx2c and form a distinct clade by SNP analysis. One point where the SNP and allelic approaches do not agree is the placement of S79 and S80 from herds E and N, respectively. These strains have only stx2c and group more closely with the major herd P clade by SNP analysis than by the allelic approach, which places them as more closely related to the Sakai-like strains. They belong to a separate clade by the classification scheme developed by Manning et al. (34), clade 6.
Virulence factor genes indicating potential high pathogenicity.
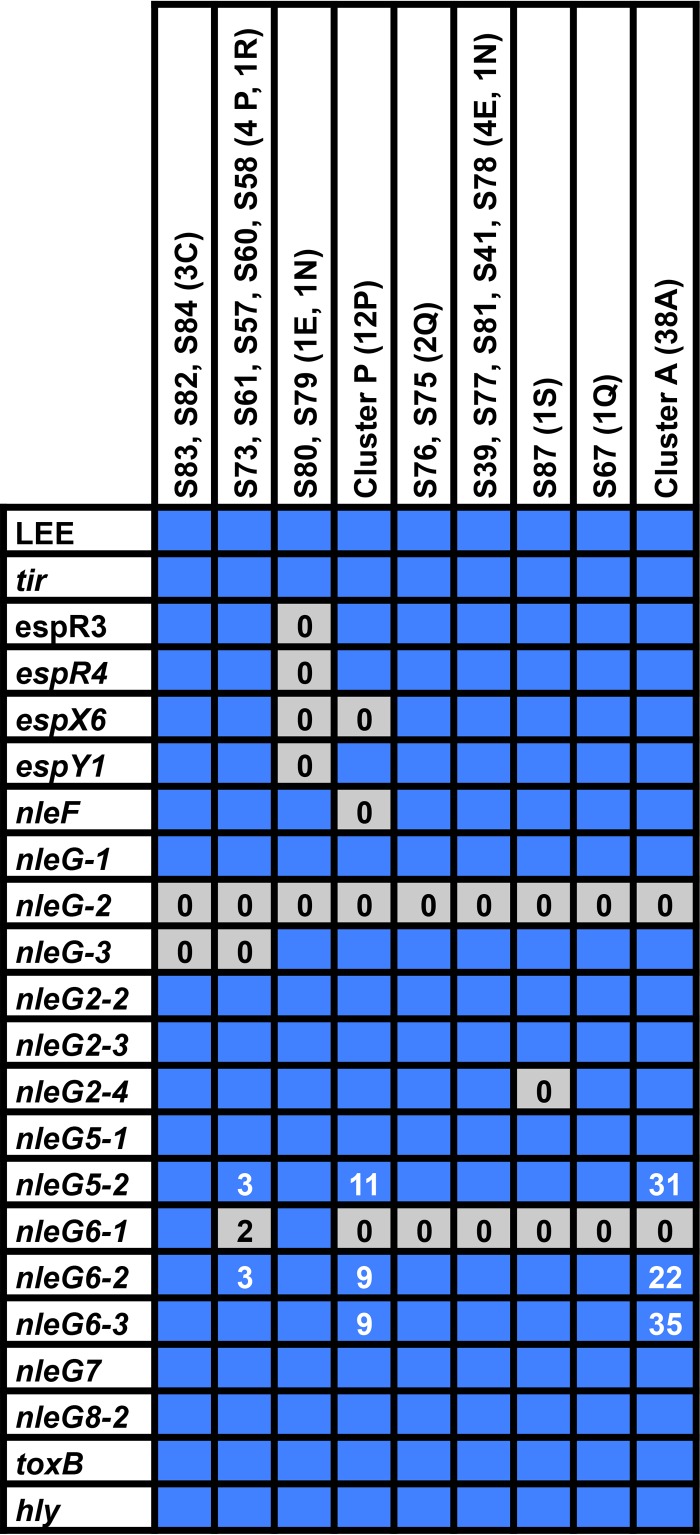
These bovine isolates contain very similar profiles of pathogenicity-linked genes. We used BLASTp to compare a database of known E. coli effectors along with some putative type III effector-like genes and other pathogenicity-related genes against our genomes (40). We used a cutoff of 90% sequence identity and 60% gene length for calling a gene present or absent in a genome. We used a process that assigned partial hits to genes if they met the sequence identity threshold, and then added those lengths up to calculate the total gene length present. All genome sequences from the isolates in this study have a locus of enterocyte effacement (LEE) and a similar suite of virulence factors, with some differences detailed below (Fig. 4; see also Data Set S2).
FIG 4.
Presence or absence of virulence genes in the genome sequences. Presence of a gene was called at 60% of the total sequence being represented in homologous sequences with 90% sequence identity. Blue squares represent presence in at least half of the isolates, while gray squares represent absence in more than half. Numbers show how many sequences had a positive call when not all sequences were positive. Additional detail and genes are presented in Data Set S2 in the supplemental material.
One particularly troublesome set of sequences for genome assembly is the nleG family of non-LEE type III secreted effector protein-encoding genes. There are several of these genes in each genome, many with a high sequence identity and a very highly conserved E3-ligase domain (41). SPAdes, the program we used to assemble these genomes, seems to have particular problems with these genes because of their repetitive nature. Approximately 1% of our contig ends fall within an nleG family gene, though they make up only 0.1% of the genomic content, showing that they are problematic for assembly (calculation shown in GitHub repository file “NleG Contig and Position Report.ipynb”). Because of this apparent issue in assembly, we have chosen not to assign some sequences, particularly potential new relatives of nleG5 and nleG6, as new effector gene sequences.
Other virulence factors showed broad conservation with a few notable exceptions. First, there were several strains for which we found ToxB-encoding sequences for only parts of the gene. We also attribute these as sequencing or assembly issues, since this loss of sequence does not seem to be inherited and toxB is a long gene (ca. 9.5 kb) with repetitive elements and, furthermore, is plasmid carried, which can affect sequencing quality. nleF encodes a non-LEE effector, and is absent from the clade including most of the strains from herd P and a single strain from herd Q. S79 and S80 have the most gene absences in our screen, missing espR3, espR4, espX6, and espY1.
These bovine isolates all contained at least one Shiga toxin gene, which are found on lysogenic Shiga toxin-converting phages. The differences in this collection of bovine stx gene types are the largest differences in pathogenic potential uncovered in this set of isolates. Isolates containing stx1 were found in herds C, P, and R, which comprise all isolates found from clade 2. All herds, and all isolates sequenced in this study, contain at least one version of the stx2 gene belonging to type 2a or 2c (Fig. 2). In some of these isolates, we were able identify the presence of multiple stx2 alleles. Individual isolates belonged to one of three types: stx1 plus stx2a, stx2a plus stx2c, or stx2c. These patterns reflect those that have been observed before using the clade designations reported by Manning and colleagues (34, 35).
Virulence plasmid pO157 and plasmid pSS17.
All of our strains had most of the genes that are found on the reference virulence plasmid pO157Sakai (see Data Set S3). We also detected most of the genes found on reference plasmid pSS17, a plasmid identified in strain SS17, in the genetic cluster containing most of the strains from herd E (see Data Set S4) (38). No significant sequence deviations were found in our sequences for these virulence plasmids.
Lineages may be persistent over months.
The persistence of individual strains within a herd of beef cattle can be weeks to months, with considerable variation observed between previous studies (42–46). We sought to gain some insight into how the persistence of populations of E. coli O157:H7 might fluctuate over time in California cow-calf herds. Specifically, we wanted to know if E. coli O157:H7 populations changed in point prevalence and composition using a second sampling months later.
Herd A from Yuba County was first sampled in May 2012 and then sampled 11 months later in April 2013. During the first sampling, 87.5% (35/40) of samples were positive for E. coli O157:H7, but only 5.2% (2/38) were positive during the second visit (Table 1). The two isolates from the second visit are nearly genetically indistinguishable from each other and are very closely related to the major lineage found during the first visit, which suggests that these isolates are clonally linked to those found 11 months earlier (Fig. 2; see also Fig. S1). This also shows that although genetically equivalent strains were found on the same farm, the point prevalence of fecal shedding by this lineage dramatically changed between sampling events. A previous study on the same herd, reported by Kondo et al. (43), showed similar variability across months, with some lineages apparently persistent across months, while others were not.
Herd E from Stanislaus County was first sampled in May 2012 and then sampled 9 months later in February 2013. The point prevalence of E. coli O157:H7 remained relatively unchanged (2/40 and 3/40, respectively), but a new lineage was found in the second sampling that was closely related to one from herd N from San Luis Obispo County. Herd Q from Tulare County was first sampled in November 2012 and then sampled 2 months later in January 2013. The point prevalence of E. coli O157:H7 was low on both dates, and a new lineage was found at the second sampling that had not been present in the first.
DISCUSSION
We have presented a region-wide effort to identify the E. coli O157:H7 strains present in California's cow-calf operations using whole-genome sequencing. We found that this region contains several different lineages of E. coli O157:H7 that are closely related to prior outbreak strains and all of which seem to have pathogenic potential based on their virulence gene composition. The overall point prevalence fecal E. coli O157:H7 in cattle from 20 cow-calf ranches was 2.7%. We did not detect positive cattle during the study in about half (55%) of these herds, possibly due to the sporadic pattern of fecal shedding among infected animals and the limiting of the cumulative sample size per herd being to approximately 80 fecal samples (47). Despite this low overall point prevalence, 2 herd visits of the 39 total (or 5.1%) exhibited a point prevalence in excess of 30%. This herd-level pattern of zero detections or very low point prevalence for fecal shedding of E. coli O157:H7 is very similar to that from an earlier study conducted between 2008 and 2010 on cow-calf operations along Central Coastal California, which also found a low overall point prevalence of 2.6% (68/2,654) for fecal E. coli O157:H7 (16), with 38% (3/8) of these herds having zero positive cattle during the 3-year duration despite much higher samples sizes (n = 138 to 489 per herd). Interestingly, this earlier study by Benjamin et al. (16) also found an occasional spike of the point prevalence of fecal E. coli O157:H7 for fecal samples collected during herd visits. For example, herd D in this earlier study spiked to 90% point prevalence from a prior point prevalence of zero, with the point prevalence from this herd returning to 0% for fecal E. coli O157:H7 on a subsequent visit. Earlier studies of fecal E. coli O157:H7 shedding in cow-calf operations in the United States and in beef herds in countries such as Scotland have found similar sporadic increases for the point prevalence of fecal E. coli O157:H7 (47, 48). This relative unpredictability in fecal shedding point prevalence at the individual animal level can result in unpredictability for the herd infection status, which creates a food safety management challenge in terms of predicting when cow-calf operations are actively shedding this bacterial pathogen.
With respect to the current study and the data observed by Benjamin et al. (16), it appears that, most of the time, California cow-calf herds pose a limited threat to nearby produce fields or to beef food safety for E. coli O157:H7 contamination (E. coli O157:H7 not detected on more than half of the farms). Obviously, other fecal pathogens may be present at a much higher prevalence (e.g., non-O157 EHEC), and individual cow-calf herds may periodically pose an elevated threat for E. coli O157:H7; hence, prudent management is always recommended. Unfortunately, these cow-calf herds appear to experience seemingly unpredictable sporadic new infections from either an undiscovered environmental point source or from events of high interanimal transmission of an endemic pathogenic strain of E. coli O157:H7 that amplify the environmental food safety risks from the herd. This pattern of variable persistence of specific strains of pathogenic E. coli O157:H7 has been observed in numerous prior studies both in the United States and internationally (27, 45, 46). Although we found several factors associated with the odds of fecal shedding of these strains of E. coli O157:H7 among California beef cattle, it is unclear whether the shift in season, the changes to the herd density, the length of the calving season, or recent access to surface sources of drinking water can explain these upward spikes in the point prevalence of cattle infection and subsequent fecal shedding. In contrast to numerous studies that found a higher prevalence of fecal shedding of E. coli O157:H7 in feedlot and dairy herds in the summer, the risk of fecal shedding in these extensive cow-calf operations was significantly lower in the spring and summer than in the winter. One speculation for this contrast is that it represents the cumulative differences in diet composition, the stocking density, the housing conditions, and the environmental bacterial bioburden between confined livestock production systems such as feedlots and dairy herds and the extensive cow-calf herds sampled in this study that were located on either rangeland or irrigated pasture at a generally much lower stocking density. The lack of elevated summertime risk for fecal shedding of E. coli O157:H7 was also observed for cow-calf herds pastured in northeast Kansas (47).
Our genomic surveys show that all E. coli O157:H7 genomes in these cow-calf herds from throughout California encode a set of pathogenicity factors that indicate they can function as human pathogens. All strains have putatively active type III secretion systems with typical E. coli O157:H7 effector repertoires and a standard complement of toxin-encoding genes.
When E. coli O157:H7 is present in a cow-calf herd, the number of distinct lineages tends to be low. On two occasions, we observed the same strain in a single herd on two different dates (A and E), which may indicate that a strain can persist in the herd for at least 1 year, but we cannot rule out strain reacquisition from an outside source. Strain replacement was also seen (herd Q). Altogether, our prevalence and genomic data, along with the findings from Benjamin et al. (16), suggest that strains of E. coli O157:H7 are infrequently acquired by these populations of beef cattle in California, though they may persist at low endemic levels once a herd becomes infected. One broad study in Scotland found only two instances of a genome digest pattern resulting from isolates from the same farm at two different time points, 4 years apart (27). Given the generally low prevalence of many strains of E. coli O157:H7 in these beef cattle herds, one cannot readily determine whether these new lineages result from a recent introduction or rather from sporadic detection of rare endemic strains, given our sample size of 40 individuals per herd visit. For example, assuming a 2.5% to 7.5% prevalence of fecal E. coli O157:H7 for most of the positive cow-calf herds in this study (Table 1), there is a 36% to 4.4% probability, respectively, of detecting no true positive samples when 40 random cattle are tested per visit (based on the binomial approximation).
This study, although broad geographically, has not captured the total diversity of E. coli O157:H7 present in the cow-calf regions of California and does not comprise data sufficient to give an estimate of how much diversity exists in these cattle, but it does indicate how such a study could be efficiently done. We found a few repeat lineages in our survey (for example, S37 from herd B is nested within the major clade from herd A), suggesting that the number of highly prevalent strains may not be unapproachably large, since we are already encountering repeat lineages with a small sample size relative to the large number of cow-calf operations in California. A broader study, including whole-genome sequencing of E. coli O157:H7 isolates from more ranches, is needed to estimate the genetic diversity of strains in the region. The results from our study suggest that the most efficient way of characterizing the diversity of a pathogen in a region would be to sequence only a few positive samples from a farm, since strain diversity on farms tends to be low, at least for E. coli O157:H7 in California cow-calf populations.
In conclusion, a diverse set of E. coli O157:H7 lineages appears to be present in California's cow-calf herds, with these analyses constrained by the limited number of positive samples available for genomic characterization. These lineages are all ostensibly pathogenic, and most are related to known outbreak strains. When present, these strains are typically present at a low prevalence, with intermittent periods of high prevalence, but seemingly in low diversity. We found some repeat lineages on geographically separated farms, and it may be important to distinguish what lineages are present broadly throughout the region and if any are more localized, both for academic and regulatory interests.
MATERIALS AND METHODS
Study population.
A statewide cross-sectional study of 20 California cow-calf operations was conducted between the spring of 2012 and fall of 2013. Operations were chosen for voluntary participation from a list of cooperating livestock operations maintained by the University of California Cooperative Extension (UCCE), focusing on counties that comprise the Central Valley and adjacent foothill regions, with an outgroup herd chosen from the Central Coastal area where a similar survey was conducted from 2008 to 2010 (16). Only grazing operations on irrigated pasture or native rangeland were considered for the study. Herds consisting of less than 20 adult breeding animals were excluded from the study. Visits were performed at key cattle gathering times during the production cycle, such as castration, branding, weaning, and pregnancy checks, or convenience samples were taken when cattle were moved to an easily accessible pasture. Fresh fecal samples were taken from cows and calves either from the rectum or from freshly voided feces off the ground. A new pair of disposable nitrile gloves was used for each sample collected. All samples were placed and stored in sterile 4-oz fecal cups until processing. Herd visits and sampling were overseen and/or performed by a veterinarian from the UC Davis Veterinary Medicine Teaching Hospital or by a UCCE advisor. Forty fecal samples were targeted for each visit, namely, 20 from cows (2 years of age or older) and 20 from calves (less than 12 months of age), but for several herd visits, only adult animals or calves were sampled due to the limited availability of animals dictated by the stage of the production cycle. Herds were visited up to three times during the sampling period during either winter (December, January, and February), spring (March, April, and May), summer (June, July, and August), or fall (September, October, and November).
The following risk factor information was recorded for each herd: (i) the total number of cows and replacement heifers in the herd, (ii) the beginning and end of the breeding season, (iii) the beginning of the calving season, (iv) the number of days from the beginning of the calving season until approximately 75% of the calves were born, (v) the weaning age, (vi) the approximate size, in acres, of calving pastures for cows and heifers, (vii) the amount of time that they stayed on the calving pastures prior to parturition, (viii) the amount of time that they stayed on calving pastures postpartum, (ix) the pasture type and the stocking density (total head/acreage) that cattle were exposed to during the 2 months prior to sampling, and (x) the type of water sources animals were exposed to during the 2 months prior to sampling. Questionnaire data were entered and stored in a computerized database.
Statistical analyses of risk factors associated with fecal shedding of E. coli O157:H7.
Logistic regression was used to characterize the association between cattle, management, and environmental risk factors and the likelihood of testing positive for fecal E. coli O157:H7. A forward-stepping algorithm was used to build the multiplicative logistic regression model, with a robust P value (adjusted for potential intraherd clustering of disease status) of ≤ 0.05 used for inclusion in the final model. All variables not included in the final analysis were reentered into the full model to test for significance. The adequacy of the final model was evaluated by comparing the full model's Akaike information criterion (AIC) against all possible nested models where one term was eliminated and by calculating the pseudo-R2 and the Hosmer-Lemeshow goodness-of-fit statistics.
Fecal isolation of presumptive E. coli O157:H7.
E. coli O157:H7 was isolated from bovine feces using methods described by Cooley et al. (49) with the following modifications. Ten grams of feces was aseptically weighed and transferred into 100-ml tryptic soy broth (TSB). The samples were incubated for 2 h at 25°C with an orbital rotation of 150 rpm and for 8 h at 42°C and 150 rpm and were finally held static overnight at 6°C. Immunomagnetic separation (IMS) was performed using an automated Dynal BeadRetriever (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions using Dynabeads with anti-E. coli O157 (Invitrogen, Carlsbad, CA, USA). Fifty microliters of the resuspended beads-phosphate-buffered saline (PBS) complex was plated and streaked for isolation on Rainbow agar (Biolog, Hayward, CA) with novobiocin (20 mg/liter; MP Biomedicals LLC, Solon, OH, USA) and tellurite (0.8 mg/liter; MP Biomedicals) (NT-Rainbow). The remaining 50 μl was plated and streaked for isolation on sorbitol MacConkey agar (BD, Sparks, MD, USA) with cefixime (0.05 mg/liter; USP, Rockville, MD, USA) and tellurite (2.5 mg/liter) (CT-SMAC). The plates were incubated for 18 to 24 h at 37°C. Two presumptive positive colonies per plate were picked and streaked on LB agar (BD, Franklin Lakes, NJ) for extraction and cryogenic storage.
Molecular confirmation of E. coli O157:H7 isolates.
DNA was extracted using a simple boiling method. Briefly, a 10-μl loopful of bacteria was swirled into 100 μl DNase free water in a microcentrifuge tube and incubated at 100°C for 20 min. After incubation, the microcentrifuge tubes were centrifuged for 10 min at 5,000 rpm. The supernatants were transferred to sterile microcentrifuge tubes. The two suspect colonies per positive sample were confirmed by PCR using primers from Paton and Paton (50). Each reaction mixture contained 48.5 μl master mixture and 1.5 μl DNA. The master mixture was composed of 1× buffer, 0.4 μM forward and reverse primers, 200 μM deoxynucleoside triphosphates (dNTPs), and 1.5 mM MgCl2, and the remaining volume was adjusted with DNase-free water to a final volume of 48.5 μl per reaction. The PCR assays were performed using an Eppendorf thermocycler (Eppendorf, Hauppauge, NY, USA) with an initial denaturation at 95°C for 1 min followed by 30 cycles of 94°C for 15 s, 55°C for 15 s, and 72°C for 1 min, with a final extension of at 72°C for 1 min. The samples were held at 4°C until they were removed from the thermocycler. PCR products were visualized on an ethidium-bromide (EtBr)-stained 2% agarose gel using UV transillumination and measured with an Invitrogen low mass ladder (Invitrogen, Carlsbad, CA, USA).
DNA sequencing.
DNA extractions from isolates of E. coli O157:H7 were performed using the QIAcube with the DNeasy blood and tissue kit according to the manufacturer's protocol (DNeasy blood and tissue—bacterial pellet—standard). Sequencing reactions were performed using an Illumina MiSeq machine using a Nextera XT DNA library preparation kit and the MiSeq reagent kit V2 500 cycle. The machine was run using paired-ends and 2 × 250 base reads. One strain was sequenced per positive sample to avoid clonally related bacteria made during the enrichment.
Sequence analysis.
Adapter trimming of raw reads was performed using the MiSeq reporter software. Trimmed reads were assembled using SPAdes genome assembler. Genome assembly quality checks were performed using Python 3.5 scripts which utilize Biopython. Phylogenetic trees based on SNP data were created using kSNP to identify SNPs, and RAxML was used to create phylogenetic distance maps. The RAxML run was called with 100 bootstraps and the GTRGAMMA substitution model. Minimum spanning trees were based on allelic data, and comparisons of gene content between isolates were performed using Ridom SeqSphere+ 3.0. In silico sequence typing was performed in SeqSphere+ using the University of Warwick E. coli MLST database. The identification of stx and other genes was done using Python 3.5, Biopython, and BLAST, with parameters specified in the results (40, 51). A previously published core genome for E. coli was used to extract core gene sequences using BLAST (36). We used the virulence factor database as a reference to identify virulence genes in the genome assemblies (52). We used GenBank accessions NC_002128 and NZ_CP008807 for pO157Sakai and pSS17 gene references, respectively (38, 53).
Clade identification.
Isolate genomes were classified as in the clade structure presented by Manning et al. (34). We used this classification because it was used in a publication that highlights a key finding in this paper (30). For this, we identified the relevant sequences used for the identification with BLAST, and then compared the results with those in the original paper.
Code availability.
The python code, in notebook format, is available at https://github.com/jnw29/2016_UCD_O157_Collab. Files referenced in the code are available upon request.
Accession number(s).
Whole-genome shotgun (WGS) assemblies from our project have been deposited at DDBJ/ENA/GenBank under accession numbers NHVM00000000 to NHYC00000000. The versions described are XXXX01000000. The sequence reads have been deposited in the NCBI Sequence Read Archive under accession numbers SRS1807737 to SRS1807821. Please refer to Table S1 in the supplemental material for specific associations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Mammel of the USFDA for providing us with the SNP matrix used to calculate clade designations.
This project was funded in part by contract U01-003-572 from the U.S. Food and Drug Administration to support the Western Center for Food Safety.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00734-17.
REFERENCES
- 1.United States Department of Agriculture. 2015. California agricultural statistics 2013 crop year. United States Department of Agriculture, National Agricultural Statistics Service, Pacific Regional Field Office, Sacramento, CA. [Google Scholar]
- 2.Persad AK, LeJeune JT. 2014. Animal reservoirs of Shiga toxin-producing Escherichia coli. Microbiol Spectr 2:EHEC-0027-2014. doi: 10.1128/microbiolspec.EHEC-0027-2014. [DOI] [PubMed] [Google Scholar]
- 3.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbishley A, Ahmad NI, Hughes K, Hutchings MR, McAteer SP, Connelley TK, Brown H, Gally DL, McNeilly TN. 2014. Strain-dependent cellular immune responses in cattle following Escherichia coli O157:H7 colonization. Infect Immun 82:5117–5131. doi: 10.1128/IAI.02462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croxen MA, Law RJ, Scholz R, Keeney KM, Wlodarska M, Finlay BB. 2013. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 7.Melton-Celsa AR. 2014. Shiga toxin (Stx) classification, structure, and function. Microbiol Spectr 2:EHEC-0024-2013. doi: 10.1128/microbiolspec.EHEC-0024-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle MP, Erickson MC. 2008. Summer meeting 2007—the problems with fresh produce: an overview. J Appl Microbiol 105:317–330. doi: 10.1111/j.1365-2672.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- 9.Henao OL, Jones TF, Vugia DJ, Griffin PM. 2015. Foodborne Diseases Active Surveillance Network—2 decades of achievements, 1996–2015. Emerg Infect Dis 21:1529–1536. doi: 10.3201/eid2109.150581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Griffin PM. 2013. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis 19:407–415. doi: 10.3201/eid1903.111866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karp DS, Gennet S, Kilonzo C, Partyka M, Chaumont N, Atwill ER, Kremen C. 2015. Comanaging fresh produce for nature conservation and food safety. Proc Natl Acad Sci U S A 112:11126–11131. doi: 10.1073/pnas.1508435112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon EB, Yaron S, Matthews KR. 2002. Transmission of Escherichia coli O157:H7 from contaminated manure and irrigation water to lettuce plant tissue and its subsequent internalization. Appl Environ Microbiol 68:397–400. doi: 10.1128/AEM.68.1.397-400.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright KM, Chapman S, McGeachy K, Humphris S, Campbell E, Toth IK, Holden NJ. 2013. The endophytic lifestyle of Escherichia coli O157:H7: quantification and internal localization in roots. Phytopathology 103:333–340. doi: 10.1094/PHYTO-08-12-0209-FI. [DOI] [PubMed] [Google Scholar]
- 14.Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J Anim Sci 85:E45–E62. doi: 10.2527/jas.2006-508. [DOI] [PubMed] [Google Scholar]
- 15.Hussein HS, Bollinger LM. 2005. Prevalence of Shiga toxin-producing Escherichia coli in beef cattle. J Food Prot 68:2224–2241. doi: 10.4315/0362-028X-68.10.2224. [DOI] [PubMed] [Google Scholar]
- 16.Benjamin LA, Jay-Russell MT, Atwill ER, Cooley MB, Carychao D, Larsen RE, Mandrell RE. 2015. Risk factors for Escherichia coli O157 on beef cattle ranches located near a major produce production region. Epidemiol Infect 143:81–93. doi: 10.1017/S0950268814000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeJeune JT, Besser TE, Rice DH, Berg JL, Stilborn RP, Hancock DD. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl Environ Microbiol 70:377–384. doi: 10.1128/AEM.70.1.377-384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanderson MW, Besser TE, Gay JM, Gay CC, Hancock DD. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet Microbiol 69:199–205. doi: 10.1016/S0378-1135(99)00106-6. [DOI] [PubMed] [Google Scholar]
- 19.Cray WC, Moon HW. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol 61:1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Döpfer D, Geue L, Schares S, Mintel B, Hoffmann B, Fischer EA. 2012. Dynamics of Shiga-toxin producing Escherichia coli (STEC) and their virulence factors in cattle. Prev Vet Med 103:22–30. doi: 10.1016/j.prevetmed.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Carlson BA, Nightingale KK, Mason GL, Ruby JR, Choat WT, Loneragan GH, Smith GC, Sofos JN, Belk KE. 2009. Escherichia coli O157:H7 strains that persist in feedlot cattle are genetically related and demonstrate an enhanced ability to adhere to intestinal epithelial cells. Appl Environ Microbiol 75:5927–5937. doi: 10.1128/AEM.00972-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams ML, Pearl DL, Bishop KE, Lejeune JT. 2013. Use of multiple-locus variable-number tandem repeat analysis to evaluate Escherichia coli O157 subtype distribution and transmission dynamics following natural exposure on a closed beef feedlot facility. Foodborne Pathog Dis 10:827–834. doi: 10.1089/fpd.2013.1484. [DOI] [PubMed] [Google Scholar]
- 23.Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat Rev Microbiol 6:904–912. doi: 10.1038/nrmicro2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omisakin F, MacRae M, Ogden ID, Strachan NJC. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl Environ Microbiol 69:2444–2447. doi: 10.1128/AEM.69.5.2444-2447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferens WA, Hovde CJ. 2011. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog Dis 8:465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chase-Topping ME, McKendrick IJ, Pearce MC, MacDonald P, Matthews L, Halliday J, Allison L, Fenlon D, Low JC, Gunn G, Woolhouse MEJ. 2007. Risk factors for the presence of high-level shedders of Escherichia coli O157 on Scottish farms. J Clin Microbiol 45:1594–1603. doi: 10.1128/JCM.01690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert LJ, Vali L, Hoyle DV, Innocent G, McKendrick IJ, Pearce MC, Mellor D, Porphyre T, Locking M, Allison L, Hanson M, Matthews L, Gunn GJ, Woolhouse ME, Chase-Topping ME. 2014. E. coli O157 on Scottish cattle farms: evidence of local spread and persistence using repeat cross-sectional data. BMC Vet Res 10:95. doi: 10.1186/1746-6148-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dewsbury DMA, Renter DG, Shridhar PB, Noll LW, Shi X, Nagaraja TG, Cernicchiaro N. 2015. Summer and winter prevalence of Shiga toxin-producing Escherichia coli (STEC) O26, O45, O103, O111, O121, O145, and O157 in feces of feedlot cattle. Foodborne Pathog Dis 12:726–732. doi: 10.1089/fpd.2015.1987. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X-S, Chase-Topping ME, McKendrick IJ, Savill NJ, Woolhouse MEJ. 2010. Spread of E. coli O157 infection among Scottish cattle farms: stochastic models and model selection. Epidemics 2:11–20. doi: 10.1016/j.epidem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A 108:20142–20147. doi: 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 32.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 33.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, Mladonicky JM, Somsel P, Rudrik JT, Dietrich SE, Zhang W, Swaminathan B, Alland D, Whittam TS. 2008. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci U S A 105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iyoda S, Manning SD, Seto K, Kimata K, Isobe J, Etoh Y, Ichihara S, Migita Y, Ogata K, Honda M, Kubota T, Kawano K, Matsumoto K, Kudaka J, Asai N, Yabata J, Tominaga K, Terajima J, Morita-Ishihara T, Izumiya H, Ogura Y, Saitoh T, Iguchi A, Kobayashi H, Hara-Kudo Y, Ohnishi M, Arai R, Kawase M, Asano Y, Asoshima N, Chiba K, Furukawa I, Kuroki T, Hamada M, Harada S, Hatakeyama T, Hirochi T, Sakamoto Y, Hiroi M, Takashi K, Horikawa K, Iwabuchi K, Kameyama M, Kasahara H, Kawanishi S, Kikuchi K, Ueno H, Kitahashi T, Kojima Y, Konishi N, et al. 2014. Phylogenetic clades 6 and 8 of enterohemorrhagic Escherichia coli O157:H7 with particular stx subtypes are more frequently found in isolates from hemolytic uremic syndrome patients than from asymptomatic carriers. Open Forum Infect Dis 1:ofu061. doi: 10.1093/ofid/ofu061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davids W, Zhang Z. 2008. The impact of horizontal gene transfer in shaping operons and protein interaction networks–direct evidence of preferential attachment. BMC Evol Biol 8:23. doi: 10.1186/1471-2148-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turabelidze G, Lawrence SJ, Gao H, Sodergren E, Weinstock GM, Abubucker S, Wylie T, Mitreva M, Shaikh N, Gautom R, Tarr PI. 2013. Precise dissection of an Escherichia coli O157:H7 outbreak by single nucleotide polymorphism analysis. J Clin Microbiol 51:3950–3954. doi: 10.1128/JCM.01930-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cote R, Katani R, Moreau MR, Kudva IT, Arthur TM, DebRoy C, Mwangi MM, Albert I, Raygoza Garay JA, Li L, Brandl MT, Carter MQ, Kapur V. 2015. Comparative analysis of super-shedder strains of Escherichia coli O157:H7 reveals distinctive genomic features and a strongly aggregative adherent phenotype on bovine rectoanal junction squamous epithelial cells. PLoS One 10:e0116743. doi: 10.1371/journal.pone.0116743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wendel AM, Johnson DH, Sharapov U, Grant J, Archer JR, Monson T, Koschmann C, Davis JP. 2009. Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August–September 2006: the Wisconsin investigation. Clin Infect Dis 48:1079–1086. doi: 10.1086/597399. [DOI] [PubMed] [Google Scholar]
- 40.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu B, Skarina T, Yee A, Jobin M-C, Dileo R, Semesi A, Fares C, Lemak A, Coombes BK, Arrowsmith CH, Singer AU, Savchenko A. 2010. NleG type 3 effectors from enterohaemorrhagic Escherichia coli are U-box E3 ubiquitin ligases. PLoS Pathog 6:e1000960. doi: 10.1371/journal.ppat.1000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rahn K, Renwick SA, Johnson RP, Wilson JB, Clarke RC, Alves D, McEwen S, Lior H, Spika J. 1997. Persistence of Escherichia coli O157:H7 in dairy cattle and the dairy farm environment. Epidemiol Infect 119:251–259. doi: 10.1017/S0950268897007929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo S, Hoar BR, Villanueva V, Mandrell RE, Atwill ER. 2010. Longitudinal prevalence and molecular typing of Escherichia coli O157:H7 by use of multiple-locus variable-number tandem-repeat analysis and pulsed-field gel electrophoresis in fecal samples collected from a range-based herd of beef cattle in California. Am J Vet Res 71:1339–1347. doi: 10.2460/ajvr.71.11.1339. [DOI] [PubMed] [Google Scholar]
- 44.Joris M-A, Verstraete K, De Reu K, De Zutter L. 2013. Longitudinal follow-up of the persistence and dissemination of EHEC on cattle farms in Belgium. Foodborne Pathog Dis 10:295–301. doi: 10.1089/fpd.2012.1277. [DOI] [PubMed] [Google Scholar]
- 45.Renter DG, Sargeant JM, Oberst RD, Samadpour M. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl Environ Microbiol 69:542–547. doi: 10.1128/AEM.69.1.542-547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liebana E, Smith RP, Batchelor M, McLaren I, Cassar C, Clifton-Hadley FA, Paiba GA. 2005. Persistence of Escherichia coli O157 isolates on bovine farms in England and Wales. J Clin Microbiol 43:898–902. doi: 10.1128/JCM.43.2.898-902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sargeant JM, Gillespie JR, Oberst RD, Phebus RK, Hyatt DR, Bohra LK, Galland JC. 2000. Results of a longitudinal study of the prevalence of Escherichia coli O157:H7 on cow-calf farms. Am J Vet Res 61:1375–1379. doi: 10.2460/ajvr.2000.61.1375. [DOI] [PubMed] [Google Scholar]
- 48.Gunn GJ, McKendrick IJ, Ternent HE, Thomson-Carter F, Foster G, Synge BA. 2007. An investigation of factors associated with the prevalence of verocytotoxin producing Escherichia coli O157 shedding in Scottish beef cattle. Vet J 174:554–564. doi: 10.1016/j.tvjl.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 49.Cooley M, Carychao D, Crawford-Miksza L, Jay MT, Myers C, Rose C, Keys C, Farrar J, Mandrell RE. 2007. Incidence and tracking of Escherichia coli O157:H7 in a major produce production region in California. PLoS One 2:e1159. doi: 10.1371/journal.pone.0001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paton AW, Paton JC. 2003. Detection and characterization of STEC in stool samples using PCR, p 45–54. In Philpott D, Ebel F (ed), Methods in molecular medicine, vol 73: E. coli Shiga toxin methods and protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 51.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han CG, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res 5:1–9. doi: 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.